Abstract
Here we describe a spectral imaging system for minimally invasive identification, localization, and relative quantification of pigments in cells and microbial communities. The modularity of the system allows pigment detection on spatial scales ranging from the single-cell level to regions whose areas are several tens of square centimeters. For pigment identification in vivo absorption and/or autofluorescence spectra are used as the analytical signals. Along with the hardware, which is easy to transport and simple to assemble and allows rapid measurement, we describe newly developed software that allows highly sensitive and pigment-specific analyses of the hyperspectral data. We also propose and describe a number of applications of the system for microbial ecology, including identification of pigments in living cells and high-spatial-resolution imaging of pigments and the associated phototrophic groups in complex microbial communities, such as photosynthetic endolithic biofilms, microbial mats, and intertidal sediments. This system provides new possibilities for studying the role of spatial organization of microorganisms in the ecological functioning of complex benthic microbial communities or for noninvasively monitoring changes in the spatial organization and/or composition of a microbial community in response to changing environmental factors.
Spectral imaging is a technique in which spectral information (i.e., the spectrum of light that is scattered from, transmitted through, or emitted by an object) is acquired at every location in an image. Since the spectral information reflects the object's identity, status, and/or composition, combining it with spatial information (i.e., the size, shape, and location of the object) enhances our ability to unravel and understand possible links between the spatial organization and functional relationships for constituents of a system. These attributes have made spectral imaging important in various areas of basic research and in industrial applications.
Generally, previously described methods concentrated either on very large scales (e.g., astronomy and satellite or airborne remote sensing of the Earth) or on very small scales (e.g., microscopic observations in medicine and microbiology). In the field of benthic microbial ecology, which is the focus of this paper, large-scale spectral imaging techniques generally aim to identify pigments or to quantify biomass concentrations in microbial communities spread over several meters to kilometers, such as intertidal flats (5, 6, 17, 20). Such techniques usually employ airborne imagers to detect reflected light in several tens of spectral bands covering visible and near-infrared regions. The spectral reflectance data are calibrated and validated by combining ground truth measurements of the parameters of interest (e.g., pigment content) with the spectral reflectance measurements obtained using single-point spectrometers, which detect the signals from regions whose areas are several square centimeters to several square decimeters. Qualitative analysis of pigments involves the fourth derivative of the reflectance spectra with respect to wavelength (5, 6), whereas for pigment quantification various approaches are employed, ranging from simple indices based on reflectances in specific bands (15, 36, 44, 51, 53, 54, 70, 75) to more sophisticated algorithms using a modified Gaussian model approach (9, 17). Although not directly an imaging method, the fluorometer described by Aberle et al. (2) can also be used for large-scale (centimeter or meter) quantification of microphytobenthos biomass and composition. This device allows discrimination between different groups of microalgae (e.g., diatoms, green algae, and cyanobacteria) based on measurement of pigment autofluorescence that is excited sequentially by several different wavelengths.
Rather than absolute quantification of the pigment content, microscale spectral imaging typically is used for noninvasively identifying pigments in single cells, for identifying cells based on their spectral fingerprint, or for determining the microscopic spatial organization of cells in mixed microbial communities (48, 52, 67, 77). Furthermore, Ainsworth et al. (3) demonstrated that spectral imaging can be used to specifically remove unwanted background autofluorescence so that selected bacterial communities stained by specific fluorescence in situ hybridization probes can be detected. Microscale spectral observation usually is performed with a confocal laser scanning microscope equipped with a spectrally resolved detector that allows three-dimensional imaging with cellular resolution (3, 48, 67), although standard white-light or epifluorescence microscopes have also been used (52, 77).
Microbial communities associated with surfaces, such as biofilms or microbial mats, are typically limited by the rate of transport of chemical substrates in the direction perpendicular to the surface, either through the diffusive boundary layer above the surface or through the community itself (39). This mass transport limitation plays a fundamental role in the way that the community as a whole functions and is spatially organized. As revealed by many confocal microscopy studies, the microscopic spatial organization of cells in such communities is complex and diverse; for example, migrating or nonmotile cells are loosely associated with each other or form tight multicellular consortia (13, 34, 49, 50). However, many microbial communities also exhibit distinctive spatial organization on a mesoscopic level, where the community structure changes from one dominant species or group to another dominant species or group over distances many times greater than a typical cell size. This patchiness occurs in both vertical and horizontal directions. An illustrative example is the pronounced vertical layering in photosynthetic microbial mats with, e.g., a zone dominated by oxygenic phototrophs at the top, anoxygenic phototrophs beneath this layer, sulfide oxidizers in a zone where oxygen and sulfide overlap, and sulfate reducers primarily at the bottom (73, 79). The spatial scale on which this structural heterogeneity occurs depends on the cell density and activity, but it most crucially depends on the fluxes of available substrates that supply the community with energy, electron donors and acceptors, and nutrients. In photosynthetic mats, distinct layers can be several tens or hundreds of micrometers (37, 45) to several decimeters (78) thick, which is the scale at which the relevant substrates on which a particular community member or group depends (e.g., light, oxygen, and sulfide) varies significantly, as revealed by microsensor measurements (23, 42, 81).
To quantify this zonation or, in general, the three-dimensional patchiness of a microbial community on a mesoscale, the community is typically sectioned horizontally, and the pigments are extracted and quantified by high-performance liquid chromatography (HPLC). Additionally, spectroscopic measurements to quantify pigments in thin sections have also been described (4, 59, 60, 76). Although useful insights into the community structure have been obtained by using this sample-sectioning approach, the major disadvantages of the technique are that samples are destroyed, the native spatial organization of the microbial community is disturbed during fixation and preparation, and the spatial resolution is limited. Less invasive, higher-resolution techniques are necessary to adequately describe small-scale heterogeneity in microbial communities. Spectral imaging systems with a spatial resolution that could be adjusted to the relevant scale would clearly be a valuable alternative. This has been achieved to some extent with confocal laser scanning microscopy (34, 50, 55), but such studies require hardware that is expensive and not portable.
The aim of the present work was to develop a measuring system that allows spectral imaging on spatial scales ranging from a few tens of micrometers to several tens of centimeters and thus facilitates minimally invasive identification and localization of pigments from the single-cell level up to the microbial community level. The main requirements for the system were portability, flexibility, simplicity of use, and reliance on commercially available and relatively low-cost components. Below we provide a detailed description of our system's hardware and software and explain typical measurement procedures. We also suggest and describe, using examples, a number of applications for the system in microbial ecology. Specifically, we used spectral imaging with single-cell resolution to identify pigment contents of cyanobacterial and algal cells originating from cultures grown in the laboratory and from a natural endolithic photosynthetic biofilm. Furthermore, we used mesoscale spectral imaging to determine the spatial organization of phototrophic groups in a natural endolithic biofilm and a laboratory-grown hypersaline microbial mat and combined this technique with oxygen imaging to investigate community structure and metabolic activity in intertidal permeable sediments.
MATERIALS AND METHODS
Hardware components and measurement configurations.
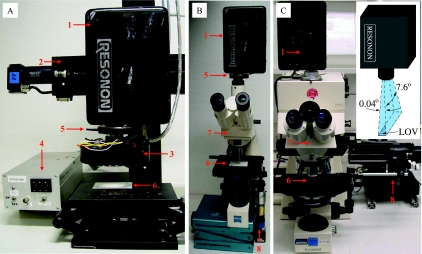
The modular spectral imaging (MOSI) system was constructed using a combination of commercially available and self-made components (Fig. 1). Spectral imaging was performed with a fire-wire hyperspectral (HS) camera (Pika II; Resonon, United States), which allowed spectral detection in 640 discrete bands with the same bandwidth (∼0.7 nm) over the wavelength range from 460 to 913 nm. The spectra were measured using a line of 480 pixels, for which we use the term “line of view” (LOV), instead of the more common “field of view” (Fig. 1C, inset). Two-dimensional spectral imaging was performed by scanning the LOV over the sample at a constant velocity. Thus, each pixel in the HS data set, referred to as the “hypercube,” had a spectral dimension in addition to two spatial dimensions.
FIG. 1.
MOSI system, showing the arrangement of the components during (A) mesoscopic and (B and C) microscopic measurements. 1, HS camera; 2, motorized linear stage; 3, LEDs with collimating optics and short-pass excitation filters; 4, LED power supply; 5, holder for the long-pass emission filter; 6, substrate for the sample; 7, microscope; 8, motor for microscope table movement. (Inset) Orientation and angular divergence (not to scale) of the LOV imaged by the camera.
For imaging of spectral reflectance and transmission, an array of light-emitting diodes (LEDs), a bare halogen bulb (Philips), or a halogen lamp with a fiber optic light guide (KL-2500; Schott) was used for sample illumination in the visible to near-infrared range (visible light, 400 to 700 nm; near-infrared light, 700 to 1,000 nm) (Table 1). For imaging of autofluorescence, high-power LEDs (Lumileds, United States) that emitted blue, green, and amber light and were equipped with 5° collimating optics (LXHL-NX05-5; Lumileds) were used as narrow-band excitation sources (Table 1). Additionally, a combination of a low-pass optical filter placed in front of the LEDs and a complementary high-pass optical filter placed in front of the HS camera (Table 1) was used to remove the longer-wavelength LED emission and thus allowed sensitive detection of autofluorescence at wavelengths greater than the cutoff wavelength (λc). LEDs were powered by a self-made power supply that provided direct current in the range from 1 to 999 mA. Alternatively, a mercury lamp (HBO 50; Zeiss) and a combination of low-pass and high-pass optical filters (Table 1) were used when autofluorescence was imaged with an epifluorescence microscope (see below).
TABLE 1.
