Abstract
To thoroughly investigate the bacterial community diversity present in a single composite sample from an agricultural soil and to examine potential biases resulting from data acquisition and analytical approaches, we examined the effects of percent G+C DNA fractionation, sequence length, and degree of coverage of bacterial diversity on several commonly used ecological parameters (species estimation, diversity indices, and evenness). We also examined variation in phylogenetic placement based on multiple commonly used approaches (ARB alignments and multiple RDP tools). The results demonstrate that this soil bacterial community is highly diverse, with 1,714 operational taxonomic units demonstrated and 3,555 estimated (based on the Chao1 richness estimation) at 97% sequence similarity using the 16S rRNA gene. The results also demonstrate a fundamental lack of dominance (i.e., a high degree of evenness), with 82% of phylotypes being encountered three times or less. The data also indicate that generally accepted cutoff values for phylum-level taxonomic classification might not be as applicable or as general as previously assumed and that such values likely vary between prokaryotic phyla or groups.
Efforts to describe bacterial species richness and diversity have long been hampered by the inability to cultivate the vast majority of bacteria from natural environments. New methods to study bacterial diversity have been developed in the last two decades (32), many of which rely on PCR-based procedures and phylogenetic comparison of 16S rRNA gene sequences. However, PCR using complex mixtures of templates (as in the case of total microbial community DNA) is presumed to preferentially amplify certain templates in the mixture (23) based on their primary sequence, percent G+C (hereafter GC) content, or other factors, resulting in so-called PCR bias. Moreover, the amplification of template sequences depends on their initial concentration and tends to skew detection toward the most abundant members of the community (23). To further complicate matters, subsequent random cloning steps on amplicon mixtures are destined to result in the detection of numerically dominant sequences, especially where relative abundance can vary over orders of magnitude. Indeed, any analysis based on random encounter is destined to primarily detect numerically dominant populations. This is especially of concern where limited sampling is performed on highly complex microbial communities exhibiting mostly even distribution of populations with only a few showing any degree of dominance, as typically perceived for soils (17). These artifacts and sampling limitations represent major hurdles in bacterial community diversity analysis, since the vast majority of bacterial diversity probably lies in “underrepresented minority” populations (24, 30). This is important because taxa that are present only in low abundance may still perform important ecosystem functions (e.g., ammonia-oxidizing bacteria). Of special concern is that biases in detection might invalidate hypothesis testing on complex communities where limited sampling is performed (5).
Recently, there has been a concerted effort toward addressing problems impeding comprehensive bacterial diversity studies (7, 13, 24, 26, 28). In recent years, studies have increased sequencing efforts, with targeted 16S rRNA gene sequence libraries approaching 2,000 clones (11) and high-throughput DNA-sequencing efforts (e.g., via 454 pyrosequencing and newer-generation high-throughput approaches) of up to 149,000 templates from one or a few samples (25, 30). These technological advances have come as researchers recognize that massive sequencing efforts are required to accurately assess the diversity of populations that comprise complex microbial communities (29, 30). Alternatively, where fully aligned sequence comparisons need to be made, novel experimental strategies that allow more-comprehensive detection of underrepresented bacterial taxa can be applied. One such approach involves the application of prefractionation of total bacterial community genomic DNA based on its GC content (hereafter GC fractionation) prior to subsequent molecular manipulations of total community DNA (14). This strategy has been successfully applied in combination with denaturing gradient gel electrophoresis (13) and 16S rRNA gene cloning (2, 21) to study microbial communities. This approach separates community genomic DNA, prior to any PCR, into fractions of similar percent GC content, effectively reducing the overall complexity of the total community DNA mixture by physical separation into multiple fractions. This facilitates PCR amplification, cloning, and detection of sequences in fractions with relatively low abundance in the community, thereby enhancing the detection of minority populations (13). Collectively, this strategy reduces the biases introduced by PCR amplification and random cloning of the extremely complex mixtures of templates of different GC content, primary sequence, and relative abundance present in total environmental genomic DNA.
Any large molecular survey that relies on sequencing further requires the analysis of large amounts of data that must be catalogued into phylogenetically relevant groups. This is usually done using high-throughput methods like RDP Classifier or Sequence Match (6) or a tree-based method like Greengenes (8) or ARB (18). Two major pitfalls that are encountered using these former approaches are the presence of huge numbers of unclassified sequences in databases and the lack of representative sequences from all phyla. This leads to most surveys having large portions of their phylotypes designated as unclassified. The latter tree-based approaches, although better suited for classification schemes, are also dependent on having a comprehensive database with well-classified sequences for reproducible results. This reproducibility becomes especially important when trying to compare data across different studies, especially those that utilize different approaches and study systems.
In the current study, we analyzed an extensive (∼5,000 clones) partial 16S rRNA gene library from a single soil sample that was generated using very general primers and GC-fractionated DNA. Total DNA was extracted from soil at a cultivated treatment plot at the National Science Foundation Long Term Ecological Research (NSF-LTER) site at the Kellogg Biological Station (KBS) in mid-Michigan (http://www.kbs.msu.edu/lter). To test the effect of GC fractionation on recovery of 16S rRNA gene sequences, we conducted a direct comparison with a nonfractionated library generated from the same soil sample. Using the GC-fractionated library, we also calculated several measures of bacterial diversity and examined the effects of sampling size and sequence length on Shannon-Weaver diversity index, Simpson's reciprocal index (1/D, where D is the probability that two randomly selected individuals from a sample belong to the same species), evenness, and Chao1 richness estimation. The results show that GC fractionation is a powerful tool to help mitigate limitations of random PCR- and cloning-based analyses of total microbial community diversity, resulting in the recovery of underrepresented taxa and, in turn, reducing the sampling size needed for accurate estimations of bacterial richness. The results also provided evidence for the need to expand the typical scale of sequence-based survey efforts, particularly in environments where evenness abounds or where minority bacterial populations may have important effects on community function and processes. We suggest that there is a need for the establishment of standardized approaches for the analysis of sequence data from community diversity studies in order to maximize data comparisons across independent studies and show examples of software programs developed to facilitate comparative analysis of large sequence datasets.
