Abstract
The ability of genetic networks to integrate multiple inputs in the generation of cellular responses is critical for the adaptation of cellular phenotype to distinct environments and of great interest in the construction of complex artificial circuits. To develop artificial genetic circuits that can integrate intercellular signaling molecules and commonly used inducing agents, we have constructed an artificial genetic AND gate based on the PluxI quorum-sensing promoter and the lac repressor. The hybrid promoter exhibited reduced basal and induced expression levels but increased expression capacity, generating clear logical responses that could be described using a simple mathematical model. The model also predicted that the AND gate's logic could be improved by altering the properties of the LuxR transcriptional activator and, in particular, by increasing its rate of transcriptional activation. Following these predictions, we were able to improve the AND gate's logic by ∼1.5-fold using a LuxR mutant library generated by directed evolution, providing the first example of the use of mutant transcriptional activators to improve the logic of a complex regulatory circuit. In addition, detailed characterizations of the AND gate's responses shed light on how LuxR, LacI, and RNA polymerase interact to activate gene expression.
Genetic regulatory systems are often composed of coupled components that integrate multiple inputs to increase the specificity of environmental responses and program complex cellular behavior (6, 21). Depending on their complexity, these regulatory systems can include components that function at the transcriptional (38) and posttranscriptional (1, 39) levels. In many systems, these components act to perform logical functions that are roughly analogous to those performed by traditional logic gates in electrical circuits and can be described using Boolean operators such as AND and OR. The prevalence of these logic gates in genetic regulatory circuits has prompted the creation of artificial circuits demonstrating diverse logical functions both to understand how logical responses are derived in natural systems and as modules for the creation of artificial circuits with increased complexities (2, 26-28, 35).
Logical AND responses are particularly common in biological networks, and studies have identified various mechanisms by which cells implement AND logic (29, 31). As part of the effort to classify these natural genetic logic gates and to generate novel gates for use in artificial genetic circuits, a variety of logical AND gates have been constructed using unique regulatory mechanisms including chemical complementation (10), posttranscriptional regulation (2), and allosteric control (31). While the complexities of these systems vary greatly, it has also been shown that AND logic can be obtained in a minimal system consisting of an individual promoter regulated by two transcription factors (11). A benefit of this simplified architecture is that it should allow for the easy construction of novel AND gates by the incorporation of additional cis-regulatory regions into target promoters. Applying this design principle to the PluxI quorum-sensing promoter from Vibrio fischeri (40), we have created a minimal AND gate incorporating a coupled activator-repressor pair by inserting a lac operator site adjacent to the PluxI regulatory region. Quorum-sensing promoters, and PluxI in particular, have been the focus of a significant amount of interest in the construction of artificial genetic circuits because of their ability to program intercellular responses (4, 24) and to synchronize cellular responses at the population level (3, 42). This interest has resulted in the generation of a small number of hybrid PluxI promoters controlled by multiple transcriptional regulators that have been used to construct a genetic pulse generator and to implement programmed pattern formation (7, 8). However, to our knowledge, the logical properties of these hybrid promoters have never been explored. In addition, screening of a combinatorial promoter library failed to identify any functional hybrid promoters containing the PluxI and lac regulatory regions (13). In this report, we demonstrate the construction, engineering, and detailed characterizations of such a genetic AND gate.
A major challenge in the construction of artificial genetic circuits is the inability to rapidly optimize circuit designs to balance the activities of the individual components and to account for the influences of cellular context on function. Overcoming these problems to create functional circuits can require multiple rounds of rational design and extensive changes to the initial circuit, including the replacement of key components. As alternatives to rational design, combinatorial methods provide a way to rapidly generate circuit diversity (17) but are ultimately limited by the small number of well-classified genetic elements (32). Another alternative for the tuning of genetic circuits is the creation and screening of mutant libraries for the key circuit components to identify mutants with properties that mathematical models predict will result in desired behaviors (18, 32). This method takes advantage of the ability of models to predict circuit behavior (16, 20) but relies on the availability of mutant libraries for the circuit components. Directed evolution is a particularly useful technique for the generation of these mutant libraries as it allows for the rapid generation of diversity that can be used to overcome limited understanding of the complex series of interactions that occur between circuit components (32, 41). Here we demonstrate the utility of mutant component libraries in the optimization of circuit function by using a simple mathematical model to identify a set of mutant LuxR proteins from an existing directed evolution library that result in enhancement of the logical responses of the AND gate.
MATERIALS AND METHODS
Plasmid construction, cell strains, and reagents.
