Abstract
The glucose dehydrogenase (GDH) from Bacillus subtilis BGSC 1A1 was cloned and functionally expressed in Escherichia coli BL21(pGDH1) and XL-1 Blue(pGDH1). Controlled permeabilization of recombinant E. coli BL21 and XL-1 Blue with EDTA-toluene under optimized conditions resulted in permeabilized cells with specific activities of 61 and 14 U/g (dry weight) of cells, respectively, for the conversion of NADP+ to NADPH upon oxidation of glucose. The permeabilized recombinant strains were more active than permeabilized B. subtilis BGSC 1A1, did not exhibit NADPH/NADH oxidase activity, and were useful for regeneration of both NADH and NADPH. Coupling of permeabilized cells of Bacillus pumilus Phe-C3 containing an NADPH-dependent ketoreductase and an E. coli recombinant expressing GDH as a novel biocatalytic system allowed enantioselective reduction of ethyl 3-keto-4,4,4-trifluorobutyrate with efficient recycling of NADPH; a total turnover number (TTN) of 4,200 mol/mol was obtained by using E. coli BL21(pGDH1) as the cofactor-regenerating microorganism with initial addition of 0.005 mM NADP+. The high TTN obtained is in the practical range for producing fine chemicals. Long-term stability of the permeabilized cell couple and a higher product concentration were demonstrated by 68 h of bioreduction of ethyl 3-keto-4,4,4-trifluorobutyrate with addition of 0.005 mM NADP+ three times; 50.5 mM (R)-ethyl 3-hydroxy-4,4,4-trifluorobutyrate was obtained with 95% enantiomeric excess, 84% conversion, and an overall TTN of 3,400 mol/mol. Our method results in practical synthesis of (R)-ethyl 3-hydroxy-4,4,4-trifluorobutyrate, and the principle described here is generally applicable to other microbial reductions with cofactor recycling.
Biocatalytic oxidoreductions are important reactions in asymmetric synthesis, and they have great potential for industrial production of enantiopure chemicals and pharmaceuticals (4, 11, 23, 24, 26). These reactions often require a stoichiometric amount of the expensive cofactor NAD(P)H or NAD(P)+, and thus practical applications require efficient recycling of the necessary cofactor (1, 5, 9, 13, 16, 17, 22, 32). In general, cofactor recycling can be achieved by coupling a desired enzymatic reaction with an additional chemical, electrochemical, photocatalytic, or enzymatic reaction, and the enzymatic method is favored (1, 5, 9, 13, 16, 17, 22, 32, 39). Enzymatic cofactor recycling can be obtained by using “coupled-substrate” (8, 12, 30, 31) and “coupled-enzyme” (14, 15, 18, 19, 20, 21, 29, 33, 34) approaches. The latter is a more general approach and utilizes the first enzyme for the desired biotransformation and the second enzyme for cofactor regeneration. Formate dehydrogenase (15, 18) and glucose dehydrogenase (GDH) (33, 34) are well-known enzymes used for regeneration of NADH and NADPH, respectively. The “coupled-enzyme” approach has been successfully used with two isolated enzymes (15, 18, 29, 33, 34) or whole cells (14, 20, 21) of a microorganism coexpressing the two necessary enzymes. While the use of isolated enzymes is costly, the use of whole cells depends on the availability of an intracellular cofactor which may be limiting and cannot be altered by addition of an extracellular cofactor. On the other hand, whole cells can be made permeable to NAD(P)H and NAD(P)+ by treatment with an organic solvent or detergent, while the enzymatic activity remains high (3, 6, 7, 27). Thus, using readily available permeabilized cells for oxidoreduction with cofactor recycling might have an advantage over using the isolated-enzyme and whole-cell approaches.
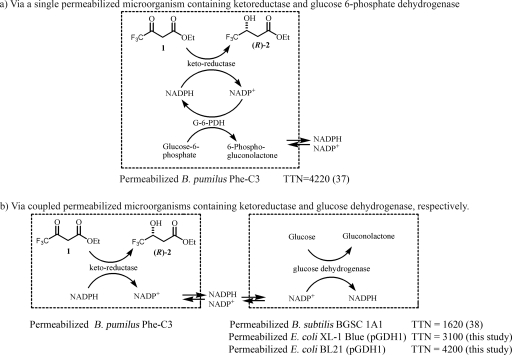
We recently discovered that Bacillus pumilus Phe-C3 containing an NADPH-dependent ketoreductase catalyzed the enantioselective reduction of ethyl 3-keto-4,4,4-trifluorobutyrate (3-ketoester 1), resulting in the corresponding (R)-ethyl 3-hydroxy-4,4,4-trifluorobutyrate [(R)-3-hydroxyester 2], a useful intermediate for the preparation of the antidepressant Befloxatone (10), with 95% enantiomeric excess (36). This strain was also found to contain an NADP+-dependent glucose-6-phosphate dehydrogenase (37). Treatment of cells of B. pumilus Phe-C3 with EDTA-toluene gave active permeabilized cells that catalyzed the bioreduction of 3-ketoester 1 and the recycling of NADPH (37). Under optimized conditions, use of permeabilized cells of B. pumilus Phe-C3 with the initial supply of glucose-6-phosphate and 0.005 mM NADP+ resulted in bioreduction of 3-ketoester 1 with recycling of NADPH 4,220 times (37) (Fig. 1). The high total turnover number (TTN) for cofactor recycling significantly reduced the cofactor cost for the bioreduction. However, the requirement for two enzymes in one microorganism and using relatively expensive glucose-6-phosphate may limit the scope of the application of the single-permeabilized-microorganism approach. To solve these problems, we recently developed a novel general approach involving the use of two permeabilized microorganisms for bioreduction with cofactor recycling (38); permeabilized cells of B. pumilus Phe-C3 containing a ketoreductase were coupled with permeabilized cells of Bacillus subtilis BGSC 1A1 containing a GDH (35) for bioreduction of 3-ketoester 1 to produce (R)-3-hydroxyester 2, which resulted in a TTN for NADPH recycling of 1,600 with initial addition of glucose and 0.01 mM NADP+. This method had several advantages. Compared with the whole-cell approach, this approach enables use of the externally added cofactor for efficient catalysis and cofactor recycling, and it allows easy substrate access and easy product release. Compared with the isolated-enzyme approach, permeabilized cells are cheap, active for a longer period, stable, and reusable, and large amounts are readily available. The coupled-permeabilized-cell approach also has great potential due to its general applicability in addition to the advantages described above. However, so far the highest TTN for NADPH recycling with this approach is only 1,620. To make this method suitable for practical synthesis of fine chemicals, the TTN of the expensive cofactor has to be increased significantly. To achieve this goal, we have been working on development of better cofactor-regenerating systems, a better permeabilized-cell couple, and efficient synthesis of (R)-3-hydroxyester 2 by the new biocatalyst system. Here we report our recent success in such development for practical bioreduction with efficient NADPH recycling.
FIG. 1.
