Abstract
Denitrification is an alternative type of anaerobic respiration in which nitrate is reduced to gaseous products via nitrite. The key step in this process is the reduction of nitrite to nitric oxide, which is catalyzed by two structurally different but functionally equivalent forms of nitrite reductase encoded by the nirK and nirS genes. Cultivation-independent studies based on these functional marker genes showed that in the environment there was a dominance of organisms with nirK and nirS genes presumably derived from organisms that have not been cultured yet. However, the phylogenetic affiliation of these organisms has not been resolved since the ability to denitrify is widespread in phylogenetically unrelated organisms. To unravel the phylogeny of the organisms from which the nitrite reductase (nirK) genes originated, one option is to use a special variant of whole-cell hybridization termed recognition of individual genes-fluorescence in situ hybridization (RING-FISH). In RING-FISH a multiply labeled transcript polynucleotide probe is used to detect a single gene on the bacterial chromosome during FISH. Here, RING-FISH was used with laboratory cultures and environmental samples, such as activated sludge. Furthermore, probe-based cell sorting using magnetic beads could also be carried out with mixtures of pure cultures, which led to effective depletion of the nirK-negative organism but capture of the nirK-positive organism, which was demonstrated by terminal restriction fragment length polymorphism analysis based on 16S rRNA genes. The results indicate that RING-FISH coupled with probe-based cell sorting could be used with environmental samples, which could provide a means for phylogenetic classification of nirK-type denitrifiers. Thus, the results of RING-FISH could increase our understanding of the phylogeny and function of denitrifying microorganisms in the environment.
Denitrification sensu stricto is the reduction of oxidized nitrogen compounds (nitrate and nitrite) with the production of nitrous oxide (N2O) and/or dinitrogen (N2). Nitrous oxide is a very potent greenhouse gas that is emitted into the atmosphere and has well-known climatic effects (10, 13). Denitrification and other nitrogen-converting processes in soils limit the availability of nitrate to plants by converting it to gaseous products and account for up to 70% of the total annual global N2O emission (8, 21). On the other hand, the conversion to dinitrogen is beneficial in wastewater treatment because it removes nitrate and thus counteracts the eutrophication of receiving water bodies.
Most of the microorganisms responsible for denitrification are facultative anaerobes that use this process as an alternative anaerobic respiration pathway for energy conservation. This ability is widespread in a broad variety of phylogenetically unrelated organisms and has been found in more than 50 genera and 130 species belonging to the Bacteria, as well as the Archaea (33, 41). A common feature of denitrifiers is the conversion of nitrite to gaseous products that cannot be further assimilated by the organisms, which distinguishes true denitrifiers from nitrate reducers. Dissimilatory nitrite reduction is catalyzed by two structurally different but functionally equivalent enzymes (16), a copper-containing nitrite reductase and a cytochrome cd1-containing nitrite reductase, which are encoded by the nirK and nirS genes, respectively. Due to the wide distribution of these genes in phylogenetically unrelated microorganisms and the resulting unsuitability of a 16S rRNA gene-based approach, nitrite reductase genes have been targeted frequently as functional marker genes for detection of denitrifiers in diverse environments. These environments included marine water columns (7, 29) and sediments (5, 25), sewage sludge (19), groundwater (39), and soil (6, 32), and the denitrifier communities were examined by using cultivation-independent PCR-based approaches. These studies revealed an enormous diversity of nitrite reductase genotypes that were only distantly related to the nitrite reductase genotypes of cultured denitrifiers. Some clusters in the nir gene trees are dominated by or consist exclusively of sequences from uncultured organisms. However, a limitation of this approach is the unresolved link between the functional gene diversity and the phylogenetic diversity of the organisms in the environment since at present it is not possible to infer phylogenetic relationships of the organisms based only on their nitrite reductase genes.
To unravel the phylogenetic affiliation of the key players in the denitrification process in the environment, two options are available. One option is to obtain isolates that are dominant in their habitats and thus relevant for nitrogen cycling in nature. However, the fraction of the environmental microbial fauna that can be cultured is generally very small (2). In addition, cultivation experiments are very labor-intensive and usually result in a limited number of isolates that are well adapted to the cultivation conditions but are often numerically and functionally irrelevant in their habitats. A different approach to obtain broader insight into the phylogenetic affiliation of members of denitrifier communities in the environment is to use a special variant of fluorescence in situ hybridization (FISH) called recognition of individual genes (RING)-FISH and to combine it with subsequent cell sorting. RING-FISH (or poly-FISH) involves using polyribonucleotide probes that are multiply labeled with several reporter molecules, and it is characterized by typical halo-shaped fluorescence signals in the periphery of the cells. These halo-shaped signals are hypothesized to occur due to folding of the single-stranded RNA-probe molecules into secondary structures (42), which results in the formation of a network of probes around the cells during whole-cell hybridization (38, 44). Polynucleotide probes were first used during in situ hybridization for identification of bacteria based on their 23S rRNA (38), but they also specifically hybridized to large-subunit rRNA in a variety of yeasts (44). Moreover, using multiply labeled 100-bp 16S rRNA-based polynucleotide probes allowed workers to identify and quantify archaeal and bacterial cells in marine plankton samples (12, 22, 31). Compared to the results obtained with conventional FISH using singly labeled oligonucleotide probes, the use of multiply labeled probes increased the intensity of the fluorescence signal and thus the sensitivity of RING-FISH between 10- and 50-fold (12, 38). Hence, the use of RING-FISH also allowed in situ detection of individual genes or gene fragments whose copy numbers in each cell were low, including genes or gene fragments in plasmids (101 to 103 copies) and even in chromosomal DNA (<10 copies) (46). For instance, the plasmid-carried beta-lactamase gene and the chromosomal glyceraldehyde-3-phosphate dehydrogenase gene in Escherichia coli, as well as the prepilin peptidase gene in Xanthomonas campestris (46), have been detected. Moreover, after hybridization of the probes to functional genes, cells can be specifically retrieved from cell mixtures (e.g., from enrichments or environmental samples) by polynucleotide probe-based cell sorting (35, 45). Retained cells can then be subjected to further analyses (e.g., amplification and cloning of their 16S rRNA genes) to determine the phylogenetic affiliations of the organisms.
