Abstract
Mycobacterium avium subsp. paratuberculosis is an important animal pathogen widely disseminated in the environment that has also been associated with Crohn's disease in humans. Three M. avium subsp. paratuberculosis genomotypes are recognized, but genomic differences have not been fully described. To further investigate these potential differences, a 60-mer oligonucleotide microarray (designated the MAPAC array), based on the combined genomes of M. avium subsp. paratuberculosis (strain K-10) and Mycobacterium avium subsp. hominissuis (strain 104), was designed and validated. By use of a test panel of defined M. avium subsp. paratuberculosis strains, the MAPAC array was able to identify a set of large sequence polymorphisms (LSPs) diagnostic for each of the three major M. avium subsp. paratuberculosis types. M. avium subsp. paratuberculosis type II strains contained a smaller genomic complement than M. avium subsp. paratuberculosis type I and M. avium subsp. paratuberculosis type III genomotypes, which included a set of genomic regions also found in M. avium subsp. hominissuis 104. Specific PCRs for genes within LSPs that differentiated M. avium subsp. paratuberculosis types were devised and shown to accurately screen a panel (n = 78) of M. avium subsp. paratuberculosis strains. Analysis of insertion/deletion region INDEL12 showed deletion events causing a reduction in the complement of mycobacterial cell entry genes in M. avium subsp. paratuberculosis type II strains and significantly altering the coding of a major immunologic protein (MPT64) associated with persistence and granuloma formation. Analysis of MAPAC data also identified signal variations in several genomic regions, termed variable genomic islands (vGIs), suggestive of transient duplication/deletion events. vGIs contained significantly low GC% and were immediately flanked by insertion sequences, integrases, or short inverted repeat sequences. Quantitative PCR demonstrated that variation in vGI signals could be associated with colony growth rate and morphology.
Mycobacterium avium subsp. paratuberculosis is a weakly gram-positive, acid-fast bacillus causing chronic enteritis, or Johne's disease (JD), in many animal species, including primates. JD is an infectious wasting condition that develops as a consequence of chronic inflammation of the gastrointestinal tract and is an important cause of economic losses associated with farm animals (17, 22). Long-term excretion by animals with subclinical or clinical infection has led to the establishment of reservoirs in many wildlife species and extensive spread into the environment and dairy products. This exposure of humans to M. avium subsp. paratuberculosis and the findings that M. avium subsp. paratuberculosis can be detected in a significant majority of patients with Crohn's disease, a chronic enteritis of humans with striking similarities to JD, suggest the potential, although still controversial, of this organism as a zoonotic agent (2, 4, 14, 18, 28).
To fully investigate these links, it is important to accurately define M. avium subsp. paratuberculosis phylogeny. Previously, three major M. avium subsp. paratuberculosis types have been classified using pulsed-field gel electrophoresis (PFGE), IS900 restriction fragment length polymorphism, PCR and restriction enzyme analysis of gyrB, denaturing gradient gel electrophoresis, and conventional culture characteristics (6, 9, 10, 16, 33, 39). These include type I (previously described as the “sheep type”), comprising pigmented and nonpigmented strains isolated from sheep in Morocco, Scotland, Iceland, South Africa, Australia, and New Zealand, strains isolated from cattle in Australia and Iceland, and some Norwegian and New Zealand caprine strains; type II (previously described as the “cattle type”), which is associated primarily with cattle but which can also be isolated from a broad range of hosts, including humans; and type III (intermediate type), which has been described for a few ovine isolates from South Africa, Canada, and Iceland and a porcine isolate from Canada, as well as caprine and bovine isolates from Spain (8). Previous comparative genomic hybridizations (CGH) between M. avium subsp. paratuberculosis strains and other related members of the M. avium complex (MAC) have demonstrated the presence of broad genomic differences called either genomic islands, comprising regions of contiguous genes (42) probably acquired as single units by horizontal transfer (1), or large sequence polymorphisms (LSPs) (31, 32, 35, 37). Some LSPs specifically associate with distinct M. avium subsp. paratuberculosis types and have indicated that M. avium subsp. paratuberculosis strains found predominantly in sheep are much more closely related to other members of the MAC than are the more commonly isolated M. avium subsp. paratuberculosis type II strains, suggesting that M. avium subsp. paratuberculosis type II strains have a reduced genomic complement (11, 24, 36).