Combinations of light sources and excitation and emission filters used for spectral imaging of reflectance, transmission, and autofluorescence of pigments in microbial communities
| Imaging | Light source
|
Excitation/emission filter
|
||||
|---|---|---|---|---|---|---|
| LED(s), lamp, or bulb | Supplier or type | Emission range or λmax (nm) | Filter | Supplier | λc (nm) | |
| Reflectance | LXHL-MWGC + ELD-740-524 | Lumileds + Roithner Lasertechnik | 450-720 (visible) + 710-770 (near infrared) | |||
| Halogen bulb | Philips | 400-1,000 (visible plus near infrared) | ||||
| Halogen lamp | Schott KL-2500 | 400-1,000 (visible plus near infrared) | ||||
| Transmission | Halogen lamp | Schott KL-2500 | 400-1,000 (visible plus near infrared) | |||
| Autofluorescence | LXHL-LR5C | Lumileds | 455 (blue) | 455DF70/510ALP | Omega Optical | 510 |
| Autofluorescence | LXHL-LM3C | Lumileds | 530 (green) | DC-Red/R-61 | Linos Photonics | 610 |
| LXHL-LL3C | Lumileds | 590 (amber) | ||||
| Autofluorescence | HBO 50 lamp | Zeiss (part of Axiophot microscope) | 450-490 (blue) | 450-490/LP520 | Zeiss (part of Axiophot microscope) | 520 |
| HBO 50 lamp | Zeiss (part of Axiophot microscope) | 535-555 (green) | BP546/LP590 | Zeiss (part of Axiophot microscope) | 590 | |
The system allowed spectral imaging on variable spatial scales, and the imaged areas were several centimeters (macroscopic), several millimeters (mesoscopic), and several tens of micrometers (microscopic) wide. The corresponding maximum theoretical resolution was equal to the LOV width divided by 480 (the number of pixels in the LOV), although the actual resolution was lower due to optical aberrations. The depth of focus depended on the imaging scale and ranged from a few micrometers in microscopic imaging to 1 to 2 millimeters in meso- and macroscopic imaging.
Macroscopic reflectance imaging and autofluorescence imaging were performed using a standard objective lens (Tamron 85089; 25 mm; 1:1.6), while an additional buffer ring (thickness, 10 mm) placed between the lens and the camera was used for mesoscopic imaging. The camera was attached to a high-precision motorized stage (VT-80; Micos, Germany), which was mounted on a stable stand, and the sample was placed ∼30 and ∼5 cm in front of the objective during macro- and mesoscopic imaging, respectively (Fig. 1A). The light source was fixed next to the objective lens to provide illumination through the space between the sample and the objective lens in these imaging modes. The angle of illumination was approximately 10° to 45° with respect to the direction of observation.
Two alternative configurations, which differed in performance and cost, were employed for microscopic spectral imaging. In the first configuration, which was used for reflectance and autofluorescence imaging in the field, the HS camera was mounted on an Axiolab microscope (Zeiss). A motorized axis (3564 K024 BC; Faulhaber, Switzerland) equipped with a 308:1 gear box (321 308:1; Faulhaber) was attached to the knob of the microscope stage to control its x-axis movement during sample scanning (Fig. 1B). The use of a long-distance objective lens (LD ACHROPLAN 20×/0.4; Zeiss) allowed direct illumination of the sample from behind the objective at an angle of ∼45°. In the second configuration, which was used for transmission and autofluorescence imaging in the lab, the HS camera was mounted on an Axiophot epifluorescence microscope (Zeiss). An image of the sample was obtained by using a ×40 or ×100 objective lens (Plan-NEOFLUAR 40×/0.75 or Plan-APOCHROMAT 100×/1.4 Oil; Zeiss) while the sample was illuminated from below during transmission imaging and from above (through the objective lens) during autofluorescence imaging (Table 1). The microscope stage was connected to a motorized positioner (VT-80 Pollux drive; Micos), whose computer-controlled movement was transduced directly to the x-axis movement of the sample during scanning (Fig. 1C).
During reflectance imaging, the sample was placed on a white standard substrate (Spectralon; Labsphere, United States), whereas transparent microscope slides with a coverslip were used for transmission imaging. In both modes, the scan covered both the sample and the white sample-free reference area, and the data set acquired was stored in one hypercube file. During meso- and macroscopic autofluorescence imaging, the sample was placed on a flat, black substrate to minimize reflection of the excitation light from the substrate. Samples on a microscope slide covered with a coverslip were employed for microscopic autofluorescence imaging.
Measurement procedure.
Spectral scanning was performed under conditions that excluded ambient light to avoid variable illumination. This was especially necessary during autofluorescence imaging due to the low intensity of the fluorescence compared to the ambient light levels. After the system was assembled in the desired configuration (i.e., for macro-, meso-, or microscopic scanning of spectral reflectance, transmission, or autofluorescence), a typical measurement involved the following steps. First, the sample was illuminated with the appropriate light source (Table 1) and moved so that the LOV covered a spatial feature with distinct spectral properties (e.g., an edge of the cell or a sediment grain). Then the LOV was continuously displayed, and the frame rate, gain, and shutter duration for the HS camera were adjusted interactively to ensure that the brightest possible image which was not saturated was obtained. After this, the LOV was focused, either by refocusing the objective lens or, preferably, by slowly moving the sample toward or away from the objective lens while observing the level of sharpness of the spatial feature seen in the LOV. After focusing, the motor was moved to the initial location of the scan, with a final check for signal oversaturation.
Then the sample was scanned using a constant velocity of the LOV relative to the sample. For an image acquisition frame rate of 3.75 frames per second, which was used in our measurements, the typical velocities were 1 to 2, 10 to 20, and 50 to 200 μm s−1 during the micro-, meso-, and macroscopic scans, respectively. Depending on the velocity and the scan range, measurement took from several tens of seconds to a few minutes, which resulted in a 50- to 200-MB hypercube. For autofluorescence scanning, the sample was illuminated with the excitation light for a few minutes prior to scanning in order to ensure that the variable fluorescence of pigments associated with reaction centers of the photosynthetic apparatus had reached a steady state (26, 27) and thus would not play a role when the fluorescence intensities in different parts of the same sample were compared.
The spectral scanning was automated by using self-made software written in Borland C++ and running on computers using Microsoft Windows. The software employed a server-client approach, in which a core client application (HSscan) used the TCP/IP protocol to send simple text-based commands to server applications controlling the motor movement (LINPOSserver) and image acquisition (FireView, supplied with the HS camera by Resonon). The nonproprietary software can be downloaded for free from the Max Planck Institute website (http://www.mpi-bremen.de/Lubos_Polerecky.html), and its basic functions are described in the supplemental material.
HS data analysis.
In this work, the spectral reflectance of a sample [R(λ)] was defined as the ratio of the intensity of light reflected from the sample placed on a perfect diffusive reflector substrate (such as the Spectralon white standard) to the intensity of light reflected from the bare substrate. Similarly, the spectral transmission of a sample [T(λ)] was defined as the ratio of the intensity of the light transmitted through the sample placed on a transparent substrate to the intensity of the light transmitted through the bare substrate. Because of normalization to the illuminating light spectrum, the mathematical methods for obtaining the information about the pigment content are equivalent for the spectral reflectance and the spectral transmission, and therefore only the method for spectral reflectance is described below. For microscopic spectral imaging of single cells, where the cells could be considered weak absorbers, the fourth derivative of spectral reflectance with respect to wavelength λ, designated R(4)(λ) (equivalent to d4R(λ)/dλ4) and referred to as the “fourth spectral derivative,” was used to determine the presence or absence of a pigment characterized by an absorption peak centered at λc, i.e., [pigment] ≈ R(4)(λc). This approach is commonly used in spectroscopy (14, 24) and is based on the reasoning described in the supplemental material. For sample images obtained on a meso- or macroscale, the exponential decay of the intensity of light propagating through the sample, governed by the Beer-Lambert law, was also taken into account. Thus, the approximate pigment content in such samples was determined by determining the fourth spectral derivative of the logarithm of reflectance, designated [log R](4)(λ) { d4log[R](λ)/dλ4}, i.e., [pigment] ≈ [log R](4)(λc) (see the supplemental material).
For interpretation of the spectral images of autofluorescence, we assumed that the concentration of a pigment characterized by an emission peak centered at λc was directly proportional to the peak intensity. Since the presence of more pigments (or different forms of pigments) in the sample generally resulted in more peaks in the emission spectra, the contribution of each pigment was determined by linear decomposition of the measured emission spectra into a sum of peaks and by considering the magnitude of each separate peak the value for the associated pigment content.
Because the strategies used to identify, localize, and quantify pigments depended on the approach used for measurement (reflectance or transmission imaging versus fluorescence imaging), were different for different community types, and were based on theoretical approaches that are typically not included in freely available software tools, new software called HS_ImAn was written in Borland C++ to facilitate HS data analysis. The basic new features offered by this software were (i) the ability to define a reference region in the HS image, in which the spectra were averaged for the vertical dimension and used as reference spectra separately for each vertical row of pixels, thus correcting for uneven sample illumination; (ii) the ability to display the reflectance [R(λc)], its logarithm [log R(λc)], and the fourth spectral derivatives {R(4)(λc) and [log R](4)(λc)} calculated from the smoothed reflectance spectrum [R(λ)] around any specific wavelength (λc) in every pixel of the image and to export this information as an eight-bit image; and (iii) the ability to convert the reflectance hypercube [R(x,y,λ)] into a hypercube of its fourth spectral derivative [R(4)(x,y,λ)]. Furthermore, the software provided functions common in other, commercially or freely available software tools, such as (i) the ability to average spectra for a selected region of interest and (ii) the ability to linearly decompose spectra in each pixel into a sum of predefined (commonly called “end member”) spectra, which was used for spectral pixel classification or pixelwise deconvolution of fluorescence emission spectra into a sum of emission peaks. The HS_ImAn software runs on Microsoft Windows computers and can be downloaded free from the Max Planck Institute website (http://www.mpi-bremen.de/Lubos_Polerecky.html), and a more detailed description of it is provided in the supplemental material.
Identification of pigments in living cells by microscopic spectral imaging.
The first application of the MOSI system involved microscopic spectral imaging of cells. A set of samples was prepared by pipetting a small drop of cyanobacterial and algal cells grown in a culture onto microscope slides. The cyanobacteria used were (i) the red filamentous organism Leptolyngbya sp. strain PCC 8103, (ii) the green filamentous Leptolyngbya-related morphotype M1C10, and (iii) the yellowish green spherical Rhabdoderma-related morphotype M7R1. The first organism was purchased from the Pasteur collection of cyanobacteria (Paris), and the other two organisms were isolated from microbial mats on intertidal flats in the Arabian Gulf (Abu Dhabi, United Arab Emirates) (1). In the present study, all organisms were maintained at 25°C with a light-dark cycle consisting of 12 h of light and 12 h of darkness (light intensity, 25 μmol photons m−2 s−1) in ASN-III B12 medium (66). The oval-shaped green algae used (species not determined) originated from Riou Mort (France) and were grown at 15°C with a light-dark cycle consisting of 12 h of light and 12 h of darkness (light intensity, 100 μmol photons m−2 s−1) in modified Bold's basal medium (74). The diatoms used (Cylindrotheca closterium CCMP1855; United States) were grown at 15°C with a light-dark cycle consisting of 12 h of light and 12 h of darkness (light intensity, ∼100 μmol photons m−2 s−1) in f/2 medium (28, 29) with additional silicate. Transmission and autofluorescence spectra for the samples were obtained immediately after sample preparation, using an Axiophot microscope and a ×40 or ×100 objective lens. The spectra were averaged for 20 to 50 pixels associated with a single cell and used to determine the cellular pigment content. For all samples, the fourth spectral derivative of transmission at wavelengths corresponding to absorption maxima of the major pigments in the cells and the magnitude of the autofluorescence peak were used to visualize the cells.
