Abstract
Many camphor-degrading bacteria that are able to transform 2-methylisoborneol (2-MIB) have been identified. Three of these strains have been examined in detail. Rhodococcus ruber T1 metabolizes camphor through 6-hydroxycamphor but converts 2-MIB to 3-hydroxy-2-MIB. Pseudomonas putida G1, which metabolizes camphor through 5-hydroxycamphor, converts MIB primarily to 6-hydroxy-2-MIB. Rhodococcus wratislaviensis DLC-cam converts 2-MIB through 5-hydroxy-2-MIB to 5-keto-2-MIB. Together, these three strains produce metabolites resulting from hydroxylation at all of the three available secondary carbons on the six-member ring of 2-MIB.
2-Methylisoborneol (2-MIB) is produced by a variety of bacteria including actinomycetes and cyanobacteria (13). When present, it gives a camphoraceous earthy off-flavor and smell to farm-raised catfish (11) or drinking water (13). Since it is not always possible to prevent 2-MIB-producing cyanobacterial blooms from occurring in ponds and reservoirs, a practical solution to the 2-MIB problem might be to remove the chemical by treatment with 2-MIB-degrading bacteria.
Several studies have demonstrated bacterial degradation of 2-MIB, although intermediates and products have in most cases not been identified and where identified have not been particularly well documented. A consortium was obtained from lakewater following enrichment with 2-MIB; several different bacteria were isolated, including a red bacterium and strains identified as Pseudomonas paucimobilis, Pseudomonas aeruginosa, Pseudomonas pseudoalcaligenes, and Pseudomonas mendocina (12). Although they were shown to degrade 2-MIB as a consortium (with no identified intermediates), none of these organisms metabolized 2-MIB in pure culture. Sand filters and derived batch bioreactors could completely remove 2-MIB (100 ng/liter) (10), while a biofilm reactor using peat fulvic acid as the primary substrate removed 44% of 2-MIB (at 100 μg/liter) (15). Two strains, a Pseudomonas sp. and an Enterobacter sp., were isolated by enrichment with 2-MIB using the backwash water from a biological filter as a source (19). The latter strain converted a small portion (probably less than 5%) of the 2-MIB to chemicals identified as the dehydration products 2-methylenebornane and 2-methylcamphene. The same products resulted from the incubation of sludge from a gravel filter with 2-MIB (18) along with some other tentatively identified intermediates; no data were provided for any of these metabolites. Relevant to the present study, Escherichia coli LE392 carrying the cam operon cloned from Pseudomonas putida G1 was able to rapidly remove 2-MIB from media (16) and clones lacking the repressor gene camR were particularly adept, removing 90 to 95% of 2-MIB (at 20 μg/ml) within 1 day. It was suggested, without accompanying data, that 2-MIB is degraded through the catabolic pathway used by strain G1 to degrade camphor.
When this work was initiated, it was expected that 2-MIB-degrading bacteria could be readily isolated by using enrichment culture with 2-MIB as the sole carbon and energy source. This simple technique with its strong selection for organisms of interest is often the starting point for studies of bacterial metabolism. However, despite numerous attempts using a variety of environmental sources, no bacteria that could grow on 2-MIB were obtained (R. Eaton, unpublished data). An alternative approach was subsequently taken in which bacteria that grow with potential 2-MIB substrate analogs, including d-camphor, were isolated by enrichment culture or obtained from existing stock cultures. This yielded several 2-MIB-transforming bacteria. Some of these will be described here.
(Part of this work was presented previously in a preliminary form at the 107th General Meeting of the American Society for Microbiology, Toronto, Ontario, Canada, 21 to 25 May 2007 [4].)
MATERIALS AND METHODS
Bacterial strains.
Pseudomonas putida G1 (9) and Rhodococcus sp. strain T1 (NCIMB 9784) (2, 8) were obtained from Peter Chapman, U.S. Environmental Protection Agency, Gulf Breeze, FL. Other strains were obtained by enrichment culture with d-camphor as the sole carbon and energy source from various environments, including activated sewage sludge from Florida Ave., New Orleans (SWS), LA, Port Allen, LA (PASS), and South Baton Rouge, LA (SBR) sewage treatment plants; water from Donner Lake, CA (DLC); and soil from beneath a ginger plant (GIN) and a dead magnolia (MAG) tree on Octavia St., New Orleans, LA, from a backyard in Midland, MI (no. 9), and from beneath a pineapple sage at Dow Gardens, Midland, MI (no. 22).