MATERIALS AND METHODS
Study site and sample collection.
Samples were collected from the KBS LTER Row-Crop Agriculture site in mid-Michigan (for an overview of that project see http://lter.kbs.msu.edu/). The current study examined the bacterial community in the replicate plots of Treatment 1 at the main experimental site, which is representative of canonical agricultural practice in the upper Midwest. The treatment consisted of conventional wheat, corn, and soybean annual rotations receiving standard levels of chemical inputs, with chisel plowing. Soil was classified as a fine-loamy, mixed, mesic Typic Hapludalfs. For this bacterial population survey, five randomly positioned, 0- to 20-cm soil cores were taken from each of six treatment replicates in July, 2004, at the height of the growing season. Each replicate treatment sample was sieved through 2-mm mesh and mixed thoroughly, providing six replicate samples. All soil samples for this study were stored on dry ice or at −70°C immediately after soil processing (i.e., sieving and mixing) prior to bacterial community DNA extraction.
DNA manipulations.
Total microbial community DNA was extracted and purified from the samples by using the large-scale direct lysis method developed by Holben (12). Equal amounts of DNA (10 μg) from each replicate sample were pooled to provide a representative sample from this treatment regimen that was subsequently fractionated based on the percent GC content of the DNA of the component populations of the community as originally described by Holben and Harris (14). Following centrifugation, the gradients were fractionated into 15 separate fractions representing percent GC contents ranging from 20 to 80% (the full range observed in the domain Bacteria) and the amount and percent GC content of the DNA at each position in the gradient were determined as described elsewhere (1). The DNA in individual fractions was desalted by using PD-10 columns (Amersham Pharmacia Biotech, Piscataway, NJ) with the manufacturer's recommended protocol. Each individual fraction was then PCR amplified independently for creation of the 16S rRNA gene clone library.
PCR conditions employed the primer pair 536f (5′-CAGCMGCCGCGGTAATWC-3′) and 907r (5′-CCGTCAATTCMTTTRAGTTT-3′) (13) and used the optimal reaction and amplification conditions described by Ishii and Fukui (16) for reducing PCR bias, namely, 50-μl volumes containing 10 pg of template DNA, 1× Taq buffer, 200 μM of each deoxynucleoside triphosphate, 25 pmol of each primer, and 1.25 U of Taq polymerase amplified for 21 cycles of 94°C for 1 min, 45°C for 1 min, and 72°C for 2 min. PCR products were cloned by using the plasmid vector pT7Blue-3 and a Perfectly Blunt cloning kit (Novagen, Inc., Madison, WI) according to the manufacturer's instructions. Plasmid clones were purified from 2-ml cultures of Escherichia coli incubated overnight at 37°C with shaking using Qiagen mini-prep kits (Qiagen, Valencia, CA) as recommended by the manufacturer. Restriction analysis using EcoRI was performed to ensure that plasmids contained correctly sized inserts. Plasmid DNA was sequenced by using the universal primer T7 and standard dideoxy sequencing conditions.
Phylogenetic placement and tree creation based on clone libraries.
All 16S rRNA gene sequences were manually trimmed of vector and primer sequence prior to alignment and analysis. Trimmed sequences were subsequently checked for chimeric character and other anomalies by using Pintail (3), and suspect sequences were excluded from further analysis, leaving 4,889 sequences to be analyzed. Multiple Fasta files were created and independently aligned in ARB (18). Alignments were performed in ARB using the Fast Aligner and at least three reference sequences for each clone from the 16S rRNA gene database PT server containing 51,024 reference sequences (http://www.arb-home.de/downloads.html). Sequences from the current study were integrated into the annotated tree based on parsimony.
Assignment to similarity-based OTUs and species richness estimators.
Prior to assignment into operational taxonomic units (OTUs), ARB-generated 16S sequence alignments were used to create Jukes-Cantor corrected distance matrices and exported. These matrices were used as input for the DOTUR program (26), which was used to calculate Simpson's and Shannon-Weaver diversity indices, Chao1 richness estimates, and OTU bins using default settings.
Comparison of GC-fractionated to nonfractionated data was performed by creating a master sequence library containing both fractionated and nonfractionated sequence libraries. Approximately 500 (487 and 490, respectively) sequences were compared for fractionated and nonfractionated libraries by comparing ∼33 sequences obtained from each of the 15 GC-based fractions of the total community to a library of 490 sequences randomly cloned from nonfractionated total community DNA from the same sample. The sequences obtained were aligned in ARB and then run through the DOTUR program. DOTUR data files were then used as input for the SONS program (27), which was used to compare OTU representation within each library.
Identification of phylum-specific taxonomic bins and OTU composition.
To identify distance score cutoff values for individual phyla, we developed the DAM (DOTUR-ARB matching) program (19), available at (http://dbs.umt.edu/research_labs/holbenlab/links.php). This allowed comparison between ARB-generated group lists and DOTUR list files created from the total data set of 4,889 sequences. The DAM program was employed to match a query list of sequence identifications (hereafter, IDs) from ARB to OTUs as determined by the DOTUR program, allowing for a user-specified range of DOTUR distance values. Querying against a DOTUR list file for each distance value in range, the program extracted only OTUs that contained one or more of the query IDs. Results were written to a file formatted as a DOTUR list file, with each line listing the DOTUR distance value, the number of matched OTUs for the prescribed distance, and a list of each bin's contents. For this study, DAM results provided the percent sequence similarity at which an ARB-generated phylum list was contained in a single DOTUR OTU.