The second sample was prepared and imaged during a field trip to Westerhöfer Creek (the Harz, Germany) in August 2006. The surface of the endolithic biofilm that formed on top of the tufa substrate in the creek was scratched off with a scalpel and diluted with creek water, and the cell suspension was pipetted onto a microscope slide. The sample was covered with a coverslip and placed on a white reference substrate, and the spectral reflectance was imaged with an Axiolab microscope using a ×20 objective lens. The spectra were averaged for 20 to 30 pixels for one cell or filament and used to determine the cellular pigment content. Additionally, the spectra were used as end members to classify the cells in the image.
Mesoscopic spectral imaging of mixed microbial communities.
The second application involved high-spatial-resolution mesoscopic imaging of pigments and the associated phototrophic groups in two benthic photosynthetic systems, an endolithic biofilm growing on tufa in Westerhöfer Creek and a hypersaline microbial mat. The tufa was gently broken so that a vertical section was reasonably flat and contained distinct colored regions. It was then placed in a petri dish, submersed in creek water, and illuminated with a combination of white and near-infrared LEDs (Table 1), and the reflectance spectra for the vertical section were imaged from a distance of ∼5 cm.
The photosynthetic microbial mat was maintained in a flowthrough mesocosm with continuous in- and outflow of hypersaline water. The mesocosm system was inoculated with mat samples obtained from hypersaline Lake Chiprana in Spain (37) and grown for 3 years in water with a salt composition similar to that of the natural lake water (seawater plus 80 g/liter MgSO4 · 7H2O). The mat was illuminated with light at an intensity of 500 μmol photons m−2 s−1 using a light regimen of 16 h of light and 8 h of darkness during incubation. An approximately 1-mm-thick 1- by 1-cm vertical section of the mat was cut with a scalpel, placed on a microscope slide, and covered with a small drop of the hypersaline water. The sample was then covered with a coverslip, which was gently pressed to ensure that the surface was flat and there was good optical contact between the mat and the coverslip. Spectral images were acquired from a distance of ∼5 cm using visible and near-infrared illumination for measurement of the reflectance and blue and amber illumination together with the appropriate filters for measurement of the autofluorescence (Table 1).
The pigment distribution was visualized using the fourth spectral derivative of log reflectance at wavelengths corresponding to the absorption maxima of the major pigments identified in the samples. Additionally, the fluorescence spectra acquired from the mat were linearly decomposed, and the magnitude of the peaks corresponding to selected pigments was also used to visualize the pigment distribution.
Macroscopic spectral imaging combined with measurement of the OCR.
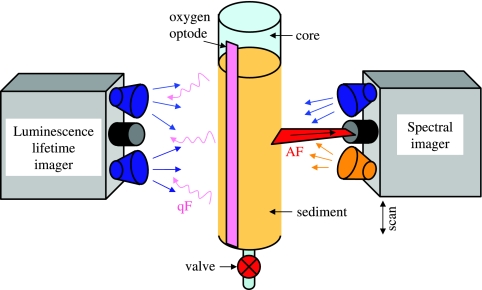
In the third application, we combined the spectral imaging method with the flowthrough method based on oxygen imaging (62) to obtain parallel measurements of pigment autofluorescence and potential oxygen consumption rate (OCR) for the same sampling core. A narrow stripe of a semitransparent planar oxygen optode (63) was attached to the inner wall of a round polycarbonate core (inside diameter, ∼3.5 cm) with water-resistant tape. Such cores were used to sample the permeable sediment from the intertidal sandflat at Janssand in the German North Sea at different mid-flat and upper-flat locations (11) in December 2006. Measurement was begun within 2 h after collection at the in situ temperature.
The core was fixed vertically on a stand, and spectral images of autofluorescence were acquired through the transparent wall of the core (Fig. 2). Two sets of scans were obtained, one from the top to the bottom and the other in the opposite direction. Each scan was done separately using blue and amber excitation (intensity, ∼100 μmol photon m−2 s−1) and the corresponding optical filters (Table 1). The scans started ∼3 min after the excitation light was switched on and covered a sediment depth of ∼8 cm at a motor speed of 80 μm s−1. The emission spectra were spectrally decomposed, and the magnitude of the chlorophyll a (Chl a) fluorescence peak at 675 nm was assumed to represent the relative Chl a concentration in each pixel of the image. The horizontal values for the images were averaged to obtain vertical profiles of relative Chl a concentrations.
FIG. 2.
Schematic diagram of a setup used for combined measurement of pigments and potential OCRs in a single sediment core. AF, autofluorescence of pigments from the microorganisms in the sediment; qF, quenchable fluorescence of the O2-sensitive fluorophore, which is detected by the luminescence lifetime imaging system (33).
To assess whether the Chl a measured in the step described above was part of a functional photosynthetic reaction center or was from damaged or dead cells, spectrally resolved kinetics of the variable fluorescence induced by blue actinic illumination were measured at three depths (0.4, 4, and 6 cm) below the sediment surface. First, the LOV of the HS camera, oriented horizontally, was positioned at a specific depth (Fig. 2), and the sediment was dark adapted for ∼15 min. Then spectral frames were recorded at a rate of 3.75 frames per second for 3 min in the dark during the first ∼5 s and then with actinic illumination (intensity, ∼300 μmol photons m−2 s−1) during the remaining time. Thus, the acquired hypercube had a spatial component representing the autofluorescence intensity along a horizontal line of pixels at a specific sediment depth and a temporal component representing the fluorescence kinetics in each pixel on the line. The emission spectra were spectrally decomposed, and the magnitudes of the Chl a fluorescence peak at 675 nm were normalized by defining the average peak magnitude measured during the first 2 s after the actinic illumination was switched on as 1, which was done for every pixel on the line. These normalized fluorescence kinetics were plotted as images (x axis, time; y axis, horizontal position). They were also horizontally averaged to obtain the fluorescence kinetics at each sediment depth.
After spectral imaging, the potential aerobic respiration rate of the microorganisms in the same sediment core (on the opposite side) (Fig. 2) was determined by the flowthrough method (62). Aerated in situ seawater was percolated through the sediment until oxic conditions were obtained throughout the core (2 to 3 min), which was checked by continuous oxygen imaging. Then the percolation was stopped, and O2 concentration images were recorded every 60 s for ∼30 min. Two-dimensional maps of the OCR throughout the core were calculated by using the initial rates of the decrease in the O2 level after the percolation was stopped, as previously described (62). Finally, the OCRs were horizontally averaged and plotted as vertical profiles.
To check for possible effects of the percolation on the transport of pigments through the sediment, spectral scanning was conducted as described above after OCR measurement. Also, to obtain an independent measure of the variability of Chl a fluorescence, a selected sediment core was, after the second spectral scan, sectioned at 1-cm intervals and the dark-adapted photosynthesis yields (Fv/Fm [Fv is the maximum variable fluorescence yield, and Fm is the maximum fluorescence yield]) were determined for homogenized sediment samples using a pulse amplitude modulated fluorometer (Diving-PAM; Walz, Germany).
RESULTS
Identification of pigments in living cells by microscopic spectral imaging.
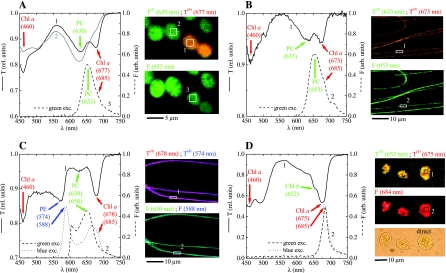
The transmission and autofluorescence spectra for single cells were clearly resolved when the Axiophot microscope was used (Fig. 3, graphs). The spectra obtained for different pixels for the same cell or for different cells of the same species in the same physiological state were very similar (data not shown), and thus only the average spectra are discussed below. The transmission and emission spectra of the green cyanobacteria (morphotypes M1C10 and M7R1) were similar (Fig. 3A and B). Valleys in the transmission spectrum around 675 and 460 nm indicated the presence of Chl a, whereas phycocyanin (PC) was detected by the presence of its characteristic absorption maximum around 630 nm (35). The emission spectra had a pronounced peak at 653 nm and a shoulder around 685 nm, which were due to PC and Chl a, respectively (32, 83). In the red filamentous cyanobacteria (Leptolyngbya sp. strain PCC 8103) phycoerythrin (PE) was the main accessory pigment, as indicated by the pronounced valley around 574 nm in the transmission spectrum (Fig. 3C) (25, 83). Chl a and PC were also detected in the filaments, as shown by the valleys around 678 and 630 nm, respectively. Compared to the spectrum for the green cyanobacteria, however, the valley around 630 nm for the red cyanobacteria was shallower than that around 678 nm (compare Fig. 3A and B with Fig. 3C), suggesting that the red cyanobacteria had a much lower PC/Chl a ratio than the green cyanobacteria. The presence of PE and PC in Leptolyngbya sp. strain PCC 8103 was also confirmed by the emission peaks around 588 and 650 nm when it was excited by blue and green light, respectively. Furthermore, when blue excitation was used, a shoulder in the emission spectrum due to Chl a was also observed around 685 nm (Fig. 3C). The major pigments detected in the green algal cells were Chl a and Chl b, as indicated by the valleys in the transmission spectra around 460 and 675 nm and around 652 nm, respectively (Fig. 3D) (31, 35). The presence of Chl a was confirmed by the emission peak at 685 nm when either blue or green excitation was used.
FIG. 3.