Chemicals and media.
2-MIB was obtained from Dalton Pharma Services, Toronto, Ontario, Canada; (1R)-(+)-camphor was from Aldrich Chemical Company, Milwaukee, WI. All other chemicals were of the highest purity available. The minimal medium was R medium (5), solidified when needed with 1.6% Noble agar (Difco).
Cultivation of organisms.
Cultures were grown in 2 ml minimal medium in 20-ml glass scintillation vials with Teflon-lined stoppers. For enrichment cultures, enough soil or sludge was added to the medium to produce a slightly turbid suspension to which 1 mg solid d-camphor was added. Bottles were incubated at 30°C with occasional shaking. When growth was noted after a few days, a loopful of culture was transferred to new medium, and when dense growth occurred, a loopful of the culture was streaked onto a minimal plate to which solid camphor was provided in the lid. Colonies were purified by transfer to and from LB agar plates (3).
For the initial screening of 2-MIB biotransformation, bacteria were inoculated into 2 ml minimal medium to which about 1 mg d-camphor was added. After overnight growth, a grain of solid 2-MIB was added and the incubation continued for 2 to 3 days. The culture was extracted with 1 ml methylene chloride, which was dried over sodium sulfate and analyzed by gas chromatography-mass spectrometry (GC-MS).
Time series biotransformations of 2-MIB.
Bacterial strains representative of three patterns of 2-MIB transformation were incubated with 2-MIB, and the conversion of 2-MIB to metabolites was recorded over time. Primarily because of differing reaction rates between strains, different experimental conditions were required for each.
Transformation of 2-MIB by Rhodococcus ruber T1.
Minimal medium (18 ml) containing 10 mM glucose and 0.025% yeast extract was inoculated with strain T1 and then divided into nine scintillation vials. To each vial was added 2 μl ethanol containing 0.75 mg camphor. After cultures were incubated at 30° C for 16 h with shaking, vial contents were pooled and diluted with an equal volume of the same medium, without camphor. Two milliliters of this culture was added to each of eight new vials. To each vial was added 2 μl ethanol containing 0.5 mg 2-MIB. Vials were then incubated with shaking at 250 rpm. At various times, individual vials were removed and extracted with 1 ml methylene chloride, which was subsequently dried over sodium sulfate. One microliter, split 1:5, was analyzed by GC-MS.
Transformation of MIB by Pseudomonas putida G1.
Minimal medium (35 ml) containing 10 mM glucose and 0.025% yeast extract was inoculated with strain G1. Two milliliters of this culture was added to each of 16 scintillation vials. To each vial was added 2 μl ethanol containing 0.75 mg camphor. Cultures were incubated at 30°C for 20 h with shaking. The contents of the vials were pooled, centrifuged (6,000 rpm for 10 min in a Sorvall SS-34 rotor), and resuspended in 24 ml of the same medium without camphor. Two milliliters of culture was added to each of 11 new vials to which 2 μl ethanol containing 0.125 mg 2-MIB was subsequently added. Vials were incubated at 30°C with shaking at 250 rpm. At various times, individual vials were removed and extracted with 2 ml methylene chloride, which was subsequently dried over sodium sulfate. One microliter was analyzed by GC-MS.
Transformation of 2-MIB by Rhodococcus wratislaviensis DLC-cam.
Minimal medium (50 ml) containing 0.1% succinate and 0.025% yeast extract was inoculated with strain DLC-cam. Two milliliters of this culture was added to each of 23 scintillation vials as well as 2 μl ethanol containing 0.75 mg camphor. Cultures were incubated at 30°C for 20 h with shaking. The contents of the vials were pooled, centrifuged (6,000 rpm for 10 min in a Sorvall SS-34 rotor), and resuspended in 30 ml of the same medium without camphor. Two milliliters of culture was added to each of 13 new vials to which 2 μl ethanol containing 0.125 mg 2-MIB was subsequently added. Vials were incubated at 30°C with shaking at 250 rpm. At various times, individual vials were removed and extracted with 1 ml methylene chloride which was subsequently dried over sodium sulfate. One microliter was analyzed by GC-MS.
To determine whether 2-MIB transformations are inducible by camphor, cells grown in the absence of camphor were incubated with 2-MIB under similar conditions. Culture densities at the beginning of time series incubations with 2-MIB were recorded at 600 nm in 1-cm cuvettes with a Perkin-Elmer Lambda 35 spectrophotometer.