In this study, we adapted the RING-FISH technique and used it for detection nirK-type nitrite reductase genes in denitrifier pure cultures, and we confirmed the specificity of the hybridization of polynucleotide probes to chromosomally located single-copy nirK genes by subsequent cell sorting using magnetic beads. The use of RING-FISH also allowed us to visualize nirK-containing microorganisms in environmental samples, such as activated sludge.
MATERIALS AND METHODS
Organisms.
The strains used as nirK-positive control organisms were Alcaligenes xylosoxidans NCIMB 11015, Hyphomicrobium aestuarii DSM 1564, Pseudomonas sp. strain G-179 (40), and Sinorhizobium meliloti Rm20115. The following strains were used as nirK-negative controls: Bacillus pumilus DSM 27, Desulfobacter postgatei DSM 2034, Hyphomicrobium zavarzinii IFAM ZV-580 (20), and Pseudomonas stutzeri JM300 (9). Most strains were cultured aerobically at 25°C in the media recommended by the culture collections or by previous workers; the only exception was D. postgatei, which was grown anaerobically under an N2-CO2 (80:20, vol/vol) atmosphere at 30°C. Bacterial strains were grown to the exponential phase (optical density at 600 nm, 0.4 to 0.6) and then inoculated into fresh growth medium (5 to 10%) and incubated again. This procedure was repeated three or four times to obtain actively growing cells in similar growth phases.
Environmental samples and enrichments.
Activated sludge was collected at the sewage treatment plant in Marburg-Cappel, Germany, in June 2007. Denitrifiers from 1 liter of activated sludge were enriched by adding (i) KNO3 to a final concentration of 5 mM, (ii) KNO3 and glucose to final concentrations of 5 mM and 5 μM, respectively, and (iii) KNO3 and methanol to final concentrations of 5 mM and 25 μM, respectively. The enrichments were incubated anaerobically at room temperature in the dark for 4 weeks. Nitrate consumption was monitored weekly using the Merckoquant nitrate test (Merck, Darmstadt, Germany), and nitrate was replenished whenever it was consumed.
Cell fixation.
Prior to cell fixation cells from pure cultures and activated sludge were treated as follows. Cells from pure cultures were collected in the exponential growth phase by centrifugation at 2,800 × g for 10 min, and each cell pellet was resuspended in phosphate-buffered saline (PBS) (137 mM NaCl, 10 mM Na2HPO4-KH2PO4, 2.7 mM KCl; pH 7.2). Cell aggregates from activated sludge (2 ml) were disrupted by pressing them through a fine needle (25 gauge; 0.5 by 25 mm) several times. Gram-negative cells and cells from environmental samples were fixed in paraformaldehyde, and gram-positive cells were fixed in 96% (vol/vol) ethanol as described previously (2).
DNA extraction and generation of polynucleotide transcript probes.
Genomic DNA from pure cultures of A. xylosoxidans, H. aestuarii, Pseudomonas sp. strain G-179, and S. meliloti was extracted with phenol-chloroform (3). Templates for in vitro transcription were generated from the DNA extracts by PCR amplification of nirK gene fragments (515 bp) using PCR conditions and primers nirK1F and nirK5R described by Braker et al. (4) with the T3 promoter sequence attached to the reverse primer to allow initiation of transcription. Amplicons of the proper size were excised from agarose (1.5%, wt/vol) gels and purified using the Wizard SV gel and PCR clean up system (Promega, Mannheim, Germany). Polynucleotide transcript probes were generated by in vitro transcription and were simultaneously labeled by incorporating either biotin-16-UTP or digoxigenin-11-UTP (Roche Molecular Diagnostics) as described by Zwirglmaier et al. (44). The concentration of the polynucleotide probes was determined using a NanoDrop instrument (Thermo Fisher Scientific, Wilmington, DE).
In situ hybridization with polyribonucleotide transcript and oligonucleotide probes.
Hybridizations were performed either with fixed cells immobilized on Teflon-coated glass slides (MAGV, Rabenau-Londorf, Germany) or with cells in solution. For hybridizations on glass slides, aliquots (5 to 10 μl) of fixed cells in suspension (approximately 5 × 104 cells), of fixed cells from enrichments, or of activated sludge were subjected to the hybridization procedure described by Zwirglmaier et al. (44). For hybridizations in solution, aliquots (30 μl) of fixed cells from pure cultures were treated as described by Zwirglmaier et al. (45). All hybridizations were performed for 24 h using a formamide concentration of 15% in the hybridization buffer and 4 and 5 μg of polynucleotide probe for hybridizations on glass slides and in solution, respectively. After hybridization in solution, cells were collected by centrifugation at 8,000 × g for 3 min, resuspended in 100 μl washing buffer (150 mM NaCl, 100 mM Tris-HCl [pH 8.0], 10% [wt/vol] sodium dodecyl sulfate; pH 7.4), and washed at 53°C for 30 min to remove the unbound polynucleotide probe. Cells were again collected by centrifugation at 8,000 × g for 3 min and resuspended in 15 μl PBS.
After hybridization with polynucleotide probes, the biotin or digoxigenin label of the polynucleotide probes was detected. Biotin-labeled probes were detected with a streptavidin-Cy3 conjugate (Sigma Aldrich), and digoxigenin-labeled probes were detected with anti-digoxigenin-Fab fragments (Roche Molecular Diagnostics) coupled to fluorescein according to the manufacturer's recommendations. All hybridization preparations were visualized by fluorescence microscopy (Axiophot; Carl Zeiss Microimaging GmbH, Göttingen, Germany) at wavelengths of 570 and 523 nm to detect the Cy3 and fluorescein labels, respectively.
In mixtures of nirK-positive and nirK-negative strains, cells were differentiated prior to detection by labeling the nirK-negative organisms by whole-cell hybridization with Cy3-labeled 16S rRNA-targeted oligonucleotide probes, as described previously (34). Cells from environmental samples were hybridized by performing whole-cell hybridization with 16S rRNA-targeted probes EUB338I to EUB338III (1, 11).