In order to identify sequences belonging to specific OTUs, a new program, DOTMAN (for “DOTUR manipulation”; available at http://dbs.umt.edu/research_labs/holbenlab/links.php), was created (19). DOTMAN queries selected OTUs (based on DOTUR bins) against a sequence database, generating FASTA files from a user-given file. To accomplish this, the program is given a range of DOTUR distance values, a DOTUR list file, and a file in FASTA format containing sequences corresponding to the IDs in the list file. For each distance value d, DOTMAN makes one FASTA file for each of the n largest OTUs. n is set by the user and is less than or equal to the total number of OTUs for a distance d.
Sample size simulations.
To explore the effects of sampling size on ecological parameters (Chao1 richness estimation, Shannon-Weaver indices, and dominance), we used EcoSim700 null model software for ecology (version 7.0) to analyze data created from the first 500, 2,000, 3,390, and 5,000 sequences contained in our library. Input files were created from OTUs that clustered with 97% similarity and were subsequently used as the data matrix for running the program.
Nucleotide sequence accession numbers.
All sequences used in this paper have been deposited in the GenBank database (accession no. EU352912 to EU357802).
RESULTS
Effect of sample size on observed and estimated richness.
Environmental rRNA gene libraries vary considerably in size but typically are of 500 sequences or less (4, 20). Although it has been shown that small sample sizes are useful for providing a “snapshot” of the predominant species (29) and they have been employed in theoretical estimates of bacterial species richness (20), there is little empirically derived data actually demonstrating the effect of sample size on ecological parameters, such as richness estimation, dominance, diversity indices, or evenness. To better understand sampling size-induced errors and to better estimate bacterial diversity in soil, we paired the additional resolving power of GC fractionation with the general utility of 16S rRNA gene clone libraries in a microbial community survey of a single soil type.
The effect of sample size was tested by creating datasets from the first 500, 2,000, 3,390, and 5,000 sequenced clones in our GC-fractionated library. Subsequent removal of anomalous and nonbacterial sequences produced sets of 487, 1,962, 3,322, and 4,889 sequences, respectively. These datasets were analyzed based on “bins” created as a function of 16S sequence similarity. Since 16S sequences are not necessarily linked to a whole-genome evolutionary or ecological context, the values chosen for binning are arbitrary and only serve the purpose of creating objectively derived bins that cluster data into a reasonable number of taxonomically related groups (10, 22). In order to facilitate comparison to prior bacterial community diversity studies, the data were grouped at multiple levels of similarity (Table 1), but discussion in this report is focused primarily on the widely utilized 97% sequence similarity level.
TABLE 1.
Effect of sample size on similarity-based OTUs, Shannon-Weaver diversity index, evenness, and richness estimation
| Sequence sample size, % similarity level | No. of unique OTUs | Shannon-Weaver index | Evenness | Richness estimatea | No. of sequences represented in top 10 OTUs
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OTU1 | OTU2 | OTU3 | OTU4 | OTU5 | OTU6 | OTU7 | OTU8 | OTU9 | OTU10 | |||||
| First 500b | ||||||||||||||
| 100 | 461 | 6.10 | 0.995 | 5,851 | 6 | 3 | 3 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| 97 | 335 | 5.62 | 0.966 | 1,020 | 12 | 8 | 7 | 7 | 7 | 7 | 5 | 5 | 5 | 5 |
| 70 | 30 | 2.94 | 0.866 | 32 | 59 | 45 | 43 | 41 | 40 | 31 | 27 | 24 | 24 | 23 |
| 55 | 6 | 0.78 | 0.434 | 6 | 379 | 51 | 43 | 6 | 5 | 3 | ||||
| 47 | 2 | 0.13 | 0.188 | 2 | 473 | 14 | ||||||||
| First 2,000c | ||||||||||||||
| 100 | 1,662 | 7.33 | 0.986 | 12,163 | 22 | 9 | 8 | 8 | 7 | 6 | 5 | 5 | 5 | 5 |
| 97 | 928 | 6.39 | 0.935 | 2,126 | 44 | 33 | 24 | 24 | 24 | 22 | 19 | 18 | 16 | 12 |
| 70 | 67 | 3.34 | 0.794 | 80 | 174 | 156 | 139 | 134 | 132 | 110 | 100 | 97 | 96 | 91 |
| 55 | 14 | 1.69 | 0.641 | 20 | 608 | 597 | 297 | 224 | 91 | 82 | 29 | 12 | 10 | 8 |
| 38 | 6 | 0.04 | 0.025 | 12 | 1,950 | 8 | 1 | 1 | 1 | 1 | ||||
| First 3,390d | ||||||||||||||
| 100 | 2,680 | 7.73 | 0.980 | 15,015 | 40 | 17 | 15 | 15 | 11 | 10 | 8 | 8 | 8 | 7 |
| 97 | 1,319 | 6.59 | 0.918 | 2,991 | 76 | 54 | 54 | 41 | 38 | 31 | 27 | 27 | 27 | 26 |
| 70 | 84 | 3.48 | 0.785 | 88 | 309 | 243 | 237 | 210 | 200 | 182 | 165 | 159 | 150 | 147 |
| 55 | 14 | 1.62 | 0.615 | 14 | 1,284 | 1,064 | 387 | 205 | 85 | 85 | 60 | 54 | 46 | 28 |
| 38 | 2 | 0.02 | 0.032 | 2 | 3,311 | 11 | ||||||||
| First 5,000e | ||||||||||||||
| 100 | 3,789 | 8.04 | 0.976 | 20,790 | 54 | 25 | 22 | 17 | 15 | 14 | 13 | 12 | 12 | 10 |
| 97 | 1,714 | 6.75 | 0.906 | 3,555 | 99 | 81 | 81 | 63 | 62 | 61 | 46 | 39 | 38 | 38 |
| 70 | 102 | 3.60 | 0.778 | 119 | 474 | 345 | 297 | 256 | 248 | 236 | 233 | 215 | 211 | 210 |
| 55 | 18 | 1.98 | 0.685 | 20 | 1,402 | 1,026 | 729 | 504 | 453 | 231 | 221 | 175 | 58 | 37 |
| 38 | 5 | 0.03 | 0.017 | 5 | 4,873 | 13 | 2 | 2 | 1 | |||||
Based on full biased corrected Chao1 richness estimates.