Bioreduction with NADPH recycling by using permeabilized microorganisms. “(37)” and “(38)” indicate references 37 and 38, respectively. OEt, OC2H5; G-6-PDH, glucose-6-phosphate dehydrogenase; 1, ethyl 3-keto-4,4,4-trifluorobutyrate; (R)-2, (R)-ethyl 3-hydroxy-4,4,4-trifluorobutyrate.
MATERIALS AND METHODS
Chemicals.
NADP+ (>99%), NAD+ (>99%), ampicillin (>99%), and ethyl 3-keto-4,4,4-trifluorobutyrate 1 (>98%) were purchased from Sigma-Aldrich. Isopropyl-β-d-thiogalactopyranoside (IPTG) (>99%) was obtained from 1st BASE. The medium components tryptone and yeast extract were purchased from Biomed Diagnostics. Ethyl 3-hydroxyl-4,4,4-trifluorobutyrate 2 was prepared by using previously described procedures (28).
Analytical method.
The assays for GDH activity and NADPH or NADH oxidase activity were carried out by using a Shimadzu UV-Vis 1700 spectrophotometer with the time scan function. The concentrations of ethyl 3-keto-4,4,4-trifluorobutyrate 1 and ethyl-3-hydroxyl-4,4,4-trifluorobutyrate 2 were analyzed by using an Agilent GC 6890 and an Agilent HP-5 column (25 m by 0.32 mm) with an inlet temperature of 290°C and a detector temperature of 310°C. The temperature program was as follows: 50°C for 8 min and then the temperature increased to 300°C at a rate of 50°C/min and kept at 300°C for 2 min. The retention times were 2.8 and 9.3 min for 3-ketoester 1, 6.2 min for 3-hydroxyester 2, and 12.5 min for n-hexadecane. The enantiomeric excess of (R)-ethyl-3-hydroxyl-4,4,4-trifluorobutyrate 2 was analyzed by using an Agilent GC 6890 with a Lipodex A column (25 m by 0.25 mm) and the following temperature program: temperature increased from 40 to 120°C at a rate of 5°C/min and then to 170°C at a rate of 45°C/min. The retention times were 11.9 min for (S)-3-hydroxyester 2 and 12.2 min for (R)-3-hydroxyester 2.
Strains and cultivation media.
B. subtilis BGSC 1A1 was obtained from the Bacillus Genetic Stock Center at The Ohio State University; Escherichia coli XL-1 Blue, E. coli BL21, and B. pumilus Phe-C3 (36) were obtained from the collections of our laboratories. Cells of B. pumilus Phe-C3 were grown in E2 medium containing glucose (1%, wt/vol) at 25°C in a 2-liter fermentor, and the cells were harvested and permeabilized by using a previously described procedure (38). E. coli XL-1 Blue(pGDH1) and BL21(pGDH1) were grown in LB medium containing ampicillin (100 μg/ml).
Genetic engineering of E. coli XL-1 Blue(pGDH1) and BL21(pGDH1).
Genomic DNA of B. subtilis BGSC 1A1 was obtained using the Trizol reagent (Sigma) according to the manufacturer's instructions. Full-length GDH was amplified by high-fidelity PCR with GDH primers. Genomic DNA was used as the template. Primer BSG-TAA1 (5′GGTAAGCTTCTCGAGTTAACCGCGG CCTGCCTG3′) and primer BSG-ATG1 (5′CAGGAATTCATACATGTATCCAGAT TTAAAAGGAA3′) were designed by using the nucleotide sequence of the GDH of B. subtilis BGSC 1A1. These primers contained restriction sites for HindIII and EcoRI, respectively. The amplification conditions used to obtain the 817-bp PCR product were 95°C for 5 min (hot start), followed by 30 cycles of 95°C for 45 s, 55°C for 45 s, and 72°C for 45 s and finally 72°C for 7 min. The GDH fragment was digested with EcoRI (Roche) and HindIII (Roche) for 90 min at 37°C and was inserted into the lacZ expression vector pUC18 using the restriction enzyme sites to form pGDH1.
E. coli XL-1 Blue cells were made chemically competent and were then transformed with pGDH1. The insertion was confirmed by plasmid prep (Qiagen) from cell culture and restriction digestion analysis using EcoRI and HindIII. The GDH protein was expressed in recombinant E. coli XL-1 Blue by using the inducible lac promoter under the control of IPTG. Similarly, plasmid pGDH1 was transferred into E. coli BL21 by the electroporation method.
Growth and GDH activity of E. coli BL21(pGDH1) and XL-1 Blue(pGDH1).
Strains BL21(pGDH1) and XL-1 Blue(pGDH1) were inoculated onto LB agar plates (10 g tryptone, 5 g yeast extract, and 5 g NaCl in 1 liter deionized water with 1.5% agar) containing ampicillin (100 μg/ml) and grown overnight at 37°C. A single colony from the LB agar plate containing each strain was inoculated into 100 ml of LB medium with ampicillin (100 μg/ml) and grown at 250 rpm and 37°C for 12 h, which resulted in optical densities at 450 nm (OD450) of 8.0 (2.2 g [dry weight] of cells per liter) for E. coli BL21(pGDH1) and 6.0 (1.7 g [dry weight] of cells per liter) for E. coli XL-1 Blue(pGDH1).
Twelve milliliters of the preculture of E. coli BL21(pGDH1) described above was added to 800 ml of LB medium containing ampicillin (100 μg/ml), and the mixture was shaken at 250 rpm and 37°C. Samples (1 ml) were taken at different time points for measurement of the OD450 and an activity test. IPTG (1 mM) was added when the OD450 reached 1.8 (0.5 g [dry weight] of cells per liter) after 2.5 h. The cells were harvested at the late exponential phase at an OD450 of 5.8 (1.6 g [dry weight] of cells per liter) after 6 h, washed with potassium phosphate (KP) buffer (5 mM; pH 7.5), and then stored in a freezer at −80°C.
Twelve milliliters of the preculture of E. coli XL-1 Blue(pGDH1) described above was added to 600 ml of LB medium containing ampicillin (100 μg/ml), and the mixture was shaken at 250 rpm and 37°C. Samples (1 ml) were taken at different time points for measurement of the OD450 and an activity test. IPTG (1 mM) was added when the OD450 reached 1.2 (0.3 g [dry weight] of cells per liter) after 3 h. The cells were harvested at the late exponential phase at an OD450 of 7.2 (2.0 g [dry weight] of cells per liter) after 10 h, washed with KP buffer (5 mM; pH 7.5), and then stored at −80°C.