To establish AND logic, all of our systems used LuxR expressed constitutively from a plasmid-borne β-lactamase promoter (Pbla-LuxR) and LacI expressed constitutively from the F′ episome of Escherichia coli strain TOP10F′ {F′ [lacIq Tn10(TetR)] mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG; Invitrogen}. Reporter plasmid PluxI-lacO-GFPmut2 was constructed by amplifying the PluxI promoter from plasmid pGFPuv (33) using primers PluxI-F(AatII), 5′-ACGGACGTCAGTCCTTTGATTCTAATAAATTGG-3′, and PluxI-lacO-R(HindIII), 5′-ATTAAGCTTGGAATTGTTATCCGCTCACAATTCCATTCGACTATAACAAACC, where the restriction digest sites are shown in bold and the wild-type lac operator is shown in italics. PluxI-lacO was then inserted into the AatII and HindIII restriction sites of plasmid PluxI-GFPmut2, completely replacing the wild-type PluxI promoter. Plasmid Pbla-LuxR was constructed by amplifying the Pbla promoter region from plasmid pUC19 using primers Pbla-F(XhoI), 5′-TATCTCGAGCAGGTGGCACTTTTCG-3′, and Pbla-R(EcoRI), 5′-GCGGAATTCTTTCAATATTATTGAAGC-3′, followed by insertion into plasmid pLuxR (33) using the XhoI and EcoRI restriction sites. Insertion of the Pbla promoter replaced the native Plac/ara-1 promoter of pLuxR. Construction of the Pbla-LuxR mutant plasmids was done in an identical manner starting with pLuxR plasmids containing the mutant proteins (33).
E. coli TOP10F′ strains were cotransformed, by electroporation, with the PluxI-lacO-GFPmut2 and the Pbla-LuxR or Pbla-LuxR mutant plasmids. Control experiments and measurement of the dose responses for the wild-type and mutant LuxR proteins used plasmid PluxI-GFPmut2 in place of PluxI-lacO- GFPmut2. For both the fluorimetry and flow cytometry measurements, cultures were grown overnight (∼15 h) from single colonies in LB medium at 37°C and 225 rpm with appropriate antibiotics for selection. Two-milliliter growth cultures were then inoculated 1:200 from the overnight cultures and induced with OHHL (3-oxohexanoyl-homoserine lactone) (0 to 3 μM) and IPTG (isopropyl-β-d-thiogalactopyranoside) (0 to 100 μM) upon reaching early exponential phase (optical density at 595 nm, ≈0.3). Following induction, cultures were grown for an additional 4 h, after which the cells were pelleted and resuspended in sterilized phosphate-buffered saline (PBS).
Fluorescence measurements.
After resuspension in PBS, 200-μl aliquots of each sample were transferred to clear-bottomed 96-well plates (Costar). Fluorescence was measured using a SPECTRAmax Gemini XS microplate spectrofluorometer (excitation, 481/emission, 507). Fluorescence values were corrected for differences in cell density using measurements of optical density at 595 nm (Bio-Tek μQuant Universal Microplate spectrophotometer). All measurements used to compare the responses of the PluxI and PluxI-lacO promoters were done on the same day. The dose-response curves for the wild-type and mutant LuxR proteins were from data collected on the same day and were representative of measurements done on multiple days. Measurements for the generation of surface plots for the mutant systems were repeated on multiple days, with an average day-to-day variation of less than 15%.
Flow cytometry.
Resuspended PBS samples were analyzed using a BD LSR II flow cytometer to measure the single-cell responses of the AND gate cultures. Twenty thousand events were collected for each sample using the low-flow setting. FCSExtract software (E. Glynn, Stowe's Institute for Medical Research) was used to convert flow cytometry standard data files to ASCII, and data analysis was done in Excel. The data were gated to a narrow range of forward- and side-scattering measurements to minimize fluorescence variability caused by debris and cell aggregates. Statistics were calculated using 10,000 events randomly selected from the gated populations.
Mathematical model.