Spectral imaging of cyanobacterial cells, including (A) Rhabdoderma-related morphotype M7R1, (B) Leptolyngbya-related morphotype M1C10, and (C) Leptolyngbya sp. strain PCC 8103, and of a green alga (D). The numbered lines in the graphs show the transmission (solid lines) and autofluorescence (dashed and dotted lines) spectra, which were obtained as averages for the corresponding areas indicated in the images on the right (pixels not belonging to the filament were not included). The arrows with corresponding central wavelengths (in nanometers) (in parentheses) indicate selected spectral features of pigments identified in the cells. The false-color images, with the cell morphology outlined, were obtained by assigning the intensities of the red, green, and blue channels to the fourth derivative of transmission [T(4)] (upper panels) and the magnitude of autofluorescence [F] (bottom panels) at the wavelengths indicated. The direct image of algal cells in panel D, in which the pigment-containing cell organelles can be discerned, is included for comparison. The spectral scans in panels A to C and in panel D were obtained using ×40 and ×100 objective lenses, respectively. The characteristics of the excitation light used are shown in Table 1. Note the different scales of the images, as indicated by the scale bars. rel., relative; exc., excitation; arb., arbitrary.
The cell morphology was clearly visible in the images obtained from the hypercubes using both the fourth-derivative approach (transmission imaging) and the autofluorescence magnitude approach (autofluorescence imaging). The oval-shaped strain M7R1 cyanobacteria were relatively large (∼5 μm), and each cell covered several hundred pixels (Fig. 3A). Single cells were well resolved even when they were close to each other. The distribution of T(4)(630) and T(4)(677) suggested that some cells had a decreased Chl a content compared to the PC content. This was clearly observed in the transmission spectra, where the valley at 677 nm was barely visible for the green cells, whereas it was pronounced for the orange cells (Fig. 3A, compare spectra 1 and 2). This difference suggested that, although cells originate from the same axenic culture, the physiological status of individual cells could differ. Spatial resolution of the red and green filamentous cyanobacteria was also possible, although their diameter (∼1 μm) covered only 3 or 4 pixels when the cells were observed with a ×40 objective lens (Fig. 3B and C). The green algal cells covered several hundred pixels, and when the ×100 objective lens was used, chloroplasts could be discerned in both fourth-derivative transmission and autofluorescence images as heterogeneities in the cellular pigment distribution (compare the pigment images with the direct image of the cells in Fig. 3D).
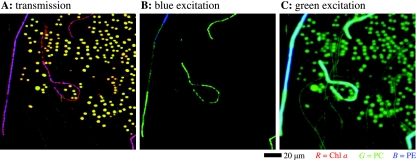
When a mixture of the three cyanobacterial species shown in Fig. 3 was scanned using a ×40 lens, the resulting images obtained from the hypercubes demonstrated that the cellular morphology and pigment content can be well resolved even for a complex mixture of cells (Fig. 4). The spectral signal for Chl a and PC from the green filaments (M1C10) was relatively weak compared to that from the M7R1 cocci, as indicated by the darker yellow and green colors of the filaments shown in Fig. 4A and C, respectively. This may have been related to the larger size of the cocci, but it also could have indicated an overall lower pigment content in the filaments. The variation in the false color of the red filamentous organism Leptolyngbya sp. strain PCC 8103 between red and purple (Fig. 4A) and green and cyan (Fig. 4B and C) suggested that there may have been heterogeneities in the PE/Chl a and PE/PC pigment ratios along the filaments, respectively.
FIG. 4.
False-color images of a mixture of cyanobacterial cells, derived from the spectral images of transmission (A) and autofluorescence (B and C) obtained with a ×40 objective lens. The mixture consisted of the same cells that were used for Fig. 3A to C, which show the corresponding transmission and emission spectra of the cells. In panel A, the intensities of the red (R), green (G), and blue (B) channels correspond to the magnitude of fourth derivative of transmission at 675, 630, and 574 nm and represent the relative distributions of the photopigments Chl a, PC, and PE, respectively. In panel B, the green and blue color intensities represent the magnitudes of autofluorescence peaks at 653 and 588 nm excited by blue light and represent the distribution of PC and PE, respectively. In panel C, the green and blue color intensities represent the magnitudes of autofluorescence peaks in the regions at 650 to 653 nm and at 604 nm excited by green light and represent the distributions of PC and PE, respectively. Note that since the long-pass filter used for autofluorescence imaging with green excitation cut off the fluorescence peak of PE at ∼588 nm, the PE distribution was approximated by the fluorescence intensity at the closest available wavelength, 604 nm (Fig. 3C, dashed line). The fuzzy appearance of the green oval-shaped cells and the cyan filaments in panel C is due to the nonlinear color mapping applied to the final image to facilitate better visibility of the darker-green filaments. The scale bar is valid for all of the images.
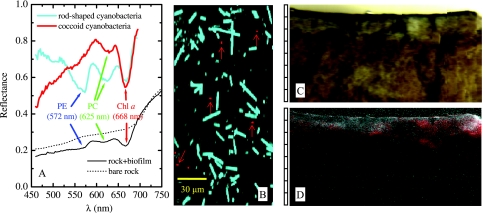
Using the Axiolab microscope with a ×20 objective lens, two major cell types characterized by distinct reflectance spectra and morphologies were identified in the endolithic biofilm (Fig. 5A and B). The rod-shaped cells had pronounced reflectance minima at 572, 625, and 668 nm (Fig. 5A, cyan line), corresponding to absorption maxima of PE, PC, and Chl a, respectively (32, 83), and thus they were putatively identified as cyanobacteria. The reflectance spectra of the small cocci (1 to 2 μm) were marked by the spectral signatures of Chl a (668 nm) and PC (∼625 nm) (Fig. 5A, red line), and these cells were probably also cyanobacteria. Using these two spectra as end members, the classified image of the cells indicated that the rod-shaped cyanobacteria were much more abundant than the cocci. This conclusion is consistent with the findings of Shiraishi et al. (72), who reported dominance of filamentous cyanobacteria with the “Phormidium incrustatum” morphotype and the presence of unicellular cyanobacteria in the creek biofilms.
FIG. 5.
Spectral imaging of an endolithic biofilm. (A) Reflectance spectra of cells in a biofilm (cyan and red lines) and of the rock substrate with (solid black line) and without (dotted line) the cells. (B) Classified HS image for cells scratched off the biofilm surface and spread on a microscope slide. Cyan and red cells (indicated by arrows) correspond to classes characterized by the cyan and red spectra in panel A, respectively. (C) Direct image of a vertical cut in the rock, approximately as it appeared to the naked eye. (D) False-color image of the same cut obtained by pixelwise assignment of the fourth derivative of the log-transferred reflectance at 668, 625, and 572 nm to the intensities of the red, green, and blue colors, respectively. The scale to the left of panels C and D is in millimeters.
Mesoscopic spectral imaging of mixed microbial communities.
To the naked eye, the cross-section of the tufa sample from Westerhöfer Creek appeared to be gray-black proximal to the surface and yellow-brown in deeper areas (Fig. 5C). Examination of the spectra of the sample revealed rather flat and featureless reflectance spectra that were associated with the underlying rock substratum to which the biofilm was attached (Fig. 5A, dotted line), while the reflectance spectra for the gray-black surface layer had pronounced minima at the same wavelengths as the minima in the reflectance spectra of the most abundant cells in the biofilm (Fig. 5A, compare the solid black and cyan lines). This shows that the spectral features found at the microscopic and mesoscopic levels directly corresponded to each other, although, due to light scattering (see Discussion), the shapes of the reflectance spectra at the levels were considerably different. The pixelwise analysis of [log R](4) at wavelengths of 572, 625, and 668 nm revealed that most of the rod-shaped cells were present close to the surface of the endolithic biofilm (maximum depth, ∼1 mm), while the cocci were also present in deeper parts (down to 1 to 3 mm) of the rock substrate (Fig. 5D, compare gray and red regions).
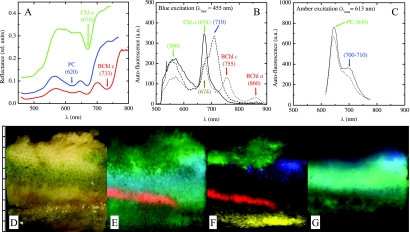
The hypersaline microbial mat had pronounced layers with distinct coloration (brown-yellow in the top 2 to 3 mm, darker green between 3 and 5 mm, and brown-purple at depths of 5 to 7 mm) (Fig. 6D) when it was observed by the naked eye with white illumination. Spectral imaging of the reflectance spectra revealed pronounced minima at wavelengths of 670, 620, and 733 nm (Fig. 6A), which corresponded to in vivo absorption maxima of Chl a, PC, and bacteriochlorophyll c (BChl c), respectively (32, 69). Since these pigments are characteristic of diatoms (Chl a), cyanobacteria (Chl a plus PC), and members of the Chloroflexaceae (BChl c), the spatial organization of these phototrophic groups in the mat could be reconstructed from [log R](4) analysis of the measured hyperspectral image (Fig. 6E). Specifically, the data suggested that the top 3-mm layer was dominated by diatoms (Fig. 6E, green region), as no spectral signature of PC was detected. Cyanobacteria dominated the mat layer at depths between 3 and 6 mm and overlapped the diatom layer at depths of 2 to 3 mm (Fig. 6E, cyan region). At the bottom of the cyanobacterial layer (depth, 6 to 7 mm) was a distinct band of BChl c-producing Chloroflexaceae (Fig. 6E, red). A valley around 670 nm observed for deeper mat layers, as indicated by the green patch at depths of 7 to 10 mm in Fig. 6E, may have been due to Chl a degradation products, which have absorption properties similar to those of Chl a in this spectral region (25, 35). This interpretation is in line with functional studies of similar mats, which have shown that such deep mat layers are permanently anoxic and highly sulfidic and do not have access to visible light that induces oxygenic photosynthesis (8).
FIG. 6.
Spectral imaging of a vertical section of a photosynthetic microbial mat. (A to C) Average reflectance (A) and autofluorescence (B and C) spectra for several selected areas in the mat. The arrows and the corresponding wavelengths (in nanometers) (in parentheses) indicate selected spectral features of pigments identified in the cells. (D) Direct image of the microbial mat, approximately as it appeared to the naked eye. (E) False-color image of the mat obtained by pixelwise assignment of the fourth derivative of the log-transferred reflectance at 733, 670, and 620 nm to the intensities of the red, green, and blue colors, respectively. (F and G) False-color images of the mat obtained using the magnitudes of the autofluorescence peaks indicated by the corresponding colors in panels B and C, respectively. The scale to the left of panels D to G is in millimeters. See text for additional explanation. rel., relative; a.u., arbitrary units.