Analysis of metabolites.
GC-MS analyses were carried out using an Agilent 6890 gas chromatograph with an HP5-ms column (30-m by 0.25-mm inside diameter, 0.25-μm film thickness) coupled to an Agilent 5973 mass selective detector. Helium (1 ml min−1) was the carrier gas, the inlet was set at 250°C, and the oven was programmed as follows: 50°C for 2 min then increasing 20°C per minute to 250°C.
For preparation of metabolites for nuclear magnetic resonance (NMR) spectroscopy, multiple incubations were carried out in 2 ml minimal medium containing a growth substrate (10 mM glucose or 0.1% succinate, 0.05% camphor, and 0.025% yeast extract) in 20-ml glass scintillation vials with Teflon-lined stoppers. After 1 day, 2-MIB was added as a solid (1 mg/vial), and after a further 2 to 4 days of incubation, cultures were extracted with methylene chloride which was dried over sodium sulfate.
The product from strain T1 was purified on activated silica gel thin-layer chromatography plates with ethyl acetate as solvent. Spots on thin-layer chromatography plates were visualized using iodine. Spots and bands were scraped off plates, and the chemicals were eluted from the silica gel with methylene chloride. The products from strains G1 and DLC-cam were purified by Flash chromatography using a Combiflash Companion (Teledyne Isco, Lincoln, NE) with a silica gel column and 70:30 toluene-ethyl acetate as the solvent. Collected fractions were analyzed by GC-MS, peak fractions were pooled, and the solvent was evaporated.
All NMR spectra were acquired at 25°C on solutions of the isolated metabolites in “vial quality” 99% deuterated dimethyl sulfoxide (DMSO-D6; Cambridge Isotope Laboratory, Andover, MA). NMR spectra of 2-MIB, 3-hydroxy-2-MIB, and 6-hydroxy-2-MIB were acquired on a Bruker AVANCE DRX 500-MHz instrument using a Bruker 5-mm z-gradient broad band probe. Standard experiments as found in the Bruker XWIN NMR pulse sequence library were employed. These included 1D proton, 1D carbon, DEPT (distortionless enhancement by polarization transfer) 45, DEPT 90, DEPT 135, HMQC (heteronuclear multiple quantum correlation), HMBC (heteronuclear multiple bond correlation), COSY (correlation spectroscopy), and NOESY (nuclear Overhauser effect spectroscopy) NMR. Spectra were processed using XWIN NMR and MestRe-C software. NMR spectra of 5-keto-2-MIB were acquired on a 400-MHz Varian INOVA Plus instrument using a 5-mm z-gradient inverse broad band probe. Additional COSY and NOESY of 5-keto-2-MIB were taken on a 700-MHz Varian INOVA Plus instrument using a 5-mm x,y,z-gradient HCN inverse probe. NMR spectra taken on the Varian instruments employed standard pulse sequences and parameter sets as found in VNMRJ 2.1D Chempack 4. In all cases, isolated metabolite structure determinations were based on molecular weights and NMR spectra. Structures were confirmed by back calculation of carbon chemical shifts using CNMR Predictor 8.08 from Advanced Chemical Development (Toronto, Canada), accessed online via the ACD I-Lab.
For the detection of acidic metabolites, cultures were acidified to pH 1, extracted with ethyl acetate which was dried over sodium sulfate and evaporated. Residues were dissolved in pyridine and derivatized with BSTFA [N,O-bis(trimethylsilyl)trifluoroacetamide] at 60°C for 30 min prior to analysis by GC-MS.
Taxonomy.
For bacterial strain identifications, 16S rRNA genes were sequenced. Total DNA was isolated from overnight LB medium (3) cultures using the Qiagen Dneasy blood and tissue kit and PCR amplified using the Phusion high-fidelity PCR kit (Finnzymes, obtained through New England Biolabs) with primers 27f and 1492r (14). These primers and additional primers for sequencing were synthesized by Integrated DNA Technologies, Skokie, IL. PCR products were purified using a Qiagen QIAquick PCR purification kit and sequenced at the ARS Mid-South Area, Stoneville, MS, by Brian Scheffler and Xiaofen Liu using six primers (27f, 530f, 1114f, 519r, 907r, and 1492r) (14). Sequence data were assembled and edited using Lasergene (DNAStar, Inc.). DNA sequence searches of the GenBank database were carried out using BLASTN (1).