A model describing the hybrid PluxI-lacO promoter was derived from mass balance equations detailing the interactions of PluxI-lacO with the LuxR and LacI transcriptional regulators. The model uses standard simplifying assumptions and has a structure similar to that of previous models based on the lac operon (35). Specifically, the following assumptions were made: (i) all reactions are at equilibrium, (ii) concentrations of LuxR and LacI are constant and independent of growth conditions, (iii) binding of LuxR and LacI to PluxI-lacO does not affect free protein concentrations, (iv) inducer concentrations are constant, and (v) LuxR and LacI bind PluxI-lacO competitively. Induction of LuxR by OHHL was assumed to follow Michaelis-Menten kinetics and has the following form: activator = [activator]/[LuxRtotal] = O/(1 + 2O), where O = [OHHL]/KA, and KA is the dissociation constant for OHHL binding to LuxR. Similarly, deactivation of LacI binding by IPTG has the following form: repressor = [LacI]/[LacItotal] = 1/(1 + I), where I = [IPTG]/KI. With these assumptions, the original set of differential equations can be simplified to the following (see also the supplemental material):
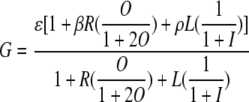 |
which describes the expression of the GFPmut2 protein (G) from PluxI-lacO with R = [LuxRtotal]/KBA and L = [LacItotal]/KBL. KBA and KBL are the dissociation constants for binding of PluxI-lacO by active LuxR and LacI, respectively. β (>1) and ρ (<1) are rate modifiers of the expression term, ɛ, for the activator- and repressor-bound states, respectively. Fitting of the model to the experimental data was done in MATLAB using the lsqcurvefit function, with a normalized root mean squared error for the best fit parameters of ∼20%. The effects of parameter variation on the responses of the model were determined by altering the best fit parameters between −25% and +25% of their initial values.
RESULTS
Design and measurement of the wild-type AND gate.
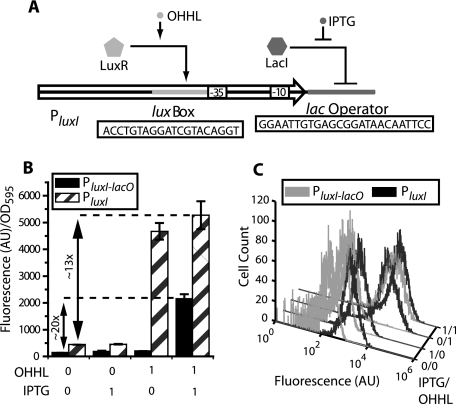
The design of our minimal AND gate focuses on altering the natural transcriptional regulatory properties of the PluxI promoter by adding a lac operator site (lacO) downstream of the native −10 site (Fig. 1A). The addition of the lac operator site results in combined transcriptional control of the hybrid PluxI-lacO promoter by LuxR, which activates transcription by binding to the lux box and recruiting RNA polymerase (RNAP) (22), and LacI, which binds to lacO and sterically prohibits binding by RNAP (34). The recruitment of RNAP by LuxR and the binding of LacI are predicted to be competitive, as studies have shown that the LuxR-RNAP complex extends ∼20 bp downstream of the PluxI promoter (37) and, thus, would overlap the lacO site in PluxI-lacO. The general design of this circuit has been proposed previously for the generation of AND logic (11) and closely resembles the natural lac operon of E. coli (25, 35).
FIG. 1.
Comparison of the hybrid PluxI-lacO promoter to PluxI. (A) Structure of the PluxI-lacO promoter. Activation occurs through binding of the LuxR-OHHL complex to the lux box while LacI binding to lacO represses transcription. (B) Promoter activities measured by expression of GFPmut2 in the presence and absence of 100 μM IPTG and 3 μM OHHL. (C) Single-cell responses of the promoters measured using flow cytometry. Inductions were performed as described for panel B. All fluorescence values are averages from four replicate cultures, with error bars representing standard deviations. AU, arbitrary units; OD595, optical density at 595 nm.
We explored the logical properties of PluxI-lacO by measuring the expression of a mutant of the green fluorescent protein (12) while controlling the activities of constitutively expressed LuxR and LacI through exogenous addition of OHHL and IPTG. LuxR is unable to bind to PluxI in the absence of OHHL, preventing transcriptional activation (37). Conversely, the affinity of LacI for the lacO site is reduced to background levels upon binding of IPTG (5). Therefore, the activity of PluxI-lacO is high in the presence of OHHL and IPTG and low when either inducer is absent (Fig. 1B). Comparison of PluxI-lacO to the native PluxI promoter shows that in addition to establishing AND logic, insertion of the lacO site also resulted in reduced total expression levels. Similar decreases in expression were also observed for cells lacking the lac repressor (data not shown), suggesting that the decreased expression resulted from altered rates of transcription from the hybrid promoter. Despite the decreased total expression level, the expression capacity, defined as the ratio between the active and inactive expression, was increased from ∼13-fold for PluxI to ∼20-fold for PluxI-lacO. Single-cell measurements confirmed the behaviors observed at the population level and showed distinct monomodal responses for each induction (Fig. 1C).