Based on 487 starting sequences.
Based on 1,962 starting sequences.
Based on 3,322 starting sequences.
Based on 4,887 starting sequences and 2 archeal sequences used as references.
A 5.1-fold increase in the number of OTUs and a 3.5-fold increase in the richness estimation were observed (at 97% sequence similarity) from the smallest to the largest data set (Table 1). Shannon diversity index values increased approximately 1.2-fold across this same span, with the Simpson's reciprocal index (1/D) increasing from 202.19 to 341.67, representing a 1.7-fold increase. In contrast, evenness estimates decreased from 0.966 to 0.906 between the smallest and largest data sets, presumably indicating that sampling was approaching a minimal saturation point where low-abundance sequences (unique in the smaller datasets) were being detected more than once.
The largest library, containing 4,889 sequences, represented the most complete survey of aligned 16S rRNA gene sequences from a single composite soil sample and was composed of 1,714 OTUs identified at 97% sequence similarity (Table 1). Projections based on this large data set predict that 3,555 different OTUs were actually present in this soil sample (Table 1) and that a GC-fractionated clone library of well over 10,000 sequences would be required to begin bordering an asymptote in the rarefaction curve.
At 97% sequence similarity, a Shannon-Weaver score of 6.75 was calculated, much greater than the values of 4.35 and 4.68 previously estimated for an Amazon and a Scottish soil, respectively (26). Further, the vast majority of bacterial taxa in the soil were present in very low numbers, producing an extremely high evenness estimate of 0.906, while only a few OTUs exhibited any numerical predominance (Table 1). Our data firmly validate the increasingly common perception (as does a recent report; see reference 29) that numerous taxa present in comparably low overall abundance comprise the bulk of the soil bacterial community.
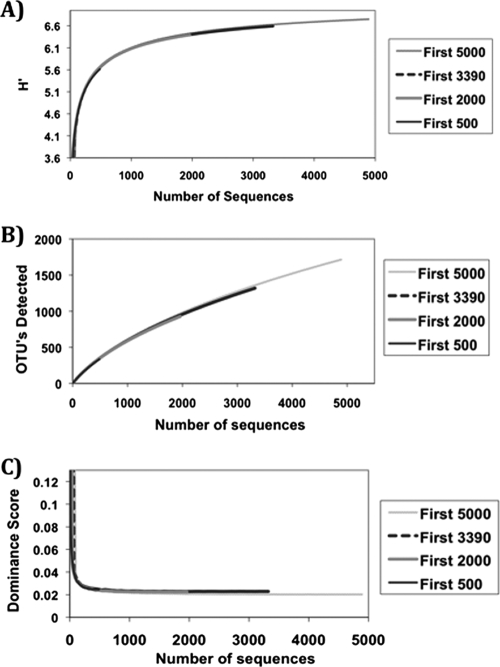
To compare common community diversity measures as a function of different sample sizes, we used EcoSim700 null model software to create species richness estimates, Shannon-Weaver diversity indices, and dominance curves. The results revealed underestimations in all three parameters when using the smaller datasets (Fig. 1). All parameters tested followed a conserved and overlapping general trend with increasing sample size, but the smaller data sets lacked sufficient sequence coverage to indicate an asymptote or to reflect end results comparable to those obtained from the larger data sets.
FIG. 1.
The effects of sampling size on estimated diversity (A), number of OTUs detected (B), and evenness (C). Iterative plots of estimated Shannon-Weaver diversity (H′), OTUs detected, and dominance score were generated for the first 500, 2,000, 3,390, and 5,000 sequences in the partial 16S gene sequence library to demonstrate the effects of sample size on each parameter. Note that the lines in each panel directly overlap as a result of this iterative process and that the trajectory of each estimation curve is extended as sample size increases.
Community composition.
To examine taxonomic representation within the community, we explored two commonly used methods of taxonomic placement for 16S rRNA gene sequence data. Sequences were first analyzed using the Classifier (version 1.0; taxonomical hierarchy release 6.0) and the SeqMatch tools (6) of the RDP. Individual sequences were considered classified only if both programs showed agreement at the phylum level. Unclassified sequences were assigned a potential placement based on Classifier. Using this method, 3,233 (66%) of the sequences were classified (Table 2) into 17 known phyla. These same sequences were also classified using ARB (18) by placement into an ARB-generated phylogenetic tree of 51,024 classified sequences. With this approach, a 33% increase in placement to known phyla was obtained, with 4,854 (99%) sequences assigned to 25 known phylogenetic groups. It is worth noting that the classification of certain groups was comparable using both methods (Table 2), but in groups with low sequence representation within databases (e.g., refer to Chlorobi, Acidobacteria, Thermomicrobia, Fibrobacteres, and candidate divisions of Table 2), the ARB-based approach allowed for more-consistent assignment of bacteria at the phylum level.
TABLE 2.