Many autofluorescence peaks were observed in the HS images when the mat was illuminated by blue light (Fig. 6B) or amber light (Fig. 6C). With blue excitation, pronounced fluorescence at 674 nm due to Chl a in diatoms and a broad emission peak at 560 nm whose origin was not identified were observed for the top 3 mm of the mat (Fig. 6F, green region), while there was a strong emission peak at 710 nm for a distinct patch at depths of 3 to 5 mm (Fig. 6F, blue region). Emission at 755 and 860 nm was observed for a distinct band at depths of 6 to 7 mm (Fig. 6F, red region), suggesting that the members of the Chloroflexaceae identified by their BChl c signature in the reflectance spectra also produced BChl a (in vivo emission peaks of BChl a and c are at ∼860 and ∼755 nm, respectively [45, 69]). The yellow patch in deeper mat layers (8 to 10 mm) (Fig. 6F) was characterized by emission at 674 nm, but there was no broad emission at 560 nm, and this may have been due to degradation products of Chl a, which have emission spectra similar to those of Chl a (25). Amber excitation resulted in a distinct emission peak around 645 nm at a depths of 3 to 6 mm (Fig. 6C and Fig. 6G, cyan region) corresponding to PC fluorescence from cyanobacteria. Additionally, a shoulder at 700 to 710 nm was locally enhanced in a patch at depths between 3 and 5 mm (Fig. 6G). This was most likely due to the same pigment that was identified by the 710-nm emission when a sample was excited by blue light (Fig. 6F, blue region), possibly indicating that a different cyanobacterial population was present. Emission at 645 nm without the additional shoulder at 700 to 710 nm was observed at depths below 6 mm (Fig. 6G, green region) and was probably due to degradation products of PC.
Thus, it can be concluded that the two spectral imaging approaches provided similar information about the spatial organization of the major phototrophic groups in the mat, although the information obtained from spectral imaging of autofluorescence was somewhat richer and more specific than the information derived from the reflectance measurements.
Macroscopic spectral imaging combined with OCR measurement.
The spectrum emitted by the microorganisms in the sediment when they were excited by blue light had a pronounced peak at 675 nm and a shoulder at ∼725 nm, and it was similar to the Chl a autofluorescence spectrum obtained for a single cell of the diatom C. closterium (Fig. 7A). The shape of the spectrum did not change with sediment depth or with the different sediment cores measured. Furthermore, no autofluorescence was detected when amber excitation was used. These data suggested that diatoms, possibly diatoms similar to C. closterium, dominated the microphytobenthic community in the sediment and that detectable amounts of cyanobacteria were not present.
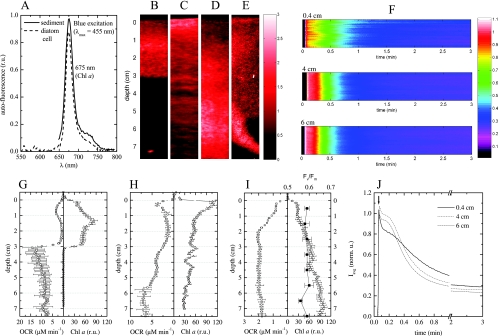
FIG. 7.
Combined spectral and oxygen imaging in permeable intertidal sediments. (A) Autofluorescence spectrum emitted by the Chl a-containing microorganisms in the sediment. The emission spectrum for a single cell of the diatom C. closterium is also shown for comparison. (B to E) Examples of Chl a distribution in the sediment cores, derived from the magnitude of the autofluorescence peak at 675 nm. (F) Slow kinetics of the variable Chl a fluorescence in a horizontal line of pixels (y coordinate) at three depths (indicated above the images) in the sediment core in panel D. The false color represents the normalized magnitude of the autofluorescence peak at 675 nm. The corresponding horizontal averages (Iavg) (i.e., averages for all pixels at a given depth) of these signals are shown in panel J (norm. u., normalized units). (G to I) The triangles indicate the depth profiles for Chl a concentrations in the sediment cores, calculated by horizontally averaging the two-dimensional distributions shown in panels B to D, although for calculation of relative units (r.u.) it was assumed that the concentrations were relatively comparable for the cores. The squares indicate vertical profiles of potential OCRs, determined by the flowthrough method, for the same cores. The filled squares in panel I indicate the depth profile for the dark-adapted photosynthesis yield of Chl a fluorescence (Fv/Fm), as measured with a pulse amplitude modulated fluorometer for the homogenized sediment samples collected at 1-cm depth intervals for the core shown in panel D.
The concentration of Chl a, as determined by the magnitude of the fluorescence peak at 675 nm, varied with depth, and Chl a was found at depths of at least 7 cm in the sediment (Fig. 7B to D), which is similar to the findings for similar sediments reported previously (10, 22). The vertical Chl a distribution was also highly variable among cores; a local maximum occurred at a depth of ∼1.5 cm (Fig. 7B and G), near the sediment surface (Fig. 7C and H), or even at a depth of ∼7 cm (Fig. 7D and I). Pronounced heterogeneity was sometimes also observed in the horizontal direction (e.g., in the vicinity of a polychaete burrow, where the pigment concentration was elevated) (Fig. 7E).
The fluorescence intensity had pronounced kinetics at all depths measured (down to 6 cm), indicating that the Chl a from which the fluorescence originated was part of a functional photosynthetic apparatus (Fig. 7F). This was confirmed by measurement of pulse amplitude modulated fluorescence for homogenized sediment sections, which showed that down to a depth of 8 cm Fv/Fm was ∼0.6, which is close to the value typical of a healthy diatom (0.65 to 0.7) (Fig. 7I) (43, 57). The kinetics of the normalized variable fluorescence depended on the depth at which it was measured, and the fluorescence for deeper sediment layers (4 and 6 cm) decayed faster than the fluorescence for the near-surface horizon (4 mm) (Fig. 7J). The horizontal variability was not pronounced (Fig. 7F). Thus, the cells at a given depth had similar light adaptations or were in a similar physiological state, but these characteristics differed with depth. The kinetics of the variable Ch a fluorescence reached a steady state after 2 to 3 min (Fig. 7J), confirming that the variability in pigment concentrations determined from spectral imaging was real and not an artifact due to temporally variable Chl a fluorescence. This was also confirmed by the fact that the spectral scans conducted from top to bottom and the spectral scans in the opposite direction yielded the same results (data not shown). Percolation of seawater through the cores during flowthrough measurement did not change the pigment distribution (data not shown).
How well the vertical distributions of Chl a and the potential OCRs were coupled depended on whether the sediments had a clear redox boundary (brown, oxidized sediment on top and black, reduced sediment underneath), which was different at different stations. In sediments with pronounced redox stratification, Chl a was detected only until the redox boundary (Fig. 7G). The OCRs were relatively low above this boundary (1 to 4 μM min−1) and increased dramatically below it (8 to 13 μM min−1) (Fig. 7G). In sediments without redox stratification, the OCRs were for the most part similar (Fig. 7G to I), although sometimes they were antiparallel (below 2 cm in Fig. 7H), to the Chl a distribution.
DISCUSSION
Spectral imaging provides specific information about the identity or composition of objects in an image. To date, such information has been used for large-scale applications, such as remote sensing for the Earth, or at the microscopic level in medicine and microbiology. Because of its construction flexibility, the MOSI system described here enables workers to bridge these spatial extremes and allows spectral imaging ranging from the single-cell level to regions whose areas are several square decimeters. A decision about the level of detail that is spectrally imaged can be made based on the spatial context in which the system of interest is to be investigated.
Spectral imaging with the spatial resolution of a few tens of micrometers is accomplished by attaching a standard 25-mm objective lens equipped with a 10-mm buffer ring to the spectral camera. In this mesoscopic imaging mode, the MOSI system can provide valuable information at scales that are relevant for the functioning of complex microbial communities, where mass transfer and light penetration control the spatial organization of benthic communities. This was illustrated by the results obtained for a photosynthetic microbial mat (Fig. 6) and a microphytobenthic population in permeable sediments (Fig. 7), where the functional zones are typically a fraction of a millimeter to a few centimeters thick.
When it is used together with a microscope, the MOSI system can also be used for spectral imaging on a microscale, which allows sensitive identification of pigments in single cells. Thus, it provides the opportunity to zoom in on specific areas of the system studied to link the pigment content and its spatial distribution in a sample revealed by mesoscopic spectral imaging to the pigment content of the cells in the sample, as demonstrated by the observations for dense phototrophic communities on tufa (Fig. 5). Furthermore, it can be used to rapidly provide basic information about a microbial system, such as the pigment content of an unknown cell or the types of pigments present in a mixed microbial community, as shown by using cells from pure and mixed cultures (Fig. 3 and 4). This is particularly useful during field studies, where existing techniques (such as confocal microscopy or HPLC) are difficult to use.
In addition to its spatial flexibility, the MOSI system has high spectral resolution (∼0.7 nm). This allows discrimination between cells whose autofluorescence peaks differ by only a few nanometers (Fig. 3). Without such resolution, it would not be possible to carry out a fourth-order derivative analysis of the reflectance and transmission spectra, which is necessary for sensitive and specific identification of pigments, as well as for discrimination between neighboring pixels in an image with different spectral features. Furthermore, the broad detection range (460 to 913 nm) of the HS camera allows detection of pigments involved in both oxygenic photosynthesis and anoxygenic photosynthesis.
The MOSI system can be used to obtain images in both reflectance-transmission and autofluorescence modes. Generally, the autofluorescence measurements provide more specific and spatially better-resolved images of pigments than the reflectance-transmission measurements. This is due mainly to the higher degree of freedom in the autofluorescence measurements (i.e., the ability to use several narrow-band excitation sources) and to the fundamental physical limitations associated with reflectance-transmission measurements due to use of spatially unconfined sample illumination. When images are obtained with a microscope, scattering of light from the cells or other particles in the sample may alter the reflectance-transmission spectrum obtained for a given pixel of the image, thus limiting the density of cells in a field of view that can be spectrally imaged with acceptable confidence. This is caused by illuminating and collecting the reflected and transmitted light for the entire sample simultaneously (i.e., by not using a focused illumination beam and a pinhole for detection, which are employed in confocal microscopy). For mesoscale reflectance measurement, the spatial overlap of microorganisms in a complex community may result in overall reflectance spectra in which, even when the fourth-derivative approach is used, discrimination between groups of microorganisms is not possible unless the spectral characteristics of the groups are considerably different in at least one distinctive spectral region. For example, the absorption peaks of Chl c at ∼635 nm and of PC at ∼625 nm are difficult to resolve. However, similar complications may occur with the autofluorescence measurements; organisms with similar emission spectra with a certain excitation light are difficult to discriminate if they overlap spatially. In such circumstances, cells from specific regions of the sample should be examined spectrally with a microscope using the MOSI system.