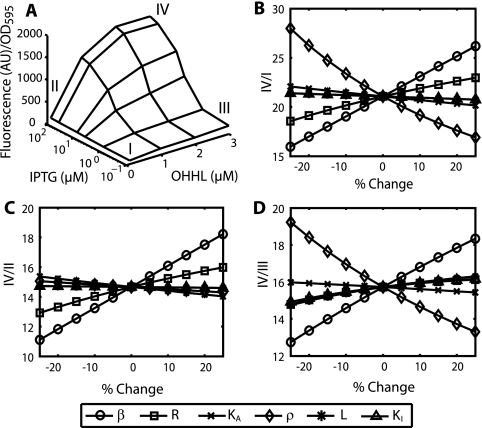
A more detailed characterization of the AND gate's responses was achieved by measuring the activation of the circuit at 24 different inducer combinations comprising four different OHHL concentrations ranging between 0 and 3 μM and six IPTG concentrations ranging between 0.1 and 100 μM (Fig. 2A). The response was dominated by a single activated plateau at high concentrations of OHHL and IPTG with an increase in expression of ∼20-fold. Higher inductions of OHHL or IPTG did not significantly increase expression. The AND gate differed in the form of its responses to induction with OHHL at set concentrations of IPTG and induction with IPTG at set OHHL concentrations. For IPTG, the response was sigmoidal (Hill coefficient of ∼1.5) with an activation threshold of ∼5 μM in the absence of OHHL. In comparison, induction with OHHL was hyperbolic (Hill coefficient of ∼1) with a measurable response occurring after 0.01 μM (see Fig. S1 in the supplemental material). Neither inducer showed a significant response in the absence of the second inducer with fluorescence levels increasing by only ∼2-fold (see Table S1 in the supplemental material). The effective activation thresholds of the AND gate decreased for induction with both IPTG and OHHL, but no significant change was observed in the shapes of the induction curves for different levels of coinduction.
FIG. 2.
Surface plot of AND gate (A) and alteration of logical properties through parameter variation (B to D). The AND gate is defined as the genetic circuit incorporating the hybrid PluxI-lacO promoter and the constitutively expressed LuxR and LacI proteins. The logical properties of the AND gate are characterized by comparing expression levels for the active state (IV) to those of each of the following inactive states: repressed basal expression (I, −IPTG/−OHHL), nonrepressed basal expression (II, +IPTG/−OHHL), and repressed active expression (III, −IPTG/+OHHL). AU, arbitrary units; OD595, optical density at 595 nm.
Prediction of critical parameters for the logical responses of the genetic AND gate.
The AND gate's responses were effectively captured by a steady-state model of the binding of LuxR and LacI to the PluxI-lacO promoter (see Materials and Methods). We used this model to explore the logical properties of the AND gate by breaking its response into three logical properties: the ratio of the activated to inactive response in the absence of both inducers and the ratios of the activated to inactive responses when each inducer is present individually (28). These states are defined in Fig. 2A. We then determined the influence of each of the model's parameters on these logical properties by varying each parameter individually from a set of best-fit values (Table 1). The results of this parameter variation are shown in Fig. 2B to D.
TABLE 1.
Best-fit parameter values for wild-type AND gate
| Parameter | Description | Value |
|---|---|---|
| ɛ | Expression rate | 131 |
| β | Expression rate modifier for active expression | 24 |
| R | Lumped LuxR term | 5 |
| KA (nM) | OHHL dissociation constant | 2,394 |
| ρ | Expression rate modifier for repressed expression | 0.66 |
| L | Lumped LacI term | 1,223 |
| KI (μM) | IPTG dissociation constant | 0.015 |
The parameter variation shows that the overall logic of the AND gate is most sensitive to the rate modifier for activated expression (β in the model) with increased transcriptional activation leading to the improvement of all three logical properties. The total levels of the LuxR protein also had positive effects on all three logical properties, though the effect on the logic between the active state and the repressed active state (IV/III) was small. In comparison to the properties of the LuxR protein, the model predicts that decreasing leaky expression from the repressed states (ρ in the model) by increasing the activity of LacI should result in substantial increases in logic between these states and the active state (IV/I and IV/III) but minimally affect the logic between the nonrepressed basal state and the active state (IV/II). The remaining parameters, describing the cellular levels of LacI and the inducer affinities of LuxR and LacI, were also shown to alter the logical properties of the AND gate, but with greatly reduced sensitivities. Using these results, we predicted that the overall logic of the AND gate could be most easily improved using LuxR mutants that showed increased rates of transcriptional activation from the PluxI promoter or that resulted in higher intracellular protein levels due to increased expression or stability.
Implementation of model predictions using LuxR mutant library.