Taxonomic classification based on multiple methods
| Taxon | ARBa | Total no. of sequences
|
% Sequence similarityd | |
|---|---|---|---|---|
| Classifiedb | Unclassifiedc | |||
| Phylum | ||||
| Acidobacteria | 955 | 88 | 41 | 38 |
| Actinobacteria | 491 | 452 | 39 | 38 |
| Bacteroidetes | 453 | 450 | 17 | 59 |
| CD OD1 | 68 | 0 | 0 | 49 |
| CD OP10 | 43 | 18 | 0 | 38 |
| CD OP11 | 14 | 6 | 6 | 27 |
| CD OP3 | 12 | 0 | 0 | 53 |
| CD TM6 | 3 | 0 | 0 | 83 |
| CD TM7 | 17 | 11 | 2 | 56 |
| CD WS1 | 2 | 0 | 0 | 99 |
| CD WS3 | 34 | 17 | 5 | 38 |
| Chamydiae | 2 | 2 | 10 | 88 |
| Chlorobi | 22 | 0 | 1 | 70 |
| Chloroflexi | 27 | 32 | 18 | 55 |
| Cyanobacteria | 12 | 6 | 57 | 38 |
| Defferibacteres | 0 | 0 | 2 | |
| Deinococcus-Thermus | 1 | 0 | 3 | 100 |
| Dictyoglomi | 0 | 0 | 21 | |
| Fibrobacteres | 11 | 0 | 0 | 60 |
| Firmicutes | 15 | 16 | 800 | 48 |
| Gemmatimonadetes | 251 | 195 | 33 | 38 |
| Lentisphaerae | 0 | 0 | 1 | |
| Nitrospira | 59 | 47 | 0 | 38 |
| Planctomycetes | 243 | 174 | 22 | 38 |
| Proteobacteria | 1,690 | 1,631 | 506 | 21 |
| Spirochaetes | 3 | 1 | 5 | 38 |
| Thermodesulfobacteria | 0 | 0 | 4 | |
| Thermomicrobia | 323 | 0 | 10 | 38 |
| Thermotogae | 0 | 0 | 1 | |
| Verrucomicrobia | 103 | 89 | 50 | 43 |
| Unclassified | 35 | 1,654 | ||
| Class | ||||
| Alphaproteobacteria | 374 | 368 | 13 | 21 |
| Betaproteobacteria | 485 | 475 | 0 | 43 |
| Deltaproteobacteria | 348 | 278 | 22 | 38 |
| Epsilonproteobacteria | 1 | 0 | 0 | 99 |
| Gammaproteobacteria | 478 | 468 | 7 | 38 |
| Unclassified Proteobacteria | 4 | |||
Classification based on ARB-generated tree. CD, candidate division.
Sequences were considered classified if assigned to the same phylum using both SeqMatch and Classifier of the RDP.
Unclassified sequences were assigned to likely phylum based on Classifier results.
Based on ARB phylum level classification.
Since the analysis reported herein was performed, a new release (34) of the Classifier tool has been made available (version 2.0; taxonomical hierarchy release 7.8). Reanalysis of our data set with this new release produced taxonomic placements that were nearly identical to those obtained with ARB for classified sequences. Despite this, Classifier was still unable to classify 1,013 (21%) of the sequences in this library.
Using DOTUR, a total of 1,405 OTUs (at 97% sequence similarity), comprising 82% of all identified OTUs, were represented three or fewer times in this 4,889-sequence library. When the data were reanalyzed to include all OTUs represented 19 or fewer times (half the value of the 10th most predominant OTU), 99% of all OTUs in the study were included in this category. This represents 83% of all sequences in the full library. In order to provide some phylogenetic context to the predominant OTU bins generated by DOTUR using the 97% similarity cutoff, we analyzed the 10 most predominant taxa, which were represented by only 99 (OTU1; Gammaproteobacteria), 81 (OTU2; Acidobacteria), 81 (OTU3; Gammaproteobacteria), 63 (OTU4; Thermomicrobia), 62 (OTU5; Betaproteobacteria), 61 (OTU6; Acidobacteria), 46 (OTU7; Thermomicrobia), 39 (OTU8; Alphaproteobacteria), 38 (OTU9; Gammaproteobacteria), and 38 (OTU10; Betaproteobacteria) sequences out of 4,889 (Tables 1 and 2).
Effect of sequence length on community analysis.
The region of the 16S rRNA gene used to generate the clone library in the current study is approximately 400 bp in length, spanning between E. coli positions 518 and 927 and encompassing two hypervariable regions (V4 and V5). Further, the highly conserved regions representing primers 536f and 907r (15) were removed prior to analysis because the minor degeneracies built into these primers potentially introduce errors into the sequences analyzed.
To test the effect that using this smaller (versus full-length) but highly variable region had on data analysis, we created a 1,184-sequence library from (nearly) full-length sequences in the ARB database. These reference sequences covered all of the phyla detected and were selected as having the greatest similarity to the sequences within our own library, thus serving as proxies to the sequences obtained in the current study. These reference sequences were analyzed separately as both full-length and truncated sequences (by trimming to match the 536-to-907 region, excluding primers) to create distance matrices at 97% sequence similarity which were used as input for the DOTUR program (26). Fairly modest differences were observed for the truncated and full-length sequences, with 911 and 1,031 OTUs identified, respectively (Table 3). Likewise, the Shannon-Weaver indices derived from truncated and full-length sequences were slightly different, being 6.65 and 6.86, respectively. A substantial effect on Chao1 richness estimation was detected however, with an almost-twofold increase in estimated OTUs when full-length sequences were used for the analysis, presumably reflecting the additional fineness of phylogenetic resolving power afforded by the additional sequence information.
TABLE 3.
Effect of fragment length on similarity-based OTU number, Shannon-Weaver diversity index, and richness estimation
| Sequence length, % similarity level | No. of unique OTUs | Shannon-Weaver index | Richness estimatea |
|---|---|---|---|
| Full lengthb | |||
| 100 | 1,183 | 7.08 | 350,169 |
| 97 | 1,031 | 6.86 | 7,452 |
| 70 | 54 | 3.01 | 67 |
| 55 | 6 | 0.67 | 6 |
| 46 | 2 | 0.01 | 2 |
| Truncatedc | |||
| 100 | 1,166 | 7.05 | 61,646 |
| 97 | 911 | 6.65 | 4,175 |
| 70 | 80 | 3.41 | 93 |
| 55 | 15 | 1.68 | 18 |
| 46 | 5 | 0.47 | 6 |
Based on full biased corrected Chao1 richness estimates.