In addition to pigment identification and localization, the MOSI system can be used for pigment quantification. In this work, we used the fourth spectral derivative of reflectance-transmission (or its logarithm) or the magnitude of the autofluorescence signal as a proxy for pigment content, which in this case were only relative measures. For absolute pigment quantification, MOSI must be combined with conventional, usually labor-intensive techniques, such as HPLC. Clearly, due to its simplicity, using spectral imaging for quantitative pigment measurement would be highly desirable, and this possibility has been extensively investigated in studies concerned with large-scale estimation of primary productivity in intertidal sediments based on spectroscopic microphytobenthos biomass quantification (15, 30, 36, 44, 53, 54, 58, 70, 75). Generally, such studies employ approaches based on simple indices derived from spectral reflectivity for a few discrete bands, which are not universally applicable as they depend on the properties of the sediment matrix, such as grain size distribution, water content, mineral composition, etc. (20). Although more sophisticated approaches have been developed (9, 17), their applicability is probably limited to systems with one or a very few predominant species of microorganisms. These complications result from a number of factors: (i) the penetration of the light that probes the system is limited to a few millimeters, making it impossible to detect pigments present at greater depths; (ii) measured reflectivity depends not only on the absorption properties of pigments present in the system but also on the absorption properties of the underlying substrate, as well as of the matrix in which the pigmented organisms are embedded, which are difficult to determine or measure in all possible habitats (e.g., sediments); and (iii) due to simultaneous absorption and intense multiple scattering of light within the matrix, the shape of the reflected light spectrum is substantially altered compared to the absorption spectrum of the absorber embedded in the matrix. These factors pose a theoretical question of how to extract information about the absorber from the signal influenced by both the absorbing and scattering components of the measured system, which has not been adequately resolved for systems such as complex communities of pigmented microorganisms embedded in a sediment matrix. The extension of the classical Koubelka-Munk theory to scattering media containing multiple absorbing species, which was developed in the context of the quality of color perception in the ink-on-paper printing industry (84-86), could provide a useful theoretical foundation for an answer to this question. However, more work is required to adapt this theory for studies of mixed microbial communities where the absorption and scattering properties of the matrix and/or the substrate of the sample are essentially unknown or are difficult to determine in a nondestructive way. Only when such work is completed will it be possible to use spectral measurements of light reflected or emitted by mixed microbial communities, such as the measurements enabled by the MOSI system, for truly quantitative, nondestructive, and universally applicable measurement of pigment content.
The MOSI system is a relatively low-cost system. A good-quality microscope and a computer with a fire-wire interface are typically available in any microbiology lab, and the main additional components include the HS camera, the motorized stage, LEDs or one of the alternative light sources listed in Table 1, a set of optical filters, a white reference standard, and a generator of power for the electrical components. These components can be purchased for a total cost that does not exceed approximately $20,000. The software used to operate the system and to analyze the data is either provided by the manufacturer (e.g., FireView provided by Resonon) or is available for free from us.
The modularity of the MOSI system means that it can be used not only in the laboratory but also during field campaigns, which is a great advantage. All the required components, except the power generator, fit into a medium-size container (∼80 by 60 by 60 cm) that can be transported in a car and can be reassembled and readjusted by one person within 1 to 2 h. Measurement requires constant illumination during reflectance-transmission imaging and low background during autofluorescence imaging. Thus, when measuring outside, workers must exclude the potentially variable or elevated ambient light during the scan, which can be done by covering the system with, e.g., a larger cardboard box. A more convenient solution is to use the system in a room (e.g., a field station) that can be darkened, although such a room is not always available during field campaigns.
Although the primary emphasis here was on methodological development, several results clearly show the potential of the MOSI system for characterizing structure-function relationships in microbial ecosystems. The observations for dense phototrophic communities at the surface of tufa corresponded well to the notion that tufa growth by calcification is controlled entirely by photosynthesis (12), where photosynthesis-coupled CO2 fixation leads to an increase in the pH and thus to calcium carbonate precipitation.
The stratification in the microbial mats shows how mass transfer and light penetration control the spatial organization of benthic communities. Diatoms are sensitive to sulfide, whereas cyanobacteria can survive highly sulfidic periods and even use sulfide as an electron acceptor (16, 21). Both types of organisms harvest visible light (primarily in the blue and red regions) for oxygenic photosynthesis, which therefore is attenuated rapidly with depth (38, 40). At night sulfide diffuses close to the surface (41, 82); thus, diatoms prefer the top zone of mats, whereas cyanobacteria also inhabit the layer underneath. Light in the near-infrared region penetrates deeper in mats (38, 40, 45), where it can be used by anoxygenic phototrophs, like members of family Chloroflexaceae, which are indeed found below the cyanobacterial layer (Fig. 6). Similar results were obtained in our recent study which investigated the link between the activity and the spatial organization of oxygenic and anoxygenic phototrophs in a hypersaline microbial mat (8).
There are open questions regarding the identity and origin of pigments corresponding to some spectral features found in a mat, such as the emission peak at 700 to 710 nm excited by both blue light and amber light or the presence of the Chl a signature at depths of >8 mm. These kinds of questions could be addressed more specifically by performing microscopic spectral imaging and independent pigment analysis (e.g., by HPLC) of cells that are obtained from particular areas in the mat identified by the mesoscopic spectral imaging approach. Such measurements were beyond the scope of the present work.
The measurements for pigments in coarse sediments revealed that diatoms seem to be rather randomly distributed in the suboxic zone and are clearly repelled by the highly reduced conditions in the deeper sediments, an adaptation which may prevent them from moving deep to reach the surface in a short time. Diatoms are highly motile, and in intertidal sediments they migrate in response to tides, surfacing during exposure and withdrawing into the sediments during high tide (18, 56, 71). Although phototrophic, they are often found several centimeters deep in sediments (18, 68) without access to light to drive photosynthesis (46, 47). At such depths, they can generate energy by respiration of O2, which can penetrate to a depth of a few centimeters in permeable sediments (11, 19, 63, 80) due to advection-driven pore water flow (64, 65). When they do not have access to O2, they must switch to anaerobic metabolism to survive. As soon as they are supplied with O2, such as during OCR measurement, they respire again. Indeed, the combined measurements for pigments and OCR are compatible with the notion that oxygen consumption is chemically controlled in strongly reduced sediments below the redox zone and is strongly linked to diatom presence in the suboxic zone. The diatoms in deeper sediment zones were alive, as the fluorescence kinetics showed that they had an intact photosystem. Thus, the possibility of degradation of decaying diatoms can be excluded, and the respiration is linked to living diatoms. Whether the diatoms themselves or diatom-associated microbial communities are responsible for the oxygen consumption cannot be assessed based on our data, but answering this question is essential for understanding the carbon cycling in benthic phototrophic communities better. Similarly, in phototrophic mats respiration appeared to be strongly dominated by phototrophs (7, 61). Since reduced sediments are very quickly oxidized when they are exposed to oxygen, it would also be interesting to assess whether diatoms could be used as an indicator of temporally averaged redox zonation of permeable sediments. The MOSI system should be a powerful tool in such studies.
In conclusion, the MOSI system described here allows nondestructive identification, localization, and relative quantification of pigments in microbial samples ranging from single cells to microbial communities. The system is relatively low cost, easy to transport and assemble, and thus suitable for field measurements. Compared to the conventional extraction techniques, measurement is fast, sensitive, nondestructive, and not prone to potential loss of fragile pigments. The system opens new possibilities for studying the role of spatial organization of microorganisms in the ecological functioning of complex microbial communities and for noninvasively monitoring changes in the spatial organization and/or composition of a microbial community in response to changing environmental factors.
Supplementary Material
Acknowledgments
We thank Georg Herz and Alfred Kutsche for construction of mechanical parts needed for the imaging system, Tim Hirzel, Casey Smith, and Rand Swanson of Resonon for excellent support and interactions during development of the measurement software, Henk Jonkers and Martin Beutler for valuable discussions, and Bo Barker Jørgensen and an anonymous reviewer for critical comments and suggestions. Katharina Kohls and Aaron Beck provided cell samples.
Financial support of this work by the European Commission (project ECODIS, project 518043) is greatly appreciated.