Cell sorting using magnetic beads.
For cell sorting experiments approximately 6 × 107 cells of mixed nirK-positive and -negative strains were used for hybridization in solution with 8 μg polynucleotide probe in 30 μl hybridization buffer. Hybridizations with polynucleotide probes were performed under conditions described above. Subsequently, cell sorting was performed in lieu of the detection procedure. For this, 100 μl of Dynabeads pan mouse immunoglobulin G with an antidigoxigenin epitope (4 × 108 beads ml−1; Invitrogen, Karlsruhe, Germany) was used. First, the beads were washed with 1 ml PBS and 0.1% bovine serum albumin (BSA) in a 1.5-ml Eppendorf tube by placing it into a MagneSphere magnetic separation stand (Promega, Mannheim, Germany) and removing the supernatant after the beads were collected on one side of the tube. Then the beads were resuspended in 100 μl PBS-0.1% (vol/vol) BSA. To 15 μl of each of the pure-culture mixtures 600 μl PBS was added, and this suspension was added to the washed beads. Cells and beads were incubated for 30 min on a rotary shaker at 100 rpm at room temperature to allow binding of cells to the beads via the digoxigenin label of the polynucleotide probe that had hybridized. Then the beads with bound cells were collected by placing the Eppendorf tube in the magnetic separation stand for 1 min, and the supernatant containing cells to which the probe had not hybridized was removed. The beads were washed four times with 1.5 ml PBS-0.1% BSA and were finally resuspended in 20 μl PBS.
T-RFLP analysis.
DNA was released from the cells bound to the beads by a freeze-thaw procedure. Then an aliquot (4 μl of a bead suspension) was subjected to PCR amplification of bacterial 16S rRNA genes using a protocol and primers described previously (3a). PCR products were purified using the Wizard SV gel and PCR clean up system (Promega). For terminal restriction fragment length polymorphism (T-RFLP) analysis aliquots (100 ng) of the purified PCR product were cleaved overnight at 37°C with 3 U of restriction endonuclease HhaI in the manufacturer's recommended reaction buffer using a reaction volume of 10 μl. After purification using Autoseq G-50 columns (Amersham Pharmacia Biotech Inc., Piscataway, NY), an aliquot (3 μl) was analyzed with a 3130 automated sequencer by comparing the sizes of the peaks to an internal standard (MapMarker 1000; 30 to 1,000 bp; BioVentures Inc., Murfreesboro, TN). The cell sorting efficiency was quantified based on the relative area of peaks indicative of a given nirK-negative pure culture by comparing the peak areas before and after cell sorting, as follows: ([relative peak area of nontarget cells before cell sorting − relative peak area of nontarget cells after cell sorting]/relative peak area of nontarget cells after cell sorting) × 100.
RESULTS
Optimization of hybridization conditions.
Polynucleotide probes were generated by in vitro transcription of a 515-bp amplicon of the nitrite reductase (nirK) gene of four denitrifier strains, A. xylosoxidans NCIMB 11015, H. aestuarii DSM 1564, Pseudomonas sp. strain G-179, and S. meliloti Rm20115. For in situ hybridizations, these probes were used either individually or as equimolar mixtures of all four probes. Initially, the optimal hybridization conditions for use of each individual probe were evaluated. Therefore, the stringency of the hybridization was varied by using different formamide concentrations in the hybridization buffer (5, 10, 15, 20, and 25%) and by estimating the number of cells that showed the halo-shaped fluorescence signal typical of RING-FISH. For all probes and nirK-type denitrifier strains, the largest fraction of cells (98 to 100% of the cells spotted on the slide) was detected using the homologous probe for each pure culture with a formamide concentration of 15% (Fig. 1; see also Fig. S1 to S4A in the supplemental material). With concentrations less than and greater than this concentration lower fractions of halos (with 5% formamide, approximately 50%; with 10% formamide, approximately 80%; with 20% formamide, approximately 60%; and with 25% formamide, approximately 10%) were observed for all probes (see Fig. S1 to S4A in the supplemental material). In addition, with a formamide concentration of 15% in the hybridization buffer none of the negative controls (B. pumilus DSM 27, D. postgatei DSM 2034, H. zavarzinii IFAM ZV-580, and P. stutzeri JM300) showed any halo-shaped fluorescence signal (see Fig. S5 in the supplemental material [data for D. postgatei not shown]). The small point-shaped fluorescent signals that occurred in large aggregates of H. zavarzinii cells were nonspecific signals that were not removed by the washing procedure. It is noteworthy that in experiments with a given stringency nirK-positive and -negative cells were hybridized on the same slide using identical solutions and hybridization conditions. The same results (i.e., almost quantitative detection and specificity of the probes with 15% formamide) were obtained when a mixture of the homologous probe and three heterologous probes was used with nirK-positive and -negative control strains (see Fig. S1 to S4A in the supplemental material). As a consequence, a formamide concentration of 15% was used to adjust the stringency conditions for all further hybridization experiments with pure cultures, as well as with environmental samples.
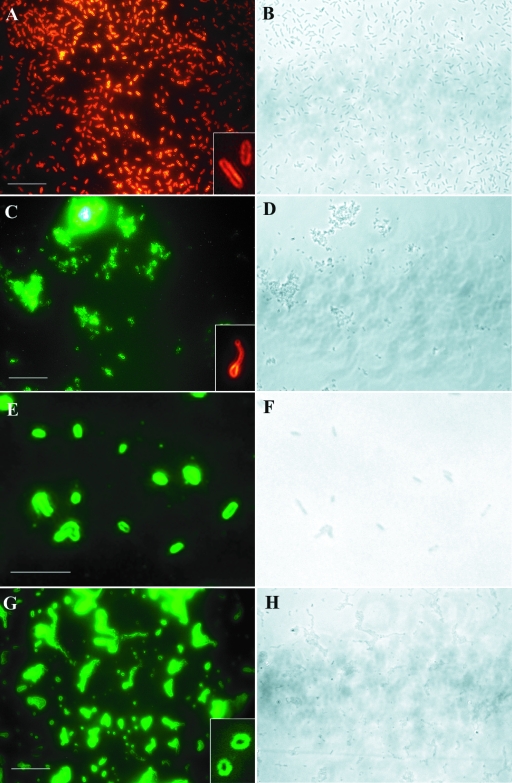
FIG. 1.