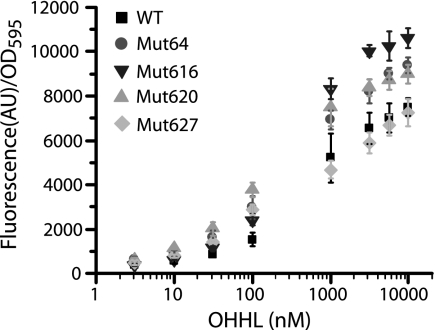
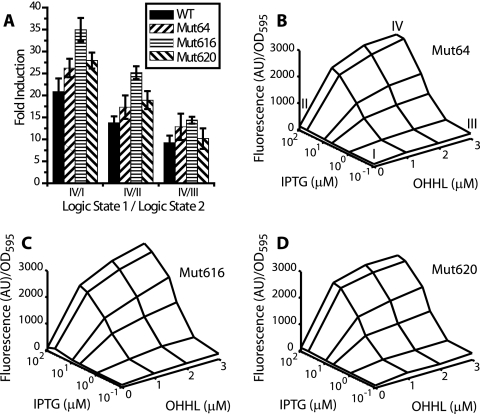
To test the model predictions, we used an existing library of four mutant LuxR proteins. These mutant proteins were generated through directed evolution and have been shown to increase the activities of LuxR at reduced concentrations of the OHHL signal molecule when incorporated into minimal positive feedback loops (33). Screening of the mutants in a simple transcriptional circuit where expression of GFPmut2 was controlled by a wild-type PluxI promoter confirmed the increased activities of the mutants at low OHHL concentrations and also showed that mut64, mut616, and mut620 increased the total expression levels from PluxI at saturating OHHL concentrations (Fig. 3). The fourth mutant, mut627, showed reduced activity compared to the wild type at saturating OHHL concentrations. Extending the definitions of the model to these simple systems, the increased expression levels from PluxI observed for mut64, mut616, and mut620 would be a result of increased transcriptional activation (β) by the mutant proteins in the limit of saturated binding to PluxI. Therefore, incorporation of any of these three mutants should result in improved logical responses for the AND gate. As predicted, the incorporation of mut64, mut616, and mut620 resulted in improved logical properties for the mutant AND gates compared to wild type (Fig. 4A). Incorporation of mut627 into the AND gate resulted in decreased logical properties (data not shown).
FIG. 3.
Dose-response curves of the wild-type (WT) and mutant LuxR proteins. Fluorescence values are averages from two replicate cultures, with error bars representing standard deviations. AU, arbitrary units; OD595, optical density at 595 nm.
FIG. 4.
Logical properties (A) and surface plots (B to D) of mutant AND gates. Error bars represent standard deviations for expression levels measured on two separate days. WT, wild type; AU, arbitrary units; OD595, optical density at 595 nm.
The surface plots for the AND gates incorporating mut64, mut616, and mut620 are shown in Fig. 4B to D. All of the mutant systems showed trends similar to that of the wild-type AND gate, retaining a hyperbolic response curve, but had increased expression levels for the OHHL-induced states (note different scale for wild-type and mutant axes). The degree of increased expression was dependent on IPTG induction, with larger relative increases occurring at IPTG concentrations above the activation threshold level. This resulted in an increased logical response between the repressed active (III) and active (IV) expression levels, as the active state had a larger relative increase in expression over the repressed active state (Fig. 4A; see also Table S1 in the supplemental material). No significant increases in expression were observed in the absence of OHHL, resulting in improved logical responses between the active (IV) and two inactive (I and II) basal expression states.
DISCUSSION
We have constructed a genetic circuit that functions as a logical AND gate using a hybrid PluxI-lacO promoter and constitutively expressed luxR and lacI genes. The circuit shows a clear logical AND response for induction with OHHL (which activates LuxR, causing expression from PluxI-lacO) and from IPTG (which inactivates LacI, thereby relieving repression of PluxI-lacO). The responses of our AND gate are well described by a simple steady-state mathematical model, which we used to explore the effects of each parameter on the logical properties of the AND gate. Using predictions from this analysis, we were able to improve the logical properties of the wild-type AND gate using a set of mutant LuxR proteins that resulted in increased expression levels from PluxI-lacO.
The logical nature of the AND gate is dependent on its ability to process signals and produce a clear “ON” response only when both inputs are present. As our design relies on the coupling of a repressor-activator pair, the main determinants of its logic are low basal expression from PluxI-lacO in the absence of activation by LuxR and strong repression of activated expression by LacI. The ability of LacI to repress activation of PluxI-lacO by LuxR is determined by interactions between LacI, LuxR, and RNAP. In the limit of independent recruitment of RNAP by LuxR and independent repression of RNAP by LacI, the response of the AND gate would be a simple product of the individual responses (11). Since the response curves for dual induction changed with induction level, as evidenced by the changing threshold levels and slopes of activation, our results indicate that binding of LuxR and recruitment of RNAP compete with the binding of LacI at PluxI-lacO. It is also seen that the binding of LuxR and LacI to PluxI-lacO is well balanced, as LacI is able to repress activation by LuxR even in the presence of saturating amounts of OHHL.