To test the GDH activity, 7-ml suspensions (10 g [dry weight] of cells per liter) of each recombinant E. coli strain in KP buffer (50 mM; pH 7.5) were passed through a homogenizer (Constant cell disruption system) twice at 20,000 lb/in2. The cell debris was removed by centrifugation at 13,000 × g at 4°C for 30 min. The supernatant was diluted 10-fold with KP buffer (50 mM; pH 7.5), and the protein concentration was determined to be 0.4 g protein/liter by using the Bradford protein content assay (2) with bovine serum albumin as a standard. To 940 μl of the prepared cell extract (CE) 50 μl of a glucose stock solution (2.0 M) and 10 μl of an NADP+ solution (0.2 M) were added. The formation of NADPH was monitored by determining the UV absorption at 340 nm at 25°C, and the concentration was calculated by using a ɛ340 of 6.22 liters mmol−1. The specific GDH activity was expressed in units/g protein, and 1 U was defined as the formation of 1 μmol NADPH/min.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed by loading 15 μl CE (0.21 g protein/liter) on a gel containing 0.1% SDS and 10% acrylamide, staining the gel with a 0.1% solution of Coomassie brilliant blue R-250 in methanol-acetic acid-water (4:1:5, vol/vol/vol), and destaining the gel by soaking it in deionized water overnight.
Preparation and GDH activity of permeabilized cells of E. coli BL21(pGDH1) and XL-1 Blue(pGDH1).
Frozen cells of E. coli BL21(pGDH1) and XL-1 Blue(pGDH1) were thawed for 2 h and resuspended in Tris-HCl buffer (100 mM; pH 8.0) to obtain a density of 10 g (dry weight) of cells per liter. For E. coli BL21(pGDH1), EDTA (10 mM) and toluene (1%, vol/vol) were added, and the mixture was shaken at 300 rpm and 25°C for 10 min. For E. coli XL-1 Blue(pGDH1), EDTA (5 mM) was added, and the mixture was shaken at 300 rpm and 25°C for 30 min and then incubated at 4°C for 1 h. Permeabilized cells were obtained by centrifugation at 4°C and 3,320 × g for 10 min, and the GDH activity assay was performed by examining the bioconversion of glucose (55 mM) with permeabilized cells (0.2 g [dry weight] of cells per liter) in Tris-HCl buffer (100 ml; pH 7.0) in the presence of NADP+ (2 mM) using a 1-ml scale at 25°C and monitoring NADPH formation at 340 nm.
Kinetics of the GDH activity of the permeabilized cells of E. coli BL21(pGDH1).
To 900 to 945 μl of permeabilized cells of E. coli BL21(pGDH1) (0.2 g [dry weight] of cells per liter) in Tris-HCl buffer (100 mM; pH 7.0) 50 μl of a glucose stock solution (2.0 M) and 5 to 50 μl of 0.02 M NADP+ or NAD+ were added. The reaction was monitored by determining the UV absorbance at 340 nm at 25°C. The initial velocities (v) were obtained from the curves for NADPH or NADH concentration versus time, and they were used to plot 1/v versus 1/[S] (where [S] is the concentration of NADP+ or NAD+) (25).
NADPH and NADH oxidase activities of the permeabilized cells of E. coli BL21(pGDH1).
Permeabilized E. coli BL21(pGDH1) cells were resuspended in 990 μl Tris-HCl buffer (100 mM; pH 7.0) to a density of 0.2 g (dry weight) of cells per liter, and 10 μl of 0.2 M NADPH or NADH was added. The concentration of NADPH or NADH was monitored by determining the UV absorption at 340 nm at 25°C.
General procedure for bioreduction of ethyl 3-keto-4,4,4-trifluorobutyrate 1 with NADPH recycling with coupled permeabilized microorganisms.
To a suspension containing permeabilized cells of B. pumilus Phe-C3 (20 to 40 g [dry weight] of cells per liter) and E. coli XL-1 Blue(pGDH1) or BL21(pGDH1) (20 to 40 g [dry weight] of cells per liter) in 10 ml Tris-HCl buffer (100 mM; pH 7.0), ethyl 3-keto-4,4,4-trifluorobutyrate 1 (110 mg; 60 mM), NADP+ (0.04 mg; 0.005 mM), and glucose (0.8 g; 440 mM) were added. The mixture was shaken at 25°C and 300 rpm, and aliquots (300 μl) were taken at different time points for gas chromatography (GC) analysis. Additional 3-keto-ester 1 (97 mg; 60 mM) was added at 5 h. More glucose (1.5 to 2.0 M) was added at several time points. Analytic samples were prepared by centrifugation, 4:1 (vol/vol) dilution, extraction with the same volume of chloroform containing 2 mM hexadecane that was used for the internal standard, and desiccation with anhydrous Na2SO4. GC analysis was used to determine the product concentrations at different time points, and the final TTN for NADPH recycling was calculated by dividing the number of moles of product formed by the number of moles of NADP+ added.
Bioreduction of ethyl 3-keto-4,4,4-trifluorobutyrate 1 with NADPH recycling 4,200 times with coupled permeabilized cells of B. pumilus Phe-C3 and E. coli BL21(pGDH1).
To a suspension of permeabilized cells of B. pumilus Phe-C3 (40 g [dry weight] of cells per liter) and E. coli BL21(pGDH1) (20 g [dry weight] of cells per liter) in 10 ml Tris-HCl buffer (100 mM; pH 7.0), ethyl 3-keto-4,4,4-trifluorobutyrate 1 (110 mg; 60 mM), NADP+ (0.04 mg; 0.005 mM), and glucose (0.8 g; 440 mM) were added. The mixture was shaken at 25°C and 300 rpm. Additional 3-ketoester 1 (97 mg; 60 mM) was added at 5 h. More glucose was added at 5 h (0.70 g; 440 mM), 10 h (0.67 g; 440 mM), 19 h (0.65 g; 440 mM), 24 h (0.16 g; 110 mM), and 29 h (0.15 g; 110 mM). At different time points, analytic samples were removed, prepared using the method described above, and analyzed by GC.
Bioreduction of ethyl 3-keto-4,4,4-trifluorobutyrate 1 with NADPH recycling for 96 h by using coupled permeabilized cells of B. pumilus Phe-C3 and E. coli BL21(pGDH1) with four additions of 0.005 mM NADP+.
The reaction procedure described above was used until 24 h. Additional NADP+ (0.005 mM) was added at 24, 48, and 72 h. More glucose was added at 24 h (0.16 g; 110 mM), 29 h (0.15 g; 110 mM), 34 h (0.57 g; 440 mM), 44 h (0.55 g; 440 mM), 48 h (0.13 g; 110 mM), 53 h (0.12 g; 110 mM), 58 h (0.48 g; 440 mM), 68 h (0.47 g; 440 mM), 72 h (0.11 g; 110 mM), 77 h (0.10 g; 110 mM), 82 h (0.38 g; 440 mM), and 92 h (0.36 g; 440 mM). At different time points, samples were removed, prepared using the method described above, and analyzed by GC. The reaction was stopped at 96 h.
RESULTS
Genetic engineering, cell growth, and GDH activity of recombinant E. coli expressing GDH.