Footnotes
Published ahead of print on 12 December 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abed, R. M. M., K. Kohls, R. Schoon, A.-K. Scherf, M. Schacht, K. A. Palinska, H. Al-Hassani, W. Hamza, J. Rullkötter, and S. Golubic. 2008. Lipid biomarkers, pigments and cyanobacterial diversity of microbial mats across intertidal flats of the arid coast of the Arabian Gulf (Abu Dhabi, UAE). FEMS Microbiol. Ecol. 65:449-462. [DOI] [PubMed] [Google Scholar]
- 2.Aberle, N., M. Beutler, C. Moldaenke, and K. H. Wiltshire. 2006. ‘Spectral fingerprinting’ for specific algal groups on sediments in situ: a new sensor. Arch. Hydrobiol. 167:575-592. [Google Scholar]
- 3.Ainsworth, T. D., M. Fine, L. L. Blackall, and O. Hoegh-Guldberg. 2006. Fluorescence in situ hybridization and spectral imaging of coral-associated bacterial communities. Appl. Environ. Microbiol. 72:3016-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Airs, R. L., and B. J. Keely. 2003. A high resolution study of the chlorophyll and bacteriochlorophyll pigment distributions in a calcite/gypsum microbial mat. Org. Geochem. 34:539-551. [Google Scholar]
- 5.Andrefouet, S., E. J. Hochberg, C. Payri, M. J. Atkinson, F. E. Muller-Karger, and H. Ripley. 2003. Multi-scale remote sensing of microbial mats in an atoll environment. Int. J. Remote Sens. 24:2661-2682. [Google Scholar]
- 6.Andrefouet, S., C. Payri, E. J. Hochberg, L. M. Che, and M. J. Atkinson. 2003. Airborne hyperspectral detection of microbial mat pigmentation in Rangiroa Atoll (French Polynesia). Limnol. Oceanogr. 48:426-430. [Google Scholar]
- 7.Bachar, A., E. Omoregie, R. de Wit, and H. M. Jonkers. 2007. Diversity and function of Chloroflexus-like bacteria in a hypersaline microbial mat: phylogenetic characterization and impact on aerobic respiration. Appl. Environ. Microbiol. 73:3975-3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bachar, A., L. Polerecky, J. Fischer, K. Vamvakopoulos, D. de Beer, and H. Jonkers. 2008. Two-dimensional mapping of photopigment distribution and activity of Chloroflexus-like bacteria in a hypersaline microbial mat. FEMS Microbiol. Ecol. 65:434-448. [DOI] [PubMed] [Google Scholar]
- 9.Barille, L., V. Meleder, J.-P. Combe, P. Launeau, Y. Rince, V. Carrere, and M. Morancais. 2007. Comparative analysis of field and laboratory spectral reflectances of benthic diatoms with a modified Gaussian model approach. J. Exp. Mar. Biol. Ecol. 343:197-209. [Google Scholar]
- 10.Billerbeck, M., H. Roy, K. Bosselmann, and M. Huettel. 2007. Benthic photosynthesis in submerged Wadden Sea intertidal flats. Estuar. Coast. Shelf Sci. 71:704-716. [Google Scholar]
- 11.Billerbeck, M., U. Werner, L. Polerecky, E. Walpersdorf, D. de Beer, and M. Huettel. 2006. Surficial and deep pore water circulation governs spatial and temporal scales of nutrient recycling in intertidal sand flat sediment. Mar. Ecol. Prog. Ser. 326:61-76. [Google Scholar]
- 12.Bissett, A., D. de Beer, R. Schoon, F. Shaaishi, A. Reimer, and G. Arp. 2008. Microbial mediation of stromatolite formation in karst-water creeks. Limnol. Oceanogr. 53:1159-1168. [Google Scholar]
- 13.Boetius, A., K. Ravenschlag, C. J. Schubert, D. Rickert, F. Widdel, A. Gieseke, R. Amann, B. B. Jørgensen, U. Witte, and O. Pfannkuche. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623-626. [DOI] [PubMed] [Google Scholar]
- 14.Butler, W. L., and D. W. Hopkins. 1970. Analysis of fourth derivative spectra. Photochem. Photobiol. 12:451-456. [Google Scholar]
- 15.Carrere, V., N. Spilmont, and D. Davoult. 2004. Comparison of simple techniques for estimating chlorophyll a concentration in the intertidal zone using high spectral-resolution field-spectrometer data. Mar. Ecol. Prog. Ser. 274:31-40. [Google Scholar]
- 16.Cohen, Y., B. B. Jørgensen, N. P. Revsbech, and R. Poplawski. 1986. Adaptation to hydrogen-sulfide of oxygenic and anoxygenic photosynthesis among cyanobacteria. Appl. Environ. Microbiol. 51:398-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Combe, J.-P., P. Launeau, V. Carrere, D. Despan, V. Meleder, L. Barille, and C. Sotin. 2005. Mapping microphytobenthos biomass by non-linear inversion of visible-infrared hyperspectral images. Remote Sens. Environ. 98:371-387. [Google Scholar]
- 18.Consalvey, M., D. M. Paterson, and G. J. C. Underwood. 2004. The ups and downs of life in a benthic biofilm: migration of benthic diatoms. Diatom Res. 19:181-202. [Google Scholar]
- 19.de Beer, D., F. Wenzhoefer, T. Ferdelman, S. E. Boehme, M. Huettel, J. E. E. van Beusekom, M. E. Bottcher, N. Musat, and N. Dubilier. 2005. Transport and mineralization rates in North Sea sandy intertidal sediments, Sylt-Romo Basin, Wadden Sea. Limnol. Oceanogr. 50:113-127. [Google Scholar]
- 20.Deronde, B., P. Kempeneers, and R. M. Forster. 2006. Imaging spectroscopy as a tool to study sediment characteristics on a tidal sandbank in the Westerschelde. Estuar. Coast. Shelf Sci. 69:580-590. [Google Scholar]
- 21.de Wit, R., W. H. M. Vanboekel, and H. van Gemerden. 1988. Growth of the cyanobacterium Microcoleus chtonoplastes on sulfide. FEMS Microbiol. Ecol. 53:203-209. [Google Scholar]
- 22.Ehrenhauss, S., U. Witte, S. L. Buhring, and M. Huettel. 2004. Effect of advective pore water transport on distribution and degradation of diatoms in permeable North Sea sediments. Mar. Ecol. Prog. Ser. 271:99-111. [Google Scholar]
- 23.Fenchel, T., and M. Kühl. 2000. Artificial cyanobacterial mats: growth, structure, and vertical zonation patterns. Microb. Ecol. 40:85-93. [DOI] [PubMed] [Google Scholar]
- 24.Fleissner, G., W. Hage, A. Hallbrucker, and E. Mayer. 1996. Improved curve resolution of highly overlapping bands by comparison of fourth-derivative curves. Appl. Spectrosc. 50:1235-1245. [Google Scholar]
- 25.French, C. S., J. H. C. Smith, H. I. Virgin, and R. L. Airth. 1956. Fluorescence-spectrum curves of chlorophylls, pheophytins, phycoerythrins, phycocyanins and hypericin. Plant Physiol. 31:369-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Govindjee. 2004. Chlorophyll fluorescence. A bit of basics and history, p. 1-42. In G. C. Papageorgiou and Govindjee (ed.), Chlorophyll fluorescence: a signature of photosynthesis. Advances in photosynthesis and respiration, vol. 19. Springer, Dordrecht, The Netherlands. [Google Scholar]
- 27.Govindjee. 1995. Sixty-three years since Kautsky: chlorophyll a fluorescence. Aust. J. Plant Physiol. 22:131-160. [Google Scholar]
- 28.Guillard, R. R. L. 1975. Culture of phytoplankton for feeding marine invertebrates, p. 22-60. In W. L. Smith and M. H. Chanley (ed.), Culture of marine invertebrate animals. Plenum Press, New York, NY.
- 29.Guillard, R. R. L., and J. H. Ryther. 1962. Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt, and Detonula confervacea (Cleve). Can. J. Microbiol. 8:229-239. [DOI] [PubMed] [Google Scholar]
- 30.Harkvoort, J. H. M., M. Heineke, K. Heymann, H. Kühl, R. Riethmüller, and G. Witte. 1998. Optical remote sensing of microphytobenthos biomass: a method to monitor tidal flat erodibility. Senckenb. Marit. 29:77-85. [Google Scholar]
- 31.Hoepffner, N., and S. Sathyendaranath. 1991. Effect of pigment composition on absorption properties of phytoplankton. Mar. Ecol. Prog. Ser. 73:11-23. [Google Scholar]
- 32.Hofstraat, J. W., W. J. M. van Zeijl, M. E. J. de Vreeze, J. C. H. Peeters, L. Peperzak, F. Colijn, and T. W. M. Rademaker. 1994. Phytoplankton monitoring by flow-cytometry. J. Plankton Res. 16:1197-1224. [Google Scholar]
- 33.Holst, G., and B. Grunwald. 2001. Luminescence lifetime imaging with transparent oxygen optodes. Sens. Actuators B Chem. 74:78-90. [Google Scholar]
- 34.Horath, T., T. R. Neu, and R. Bachofen. 2006. An endolithic microbial community in dolomite rock in central Switzerland: characterization by reflection spectroscopy, pigment analyses, scanning electron microscopy, and laser scanning microscopy. Microb. Ecol. 51:353-364. [DOI] [PubMed] [Google Scholar]
- 35.Jeffrey, S. W., R. F. C. Mantoura, and S. W. Wright (ed.). 1997. Phytoplankton pigments in oceanography: guidelines to modern methods. UNESCO Publishing, Paris, France.
- 36.Jesus, B., C. R. Mendes, V. Brotas, and D. M. Paterson. 2006. Effect of sediment type on microphytobenthos vertical distribution: modelling the productive biomass and improving ground truth measurements. J. Exp. Mar. Biol. Ecol. 332:60-74. [Google Scholar]
- 37.Jonkers, H. M., R. Ludwig, R. De Wit, O. Pringault, G. Muyzer, H. Niemann, N. Finke, and D. De Beer. 2003. Structural and functional analysis of a microbial mat ecosystem from a unique permanent hypersaline inland lake: ‘La Salada de Chiprana’ (NE Spain). FEMS Microbiol. Ecol. 44:175-189. [DOI] [PubMed] [Google Scholar]
- 38.Jørgensen, B. B. 1989. Light penetration, absorption, and action spectra in cyanobacterial mats, p. 123-137. In Y. Cohen and E. Rosenberg (ed.), Microbial mats: physiological ecology of benthic microbial communities. ASM Press, Washington, DC.