RESULTS AND DISCUSSION
Several bacterial strains that can transform 2-MIB were obtained from existing stocks of camphor-degrading bacteria and by new selective enrichments with camphor. These bacteria have been characterized by the types of metabolites produced from 2-MIB and with regard to their 16S rRNA gene sequences. Screening of all bacterial isolates for transformation of 2-MIB indicated that camphor-degrading organisms yield three patterns of metabolites. Representative strains for each pattern group were selected and subsequently studied in detail.
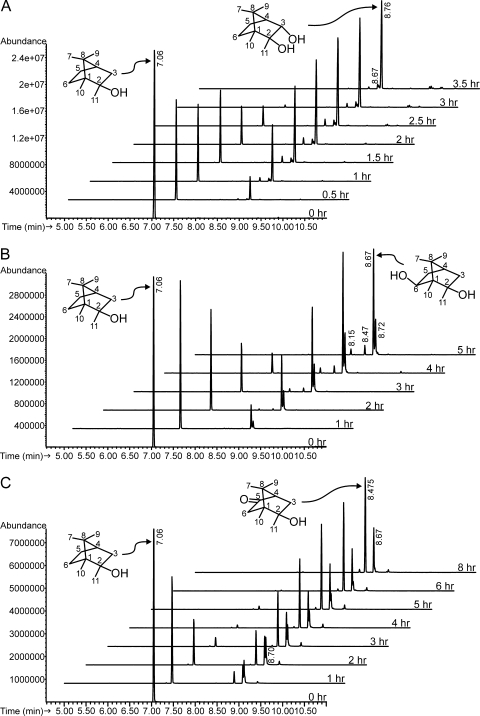
The first group consists of two strains, Rhodococcus ruber T1 (NCIMB 9784; GenBank accession no. EU445342) and a related R. ruber strain, 9camb (GenBank accession no. EF151233). Strain T1 has been studied previously (2, 8) and has been shown to metabolize camphor by a cytochrome P450camr-catalyzed hydroxylation at carbon 6 (8) followed by a dehydrogenation to give 2,6-diketocamphane. When incubated with 2-MIB, strain T1 converted it to a major product (91%) which eluted from the GC at 8.76 min (Fig. 1A) along with minor products at 8.70 and 8.67 min. The mass spectrum of the major product (Table 1) has a small, barely visible molecular ion at m/z 184 (0.4% relative abundance) and prominent ions at m/z 166 ([M − H2O]+) and 151 ([M − H2O − CH3]+). NMR spectroscopy experiments (Table 2) carried out with this chemical demonstrated that it is trans-3-hydroxy-2-MIB (trans-2,3-dihydroxy-2-methylbornane). NOESY and COSY spectra and JHH couplings (not shown) were used to assign methyl 7 and methyl 9 and to determine the exo and endo assignments for the protons at positions 3, 5, and 6. A NOESY cross peak observed between methyl 9 and the position 3 proton helped to confirm the 3 position hydroxyl orientation as endo. In the HMBC spectrum, 2-OH × C-1, 2-OH × C-2, 3-OH × C-4, and 3-OH × C-3 cross peaks were observed, confirming the position of hydroxylation and allowing assignment of hydroxyl peaks. A COSY cross peak between 3-OH and the 3-exo proton was also observed. Observed and calculated carbon chemical shifts are in close agreement.
FIG. 1.
Biotransformations of 2-MIB by camphor-grown bacteria. Time series incubations were carried out as described in Materials and Methods. Samples were taken at the beginning and at time intervals, extracted with methylene chloride, and analyzed by GC-MS. (A) R. ruber T1. The optical density at 600 nm (OD600) of strain T1 at the beginning was 2.5. In an identical experiment using a similar density of strain T1 grown in the absence of camphor, no 2-MIB had been transformed after 3.5 h. (B) P. putida G1. The OD600 of strain G1 at the beginning was 3.7. In an identical experiment using a similar density of strain G1 grown in the absence of camphor, less than 20% of 2-MIB had been consumed after 5 h. (C) R. wratislaviensis DLC-cam. The OD600 of strain DLC-cam at the beginning was 1.3. In an identical experiment using a similar density of strain DLC-cam grown in the absence of camphor, no 2-MIB had been transformed after 6 h.
TABLE 1.