Based on 1,184 full-length sequences.
Based on 1,184 truncated sequences. Truncations were created by deleting the upstream base pair region from the E. coli consensus position 536 and downstream of consensus position 906.
We further tested the effect of sequence length on outcome by comparing taxonomic placements based on full-length versus truncated sequences and found that comparable results were obtained for both. In 1,166 of the 1,184 cases (98.5%), congruent taxonomic assignments were obtained with the truncated sequences, while in only 16 cases (1.5%) did the additional sequence information result in a different taxonomic assignment (Table 3). Phylum-level classification based on ARB-based tree generation was highly reproducible independent of fragment length (Table 4). Collectively, this suggests that the ∼400-bp V4-V5 region examined for our survey, which is readily obtained from a single dideoxy sequence reaction, is sufficient to provide reliable phylogenetic placement at phylum and higher-order levels. The effect of sequence length on finer-level placement (genus and species) was not examined, being outside the context of the current study.
TABLE 4.
Effect of sequence length on taxonomic placement and distance based on ARB alignment
| Taxona | Full-length
|
Truncated
|
||
|---|---|---|---|---|
| % Sequence similarity | No. of sequences | % Sequence similarity | No. of sequences | |
| Phylum | ||||
| Acidobacteria | 58 | 176 | 46 | 175 |
| Bacteroidetes | 52 | 106 | 46 | 107 |
| CD OD1 | 57 | 20 | 46 | 20 |
| Gemmatimonadetes | 75 | 42 | 51 | 41 |
| Planctomycetes | 46 | 88 | 39 | 88 |
| Class | ||||
| Betaproteobacteria | 61 | 141 | 54 | 144 |
CD, candidate division.
Assignment of cutoff values for phylogenetic clusters.
Previous publications have suggested that sequences sharing >60 to 80% identity likely belong to the same phylum (26, 29). Using this guideline, we empirically assessed the feasibility of employing such a “universal” cutoff value for phylum-level discrimination and the effect of using truncated versus full-length sequences to determine cutoff values. To accomplish this, we developed and applied the DAM program, which matches a list of query sequences (belonging to a discrete group [i.e., phylum-level cluster] as determined by ARB) to a distance matrix-determined OTU group encompassing all sequences in a given query list (as created by the DOTUR program). This allowed determination of the percent sequence similarity at which groupings of sequences were identified as discrete OTUs. For this exercise, phylum-level groups with both large and smaller numbers of sequences were compared again using the ARB-derived full-length and truncated sequences described above, as well as the KBS-LTER study data set. The results showed that no single, consistent consensus value can be used as a phylum-level cutoff point across all taxa (Tables 1 and 4). With our own large data set, we annotated 102 separate groups at 70% sequence similarity, greatly exceeding estimates of 36 to 52 extant bacterial phyla suggested by Rappé and Giovannoni (24). At 55% sequence similarity, 18 groups were defined, in line with the number of phyla expected to be in soil (Table 1).
Comparison of fractionated versus nonfractionated DNA libraries.
In order to clearly test the effect of GC fractionation on the recovery of low-abundance sequences in the complex mixture of bacterial community DNA, a direct comparison was made between 16S rRNA gene clone libraries generated with and without the use of GC fractionation. No substantive differences in phylum or genus level community composition were detected (see the supplemental material). However, when these aligned sequences were analyzed using the DOTUR and EcoSim programs, a species detection (also known as rarefaction) curve of OTUs detected at 97% sequence similarity indicated a higher rate of recovery of new phylotypes for the GC-fractionated library (Table 5; see the supplemental material). In addition, the values for the Shannon-Weaver diversity index, evenness, and Chao1 richness estimation were all higher for the GC-fractionated DNA (Table 5).
TABLE 5.
Effect of GC fractionation on similarity-based OTU numbers, Shannon-Weaver diversity indices, and richness estimates
| Fractionation status | No. of unique OTUs | Shannon-Weaver index | Evenness | Richness estimate | % of shared OTUsc |
|---|---|---|---|---|---|
| GC fractionateda | 335 | 5.62 | 0.966 | 1,020 | 64 |
| Not GC fractionatedb | 301 | 5.45 | 0.954 | 780 | 74 |
Based on 487 starting sequences.
Based on 490 starting sequences.
OTUs identified in both libraries.
To compare community composition and classification results for the above-mentioned libraries, the data were analyzed using the SONS program (27), which was designed to compare OTUs between libraries in order to establish patterns of community membership and structure based on sequence comparisons. This analysis indicated that GC fractionation facilitated the detection of a higher number of OTUs, both shared and unique, from the same soil bacterial community (Table 5; see the supplemental material).
Community composition.
We relied on a tree-based approach utilizing an ARB-annotated (18) sequence library into which our sequences were placed for assignment of phylogenies. Essentially all of the sequences in the study were assigned into 25 known phyla by this approach, with just 35 of the 4,898 sequences not assignable to any known phylum or group (Table 2). The most predominant phylum in this soil was the Proteobacteria, which comprised 35% of the sequence library, followed by the Acidobacteria with 20% of the total. Six other phyla, including Actinobacteria, Bacteroidetes, Thermomicrobia, Gemmatimonadetes, Planctomycetes, and Verrucomicrobia, averaged 7% representation. The remaining phyla were represented by numbers of phylotypes totaling <2% of the total library.