In obtaining improved logical properties through implementation of the LuxR mutants, we have shown that it is possible to rationally optimize the AND gate by altering the activity of one of its transcription factors. This is an important result, as optimization of the AND gate's logic requires increasing the relative expression level of the AND response while maintaining the balance of the two regulatory factors so that one regulator does not become dominant. This was achieved by increasing the activated expression levels from the PluxI-lacO promoter without significantly increasing expression of the inactive or repressed active expression states. In showing that the lac repressor was able to adequately repress the increased activities of the LuxR mutants, our results agree with those of previous studies that have shown that repression dominates activation at combinatorial promoters (9, 13).
Along with their logical properties, the differences between the responses of the genetic AND gates to OHHL and IPTG are important determinants for how the systems will behave when incorporated into genetic networks. In sigmoidal responses, such as those observed for the responses of the AND gates to IPTG, ultrasensitivity to the input allows for the filtering of noise in genetic networks (36) and can create bistability (15, 30). These properties are critical in the construction of artificial genetic networks because the individual components must be able to reliably process noisy inputs and produce specific outputs for the next component in the network. In comparison, hyperbolic or graded responses such as those observed for induction of the AND gates with OHHL are susceptible to noise, as small changes to the input can result in responses that are intermediate to the “on” or “off” states. These intermediate responses negatively impact the robustness of a network because large variations in the output of a component could result in the failure of other network components. Therefore, in comparison to our current AND gate, which shows only weak ultrasensitivity for induction with IPTG, an ideal AND gate would have highly ultrasensitive responses to both of its inputs. Despite this apparent limitation of our genetic AND gates, the effects of noise on the functionality of the gates are expected to be minimized by the clear distinctions between the logical states.
The improvement of the AND gate's logical properties was aided by our previous work with the LuxR protein, as it allowed us to use our small existing library of LuxR mutants to identify mutants that could increase the logical properties of the AND gate. In identifying these mutants, it is noted that the original directed evolution library was designed and screened for mutants with general increases in activity and not specific properties. The resulting diversity of the recovered mutants allowed us to rescreen them to select those with the desired property of increased activities in the presence of saturating OHHL concentrations and made it unnecessary to create a new mutant library. While it is predicted that the increased activities of the LuxR mutants, and the resulting increased logical properties of the AND gate, are largely a result of increased rates of transcriptional activation, the current classification of the mutant proteins does not allow for identification of the exact properties of each of the mutants. Therefore, it is possible that additional properties of the LuxR mutants, such as increased expression levels or increased affinity for OHHL, could contribute to the increased logical properties of the mutant AND gates.
The incorporation of the LuxR mutant proteins resulted in a maximum improvement of logic for the mutant AND gates of ∼1.5-fold compared to the wild type. Similar alterations of logic have been achieved in studies of the lac operon by introducing multiple mutations in the cis-regulatory region of the lac promoter (28). In comparison to the wild-type promoter, which exhibited a response that was partly OR-like, the mutated promoters displayed pure AND logic. The engineering of ribosome binding sites has also been used to improve the output of a logic AND gate (2), though this approach is unlikely to enhance the properties of a minimal AND gate, as both basal and maximal expression levels will be altered concomitantly. In comparison to these studies, which focus on the alteration of the cis-regulatory region, our results show, for the first time, that circuit function can be rapidly and rationally improved by using mutant regulators generated by directed evolution. The advantages of this approach are that relatively complex interactions, such as those that occur for LuxR in our genetic AND gate, can be altered rapidly without a complete knowledge of the individual interactions. The ability to rapidly optimize components to fit specified designed parameters will be of increasing importance as the complexity of artificial networks increases and each component is subject to an increasing number of interactions. Therefore, the method that we illustrate here will be of great utility in the creation and optimization of components that are to be incorporated into complex genetic circuits.
Finally, using combinatorial techniques, it has been shown that distinct cis operator regions can be joined to quickly form promoters with complex regulatory mechanisms and demonstrating AND or OR logic (13, 23). Components necessary for more diverse logical responses including NOR, XOR, and EQU have also been identified in a number of theoretical studies (11, 19). Combined with our results, it is apparent that the construction and optimization of a diverse array of logical genetic circuits could be achieved in a straightforward manner. Using this design principle, a number of AND gate devices can be constructed to simultaneously monitor various environmental cues, such as pH, osmolarity, temperature, and light. Moreover, thresholds of AND gates can be fine-tuned by engineering individual components with directed evolution to accommodate a specific application. The extension of these techniques to additional quorum-sensing promoters would be of particular interest, as the natural diversity and intercellular signaling of quorum-sensing systems would allow for the creation of complex multicomponent networks that function at the population level. Specificity in each of the individual components could be achieved using independent quorum-sensing systems (e.g., LuxI/R and RhlI/R) each coupled to a unique repressor. In addition, it has been shown that quorum-sensing activators can be converted into repressors by repositioning their binding sites to overlap the RNAP binding region (14). This flexibility would allow for the construction of novel logic circuits incorporating both activators and repressors that are capable of processing intercellular signals and help advance the construction of more complex circuits.