- 39.Jørgensen, B. B., and D. J. D. Marais. 1990. The diffusive boundary-layer of sediments—oxygen microgradients over a microbial mat. Limnol. Oceanogr. 35:1343-1355. [DOI] [PubMed] [Google Scholar]
- 40.Jørgensen, B. B., and D. C. Nelson. 1988. Bacterial zonation, photosynthesis, and spectral light-distribution in hot-spring microbial mats of Iceland. Microb. Ecol. 16:133-147. [DOI] [PubMed] [Google Scholar]
- 41.Jørgensen, B. B., N. P. Revsbech, T. H. Blackburn, and Y. Cohen. 1979. Diurnal cycle of oxygen and sulfide microgradients and microbial photosynthesis in a cyanobacterial mat sediment. Appl. Environ. Microbiol. 38:46-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jørgensen, B. B., N. P. Revsbech, and Y. Cohen. 1983. Photosynthesis and structure of benthic microbial mats: microelectrode and SEM studies of four cyanobacterial communities. Limnol. Oceanogr. 28:1075-1093. [Google Scholar]
- 43.Kromkamp, J., C. Barranguet, and J. Peene. 1998. Determination of microphytobenthos PSII quantum efficiency and photosynthetic activity by means of variable chlorophyll fluorescence. Mar. Ecol. Prog. Ser. 162:45-55. [Google Scholar]
- 44.Kromkamp, J. C., E. P. Morris, R. M. Forster, C. Honeywill, S. Hagerthey, and D. M. Paterson. 2006. Relationship of intertidal surface sediment chlorophyll concentration to hyperspectral reflectance and chlorophyll fluorescence. Estuar. Coasts 29:183-196. [Google Scholar]
- 45.Kühl, M., and T. Fenchel. 2000. Bio-optical characteristics and the vertical distribution of photosynthetic pigments and photosynthesis in an artificial cyanobacterial mat. Microb. Ecol. 40:94-103. [DOI] [PubMed] [Google Scholar]
- 46.Kühl, M., and B. B. Jørgensen. 1994. The light-field of microbenthic communities—radiance distribution and microscale optics of sandy coastal sediments. Limnol. Oceanogr. 39:1368-1398. [Google Scholar]
- 47.Kühl, M., C. Lassen, and B. B. Jørgensen. 1994. Light penetration and light-intensity in sandy marine-sediments measured with irradiance and scalar irradiance fiberoptic microprobes. Mar. Ecol. Prog. Ser. 105:139-148. [Google Scholar]
- 48.Larson, C., and S. I. Passy. 2005. Spectral fingerprinting of algal communities: a novel approach to biofilm analysis and biomonitoring. J. Phycol. 41:439-446. [Google Scholar]
- 49.Lawrence, J. R., and T. R. Neu. 2003. Microscale analyses of the formation and nature of microbial biofilm communities in river systems. Rev. Environ. Sci. Biotechnol. 2:85-97. [Google Scholar]
- 50.Macedo, A. J., U. Kuhlicke, T. R. Neu, K. N. Timmis, and W.-R. Abraham. 2005. Three stages of a biofilm community developing at the liquid-liquid interface between polychlorinated biphenyls and water. Appl. Environ. Microbiol. 71:7301-7309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meleder, V., L. Barille, P. Launeau, V. Carrere, and Y. Rince. 2003. Spectrometric constraint in analysis of benthic diatom biomass using monospecific cultures. Remote Sens. Environ. 88:386-400. [Google Scholar]
- 52.Miyazawa, K., K. Kobayashi, S. Nakauchi, and A. Hiraishi. 2005. In situ detection and identification of microorganisms at single colony resolution using spectral imaging technique, p. 419-428. In H. Kalviainen (ed.), Lecture notes in computer science, vol. 3540. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 53.Murphy, R. J., M. G. Tolhurst, M. G. Chapman, and A. J. Underwood. 2005. Estimation of surface chlorophyll-a on an emersed mudflat using field spectrometry: accuracy of ratios and derivative-based approaches. Int. J. Remote Sens. 26:1835-1859. [Google Scholar]
- 54.Murphy, R. J., A. J. Underwood, M. H. Pinkerton, and P. Range. 2005. Field spectrometry: new methods to investigate epilithic micro-algae on rocky shores. J. Exp. Mar. Biol. Ecol. 325:111-124. [Google Scholar]
- 55.Neu, T. R., S. Woelfl, and J. R. Lawrence. 2004. Three-dimensional differentiation of photo-autotrophic biofilm constituents by multi-channel laser scanning microscopy (single-photon and two-photon excitation). J. Microbiol. Methods 56:161-172. [DOI] [PubMed] [Google Scholar]
- 56.Palmer, J. D., and F. E. Round. 1967. Persistent vertical-migration rhythms in benthic microflora. VI. The tidal and diurnal nature of the rhythm in the diatom Hantzschia virgata. Biol. Bull. 132:44-55. [Google Scholar]
- 57.Parkhill, J.-P., G. Maillet, and J. J. Cullen. 2001. Fluorescence-based maximal quantum yield for PSII as a diagnostic of nutrient stress. J. Phycol. 37:517-529. [Google Scholar]
- 58.Paterson, D. M., K. H. Wiltshire, A. Miles, J. Blackburn, I. Davidson, M. G. Yates, S. McGrorty, and J. A. Eastwood. 1998. Microbiological mediation of spectral reflectance from intertidal cohesive sediments. Limnol. Oceanogr. 43:1207-1221. [Google Scholar]
- 59.Pierson, B. K., and M. N. Parenteau. 2000. Phototrophs in high iron microbial mats: microstructure of mats in iron-depositing hot springs. FEMS Microbiol. Ecol. 32:181-196. [DOI] [PubMed] [Google Scholar]
- 60.Pierson, B. K., V. M. Sands, and J. L. Frederick. 1990. Spectral irradiance and distribution of pigments in a highly layered marine microbial mat. Appl. Environ. Microbiol. 56:2327-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Polerecky, L., A. Bachar, R. Schoon, M. Grinstein, B. B. Jørgensen, D. de Beer, and H. Jonkers. 2007. Contribution of Chloroflexus respiration to oxygen cycling in a hypersaline microbial mat from Lake Chiprana, Spain. Environ. Microbiol. 9:2007-2024. [DOI] [PubMed] [Google Scholar]
- 62.Polerecky, L., U. Franke, U. Werner, B. Grunwald, and D. de Beer. 2005. High spatial resolution measurement of oxygen consumption rates in permeable sediments. Limnol. Oceanogr. Methods 3:75-85. [Google Scholar]
- 63.Precht, E., U. Franke, L. Polerecky, and M. Huettel. 2004. Oxygen dynamics in permeable sediments with wave-driven pore water exchange. Limnol. Oceanogr. 49:693-705. [Google Scholar]
- 64.Precht, E., and M. Huettel. 2003. Advective pore-water exchange driven by surface gravity waves and its ecological implications. Limnol. Oceanogr. 48:1674-1684. [Google Scholar]
- 65.Precht, E., and M. Huettel. 2004. Rapid wave-driven advective pore water exchange in a permeable coastal sediment. J. Sea Res. 51:93-107. [Google Scholar]
- 66.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1-61. [Google Scholar]
- 67.Roldan, M., F. Thomas, S. Castel, A. Quesada, and M. Hernandez-Marine. 2004. Noninvasive pigment identification in single cells from living phototrophic biofilms by confocal imaging spectrofluorometry. Appl. Environ. Microbiol. 70:3745-3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Round, F. E., and J. D. Palmer. 1966. Persistent, vertical migration rhythms in benthic microflora. II. Field and laboratory studies on diatoms from the banks of the River Avon. J. Mar. Biol. Assoc. UK 46:191-214. [Google Scholar]
- 69.Saga, Y., and H. Tamiaki. 2004. Fluorescence spectroscopy of single photosynthetic light-harvesting supramolecular systems. Cell Biochem. Biophys. 40:149-165. [DOI] [PubMed] [Google Scholar]
- 70.Serodio, J., H. Coelho, S. Vieira, and S. Cruz. 2006. Microphytobenthos vertical migratory photoresponse as characterised by light-response curves of surface biomass. Estuar. Coast. Shelf Sci. 68:547-556. [Google Scholar]
- 71.Serodio, J., J. M. daSilva, and F. Catarino. 1997. Nondestructive tracing of migratory rhythms of intertidal benthic microalgae using in vivo chlorophyll a fluorescence. J. Phycol. 33:542-553. [Google Scholar]
- 72.Shiraishi, F., A. Bissett, D. de Beer, A. Reimer, and G. Arp. 2008. Photosynthesis, respiration and exopolymer calcium-binding in biofilm calcification (Westerhöfer and Deinschwanger Creek, Germany). Geomicrobiol. J. 25:83-94. [Google Scholar]
- 73.Stal, L. J. 1995. Physiological ecology of cyanobacteria in microbial mats and other communities. New Phytol. 131:1-32. [DOI] [PubMed] [Google Scholar]
- 74.Stein, J. R. (ed.) 1973. Handbook of phycological methods: culture methods and growth measurements. Cambridge University Press, Cambridge, United Kingdom.
- 75.Stephens, F. C., E. M. Louchard, R. P. Reid, and R. A. Maffione. 2003. Effects of microalgal communities on reflectance spectra of carbonate sediments in subtidal optically shallow marine environments. Limnol. Oceanogr. 48:535-546. [Google Scholar]
- 76.Sugiura, M., M. Takano, S.-I. Kawakami, K. Toda, and S. Hanada. 2001. Application of a portable spectrophotometer to microbial mat studies: temperature dependence of the distribution of Cyanobacteria and photosynthetic bacteria in hot spring water. Microbes Environ. 16:255-261. [Google Scholar]
- 77.Sunamura, M., A. Maruyama, T. Tsuji, and R. Kurane. 2003. Spectral imaging detection and counting of microbial cells in marine sediment. J. Microbiol. Methods 53:57-65. [DOI] [PubMed] [Google Scholar]
- 78.Trichet, J., C. Defarge, J. Tribble, G. Tribble, and F. Sansone. 2001. Christmas Island lagoonal lakes, models for the deposition of carbonate-evaporite-organic laminated sediments. Sediment. Geol. 140:177-189. [Google Scholar]
- 79.van Gemerden, H. 1993. Microbial mats: a joint venture. Mar. Geol. 113:3-25. [Google Scholar]
- 80.Werner, U., M. Billerbeck, L. Polerecky, U. Franke, M. Huettel, J. E. E. van Beusekom, and D. de Beer. 2006. Spatial and temporal patterns of mineralization rates and oxygen distribution in a permeable intertidal sand flat (Sylt, Germany). Limnol. Oceanogr. 51:2549-2563. [Google Scholar]
- 81.Wieland, A., and M. Kühl. 2006. Regulation of photosynthesis and oxygen consumption in a hypersaline cyanobacterial mat (Camargue, France) by irradiance, temperature and salinity. FEMS Microbiol. Ecol. 55:195-210. [DOI] [PubMed] [Google Scholar]
- 82.Wieland, A., J. Zopfi, M. Benthien, and M. Kühl. 2005. Biogeochemistry of an iron-rich hypersaline microbial mat (Camargue, France). Microb. Ecol. 49:34-49. [DOI] [PubMed] [Google Scholar]
- 83.Wood, A. M., P. K. Horan, K. Muirhead, D. A. Phinney, C. M. Yentsch, and J. B. Waterbury. 1985. Discrimination between types of pigments in marine Synechococcus spp. by scanning spectroscopy, epifluorescence microscopy, and flow cytometry. Limnol. Oceanogr. 30:1303-1315. [Google Scholar]
- 84.Yang, L., and B. Kruse. 2004. Revised Kubelka-Munk theory. I. Theory and application. J. Opt. Soc. Am. A Opt. Image Sci. Vision 21:1933-1941. [DOI] [PubMed] [Google Scholar]
- 85.Yang, L., B. Kruse, and S. J. Miklavcic. 2004. Revised Kubelka-Munk theory. II. Unified framework for homogeneous and inhomogeneous optical media. J. Opt. Soc. Am. A Opt. Image Sci. Vision 21:1942-1952. [DOI] [PubMed] [Google Scholar]
- 86.Yang, L., and S. J. Miklavcic. 2005. Revised Kubelka-Munk theory. III. A general theory of light propagation in scattering and absorptive media. J. Opt. Soc. Am. A Opt. Image Sci. Vision 22:1866-1873. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.