Detection of nitrite reductase (nirK) genes in nirK-positive pure cultures by RING-FISH. Cells were hybridized with the homologous nirK polynucleotide probes and labeled with Cy3 (red fluorescence) or fluorescein (green fluorescence). (A and B) A. xylosoxidans NCIMB 11015. (C and D) H. aestuarii DSM 1564. (E and F) Pseudomonas sp. strain G-179. (G and H) S. meliloti Rm20115. The insets show the halo shape of the fluorescence signal. Panels B, D, F, and H are phase-contrast images. Bars = 10 μm.
Mixtures of nirK-positive and -negative strains.
RING-FISH for detection of nirK-type denitrifiers was also used for hybridizations with mixtures of one nirK-positive strain and one nirK-negative strain by using the polynucleotide probes individually or equimolar mixtures of all four probes. This was done by mixing cells of morphologically distinct strains (e.g., A. xylosoxidans and H. zavarzinii strains or H. aestuarii and P. stutzeri strains) or, alternatively, by mixing cells of strains with similar morphologies. In the latter case nondenitrifiers were discriminated from nirK-type denitrifiers by using specific oligonucleotide probes (LGC0355 for B. pumilus, which is specific for Firmicutes [17]; Hypho-1241 for H. zavarzinii ZV-580, which is specific for Hyphomicrobium spp. [24]; and GAM42a for P. stutzeri, which is specific for Gammaproteobacteria [27]) to counterstain nondenitrifier cells. Most possible combinations of nirK-positive and -negative pure cultures were tested; the exceptions were combinations with D. postgatei due to the high background fluorescence of this strain. RING-FISH labeled the nirK-type denitrifiers exclusively, while hybridizations using oligonucleotide probes stained only the nirK-negative control strains. For instance, when mixtures of the nirK-type denitrifier A. xylosoxidans and the nondenitrifier B. pumilus or H. zavarzinii were used, the denitrifier showed the halo-shaped RING-FISH fluorescence signal, while the nondenitrifiers were detected by the specific oligonucleotide probes (Fig. 2). No difference in specificity was observed when the homologous probes were used individually or in combination with the three heterologous probes.
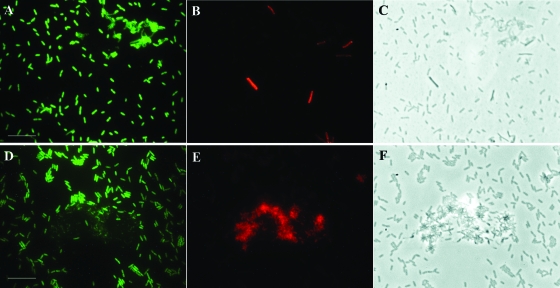
FIG. 2.
Discrimination of nirK-type denitrifiers and nondenitrifiers in mixtures of pure cultures by RING-FISH. Denitrifier cells were hybridized with nirK polynucleotide probes, and nondenitrifiers were counterstained by FISH using Cy3-labeled oligonucleotide probes. (A to C) Mixture of the denitrifier A. xylosoxidans NCIMB 11015 and the nondenitrifier B. pumilus DSM 27. (D to F) Mixture of the denitrifier A. xylosoxidans NCIMB 11015 and the nondenitrifier H. zavarzinii ZV-580. The organisms were labeled with fluorescein after RING-FISH (A and D) and with Cy3 after FISH (B and E). Panels C and F are phase-contrast images. Bars = 10 μm.
Cell sorting.
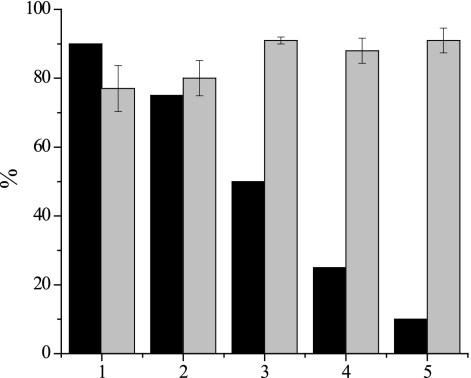
Hybridization using nirK polynucleotide probes was used as the initial step for sorting cells in mixtures of nirK-positive and -negative strains. Generally, cell sorting is performed with cells in suspension, and it was done with a mixture of four probes in this case. Given that the specificity and efficiency were reproducible for hybridizations performed on glass slides, as well as in solution, hybridizations were carried out with mixtures of two cultures, a nirK-positive pure culture and a nirK-negative pure culture. The mixtures included most of the possible combinations of nirK-positive and -negative pure cultures used in this study. They were prepared with nirK-negative cells comprising a large fraction of the total cell count (between 75 and 91%) and the nirK-positive organism comprising the remaining fraction (Table 1 and Fig. 3). Cell sorting was achieved by magnetically capturing cells that had hybridized with digoxigenin-labeled polynucleotide probes and by removing the supernatant containing unbound cells and thereby supposedly depleting nirK-negative cells. Then 16S rRNA genes were amplified from cells coupled to the beads, and the amplicons were analyzed by T-RFLP analysis. For the T-RFLP analysis the restriction endonuclease HhaI was used, which resulted in terminal restriction fragments that were of distinct lengths for the nirK-positive and -negative pure cultures in the mixture. Terminal restriction fragment lengths (the theoretical lengths for nirK-positive organisms were 565 bp for A. xylosoxidans, 336 bp for H. zavarzinii, and 56 bp for Pseudomonas sp. and S. meliloti; the theoretical lengths for nirK-negative organisms were 240 bp for B. pumilus, 206 bp for P. stutzeri, and 97 bp for D. postgatei) were determined theoretically and experimentally. Some bacterial strains have different 16S rRNA gene copy numbers, which may result in different relative peak areas even if equal amounts of cells are used. Hence, the efficiency of cell sorting (the fraction in which the nirK-negative organism was depleted from the mixture) was estimated by calculating the ratio of the relative areas of the peaks specific for the organisms before and after cell sorting. For all mixtures of strains used here effective cell sorting was obtained independent of the pure cultures chosen. The efficiency of depletion of the nirK-negative strain in the mixtures varied from 92 to 100% and thus was generally high. It was also largely independent of the ratios of nirK-positive and -negative cells, as observed, for instance, when 90, 75, 50, 25, and 10% D. postgatei was mixed with the nirK-type denitrifier A. xylosoxidans (Fig. 3). The efficiency of depletion for these mixtures was around 90% if the fraction of nirK-positive cells was higher than 50%. Thus, it was in a range (>92%) similar to the range observed for all other mixtures. However, in contrast to our initial attempts, in this experiment an exceptional lower efficiency of depletion (approximately 80%) was observed for mixtures of these two strains when nirK-negative cells accounted for more than 50% of all cells.