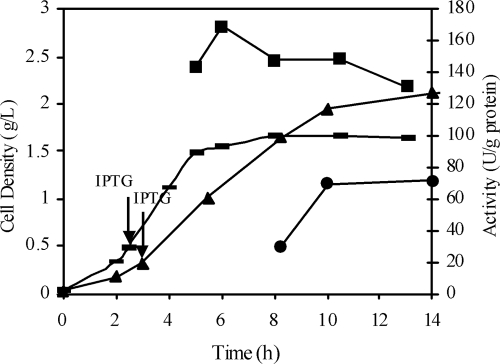
Primers BSG-TAA1 and BSG-ATG1 were designed to clone the gdh gene of B. subtilis BGSC 1A1 based on the known gene sequence (35). The full-length gdh gene was amplified by high-fidelity PCR with these primers. The gdh gene fragment was digested with EcoRI (Roche) and HindIII (Roche) and inserted into the lacZ expression vector pUC18. The plasmid formed, pGDH1, was then transformed into E. coli XL-1 Blue and BL21. The two recombinant E. coli strains were grown in LB medium containing ampicillin (100 μg/ml) at 37°C, and their GDHs were induced by addition of 0.5 to 1.0 mM IPTG after the lag phase. Typical growth curves are shown in Fig. 2. E. coli BL21(pGDH1) grew faster at the beginning and reached the stationary phase at 5 h, when the cell density was 1.5 g (dry weight) of cells per liter. On the other hand, E. coli XL-1 Blue(pGDH1) grew slower at the beginning and reached the stationary phase at 10 h, when the cell density was 2.0 g (dry weight) of cells per liter.
FIG. 2.
Growth and GDH activities of E. coli XL-1 Blue(pGDH1) and E. coli BL21(pGDH1). Symbols: ▴, E. coli XL-1 Blue(pGDH1) cell growth;  , E. coli BL21(pGDH1) cell growth; •, GDH activity of E. coli XL-1 Blue(pGDH1) CE; ▪, GDH activity of E. coli BL21(pGDH1) CE.
, E. coli BL21(pGDH1) cell growth; •, GDH activity of E. coli XL-1 Blue(pGDH1) CE; ▪, GDH activity of E. coli BL21(pGDH1) CE.
The GDH activities at different time points were determined by removing samples from the E. coli cultures, preparing cell-free extract, determining the protein concentrations with a bioassay (2), and examining the GDH activities (33) by adding 1% glucose and 2 mM NADP+ to the CE and monitoring the NADPH formation by determining the UV absorption at 340 nm. As shown in Fig. 2, the highest activities were 170 U/g protein at 6 h for E. coli BL21(pGDH1) and 70 U/g protein at 10 h for E. coli XL-1 Blue(pGDH1).
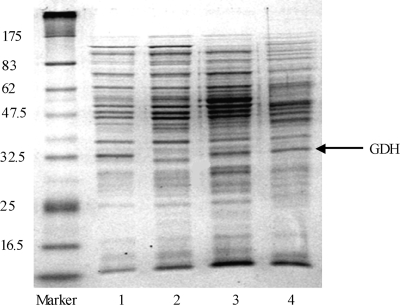
SDS-PAGE (Fig. 3) revealed the proteins in the CE of E. coli BL21(pGDH1) (lane 1), E. coli BL21(pUC18) (lane 2), E. coli XL-1 Blue(pGDH1) (lane 3), and B. subtilis BGSC 1A1 (lane 4). The GDH band was clearly visible for the two pGDH1-containing recombinant strains, and E. coli BL21(pGDH1) showed a higher level of expression of GDH than E. coli XL-1 Blue(pGDH1).
FIG. 3.
SDS-PAGE of E. coli BL21(pGDH1) (lane 1), E. coli BL21(pUC18) (lane 2), E. coli XL-1 Blue(pGDH1) (lane 3), and B. subtilis BGSC 1A1 (lane 4).
Preparation and GDH activity of permeabilized cells of E. coli recombinants expressing GDH.
The GDH activity of whole cells was examined by adding 1% glucose and 2 mM NADP+ to a cell suspension in KP buffer (0.2 g [dry weight] of cells per liter) and monitoring the NADPH formation by determining the UV absorption at 340 nm. Fresh cells of E. coli BL21(pGDH1) showed no activity in this assay. However, frozen and thawed cells exhibited an activity of 25 U/g (dry weight) of cells, indicating that there was partial permeabilization of cells. Further freeze-thaw treatments resulted in increased activity of the cells after each cycle, and the activity was 45 U/g (dry weight) of cells after five cycles. However, the activity decreased to 39 U/g (dry weight) of cells after six treatments. Nevertheless, the CE of the cells treated six times showed full activity (170 U/g protein), indicating that the GDH was stable under the treatment conditions and the cell membranes were not fully permeabilized for cofactor across.
To prepare more active permeabilized cells of E. coli BL21(pGDH1), treatment with a surfactant and an organic solvent was examined. The cells were incubated with EDTA (5 to 10 mM) containing toluene (0 to 1%) at 25°C and 300 rpm for 10 to 30 min, and then the permeabilized cells were harvested by centrifugation and the GDH activity was determined. As shown in Table 1, permeabilized cells prepared by incubation with 10 mM EDTA containing 1% toluene for 10 min exhibited the highest level of activity, 61 U/g (dry weight) of cells, which was higher than the level of activity obtained after five freeze-thaw treatments, 45 U/g (dry weight) of cells.
TABLE 1.
Preparation conditions and GDH activities of permeabilized cells of E. coli XL-1 Blue(pGDH1) and BL21(pGDH1)
| E. coli cells | EDTA concn (mM) | Toluene concn (%, vol/vol) | Time (min) | Activity (U/g [dry wt] of cells) |
|---|---|---|---|---|
| BL21(pGDH1) | 5 | 0 | 30a | 23 |
| BL21(pGDH1) | 10 | 0 | 30a | 24 |
| BL21(pGDH1) | 5 | 1 | 30a | 24 |
| BL21(pGDH1) | 5 | 0 | 10b | 26 |
| BL21(pGDH1) | 10 | 0 | 10b | 38 |
| BL21(pGDH1) | 10 | 1 | 10b | 61 |
| XL-1 Blue(pGDH1) | 5 | 0 | 30a | 14 |
Samples were shaken at 25°C and 300 rpm for 30 min and then incubated on ice for 1 h.
Samples were shaken at 25°C and 300 rpm for 10 min without treatment on ice.
Similarly, EDTA (5 to 10 mM) and toluene (0 to 1%) were used for treatment of cells of E. coli XL-1 Blue(pGDH1). The best permeabilization conditions were treatment with 5 mM EDTA without toluene, which resulted in a GDH activity of 14 U/g (dry weight) of cells.
GDH kinetics and NAD(P)H oxidase activity of E. coli BL21(pGDH1).
To investigate the cofactor dependence and the affinity of NADP+ or NAD+ for GDH in permeabilized E. coli BL21(pGDH1) cells, a set of activity assays were performed with NADP+ or NAD+ at different concentrations (0.1 to 1.0 mM) using a suspension of permeabilized cells (0.2 g [dry weight] of cells per liter) in Tris-HCl buffer (100 mM; pH 7.0) containing glucose (55 mM) at 25°C. The reaction was monitored by determining the UV absorption at 340 nm. The initial velocities at different concentrations of NADP+ or NAD+ were used to plot 1/v versus 1/[S] (25), which resulted in a Km of 0.90 mM and a Vmax of 130 U/g (dry weight) of cells for NADP+ and a Km of 1.4 mM and a Vmax of 76 U/g (dry weight) of cells for NAD+ for the permeabilized cells. Such permeabilized cells could be used for regeneration of NADPH and NADH. The Vmax/Km value, which indicates enzyme efficiency, was 2.7-fold higher for NADPH regeneration than for NADH regeneration.