Supplementary Material
Acknowledgments
This work was supported by grant CBET-0747728 from the National Science Foundation and grant IRG 93-033 from the American Cancer Society.
Footnotes
Published ahead of print on 5 December 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alifano, P., C. B. Bruni, and M. S. Carlomagno. 1994. Control of messenger-RNA processing and decay in prokaryotes. Genetica 94:157-172. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, J. C., C. A. Voigt, and A. P. Arkin. 2007. Environmental signal integration by a modular AND gate. Mol. Syst. Biol. 3:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrianantoandro, E., S. Basu, D. K. Karig, and R. Weiss. 2006. Synthetic biology: new engineering rules for an emerging discipline. Mol. Syst. Biol. 2:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balagadde, F. K., H. Song, J. Ozaki, C. H. Collins, M. Barnet, F. H. Arnold, S. R. Quake, and L. You. 2008. A synthetic Escherichia coli predator-prey ecosystem. Mol. Syst. Biol. 4:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barkley, M. D., and S. Bourgeois. 1980. Repressor recognition of operator and effectors, p. 177-220. In J. H. Miller and W. S. Reznikoff (ed.), The operon. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 6.Barnard, A., A. Wolfe, and S. Busby. 2004. Regulation at complex bacterial promoters: how bacteria use different promoter organizations to produce different regulatory outcomes. Curr. Opin. Microbiol. 7:102-108. [DOI] [PubMed] [Google Scholar]
- 7.Basu, S., Y. Gerchman, C. H. Collins, F. H. Arnold, and R. Weiss. 2005. A synthetic multicellular system for programmed pattern formation. Nature 434:1130-1134. [DOI] [PubMed] [Google Scholar]
- 8.Basu, S., R. Mehreja, S. Thiberge, M. T. Chen, and R. Weiss. 2004. Spatiotemporal control of gene expression with pulse-generating networks. Proc. Natl. Acad. Sci. USA 101:6355-6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bintu, L., N. E. Buchler, H. G. Garcia, U. Gerland, T. Hwa, J. Kondev, T. Kuhlman, and R. Phillips. 2005. Transcriptional regulation by the numbers: applications. Curr. Opin. Genet. Dev. 15:125-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bronson, J. E., W. W. Mazur, and V. W. Cornish. 2008. Transcription factor logic using chemical complementation. Mol. Biosyst. 4:56-58. [DOI] [PubMed] [Google Scholar]
- 11.Buchler, N. E., U. Gerland, and T. Hwa. 2003. On schemes of combinatorial transcription logic. Proc. Natl. Acad. Sci. USA 100:5136-5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 13.Cox, R. S., M. G. Surette, and M. B. Elowitz. 2007. Programming gene expression with combinatorial promoters. Mol. Syst. Biol. 3:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Egland, K. A., and E. P. Greenberg. 2000. Conversion of the Vibrio fischeri transcriptional activator, LuxR, to a repressor. J. Bacteriol. 182:805-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrell, J. E., and W. Xiong. 2001. Bistability in cell signaling: how to make continuous processes discontinuous, and reversible processes irreversible. Chaos 11:227-236. [DOI] [PubMed] [Google Scholar]
- 16.Gardner, T. S., C. R. Cantor, and J. J. Collins. 2000. Construction of a genetic toggle switch in Escherichia coli. Nature 403:339-342. [DOI] [PubMed] [Google Scholar]
- 17.Guet, C. C., M. B. Elowitz, W. H. Hsing, and S. Leibler. 2002. Combinatorial synthesis of genetic networks. Science 296:1466-1470. [DOI] [PubMed] [Google Scholar]
- 18.Haseltine, E. L., and F. H. Arnold. 2007. Synthetic gene circuits: design with directed evolution. Annu. Rev. Biophys. Biomol. Struct. 36:1-19. [DOI] [PubMed] [Google Scholar]
- 19.Hermsen, R., S. Tans, and P. R. ten Wolde. 2006. Transcriptional regulation by competing transcription factor modules. PLoS Comput. Biol. 2:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isaacs, F. J., J. Hasty, C. R. Cantor, and J. J. Collins. 2003. Prediction and measurement of an autoregulatory genetic module. Proc. Natl. Acad. Sci. USA 100:7714-7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Istrail, S., and E. H. Davidson. 2005. Logic functions of the genomic cis-regulatory code. Proc. Natl. Acad. Sci. USA 102:4954-4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, D. C., A. Ishihama, and A. M. Stevens. 2003. Involvement of region 4 of the sigma(70) subunit of RNA polymerase in transcriptional activation of the lux operon during quorum sensing. FEMS Microbiol. Lett. 228:193-201. [DOI] [PubMed] [Google Scholar]