DISCUSSION
Based on the results presented herein, this agricultural soil bacterial community was empirically demonstrated to be a highly complex assemblage with extremely broad evenness. Such a community composition requires vast sequencing efforts to even approach onefold coverage of richness and to obtain reliable results for traditional ecological parameters originally developed for the analysis of many metazoan communities. One way to mitigate sample size requirements for complete coverage of community diversity is to reduce sample complexity and disparity in abundance between taxa by prefractionation of community DNA, using methods such as GC fractionation. Using this method, 1,714 OTUs were detected at a sequence similarity level cutoff of 97% (representing a new OTU for every 2.9 sequences acquired), with an estimated 3,555 OTUs present. These values are potentially underestimations due to the focus on an ∼400-bp hypervariable region within the 16S gene, with the corrected richness estimation for full-length sequences approaching 6,500 OTUs (based on the twofold increase detected in our data) (Table 3). Compared to the results of other, conventional 16S rRNA gene clone library-based soil studies (26), our library exhibits an ∼1.6-fold increase in the Shannon-Weaver diversity index, most likely due to the 50-fold increase in sample size and DNA prefractionation approach employed. This is the highest index reported to date for a bacterial community and presumably reflects the additional resolution afforded by the unique combination of existing and novel approaches employed.
While it may seem intuitive even in the absence of empirical data as presented here, the comparison of different-sized libraries from the same sample clearly demonstrates that for highly complex bacterial communities, such as those typically found in surface soils, rich sampling of 16S rRNA gene sequences (i.e., several thousand) is necessary to obtain a robust measure or estimation of community diversity parameters. This is especially true where even near saturation of sampling curves is not feasible or is seemingly impossible due to large numbers of taxa exhibiting high degrees of evenness, or where theoretical estimates based on sample sizes under 1,000 do not appear to be accurate (e.g., asymptotic behavior is not yet apparent in a sampling curve). The importance of at least approaching sampling saturation is supported by a recent publication indicating that surveys missing or ignoring a small subset (e.g.<10%) of species result in minimal loss of information but that more-extensive gaps substantially increase rates of information loss (33).
To directly compare the effectiveness of GC fractionation for sampling coverage, we compared our results to a nonfractionated 500-clone library from the same soil sample, which produced lower recovery of OTUs, as well as a lower Shannon diversity index and less evenness. The main benefit of GC fractionation prior to PCR amplification is the reduction in DNA complexity within each fraction which allows underrepresented sequences to be detected more readily than in a random survey. This resulted in a higher recovery rate for minority species and more-even detection of total diversity, thereby reducing the required survey size needed to approach complete coverage of the entire bacterial community.
The low and variable levels of sequence similarity required to sort this large group of sequences into phylum-level bins comparable to those suggested by other soil microbiological studies suggests that having a universally applied phylum-level cutoff is impractical and would not apply across the full range of known bacterial taxa. Additionally, the sample size (number of sequences within a given group) showed no correlation with the percent sequence similarity required for clustering, suggesting that there are actual differences in the degree of 16S sequence variation between different phyla. This observation potentially represents different evolutionary strategies between phyla at the molecular level where ribosomal-gene sequence conservation is concerned.
When full-length sequence data were compared to those for the 400-bp region of focus in our library, a 1.4-fold increase in sequence similarity at the phylum level was observed. We suggest that this is explained by the hypervariable nature of the 536f-907r-sequenced region as mentioned above, especially given that the conserved primer regions at each end were removed prior to analysis. The inclusion of nearly full-length sequences in the comparison would introduce several additional highly conserved areas into the analyses and thus lower the overall variation observed between longer 16S rRNA gene sequences. Contrary to what was observed for percent sequence similarity, phylum-level classification based on ARB-based tree generation was highly reproducible independent of fragment length, as shown in Table 4.
Based on the data presented here, we suggest that GC fractionation or other prefractionation approaches for reducing complexity within total community DNA prior to PCR and cloning are useful for DNA-based phylogenetic surveys of microbial community diversity. We further suggest that, even with prefractionation of community DNA, 16S rRNA gene clone libraries of at least 2,000 sequences are required to achieve reliable results for estimating ecological parameters, such as richness, evenness, and diversity, for complex bacterial communities such as those typically found in surface soils. The results also validate the use of the 536f-907r primer set for rapid and relatively inexpensive analysis of total bacterial diversity based on single, unidirectional sequence reads that support binning into a reasonable number of OTUs. This strategy provided sufficient resolution for the analyses described herein. However, analysis of full-length or nearly full-length sequences is highly recommended where phylogenetic placement at the genus or near-species level is desired. The determination of evolutionary relatedness between organisms requires the use of large stretches of genetic information. This is especially true for highly conserved genes, such as the 16S rRNA gene (10).
Wherever possible, phylogenetic surveys should use large library sizes and scrutinize data using multiple taxonomic tools. As part of our study, we used methods from the study of Thompson et al. (31) (Clustal W alignments) and MUSCLE software (9), which produced datasets with similar numbers of OTUs, Chao1 richness estimates, and other diversity parameters. However, phylogenetic trees generated from those approaches did not produce coherent clustering with phylogenetic assignments using RDP tools (not shown). In contrast, phylogenetic trees generated using ARB alignments were reproducible and provided consistent phylogenetic placement with the RDP toolset.
The continued use of nucleic acid sequence-based phylogenetic approaches will yield more information, providing additional insights into the effectiveness and validity of current phylogenetic classification strategies and whether they reflect fundamental biological properties. Continued evolution of this general approach should come with the development of a common platform for data acquisition and analysis, which would allow for microbial community comparisons across multiple studies. Special focus should be given to a universal set of rules for assigning bacterial phylogenies. Although our data clearly suggest that there is no universal cutoff value for phylum assignment, it does not provide enough insight to suggest a specific number of phyla in our sample based strictly on sequence similarity, nor does it suggest phylum-specific cutoff values which might come from a more-complete integration of all data in reliably assigned phylotypes present in extant databases. The fact that sequences in the current study were only reliably affiliated to higher-order phylogenetic groups (phylum level and higher) highlights the need to develop a clearer definition for bacterial phylogenetic assignments at the genus and species level that are based on more than just single 16S rRNA gene sequences. In closing, we suggest that additional studies are needed to explore the extent of taxonomic variance within and between phylogenetic groups to provide additional ecological and biological context that will underpin bacterial community diversity studies into the future.