To examine the NADH or NADPH oxidase activity of the permeabilized cells of E. coli recombinants, oxidation of NADH or NADPH was performed using cell suspensions (0.2 g [dry weight] of cells per liter) in Tris-HCl buffer (100 mM; pH 7.0) in the presence of NADPH (2 mM) or NADH (2 mM) at 25°C for 30 min. The NADPH or NADH concentration was determined by using the UV absorbance at 340 nm. In both cases, no NADPH or NADH was consumed, suggesting that the permeabilized E. coli recombinant cells did not contain NADPH or NADH oxidase.
Coupling of permeabilized cells of B. pumilus Phe-C3 and recombinant E. coli expressing GDH for bioreduction of 3-keto-4,4,4-trifluorobutyrate 1 with NADPH recycling.
The potential for using permeabilized cells of E. coli BL21(pGDH1) or XL-1 Blue(pGDH1) for cofactor recycling was examined by studying the enantioselective reduction of 3-ketoester 1 to the corresponding (R)-3-hydroxyester 2. Previously, the best TTN obtained for NADPH recycling during reduction was 1,600 when permeabilized cells of B. pumilus Phe-C3 and B. subtilis BGSC 1A1 were used at concentrations of 20 and 40 g (dry weight) of cells per liter, respectively, along with initial addition of 0.01 mM NADP+ (38). For comparison, 0.005 mM NADP+ was used for the same biotransformation, which resulted in the same TTN shown in Table 2. Coupling of permeabilized cells of E. coli XL-1 Blue(pGDH1) with an activity of 14 U/g (dry weight) of cells (40 g [dry weight] of cells per liter) with permeabilized cells of B. pumilus Phe-C3 (20 g [dry weight] of cells per liter) for bioreduction of 3-ketoester 1 with an initial NADP+ concentration of 0.005 mM at 25°C and 300 rpm for 30 h resulted in a TTN of 3,100.
TABLE 2.
Coupled permeabilized cells of B. pumilus Phe-C3 and a cofactor-regenerating microorganism for bioreduction of ethyl 3-keto-4,4,4-trifluorobutyrate 1 with NADPH recycling
| 3-Ketoester 1 concn (mM)a | NADP+ concn (mM) | Glucose concn (mM)b | B. pumilus Phe-C3 concn (g [dry wt] of cells/liter) | B. subtilis BGSC 1A1 concn (g [dry wt] of cells/liter) | E. coli XL-1 Blue(pGDH1) concn (g [dry wt] of cells/liter) | E. coli BL21(pGDH1) concn (g [dry wt] of cells/liter) | Time (h) | (R)-3-Hydroxyester 2 concn (mM) | TTN |
|---|---|---|---|---|---|---|---|---|---|
| 60 + 60 | 0.005 | 440 | 20 | 40 | 43 | 8.0 | 1,600 | ||
| 60 + 60 | 0.005 | 440 | 20 | 40 | 43 | 15.5 | 3,100 | ||
| 60 + 60 | 0.005 | 440 | 20 | 30c | 39 | 15.0 | 3,000 | ||
| 60 + 60 | 0.005 | 440 | 20 | 40c | 39 | 16.0 | 3,200 | ||
| 60 + 60 | 0.005 | 440 | 30 | 30c | 39 | 17.5 | 3,700 | ||
| 60 + 60 | 0.005 | 440 | 40 | 20d | 34 | 21.0 | 4,200 | ||
| 60 + 60 | 0.001 | 440 | 40 | 20d | 34 | 25.0 | 2,500 |
3-Ketoester 1 was added at the beginning (60 mM) and at 5 h (60 mM).
More glucose (1.5 to 2.0 M) was supplied during the reaction.
Permeabilized cells with an activity of 24 U/g (dry weight) of cells were used.
Permeabilized cells with an activity of 61 U/g (dry weight) of cells were used.
Permeabilized cells of E. coli BL21(pGDH1) with an activity of 24 U/g (dry weight) of cells, prepared by treatment with EDTA (10 mM), were then examined for cofactor regeneration. Coupling of B. pumilus Phe-C3 (20 g [dry weight] of cells per liter) with E. coli BL21(pGDH1) (30 g [dry weight] of cells per liter) led to recycling of NADPH 3,000 times in the bioreduction. Increasing the density of E. coli BL21(pGDH1) to 40 g (dry weight) of cells per liter resulted in a slight increase in the TTN from 3,000 to 3,200. Increasing the amount of the reducing enzyme had more influence on the cofactor TTN; bioreduction of 3-ketoester 1 with 30 g (dry weight) of cells per liter of B. pumilus Phe-C3 and 30 g (dry weight) of cells per liter of E. coli BL21(pGDH1) afforded a TTN of 3,700 for the recycling of NADPH.
Finally, permeabilized cells of E. coli BL21(pGDH1) with an activity of 61 U/g (dry weight) of cells, prepared by treatment with 10 mM EDTA containing 1% toluene, were used for the coupled-permeabilized-cell approach. Since these cells had very high activity for NADPH regeneration, they were used at a concentration of 20 g (dry weight) of cells per liter along with 40 g (dry weight) of B. pumilus Phe-C3 cells per liter for bioreduction of 3-ketoester 1. After initial addition of NADP+ (0.005 mM) and glucose (440 mM), bioreduction for 34 h resulted in a TTN of 4,200 for NADPH recycling.
Long-term bioreduction of ethyl 3-keto-4,4,4-trifluorobutyrate 1 with efficient NADPH recycling by the coupled-permeabilized-cell approach with addition of NADP+ multiple times.
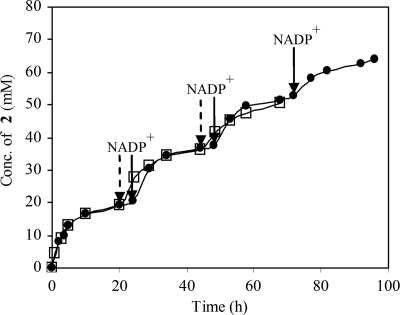
Bioreduction of 60 mM 3-ketoester 1 with permeabilized cells of B. pumilus Phe-C3 (40 g [dry weight] of cells) and E. coli BL21(pGDH1) (20 g [dry weight] of cells) was performed for 68 h with three additions of 0.005 mM NADP+. As shown in Fig. 4, the reaction rate was high at beginning, and 16.5 mM (R)-3-hydroxyester 2 was produced in the first 10 h. From 10 to 20 h, the reaction rate decreased. Nevertheless, the product concentration was 19.0 mM in the first cycle, with a TTN of 3,800. Addition of another 0.005 mM NADP+ at 20 h resulted in fast conversion of 3-ketoester 1 to (R)-3-hydroxyester 2. The findings for the second and third cycles are similar to the findings for the first cycle. As shown in Table 3, (R)-3-hydroxyester 2 with an enantiomeric excess of 95% was formed at 36.0 mM with an overall cofactor TTN of 3,600 at 44 h and at 50.5 mM with an overall TTN of 3,400 and 84% conversion at 68 h. In another experiment, the bioreduction reaction was performed with 120 mM 3-ketoester 1 for 96 h with four additions of 0.005 mM NADP+. Figure 4 shows that the curves for two cases in the first 68 h were nearly the same. At 72 h the concentration of (R)-3-hydroxyester 2 formed was 52.5 mM, and the overall TTN was 3,500. Addition of 0.005 mM NADP+ at this time point accelerated bioreduction again, and the product concentration was 64.0 mM at 96 h with an overall TTN of 3,200.