- 23.Kinkhabwala, A., and C. C. Guet. 2008. Uncovering cis regulatory codes using synthetic promoter shuffling. PLoS ONE 3:e2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi, H., M. Kaern, M. Araki, K. Chung, T. S. Gardner, C. R. Cantor, and J. J. Collins. 2004. Programmable cells: interfacing natural and engineered gene networks. Proc. Natl. Acad. Sci. USA 101:8414-8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhlman, T., Z. Zhang, M. H. Saier, Jr., and T. Hwa. 2007. Combinatorial transcriptional control of the lactose operon of Escherichia coli. Proc. Natl. Acad. Sci. USA 104:6043-6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mangan, S., and U. Alon. 2003. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. USA 100:11980-11985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mangan, S., A. Zaslaver, and U. Alon. 2003. The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. J. Mol. Biol. 334:197-204. [DOI] [PubMed] [Google Scholar]
- 28.Mayo, A. E., Y. Setty, S. Shavit, A. Zaslaver, and U. Alon. 2006. Plasticity of the cis-regulatory input function of a gene. PLoS Biol. 4:555-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukhopadhyay, A., R. Gao, and D. G. Lynn. 2004. Integrating input from multiple signals: the VirA/VirG two-component system of Agrobacterium tumefaciens. Chembiochem 5:1535-1542. [DOI] [PubMed] [Google Scholar]
- 30.Ozbudak, E. M., M. Thattai, H. N. Lim, B. I. Shraiman, and A. van Oudenaarden. 2004. Multistability in the lactose utilization network of Escherichia coli. Nature 427:737-740. [DOI] [PubMed] [Google Scholar]
- 31.Prehoda, K. E., J. A. Scott, R. D. Mullins, and W. A. Lim. 2000. Integration of multiple signals through cooperative regulation of the N-WASP-Arp2/3 complex. Science 290:801-806. [DOI] [PubMed] [Google Scholar]
- 32.Sayut, D. J., P. K. R. Kambam, and L. Sun. 2007. Engineering and applications of genetic switches. Mol. Biosyst. 3:835-840. [DOI] [PubMed] [Google Scholar]
- 33.Sayut, D. J., Y. Niu, and L. Sun. 2006. Construction and engineering of positive feedback loops. ACS Chem. Biol. 1:692-696. [DOI] [PubMed] [Google Scholar]
- 34.Schlax, P. J., M. W. Capp, and T. M. Record, Jr. 1995. Inhibition of transcription Initiation by lac repressor. J. Mol. Biol. 245:331-351. [DOI] [PubMed] [Google Scholar]
- 35.Setty, Y., A. E. Mayo, M. G. Surette, and U. Alon. 2003. Detailed map of a cis-regulatory input function. Proc. Natl. Acad. Sci. USA 100:7702-7707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thattai, M., and A. van Oudenaarden. 2002. Attenuation of noise in ultrasensitive signaling cascades. Biophys. J. 82:2943-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urbanowski, A. L., C. P. Lostroh, and E. P. Greenberg. 2004. Reversible acyl-homoserine lactone binding to purified Vibrio fischeri LuxR protein. J. Bacteriol. 186:631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wall, M. E., W. S. Hlavacek, and M. A. Savageau. 2004. Design of gene circuits: lessons from bacteria. Nature 5:34-42. [DOI] [PubMed] [Google Scholar]
- 39.Wassarman, K. M. 2002. Small RNAs in bacteria: diverse regulators of gene expression in response to environmental changes. Cell 109:141-144. [DOI] [PubMed] [Google Scholar]
- 40.Waters, C. M., and B. L. Bassler. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21:319-346. [DOI] [PubMed] [Google Scholar]
- 41.Yokobayashi, Y., R. Weiss, and F. H. Arnold. 2002. Directed evolution of a genetic circuit. Proc. Natl. Acad. Sci. USA 99:16587-16591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.You, L., R. S. Cox III, R. Weiss, and F. H. Arnold. 2004. Programmed population control by cell-cell communication and regulated killing. Nature 428:868-871. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.