Supplementary Material
Acknowledgments
Funding for this project was provided by the United States Department of Agriculture, National Research Initiative (USDA-CSREES grant no. 2004-03501).
Soil samples for this project were graciously provided by the KBS-LTER. We also gratefully acknowledge the Ribosomal Database Project (RDP-II) for aid in the chimera check process, as well as Linda Schimmelpfennig and Tara Westlie for technical assistance in performing GC fractionations, cloning, and sequence processing. We thank Jared Rapp for aiding in computer-based data analysis.
Footnotes
Published ahead of print on 14 November 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Apajalahti, J. H. A., A. Kettunen, M. R. Bedford, and W. E. Holben. 2001. Percent G+C profiling accurately reveals diet-related differences in the gastrointestinal microbial community of broiler chickens. Appl. Environ. Microbiol. 67:5656-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apajalahti, J. H. A., H. Kettunen, A. Kettunen, W. E. Holben, P. H. Nurminen, N. Rautonen, and M. Mutanen. 2002. Culture-independent microbial community analysis reveals that inulin in the diet primarily affects previously unknown bacteria in the mouse cecum. Appl. Environ. Microbiol. 68:4986-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724-7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72:5734-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brose, U., N. D. Martinez, and R. J. Williams. 2003. Estimating species richness: sensitivity to sample coverage and insensitivity to spatial patterns. Ecology 84:2364-2377. [Google Scholar]
- 6.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, A. M. Bandela, E. Cardenas, G. M. Garrity, and J. M. Tiedje. 2007. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35:D169-D172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curtis, T. 2006. Microbial ecologists: it's time to “go large”. Nature 4:488. [DOI] [PubMed] [Google Scholar]
- 8.DeSantis, T. Z., P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu, and G. L. Andersen. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanage, W. P., C. Fraser, and B. G. Spratt. 2006. Sequences, sequence clusters and bacterial species. Philos. Trans. R. Soc. B 361:1917-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartmann, M., and F. Widmer. 2006. Community structure analyses are more sensitive to differences in soil bacterial communities than anonymous diversity indices. Appl. Environ. Microbiol. 72:7804-7812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holben, W. E. 1997. Isolation and purification of bacterial community DNA from environmental samples, p. 431-436. In C. J. Hurst et al. (ed.), Manual of environmental microbiology, 1st ed. ASM Press, Washington, DC.
- 13.Holben, W. E., K. P. Feris, A. Kettunen, and J. H. A. Apajalahti. 2004. GC fractionation enhances microbial community diversity assessment and detection of minority populations of bacteria by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 70:2263-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holben, W. E., and D. Harris. 1995. DNA-based monitoring of total bacterial community structure in environmental samples. Mol. Ecol. 4:627-631. [DOI] [PubMed] [Google Scholar]
- 15.Holben, W. E., P. Williams, M. A. Gilbert, M. Saarinen, L. K. Särkilahti, and J. H. Apajalahti. 2002. Phylogenetic analysis of intestinal microflora indicates a novel mycoplasma phylotype in farmed and wild salmon. Microb. Ecol. 44:175-185. [DOI] [PubMed] [Google Scholar]
- 16.Ishii, K., and M. Fukui. 2001. Optimization of annealing temperature to reduce bias caused by a primer mismatch in multitemplate PCR. Appl. Environ. Microbiol. 67:3753-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janssen, P. H. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72:1719-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morales, S. E., T. Cosart, J. V. Johnson, and W. E. Holben. 25 July 2008. Supplemental programs for enhanced recovery of data from the DOTUR application. J. Microbiol. Methods 75:572-575. [DOI] [PubMed] [Google Scholar]
- 20.Narang, R., and J. Dunbar. 2004. Modeling bacterial species abundance from small community surveys. Microb. Ecol. 47:396-406. [DOI] [PubMed] [Google Scholar]
- 21.Nusslein, K., and J. M. Tiedje. 1998. Characterization of the dominant and rare members of a young Hawaiian soil bacterial community with small-subunit ribosomal DNA amplified from DNA fractionated on the basis of its guanine and cytosine composition. Appl. Environ. Microbiol. 64:1283-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oren, A. 2004. Prokaryote diversity and taxonomy: current status and future challenges. Philos. Trans. R. Soc. B 359:623-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rappé, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Ann. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 25.Roesch, L. F. W., R. R. Fulthorpe, A. Riva, G. Casella, A. K. M. Hadwin, A. D. Kent, S. H. Daroub, F. A. O. Camargo, W. G. Farmerie, and E. W. Triplett. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1:283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schloss, P. D., and J. Handelsman. 2006. Introducing SONS, a tool for operational taxonomic unit-based comparisons of microbial community memberships and structures. Appl. Environ. Microbiol. 72:6773-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schloss, P. D., and J. Handelsman. 2004. Status of the microbial census. Microbiol. Mol. Biol. Rev. 68:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schloss, P. D., and J. Handelsman. 2006. Toward a census of bacteria in soil. PLoS Comput. Biol. 2(7):e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sogin, M. L., H. G. Morrison, J. A. Huber, D. M. Welch, S. M. Huse, P. R. Neal, J. M. Arrieta, and G. J. Herndl. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. USA 103:12115-12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tiedje, J. M., S. Asuming-Brempong, K. Nusslein, T. L. Marsh, and S. J. Flynn. 1999. Opening the black box of soil microbial diversity. Appl. Soil Ecol. 13:109-122. [Google Scholar]
- 33.Vellend, M., P. L. Lilley, and B. M. Starzomski. 2007. Using subsets of species in biodiversity surveys. J. Appl. Ecol. 45:161-169. [Google Scholar]
- 34.Wang, Q., G. M. Garrity, J. M. Tiedje, and J. R. Cole. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.