FIG. 4.
Product formation during bioreduction of ethyl 3-keto-4,4,4-trifluorobutyrate (compound 1) by using coupled permeabilized cells with 0.005 mM NADP+ added at different time points. •, B. pumilus Phe-C3 (40 g [dry weight] of cells per liter) and E. coli BL21(pGDH1) (20 g [dry weight] of cells per liter; 61 U/g [dry weight] of cells) with 120 mM 3-ketoester 1; □, B. pumilus Phe-C3 (40 g [dry weight] of cells per liter) and E. coli BL21(pGDH1) (20 g [dry weight] of cells per liter; 61 U/g [dry weight] of cells) with 60 mM 3-ketoester 1. 2, (R)-ethyl 3-hydroxy-4,4,4-trifluorobutyrate.
TABLE 3.
Product formation for bioreduction of ethyl 3-keto-4,4,4-trifluorobutyrate 1 with coupled permeabilized cells
| B. pumilus Phe-C3 concn (g [dry wt] of cells/liter) | E. coli BL21 concn (g [dry wt] of cells/liter)a | Ethyl 3-keto-4,4,4-triflurobutyrate concn (mM) | Glucose concn (mM) | Activity (U/g [dry wt] of cells)b | Time (h) | NADP+ concn (mM) | (R)-Ethyl 3-hydroxy-4,4,4-triflurobutyrate concn (mM) | % Conversion | TTN |
|---|---|---|---|---|---|---|---|---|---|
| 40 | 20 | 60 | 440 | 1.3 | 0 | 0.005 | |||
| 20 | +0.005c | 19.0 | 32 | 3,800 | |||||
| 44 | +0.005 | 36.0 | 60 | 3,600 | |||||
| 68 | 50.5 | 84 | 3,400 | ||||||
| 40 | 20 | 60 + 60d | 440 | 1.3 | 0 | 0.005 | |||
| 24 | +0.005 | 20.0 | 17 | 4,000 | |||||
| 48 | +0.005 | 38.0 | 31 | 3,800 | |||||
| 72 | +0.005 | 52.5 | 44 | 3,500 | |||||
| 96 | 64.0 | 53 | 3,200 |
Permeabilized E. coli BL21(pGDH1) cells with an activity of 61 U/g (dry weight) of cells were used.
Activity was determined over the first 1 h.
+0.005 indicates that 0.005 mM NADP+ was added at the time indicated.
Ethyl 3-keto-4,4,4-trifluorobutyrate 1 was added at the beginning (60 mM) and at 5 h (60 mM).
DISCUSSION
Previously, permeabilized cells of B. subtilis BGSC 1A1 containing GDH were used for regeneration of NADPH in the coupled-permeabilized-cell approach (38). The GDH activity was, however, only about 5.0 U/g (dry weight) of cells (38), which is the limiting factor for obtaining a high cofactor TTN. In addition, this strain was found to have NADPH oxidase activity which competed with the NADPH-dependent bioreduction. Thus, our first goal was to engineer a recombinant strain expressing GDH from B. subtilis BGSC 1A1 that had higher GDH activity and no NADPH oxidase activity. Cloning and expression of GDH in two E. coli recombinants were successful, as demonstrated by the GDH bands in SDS-PAGE gels, as well as the observed GDH activities of the two E. coli strains. The conditions for cell growth and GDH expression were examined. Temperatures ranging from 20 to 37°C and IPTG concentrations ranging from 0.5 to1.0 mM did not influence the GDH activity and expression, and induction at an early stage was necessary. For both E. coli strains, high activity in the stationary phase was observed over a period of 4 to 6 h (Fig. 2). The GDH activities obtained for E. coli BL21(pGDH1) and XL-1 Blue(pGDH1) (170 and 72 U/g protein, respectively) are much higher than that of the wild-type Bacillus strain.
The preparation of permeabilized cells with high activity is a very important step toward successful application of the whole concept. In the case of E. coli BL21(pGDH1), a simple freeze-thaw treatment partially permeabilized the cells. However, five or six freeze-thaw treatments resulted in cells with a GDH activity of 39 to 45 U/g (dry weight) of cells, which is much lower than the theoretical value; the CE of the resulting cells had an activity of 170 U/g protein. Based on the assumption that 50% of the dry weight of cells is the weight of the total protein, the highest possible activity of permeabilized cells is 85 U/g (dry weight) of cells. Treatment of cells with a surfactant and an organic solvent turned out to be a better method. Under optimized conditions, the prepared permeabilized cells had an activity of 61 U/g (dry weight) of cells, which is higher than the activity obtained by freeze-thaw treatment and close to the theoretic maximum for the cells. The amount of toluene used was very critical. Use of more than 1% toluene decreased the activity. Also, the incubation time was important; 10 min was better than 30 min. In the case of E. coli XL-1 Blue(pGDH1), although many conditions were examined, the permeabilized cells had rather low GDH activity (14 U/g [dry weight] of cells). Nevertheless, this activity was more than double the activity of the permeabilized cells of the wild-type Bacillus strain.
The more active permeabilized cells of an E. coli recombinant were suitable catalysts for the regeneration of NADPH, as well as NADH. In addition, they contained no NADPH/NADH oxidase activity. With permeabilized cells of B. subtilis BGSC 1A1 as the cofactor regeneration microorganism, a TTN of 1,600 was obtained for bioreduction of 3-ketoester 1 with initial addition of 0.01 mM NADP+ (38). The TTN could not be increased by adding a smaller amount of cofactor, such as 0.005 mM NADP+ (Table 2). However, it increased significantly to 3,100 when E. coli XL-1 Blue(pGDH1) with an activity of 14 U/g (dry weight) of cells was used instead of the wide-type strain. A further increase in the GDH activity when even more active cells of E. coli BL21(pGDH1) were used did not increase the TTN much (Table 2). Obviously, the limiting factor was the inefficiency of the reducing microorganism. Increasing the cell density of B. pumilus Phe-C3 to 40 g (dry weight) of cells per liter and coupling with E. coli BL21(pGDH1) (20 g [dry weight] of cells per liter) with an activity of 61 U/g (dry weight) of cells led to an increase in the TTN for NADPH recycling to 4,200. This is a very significant improvement in the cofactor TTN for the coupled-permeabilized-cell approach; considering the price of NADP+ (6 to 7 euros/g), recycling of NADPH 4,200 times significantly reduced the cofactor cost and made the process practical for producing final chemicals and pharmaceutical intermediates. The high TTN obtained for the bioreduction of 3-ketoester 1 with the two-permeabilized-organism approach is similar to the TTN obtained for the same reduction with a single permeabilized microorganism with two necessary enzymes (37). This indicates that the cofactor can easily move between the two types of permeabilized microbial cells and provides the same catalytic efficiency as it does in a single permeabilized cell. In the current system, NADP+ was used at a concentration of 0.005 mM, which is far below the Km. Increasing the NADP+ concentration to 0.01 mM should have resulted in doubling of the rate for the regeneration of NADPH. However, the rate of reduction increased only slightly (21 versus 25 mM product [Table 3]), and the TTN of cofactor recycling was significantly reduced. This was due mainly to the low activity of the reductase-containing microorganism. Thus, the system could be further improved by using a highly active reducing microorganism.