GC-MS data for 2-MIB and its bacterial transformation productsa
| Strain (product retention time in min) | m/z of major ion peaks (% intensity) |
|---|---|
| [2-MIB] (7.05) | 168 (3), 153 (1.5), 150 (6), 135 (12), 125 (3), 121 (4), 110 (15), 108 (24), 106 (21), 95 (100) |
| T1b (8.76) | 184 (0.4), 166 (13), 151 (35), 148 (10), 139 (10), 123 (93), 108 (30), 95 (73), 85 (34), 74 (100) |
| T1 (8.67) | 169 (100), 151 (28), 133 (4.5), 125 (22), 123 (36), 109 (85), 101 (21), 93 (45) |
| G1b (8.67) | 169 (100), 151 (29), 133 (5), 125 (20), 123 (34), 109 (77), 101 (19), 93 (41) |
| DC (8.67) | 169 (100), 151 (28), 133 (5), 125 (20), 123 (35), 109 (78), 101 (20), 93 (42) |
| T1 (8.70) | 184 (2.6), 169 (13), 166 (17), 151 (65), 133 (7), 123 (44), 109 (100), 108 (79), 95 (37), 93 (51), 81 (27) |
| DC (8.70) | 184 (2.3), 169 (3), 166 (17), 151 (62), 133 (6), 123 (43), 109 (100), 108 (77), 95 (35), 93 (51), 81 (28) |
| G1 (8.72) | 169 (4), 166 (6), 151 (59), 133 (11), 123 (63), 109 (98), 108 (100), 95 (17), 93 (44), 81 (23) |
| DCb (8.475) | 182 (12), 167 (6), 164 (3), 149 (4), 139 (15), 124 (100), 121 (15), 109 (35) |
| G1 (8.47) | 182 (13), 167 (4), 164 (3), 149 (5), 139 (13), 124 (100), 121 (13), 109 (33) |
| G1 (8.15) | 182 (4), 167 (100), 149 (4), 139 (2), 125 (11), 124 (13), 121 (13), 109 (41), 107 (17) |
Spectra were taken at the chromatographic peaks shown in Fig. 1A (strain T1), B (strain G1), and C (strain DLC-cam [DC]).
Product purified and analyzed by NMR spectroscopy.
TABLE 2.
NMR spectroscopy data for 2-MIB and 2-MIB biotransformation productsa
| C/H | 2-MIB
|
T1 metabolite 3-OH-2-MIB
|
G1 metabolite 6-OH-2-MIB
|
DLC-cam metabolite 5-keto-2-MIB
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Type | δ 1H | δ 13C | Type | δ 1H | δ 13C | Type | δ 1H | δ 13C | Type | δ 1H | δ 13C | |
| 1 | Q | 52.8 | Q | 52.1 | Q | 56 | Q | 52.8 | ||||
| 2 | Q | 77.6 | Q | 79.3 | Q | 77 | Q | 76.5 | ||||
| 3-exo | CH2 | 1.91 | 47.5 | CH | 4.03 | 79.2 | CH2 | 1.86 | 46.4 | CH2 | 2.16 | 41.5 |
| 3-endo | 1.22 | 1.11 | 1.41 | |||||||||
| 4 | CH | 1.58 | 44.9 | CH | 1.6 | 50 | CH | 1.66 | 45 | CH | 2.04 | 60.6 |
| 5-exo | CH2 | 1.59 | 26.7 | CH2 | 1.64 | 17 | CH2 | 1.55 | 39 | C=O | 215.5 | |
| 5-endo | 0.94 | 1.25 | 1.49 | |||||||||
| 6-exo | CH2 | 1.29 | 31 | CH2 | 1.24 | 31.2 | CH | 72.5 | CH2 | 2.05 | 45.2 | |
| 6-endo | 1.29 | 1.24 | 3.7 | 1.84 | ||||||||
| 7 | CH3 | 0.78 | 21.3 | CH3 | 0.82 | 21.2 | CH3 | 0.99 | 22.8 | CH3 | 0.85 | 21.6 |
| 8 | Q | 48.5 | Q | 45.4 | Q | 48.3 | Q | 48 | ||||
| 9 | CH3 | 1.04 | 21.6 | CH3 | 1.1 | 21.4 | CH3 | 1.05 | 23.3 | CH3 | 1.17 | 20.1 |
| 10 | CH3 | 0.74 | 10.2 | CH3 | 0.74 | 11 | CH3 | 0.77 | 6.02 | CH3 | 0.89 | 9.6 |
| 2-CH3 | CH3 | 1.11 | 27.1 | CH3 | 0.98 | 20.8 | CH3 | 1.07 | 26.7 | CH3 | 1.14 | 26.2 |
| 2-OH | 3.96 | 3.99 | 4.5 | 4.52 | ||||||||
| 3-OH | 4.47 | |||||||||||
| 6-OH | 3.96 | |||||||||||
Molecules with numbered carbons are shown in Fig. 1. These data are from 1D carbon and 1D proton spectra. DEPT 45, DEPT 90, and DEPT 135 spectra were used to define carbon types as methyl, methylene, methine, or quaternary. Additional HMQC, HMBC, COSY, and NOESY spectra contributed to assignments.