TABLE 1.
Cell depletion efficiency of nirK-negative strains in mixtures of nirK-positive and -negative strains after cell sorting
| nirK-positive strain | % nirK-negative cells in all cells/% depletion of nirK-negative strain with:
|
|||
|---|---|---|---|---|
| B. pumilus DSM 27 | D. postgatei DSM 2034 | H. zavarzinii IFAM- ZV580 | P. stutzeri JM300 | |
| A. xylosoxidans NCIMB 11015 | 75/99.9 | 80/99.8 | NDa | 80/98.6 |
| H. aestuarii DSM 1564 | 75/100 | ND | ND | 91/98.6 |
| Pseudomonas sp. strain G-179 | ND | 83/100 | 75/100 | 83/92.5 |
| S. meliloti Rm20115 | ND | 83/100 | 91/99.6 | ND |
ND, not determined.
FIG. 3.
Cell sorting efficiency for mixtures of the nirK-type denitrifier A. xylosoxidans NCIMB 11015 and the nondenitrifier D. postgatei DSM 2034. Depletion of D. postgatei was determined by T-RFLP analysis of PCR-amplified 16S rRNA genes of cells captured after hybridization with nirK polynucleotide probes. Black bars, fractions of the nondenitrifier in mixtures of denitrifier and nondenitrifier pure cultures; gray bars, fractions of the nondenitrifier depleted from mixtures. The error bars indicate standard deviations (n = 3).
Activated sludge.
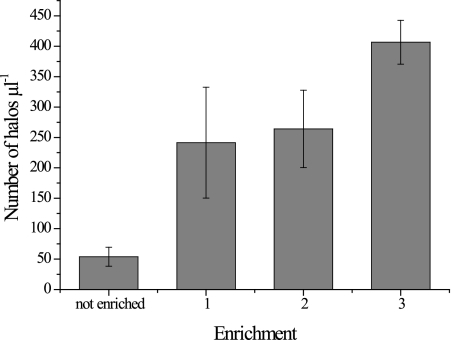
Denitrifiers were also detected in activated sludge by using RING-FISH to target nirK genes. Halo-shaped signals in the periphery of mainly rod-shaped organisms were detected for only few cells in the floc (Fig. 4). Additional evidence for successful use of RING-FISH to detect nirK-type denitrifiers in environmental samples was provided by the detection of increased numbers of halo-shaped signals after enrichment of denitrifiers in activated sludge. Enrichments were grown with nitrate as an electron acceptor either using the prevalent carbon sources or after addition of glucose or methanol as an additional carbon source. For RING-FISH with activated sludge and enriched sludge, a mixture of nirK polynucleotide probes for all four nirK-type denitrifiers was used. In fresh sludge (1 μl) an average of 54 halos were detected using either the digoxigenin-anti-digoxigenin antibody with the fluorescein label or the biotin-streptavidin conjugate labeled with Cy3 (Fig. 5). After 4 weeks of enrichment, the number of halos increased between five- and ninefold. A fivefold increase in the number of halos was observed for the enrichment with nitrate only, and a ninefold increase occurred in the enrichment supplemented with methanol.
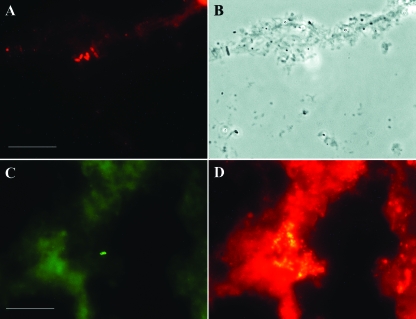
FIG. 4.
Detection of nirK-type denitrifiers in activated sludge by using RING-FISH with nirK polynucleotide probes. (A and C) Halo-shaped fluorescence signals of Cy3-labeled denitrifiers. (B) Floc in phase-contrast micrograph. (D) Bacterial cells in sludge stained by oligonucleotide probes EUB338I to EUB338III. Bars = 10 μm.
FIG. 5.
Enrichment of nirK-type denitrifiers in activated sludge detected by RING-FISH using nirK polynucleotide probes. Denitrifiers were visualized in nonenriched sludge and in sludge enriched anaerobically for 4 weeks. Bar 1, sludge supplemented with 5 mM nitrate; bar 2, sludge supplemented with 5 mM KNO3 and 5 μM glucose; bar 3, sludge supplemented 5 mM KNO3 and 25 μM methanol. The error bars indicate standard deviations (n = 2).
DISCUSSION
Evaluation of optimum hybridization conditions.
In this study, we adapted use of polyribonucleotide probes to specifically label and detect nirK-type denitrifiers based on their nirK genes. These genes are chromosomal single-copy genes coding for copper-containing nitrite reductase that are commonly used as functional marker genes to detect this type of denitrifier in environmental samples by cultivation-independent approaches (25, 29, 32, 36). Initially, the hybridization conditions were optimized using nirK-positive and -negative pure cultures to obtain specific and sensitive results. Polynucleotide probes were generated for four denitrifiers whose partial nirK sequences were 72 to 85% identical to each other and clustered with the sequences of other cultured denitrifiers in the nirK gene tree (see Fig. S6 in the supplemental material). The probes comprised a 515-bp fragment of the nirK gene, which was within the range of lengths (50 to 800 nucleotides) that were found to work for RING-FISH (44). In previous studies halo formation was observed only at high probe concentrations (200 to 250 ng/μl) (44), which is also consistent with the concentrations used here (300 and 160 ng/μl for hybridization on glass slides and in solution, respectively).