Long-term stability of the permeabilized- cell couple and a higher product concentration were observed for 68- and 96-h reductions of 3-ketoester 1 with three and four additions of 0.005 mM NADP+, respectively. In each cycle of bioreduction shown in Fig. 4, the bioreduction was quite fast in the first 10 h and decreased from 10 h to 20 to 24 h, which may have been due to decomposition of the cofactor, since the half-life of NADPH in the buffer was 24 h (34). The permeabilized cells showed long-term stability, and they were still active after 96 h. The concentration of (R)-3-hydroxyester 2 was 50.5 mM at 68 h, with an overall TTN of 3,400 and 84% conversion. This is a very good result. In a comparison experiment, bioreduction of 60 mM 3-ketoester 1 with resting cells of B. pumilus Phe-C3 (40 g [dry weight] of cells per liter) for 25 h resulted in only 20.5 mM (R)-ethyl 3-hydroxy-4,4,4-trifluorobutyrate 2 with 34% conversion.
In conclusion, the GDH of B. subtilis BGSC 1A1 was cloned and functionally expressed in E. coli BL21(pGDH1), and the activity was 170 U/g protein. The recombinant strain was successfully permeabilized with EDTA-toluene to obtain an activity of 61 U/g (dry weight) of cells for NADPH regeneration. The permeabilized cells exhibited much higher GDH activity and no NADPH or NADH oxidase activity, which was an advantage compared with the wild-type strain. They were suitable for regeneration of both NADPH and NADH. Coupling of permeabilized cells of B. pumilus Phe-C3 and E. coli BL21(pGDH1) for the reduction of ethyl 3-keto-4,4,4-trifluorobutyrate 1 resulted in a TTN of 4,200 for NADPH recycling, which is 2.6 times greater than the TTN obtained by using B. subtilis BGSC 1A1. The high TTN is in the range which is practical for the synthesis of fine chemicals. Bioreduction of ethyl 3-keto-4,4,4-trifluorobutyrate 1 using coupled permeabilized cells with three additions of 0.005 mM NADP+ resulted in 50.5 mM (R)-ethyl 3-hydroxy-4,4,4-trifluorobutyrate 2 with 95% enantiomeric excess and 84% conversion with an overall TTN of 3,400 for NADPH recycling. These results demonstrated the high stability and productivity of the new biocatalyst system and a practical method for synthesis of (R)-ethyl 3-hydroxy-4,4,4-trifluorobutanoate 2. Our method could be applicable to other microbial reduction reactions with cofactor recycling.
Acknowledgments
Financial support from the Singapore-MIT Alliance through the Flagship Research Projects in Chemical and Pharmaceutical Engineering Program is acknowledged.
Footnotes
Published ahead of print on 1 December 2008.
REFERENCES
- 1.Adlercreutz, P. 1996. Cofactor regeneration in biocatalysis in organic media. Biocatal. Biotransform. 14:1-30. [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Canovas, M., T. Torroglosa, and J. L. Iborra. 2005. Permeabilization of Escherichia coli cells in the biotransformation of trimethylammonium compounds into l-carnitine. Enzyme Microb. Tech. 25:300-308. [Google Scholar]
- 4.Chang, D., J. Zhang, B. Witholt, and Z. Li. 2004. Chemical and enzymatic synthetic methods for asymmetric oxidation of the C-C double bond. Biocatal. Biotransform. 22:113-131. [Google Scholar]
- 5.Chenault, H. K., and G. M. Whitesides. 1987. Regeneration of nicotinamide cofactors for use in organic synthesis. Appl. Biochem. Biotechnol. 14:147-197. [DOI] [PubMed] [Google Scholar]
- 6.Denyer, S. P., and J.-Y. Maillard. 2002. Cellular impermeability and uptake of biocides and antibiotics in gram-negative bacteria. J. Appl. Microbiol. Symp. Suppl. 92:35S-45S. [PubMed] [Google Scholar]
- 7.De Smet, M. J., J. Kingma, and B. Witholt. 1978. The effect of toluene on the structure and permeability of the outer and cytoplasmic membranes of Escherichia coli. Biochim. Biophys. Acta 506:64-80. [DOI] [PubMed] [Google Scholar]
- 8.Eckstein, M., M. V. Filho, A. Liese, and U. Kragl. 2004. Use of an ionic liquid in a two-phase system to improve an alcohol dehydrogenase catalysed reduction. Chem. Commun. 2004:1084-1085. [DOI] [PubMed] [Google Scholar]
- 9.Eckstein, M., T. Daussmann, and U. Kragl. 2004. Recent developments in NAD(P)H regeneration for enzymatic reductions in one- and two-phase systems. Biocatal. Biotransform. 22:89-96. [Google Scholar]
- 10.Emilien, G. 1999. Befloxatone. Idrugs 2:247-253. [PubMed] [Google Scholar]
- 11.Faber, K. 1997. Biotransformations in organic chemistry, p. 160-200. Springer, Berlin, Germany.