The second metabolite pattern group is represented by Pseudomonas putida G1, previously shown to initiate camphor metabolism with a cytochrome P450cam-catalyzed hydroxylation at carbon 5 (9) followed by dehydrogenation to give 2,5-diketocamphane. When strain G1 was incubated with 2-MIB (Fig. 1B), it gave a major product eluting at 8.67 min (61% of total) and minor products at 8.15 min (4%), 8.47 min (9%), and 8.72 min (25%). The mass spectrum of the major product (Table 1) does not show a molecular ion and has a base peak at m/z 169 (proposed [M − CH3]+). The product was purified and analyzed by NMR spectroscopy (Table 2), which led to its identification as exo-6-hydroxy-2-methylisoborneol (2,6-dihydroxy-2-methylbornane). NOESY cross peaks with the 2-methyl and 5-endo protons and stronger JHH coupling to the 5-endo proton relative to the 5-exo proton confirmed the orientation of the 6 position proton as endo. Observed and calculated carbon chemical shifts are in close agreement.
Several camphor-degrading Pseudomonas strains produced these four metabolites from 2-MIB, including three P. putida strains in addition to strain G1 (GenBank accession no. EU445344), SBR-camc (GenBank accession no. EU446283), GIN-cama (GenBank accession no. EU446284), and PASS2-cam (GenBank accession no. EU446288); five P. pseudoalcaligenes strains, SWS3-camc (GenBank accession no. EF151237), PASS-camb (GenBank accession no. EU445341), PASS3-cam (GenBank accession no. EU043332), MAG-camb (GenBank accession no. EF151236), and SBR-camb (GenBank accession no. EF151241); and two more distant Pseudomonas sp. strains, SBR3-cam (GenBank accession no. EU043315) and PASS-cama (GenBank accession no. EF151239).
The third metabolite pattern resulted from transformation of 2-MIB by Rhodococcus wratislaviensis DLC-cam (GenBank accession no. EU043327) (Fig. 1C) and by a close relative, Rhodococcus opacus 22-cam (GenBank accession no. EU445340) (6). Here, significant hydroxylation by strain DLC-cam occurred at both C-5 (8.7 min) and C-6 (8.67 min, 31%). C-6 hydroxylation yielded a product with mass spectrum identical to that of the major product formed by strain G1 (Table 1), while the putative product of C-5 hydroxylation was further oxidized to the corresponding ketone (8.47 min, 69%) (Table 1). This ketone, purified and analyzed by NMR spectroscopy (Table 2), was identified as 5-keto-2-MIB. HMBC cross peaks were observed between the carbonyl carbon at 215.5 ppm and the 3-exo, 3-endo, 4,6-exo, and 6-endo protons (data not shown), while calculated carbon chemical shifts are in agreement with those observed in the spectra. Particularly notable in this regard are the carbon chemical shifts of positions C-1 and C-4, which confirm that the carbonyl position is 5 and not 6.