The halo-shaped fluorescence signal is believed to result from a network of probes formed in the periphery of the cell that is anchored via hybridization of a single probe molecule to a gene on the chromosome (44). Network formation depends on the tendency of the single-stranded RNA probes to fold into secondary structures under certain hybridization conditions (44). As indicated by in silico analysis of the nirK polynucleotide probe sequences using RNAdraw V1.1b2 (www.rnadraw.com/), all probes had low energy values (the ΔG values were between −69 and −88 kcal/mol with 15% formamide) and thus had a strong tendency to form secondary structures (see Fig. S1 to S4 in the supplemental material). The optimum stringency conditions for specific hybridizations were evaluated empirically by varying the formamide concentration in the hybridization buffer. Highly efficient hybridization of the probes was observed when a hybridization temperature of 53°C and 15% formamide in the hybridization buffer were used. The hybridization efficiency was independent of the use of an individual homologous probe or an equimolar mixture of probes for all four strains. Consistently, quantitative hybridization was observed only for cells in the exponential growth phase, indicating that there may have been differences in the cell wall structure or DNA accessibility during different developmental stages for the individual cells (43, 46). However, only the basic RING-FISH protocol was used without any modifications to further permeabilize the cell walls of the organisms, such as treatments with lysozyme, lysostaphin, or mutanolysin, as suggested recently (14). Under stringency conditions that were lower and higher than the optimum conditions fewer nirK-positive cells were labeled. At least for Pseudomonas sp. and S. meliloti this finding agrees with the significant shift in the formation of specific secondary structures in silico when the formamide concentration was increased from 15 to 20% (see Fig. S1 to S4 in the supplemental material). Generally, the lowest hybridization efficiency with 25% formamide correlated with the most condensed secondary structures of the mRNA molecules, suggesting that these structures are not accessible for network formation.
Specificity of hybridizations.
The specificity of the polynucleotide probes was confirmed by RING-FISH and nirK polynucleotide probe-based cell sorting of nirK-positive and -negative pure cultures. Under optimum hybridization conditions probes exclusively labeled the nirK-positive cells, but with lower formamide concentrations nonspecific fluorescence signals of nirK-negative cells were also detected during RING-FISH, although this was never observed for B. pumilus, probably due to impermeability of the gram-positive cell wall. However, nonspecific hybridizations with the probes did not result in the typical halo-shaped signal; rather, the signals resembled signals obtained by conventional oligo-FISH and thus stained the entire cell. This suggests that no probe network was formed in the periphery of the cells. The specificity of the signals was not affected by using either directly labeled fluorescent probes or the indirect method that included a separate step to detect digoxigenin and biotin reporter molecules with a fluorescently labeled anti-digoxigenin antibody and streptavidin conjugate, respectively. No signals were observed when the conjugates were used without prior hybridization of the probes, thus confirming that the signal was dependent on previous hybridization of the probes.
Since the indirect detection method and polynucleotide probe-based cell sorting share initial steps, we focused on using the latter approach. The results of cell sorting experiments using magnetic beads and subsequent analysis of the 16S rRNA genes of denitrifier-nondenitrifier mixtures also indicated the specificity of the hybridization. Generally, high efficiency (80 to 100%) was observed for depletion of the nirK-negative strain from mixtures, which was largely independent of the ratio of nirK-positive and -negative cells in the mixture, indicating that there was successful enrichment of denitrifiers based on their nirK genes. We initially attempted to sort cells based on the hybridization of the nirK polynucleotide probe network to DNA-coated microplates, as described previously by Zwirglmaier et al. (45). However, the specificity of cell separation was significantly lower using this technique compared to the specificity of the very specific depletion of nirK-negative cells using digoxigenin as a reporter molecule and magnetic beads to capture cells to whose DNA the probes had hybridized.
Finally, hybridization of sense polynucleotide probes to mRNA was not tested because cultures were kept under nondenitrifying conditions. With few exceptions (23), the denitrification process is triggered by the absence of oxygen and the presence of oxidized nitrogen compounds; thus, expression of denitrification genes is unlikely and also should not hamper the enrichment of nirK-type denitrifiers from environmental samples by probe-based cell capture.
Detection of nirK-positive organisms from environmental samples.
There was effective and specific detection of nirK genes in pure cultures, as shown above, and nirK-type denitrifiers were also detected in environmental samples (activated sludge) by RING-FISH. RING-FISH was used directly with fixed activated sludge, and cells with the typical peripheral halo-shaped fluorescence signal surrounded by the vast majority of other (bacterial) cells not showing any fluorescence were detected in the flocs. The potential ability to detect nirK-type denitrifiers by RING-FISH was further shown by the results obtained with 25 nirK clones that were obtained from the activated sludge sample. The sequences clustered exclusively in the vicinity of the probe sequences in the nirK gene tree; hence, the results indicate that denitrifiers with similar genotypes were dominant in this habitat (see Fig. S6 in the supplemental material). Moreover, increased numbers of cells with halos after enrichment for denitrifiers by addition of the electron acceptor nitrate or nitrate and glucose or methanol as an additional carbon source provided further evidence that there was specific detection of nirK-type denitrifiers in the sludge. The highest numbers of halos were found in the methanol-fed enrichment. Methanol is frequently used in sewage treatment systems to enhance denitrification rates due to its low cost, and it has been found to cause shifts in the overall denitrifier community and, more specifically, in the nirK-type denitrifier community (15, 18). In contrast to previous cultivation-based work (28, 37), these more recent studies suggest that there is dominance of populations other than Hyphomicrobium spp. and Paracoccus spp. that are specialized to utilize this C1 compound. On the other hand, in another study the workers retrieved nirK sequences related to the sequences of H. zavarzinii and Rhizobium hedysari from the active methanol-assimilating nitrate-reducing bacterial populations in activated sludge (30). However, it is important to state that based on separate analyses of nirK or 16S rRNA gene phylogeny it is premature to reach conclusions concerning the phylogenetic affiliation of denitrifiers or to infer that microorganisms are able to denitrify, respectively. Thus, hybridization using polynucleotide probes combined with conventional oligo-FISH or nirK-based cell sorting with subsequent 16S rRNA gene analysis has the potential to help resolve the unknown link between functional diversity and phylogenetic diversity. Polynucleotide probes are less specific than oligonucleotides and allow discrimination of prokaryotic groups separated by large evolutionary distances. For instance, archaeal and bacterial groups that are less than 75 and 70% related based on their 16 and 23S rRNA genes, respectively, could be discriminated (26). This agrees with our findings. Analyses with the nirK polynucleotide probes used in this study did not detect pure cultures with more distantly related nirK genes (<72%) by RING-FISH (unpublished data). This suggests that nirK polynucleotide probes specific for different clusters in the gene tree could be designed, allowing more focused analysis of the genetic background. However, two prerequisites for successful cell sorting for specifically retrieving microorganisms from environmental samples based on detection of functional low-copy-number genes are breaking up cell aggregates and improving the accessibility of the probes to target organisms (for instance, by using special pretreatment protocols) (14).