- 12.Filho, M. V., T. Stillger, M. Muller, A. Liese, and C. Wandrey. 2003. Is log P a convenient criterion to guide the choice of solvents for biphasic enzymatic reactions? Angew. Chem. Int. Ed. 42:2993-2996. [DOI] [PubMed] [Google Scholar]
- 13.Goldberg, K., K. Schroer, S. Lütz, and A. Liese. 2007. Biocatalytic ketone reduction—a powerful tool for the production of chiral alcohols. Part I. Processes with isolated enzymes. Appl. Microbiol. Biotechnol. 76:237-248. [DOI] [PubMed] [Google Scholar]
- 14.Groeger, H., C. Rollmann, F. Chamouleau, I. Sebastien, O. May, W. Wienand, and K. Drauz. 2007. Enantioselective reduction of 4-fluoroacetophenone at high substrate concentration using a tailor-made recombinant whole-cell catalyst. Adv. Synth. Catal. 349:709-712. [Google Scholar]
- 15.Groeger, H., W. Hummel, C. Rollmann, F. Chamouleau, H. Husken, H. Werner, C. Wunderlich, K. Aboktse, K. Drauz, and S. Buchholz. 2004. Preparative asymmetric reduction of ketones in a biphasic medium with an (S)-alcohol dehydrogenase under in situ-cofactor-recycling with a formate dehydrogenase. Tetrahedron 60:633-640. [Google Scholar]
- 16.Hollmann, F., and A. Schmid. 2004. Electrochemical regeneration of oxidoreductases for cell-free biocatalytic redox reactions. Biocatal. Biotransform. 22:63-88. [Google Scholar]
- 17.Hummel, W. 1999. Large-scale application of NAD(P)-dependent oxidoreductases: recent developments. TIBTECH 17:487-492. [DOI] [PubMed] [Google Scholar]
- 18.Hummel, W., K. Abokitse, K. Drauz, C. Rollmann, and H. Groeger. 2003. Towards a large-scale asymmetric reduction process with isolated enzymes: expression of an (S)-alcohol dehydrogenase in E. coli and studies on the synthetic potential of this biocatalyst. Adv. Synth. Catal. 345:153-159. [Google Scholar]
- 19.Johannes, T. W., R. D. Woodyer, and H. Zhao. 2007. Efficient regeneration of NADPH using an engineered phosphate dehydrogenase. Biotechnol. Bioeng. 96:18-26. [DOI] [PubMed] [Google Scholar]
- 20.Kataoka, M., K. Yamamoto, H. Kawabata, M. Wada, K. Kita, H. Yanase, and S. Shimizu. 1999. Stereoselective reduction of ethyl 4-chloro-3-oxobutanoate by Escherichia coli transformant cells coexpressing the aldehyde reductase and glucose dehydrogenase genes. Appl. Microbiol. Biotechnol. 51:486-490. [DOI] [PubMed] [Google Scholar]
- 21.Kizaki, N., Y. Yasohara, J. Hasegawa, M. Wada, M. Kataoka, and S. Shimizu. 2001. Synthesis of optically pure ethyl (S)-4-chloro-3-hydroxybutanoate by Escherichia coli transformant cells coexpressing the carbonyl reductase and glucose dehydrogenase genes. Appl. Microbiol. Biotechnol. 55:590-595. [DOI] [PubMed] [Google Scholar]
- 22.Kroutil, W., H. Mang, K. Edegger, and K. Faber. 2004. Recent advances in the biocatalytic reduction of ketones and oxidation of sec-alcohols. Curr. Opin. Chem. Biol. 8:120-126. [DOI] [PubMed] [Google Scholar]
- 23.Li, Z., J. B. van Beilen, W. A. Duetz, A. Schmid, A. de Raadt, H. Griengl, and B. Witholt. 2002. Oxidative biotransformations using oxygenases. Curr. Opin. Chem. Biol. 6:136-144. [DOI] [PubMed] [Google Scholar]
- 24.Liese, A., K. Seelbach, and C. Wandrey. 2000. Industrial biotransformation. Wiley-VCH, Weinheim, Germany.
- 25.Lineweaver, H., and D. Burk. 1934. The determination of enzyme dissociation constants. J. Am. Chem. Soc. 56:658-666. [Google Scholar]
- 26.Nakamura, K., and T. Matsuda. 2002. Enzyme-catalyzed reduction reactions, p. 991-1047. In K. Drauz and H. Waldmann (ed.), Enzyme catalysis in organic synthesis, vol. III. Wiley-VCH, Weinheim, Germany. [Google Scholar]
- 27.Ni, Y., and R. R. Chen. 2004. Accelerating whole-cell biocatalysis by reducing outer membrane permeability barrier. Biotechnol. Bioeng. 87:804-811. [DOI] [PubMed] [Google Scholar]
- 28.Rueping, M., M. Albert, and D. Seebach. 2004. On the structure of PHB (= poly[(R)-3-hydroxybutanoic acid]) in phospholipid bilayers: preparation of trifluoromethyl-labeled oligo[(R)-3-hydroxybutanoic acid] derivatives. Helv. Chim. Acta 87:2473-2486. [Google Scholar]
- 29.Shorrock, V. J., M. Chartrain, and J. M. Woodley. 2004. An alternative bioreactor concept for application of an isolated oxidoreductase for asymmetric ketone reduction. Tetrahedron 60:781-788. [Google Scholar]
- 30.Stampfer, W., B. Kosjek, C. Moitzi, W. Kroutil, and K. Faber. 2002. Biocatalytic asymmetric hydrogen transfer. Angew. Chem. Int. Ed. 41:1014-1017. [DOI] [PubMed] [Google Scholar]
- 31.Stampfer, W., K. Edegger, B. Kosjek, K. Faber, and W. Kroutil. 2004. Simple biocatalytic access to enantiopure (S)-1-heteroarylethanols employing a microbial hydrogen transfer reaction. Adv. Synth. Catal. 346:57-62. [Google Scholar]
- 32.van der Donk, W. A., and H. Zhao. 2003. Recent developments in pyridine nucleotide regeneration. Curr. Opin. Biotechnol. 14:421-426. [DOI] [PubMed] [Google Scholar]
- 33.Wong, C. H., D. G. Drueckhammer, and H. M. Sweers. 1985. Enzymatic vs. fermentative synthesis: thermostable glucose dehydrogenase catalyzed regeneration of NAD(P)H for use in enzymatic synthesis. J. Am. Chem. Soc. 107:4028-4031. [Google Scholar]
- 34.Wong, C. H., and G. M. Whitesides. 1981. Enzyme-catalyzed organic synthesis: NAD(P)H cofactor regeneration by using glucose-6-phosphate and the glucose-5-phosphate dehydrogenase from Leuconostoc mesenteroides. J. Am. Chem. Soc. 103:4890-4899. [Google Scholar]
- 35.Yamamoto, H., A. Matsuyama, and Y. Kobayashi. 2003. Synthesis of ethyl (S)-4-chloro-3-hydroxybutanoate using fabG homologues. Appl. Microbiol. Biotechnol. 61:133-139. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, J., W. A. Duetz, B. Witholt, and Z. Li. 2004. Rapid identification of new bacterial alcohol dehydrogenases for (R)- and (S)-enantioselective reduction of β-ketoesters. Chem. Commun. 2004:2120-2121. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, J., B. Witholt, and Z. Li. 2006. Efficient NADPH recycling in enantioselective bioreduction of a ketone with permeabilized cells of a microorganism containing a ketoreductase and a glucose 6-phosphate dehydrogenase. Adv. Synth. Catal. 348:429-433. [Google Scholar]
- 38.Zhang, J., B. Witholt, and Z. Li. 2006. Coupling of permeabilized microorganisms for efficient enantioselective reduction of ketone with cofactor recycling. Chem. Commun. 2006:398-400. [DOI] [PubMed] [Google Scholar]
- 39.Zhao, H., and W. A. van der Donk. 2003. Regeneration of cofactor for use in biocatalysis. Curr. Opin. Biotechnol. 14:1-7. [DOI] [PubMed] [Google Scholar]