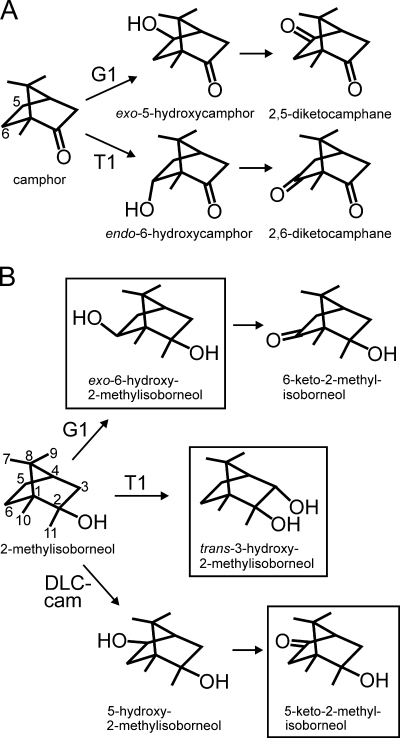
R. ruber T1 metabolizes camphor by hydroxylation at carbon 6 followed by dehydrogenation to give 2,6-diketocamphane (Fig. 2A) (2, 8), while P. putida G1 hydroxylates camphor at carbon 5; dehydrogenation yields 2,5-diketocamphane (8). Camphor metabolism has not been studied in R. wratislaviensis DLC-cam. Although 2-MIB and camphor share the bornane carbon skeleton, camphor-degrading bacteria did not oxidize 2-MIB with the same regioselectivity as they oxidize camphor (Fig. 2B). The major product of 2-MIB oxidation by R. ruber T1 was 3-hydroxy-2-MIB, not 6-hydroxy-2-MIB, while the major oxidation product of P. putida G1 was 6-hydroxy-2-MIB, not 5-hydroxy-2-MIB. Previous studies (17) with the P. putida G1 cytochrome P450 camphor hydroxylase have shown a requirement for hydrogen bonding through the carbonyl of camphor and other specific substrate-enzyme interactions for regiospecific hydroxylation, such as occur with the natural substrate camphor. Thus, camphor substrate analogs, including thiocamphor (which gave 5-exo-hydroxythiocamphor [64%], 6-exo-hydroxythiocamphor [34%], and 3-exo-hydroxythiocamphor [2%]) and norcamphor (which gave 5-exo-hydroxynorcamphor [45%], 6-exo-hydroxynorcamphor [47%], and 3-exo-hydroxynorcamphor [8%]), showed little hydroxylation regiospecificity (17).
FIG. 2.
Initial steps (A) in the metabolism of camphor by R. ruber T1 and P. putida G1 and (B) proposed in the biotransformation of 2-MIB by strains T1, G1, and DLC-cam. Compounds in boxes were identified by a combination of GC-MS and NMR spectroscopy.
Although not the major products, the camphor-analogous metabolites were among the minor products. In addition to 3-hydroxy-2-MIB (Fig. 1A; 8.76 min), R. ruber T1 gave small amounts of 6-hydroxy-2-MIB (Fig. 1A; 8.67 min) and 5-hydroxy-2-MIB (Fig. 1A; 8.70 min). P. putida G1 hydroxylated 2-MIB, not only at carbon 6 (Fig. 1B; 8.67 min) but also at carbon 5 (Fig. 1B; 8.72 min), and dehydrogenated some of each product to 5-keto-2-MIB (Fig. 1B, 8.47 min) and, probably, 6-keto-2-MIB (Fig. 1B, 8.15 min). The identity of the latter compound was suggested by a comparison of its mass spectrum with that of 6-hydroxy-2-MIB produced by strain G1 (Table 1), which has a similar appearance but with ions two mass units smaller and a molecular ion of m/z 182. R. wratislaviensis DLC-cam gave products hydroxylated at carbons 5 (Fig. 1C; 8.70 min) and 6 (Fig. 1C; 8.67 min) and converted the 5-hydroxy-2-MIB product further to 5-keto-2-MIB (Fig. 1C; 8.475 min).
While it is possible that the minor camphor-analogous metabolites were produced in greater amounts but rapidly metabolized, it is also likely that, if that occurred, some other downstream metabolite would accumulate. Thus, the camphor intermediate, 2,6-diketocamphane (6-oxocamphor), is metabolized in strain T1 by a β-diketone-hydrolase, for which 6-keto-2-MIB is not likely to be a substrate (7). In addition, no acidic 2-MIB metabolites were detected from any of the strains.
Together, the three groups of camphor-degrading bacteria are capable of hydroxylating 2-MIB at all three secondary carbons of the six-member ring as well as further oxidation reactions. The purpose of this research has been to find and develop bacterial strains that can eliminate the earthy, musty off-flavor of 2-MIB. Although the strains described here carry out the desired activity, prior to their use some modification may be required, notably the isolation of constitutive mutants, which would eliminate the current need for an inducer. While the oxidation products have not been subjected to a quantitative taste and odor test, they are less volatile than 2-MIB and even if they retained the 2-MIB odor, it would be expected to be less intense than that of 2-MIB. Preliminary evaluations indicate that the metabolites seem to lack any of the 2-MIB odor (Eaton, unpublished).