In summary, RING-FISH and polynucleotide probe-based cell sorting were successfully adapted to specifically and effectively detect nirK-type denitrifiers in pure cultures and also in environmental samples. Thus, a method using specific probes to selectively enrich these organisms is now available to obtain more insight into the phylogenetic affiliation of denitrifiers that are numerically dominant and thus relevant for denitrification in the environment. However, we believe that the entire procedure (RING-FISH combined with subsequent cell sorting) needs to be refined further for specific and more quantitative retrieval of denitrifiers from complex samples.
Supplementary Material
Acknowledgments
H. aestuarii DSM 1564 and H. zavarzinii ZV-580 were provided by C. G. Gliesche, and Pseudomonas sp. strain G-179 and P. stutzeri JM300 were obtained from J. M. Tiedje.
This work was supported by grants from the Max Planck Society (Munich, Germany).
Footnotes
Published ahead of print on 12 December 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, E. Kinston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1991. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
- 3a.Braker, G., H. L. Ayala-del-Río, A. H. Devol, A. Fesefeldt, and J. M. Tiedje. 2001. Community structure of denitrifiers, Bacteria, and Archaea along redox gradients in Pacific Northwest marine sediments by terminal restriction fragment length polymorphism analysis on amplified nitrite reductase (nirS) and 16S rRNA genes. Appl. Environ. Microbiol. 67:1893-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braker, G., A. Fesefeldt, and K.-P. Witzel. 1998. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 64:3769-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braker, G., J. Zhou, L. Wu, A. H. Devol, and J. M. Tiedje. 2000. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific Northwest marine sediment communities. Appl. Environ. Microbiol. 66:2096-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bremer, C., G. Braker, D. Matthies, A. Reuter, C. Engels, and R. Conrad. 2007. Impact of plant functional group, plant species, and sampling time on the composition of nirK-type denitrifier communities in soil. Appl. Environ. Microbiol. 73:6876-6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castro-González, M., G. Braker, L. Farías, and O. Ulloa. 2005. Communities of nirS-type denitrifiers in the water column of the oxygen minimum zone in the eastern South Pacific. Environ. Microbiol. 7:1298-1306. [DOI] [PubMed] [Google Scholar]
- 8.Conrad, R. 1996. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol. Rev. 60:609-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coyne, M. S., A. Arunakumari, B. A. Averill, and J. M. Tiedje. 1989. Immunological identification and distribution of dissimilatory heme cd1 and nonheme copper nitrite reductases in denitrifying bacteria. Appl. Environ. Microbiol. 55:2924-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crutzen, P. J. 1970. Influence of nitrogen oxides on atmospheric ozone content. Q. J. R. Meteorol. Soc. 96:320. [Google Scholar]
- 11.Daims, H., A. Bruhl, R. Amann, K.-H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 12.DeLong, E. F., L. T. Taylor, T. L. Marsh, and C. M. Preston. 1999. Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl. Environ. Microbiol. 65:5554-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dickinson, R. E., and R. J. Cicerone. 1986. Future global warming from atmospheric trace gases. Nature 319:109-115. [Google Scholar]
- 14.Fichtl, K. M. 2005. Polynucleotide probe based enrichment of bacterial cells: development of probes for species of clinical relevance. Ph.D. thesis. Technical University of Munich, Munich, Germany.
- 15.Ginige, M. P., P. Hugenholtz, H. Daims, M. Wagner, J. Keller, and L. L. Blackall. 2004. Use of stable-isotope probing, full-cycle rRNA analysis, and fluorescence in situ hybridization-microautoradiography to study a methanol-fed denitrifying microbial community. Appl. Environ. Microbiol. 70:588-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glockner, A. B., A. Jüngst, and W. G. Zumft. 1993. Copper-containing nitrite reductase from Pseudomonas aureofaciens is functional in a mutationally cytochrome cd1-free background (NirS−) of Pseudomonas stutzeri. Arch. Microbiol. 160:18-26. [DOI] [PubMed] [Google Scholar]
- 17.Hallberg, K. B., K. Coupland, S. Kimura, and D. B. Johnson. 2006. Macroscopic streamer growths in acidic, metal-rich mine waters in North Wales consist of novel and remarkably simple bacterial communities. Appl. Environ. Microbiol. 72:2022-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hallin, S., and M. Pell. 1998. Metabolic properties of denitrifying bacteria adapting to methanol and ethanol in activated sludge. Water Res. 32:13-18. [Google Scholar]
- 19.Hallin, S., I. N. Throbäck, J. Dicksved, and M. Pell. 2006. Metabolic profiles and genetic diversity of denitrifying communities in activated sludge after addition of methanol or ethanol. Appl. Environ. Microbiol. 72:5445-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirsch, P. 1989. Genus Hyphomicrobium Stutzer and Hartleb 1898, 76AL, p. 1897-1904. In J. T. Staley, M. P. Bryant, N. Pfennig, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology. Williams & Wilkins, Baltimore, MD.