Footnotes
Published ahead of print on 5 December 2008.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Meyers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Chapman, P. J., G. Meerman, I. C. Gunsalus, R. Srinivasan, and K. L. Rinehart, Jr. 1966. A new acyclic acid metabolite in camphor oxidation. J. Am. Chem. Soc. 88:618-619. [Google Scholar]
- 3.Davis, R. W., D. Botstein, and Roth. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 4.Eaton, R., and P. Sandusky. 2007. Biotransformations of 2-methylisoborneol by terpene-degrading bacteria, abstr. Q-125, p. 560. Abstr. 107th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 5.Eaton, R. W. 2001. Plasmid-encoded phthalate catabolic pathway in Arthrobacter keyseri 12B. J. Bacteriol. 183:3689-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodfellow, M., J. Chun, E. Stackebrandt, and R. M. Kroppenstedt. 2002. Transfer of Tsukamurella wratislaviensis Goodfellow et al. to the genus Rhodococcus as Rhodococcus wratislaviensis comb. nov. Int. J. Syst. Evol. Microbiol. 52:749-755. [DOI] [PubMed] [Google Scholar]
- 7.Grogan, G., J. Graf, A. Jones, S. Parsons, N. J. Turner, and S. L. Flitsch. 2001. An asymmetric enzyme-catalyzed retro-Claisen reaction for the desymmetrization of cyclic β-diketones. Angew. Chem. Int. Ed. 40:1111-1114. [PubMed] [Google Scholar]
- 8.Grogan, G., G. A. Roberts, S. Parsons, N. J. Turner, and S. L. Flitsch. 2002. P450camr, a cytochrome P450 catalysing the stereospecific 6-endo-hydroxylation of (1R)-(+)-camphor. Appl. Microbiol. Biotechnol. 59:449-454. [DOI] [PubMed] [Google Scholar]
- 9.Gunsalus, I. C., and V. P. Marshall. 1971. Monoterpene dissimilation: chemical and genetic models. Crit. Rev. Microbiol. December:291-310. [Google Scholar]
- 10.Ho, L., D. Hoefel, F. Bock, C. P. Saint, and G. Newcombe. 2007. Biodegradation of 2-methylisoborneol (MIB) and geosmin through sand filters and in bioreactors. Chemosphere 66:2210-2218. [DOI] [PubMed] [Google Scholar]
- 11.Howgate, P. 2004. Tainting of farmed fish by geosmin and 2-methyl-iso-borneol: a review of sensory aspects and of uptake/depuration. Aquaculture 234:155-181. [Google Scholar]
- 12.Izaguirre, G., R. L. Wolfe, and E. G. Means III. 1988. Degradation of 2-methylisoborneol by aquatic bacteria. Appl. Environ. Microbiol. 54:2424-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jüttner, F., and S. B. Watson. 2007. Biochemical and ecological control of geosmin and 2-methylisoborneol in source waters. Appl. Environ. Microbiol. 73:4395-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics, John Wiley & Sons, New York, NY.
- 15.Namkung, E., and B. E. Rittmann. 1987. Removal of taste- and odor-causing compounds by biofilms grown on humic substances. Am. Water Works Assoc. J. 79:107-112. [Google Scholar]
- 16.Oikawa, E., A. Shimizu, and Y. Ishibashi. 1995. 2-Methylisoborneol degradation by the cam operon from Pseudomonas putida PpG1. Water Sci. Technol. 31:79-86. [Google Scholar]
- 17.Raag, R., and T. L. Poulos. 1991. Crystal structures of cytochrome P-450CAM complexed with camphane, thiocamphor, and adamantane: factors controlling P-450 substrate hydroxylation. Biochemistry 30:2674-2684. [DOI] [PubMed] [Google Scholar]
- 18.Sumitomo, H. 1992. Biodegradation of 2-methylisoborneol by gravel sand filtration. Water Sci. Technol. 25:191-198. [Google Scholar]
- 19.Tanaka, A., T. Oritani, F. Uehara, A. Saito, H. Kishita, Y. Niizeki, H. Yokota, and K. Fuchigami. 1996. Biodegradation of a musty odour component, 2-methylisoborneol. Water Res. 30:759-761. [Google Scholar]