- 21.Intergovernmental Panel on Climate Change. 2001. Climate change 2001: a scientific basis. Cambridge University Press, Cambridge, United Kingdom.
- 22.Karner, M. B., E. F. DeLong, and D. M. Karl. 2001. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409:507-510. [DOI] [PubMed] [Google Scholar]
- 23.Knowles, R. 1982. Denitrification. Microbiol. Rev. 46:43-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Layton, A. C., P. N. Karanth, C. A. Lajoie, A. J. Meyers, I. R. Gregory, R. D. Stapleton, D. E. Taylor, and G. S. Sayler. 2000. Quantification of Hyphomicrobium populations in activated sludge from an industrial wastewater treatment system as determined by 16S rRNA analysis. Appl. Environ. Microbiol. 66:1167-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, X. D., S. M. Tiquia, G. Holguin, L. Y. Wu, S. C. Nold, A. H. Devol, K. Luo, A. V. Palumbo, J. M. Tiedje, and J. Z. Zhou. 2003. Molecular diversity of denitrifying genes in continental margin sediments within the oxygen-deficient zone off the Pacific coast of Mexico. Appl. Environ. Microbiol. 69:3549-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludwig, W., and K.-H. Schleifer. 1994. Bacterial phylogeny based on 16S and 23S rRNA sequence analysis. FEMS Microbiol. Rev. 15:155-173. [DOI] [PubMed] [Google Scholar]
- 27.Manz, W., R. I. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 28.Neef, A., A. Zaglauer, H. Meier, R. I. Amann, H. Lemmer, and K.-H. Schleifer. 1996. Population analysis in a denitrifying sand filter: conventional and in situ identification of Paracoccus spp. in methanol-fed biofilms. Appl. Environ. Microbiol. 62:4329-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oakley, B. B., C. A. Francis, K. J. Roberts, C. A. Fuchsman, S. Srinivasan, and J. T. Staley. 2007. Analysis of nitrite reductase (nirK and nirS) genes and cultivation reveal depauperate community of denitrifying bacteria in the Black Sea suboxic zone. Environ. Microbiol. 9:118-130. [DOI] [PubMed] [Google Scholar]
- 30.Osaka, T., S. Yoshie, S. Tsuneda, A. Hirata, N. Iwami, and Y. Inamori. 2006. Identification of acetate- or methanol-assimilating bacteria under nitrate-reducing conditions by stable-isotope probing. Microb. Ecol. 52:253-266. [DOI] [PubMed] [Google Scholar]
- 31.Pernthaler, A., C. M. Preston, J. Pernthaler, E. F. DeLong, and R. Amann. 2002. Comparison of fluorescently labeled oligonucleotide and polynucleotide probes for the detection of pelagic marine bacteria and archaea. Appl. Environ. Microbiol. 68:661-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Priemé, A., G. Braker, and J. M. Tiedje. 2002. Diversity of nitrite reductase (nirK and nirS) gene fragments in forested upland and wetland soils. Appl. Environ. Microbiol. 68:1893-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shapleigh, J. P. 2006. The denitrifying prokaryotes, p. 769-792. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes. Springer, New York, NY.
- 34.Snaidr, J., R. Amann, I. Huber, W. Ludwig, and K.-H. Schleifer. 1997. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl. Environ. Microbiol. 63:2884-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoffels, M., W. Ludwig, and K.-H. Schleifer. 1999. rRNA probe-based cell fishing of bacteria. Environ. Microbiol. 1:259-271. [DOI] [PubMed] [Google Scholar]
- 36.Throbäck, I. N., K. Enwall, Å. Jarvis, and S. Hallin. 2004. Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol. Ecol. 49:401-417. [DOI] [PubMed] [Google Scholar]
- 37.Timmermans, P., and A. van Haute. 1983. Denitrification with methanol. Fundamental study of the growth and denitrification capacity of Hyphomicrobium sp. Water Res. 17:1249-1255. [Google Scholar]
- 38.Trebesius, K., R. Amann, W. Ludwig, K. Muhlegger, and K.-H. Schleifer. 1994. Identification of whole fixed bacterial-cells with nonradioactive 23S ribosomal-RNA-targeted polynucleotide probes. Appl. Environ. Microbiol. 60:3228-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan, T., M. W. Fields, L. Wu, Y. Zu, J. M. Tiedje, and J. Zhou. 2003. Molecular diversity and characterization of nitrite reductase gene fragments (nirK and nirS) from nitrate- and uranium-contaminated groundwater. Environ. Microbiol. 5:13-24. [DOI] [PubMed] [Google Scholar]
- 40.Ye, R. W., M. R. Fries, S. G. Bezborodnikov, B. A. Averill, and J. M. Tiedje. 1993. Characterization of the structural gene encoding a copper-containing nitrite reductase and homology of this gene to DNA of other denitrifiers. Appl. Environ. Microbiol. 59:250-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zwirglmaier, K. 2005. Fluorescence in situ hybridisation (FISH)—the next generation. FEMS Microbiol. Lett. 246:151-158. [DOI] [PubMed] [Google Scholar]
- 43.Zwirglmaier, K., K. Fichtl, and W. Ludwig. 2005. In situ functional gene analysis: recognition of individual genes by fluorescence in situ hybridization. Methods Enzymol. 397:338-351. [DOI] [PubMed] [Google Scholar]
- 44.Zwirglmaier, K., W. Ludwig, and K.-H. Schleifer. 2003. Improved fluorescence in situ hybridization of individual microbial cells using polynucleotide probes: the network hypothesis. Syst. Appl. Microbiol. 26:327-337. [DOI] [PubMed] [Google Scholar]
- 45.Zwirglmaier, K., W. Ludwig, and K.-H. Schleifer. 2004. Improved method for polynucleotide probe-based cell sorting, using DNA-coated microplates. Appl. Environ. Microbiol. 70:494-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zwirglmaier, K., W. Ludwig, and K.-H. Schleifer. 2004. Recognition of individual genes in a single bacterial cell by fluorescence in situ hybridization—RING-FISH. Mol. Microbiol. 51:89-96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.