This work addresses these analyses in depth with the development of a microarray that comprises optimized 60-mer oligonucleotide reporters designed to represent the gene contents of the sequenced genomes of two closely related members of the MAC, M. avium subsp. paratuberculosis K-10 and Mycobacterium avium subsp. hominissuis strain 104. We describe the use of this array (designated the MAPAC array) to characterize a range of M. avium subsp. paratuberculosis genomotypes, particularly focusing upon M. avium subsp. paratuberculosis type I and III strains from various animal hosts. The study highlights the close relation of M. avium subsp. paratuberculosis type I and III strains while showing significant genomic deletions, similarities to M. avium subsp. hominissuis 104, and variations in gene copy number of low-GC% gene regions, which we suggest may contribute to host preferences and variations in epidemiological spread observed for these M. avium subsp. paratuberculosis types.
MATERIALS AND METHODS
Mycobacterial strains.
Reference strains used in this study included M. avium subsp. paratuberculosis K-10 (ATCC BAA-968) and M. avium subsp. hominissuis 104 (a kind gift from Marcel Behr, Canada). Reference strain M. avium subsp. paratuberculosis K-10 is M. avium subsp. paratuberculosis type II, originally isolated from a cow with JD (23), and M. avium subsp. hominissuis 104 is a Mycobacterium avium subsp. hominissuis serotype 4 strain originally isolated from an AIDS patient (21). M. avium subsp. paratuberculosis strains from various regions in Spain, Scotland, and Denmark (Table 1) were isolated as previously described by de Juan et al. (9) and Stevenson et al. (39). Primary cultures were incubated at 37°C for up to 10 months on Middlebrook 7H11 agar supplemented with Selectatabs (amphotericin B, polymyxin B, carbenicillin, and trimethoprim; code MS 24; MAST Laboratories, Ltd., Merseyside, United Kingdom), 10% Middlebrook oleic acid-albumin-dextrose-catalase enrichment medium (Difco, Surrey, United Kingdom), and 2 μg per ml of mycobactin J (Allied Monitor, Fayette, MO) per ml. For pigmented strains, cultures additionally contained 20% (vol/vol) heat-inactivated newborn calf serum, 2.5% (vol/vol) glycerol, and 2 mM asparagine. The study also included a sample of DNA extracted from an M. avium subsp. paratuberculosis isolate labeled 6760B (S1 restriction fragment length polymorphism profile [7]), originally isolated from a sheep in New Zealand.
TABLE 1.
Origins of M. avium subsp. paratuberculosis isolates
| Strain(s) | Host breed | Origin | PFGE type |
|---|---|---|---|
| Isolates used for MAPAC array (n = 12) | |||
| M189 | Finn sheep | Scotland (central) | I pigmented |
| 213G | Sheep | Scotland (Shetland) | I pigmented |
| 6760B | Sheep | New Zealand | I |
| 896 | Bullfighting cattle | Spain (north central) | II |
| CAM20, CAM84 | Guadarrama goat | Spain (central) | II |
| 574 | Murciano-Granadina goat | Spain (south central) | II |
| 619, 841 | Bullfighting cattle | Spain (south central) | III |
| CAM38, CAM86, CAM87 | Guadarrama goat | Spain (central) | III |
| Additional isolates used for PCR screening (n = 66) | |||
| 21P | Sheep | Denmark (Faroe Islands) | I pigmented |
| 208G, 235G | Sheep | Scotland (Shetland) | I pigmented |
| 813, 940, MI05/02938-2 | Bullfighting cattle | Spain (north central) | II |
| D206 | Fallow deer | Spain (south) | II |
| 172 | Goat | Unknown | II |
| 232, 388, 416, 417, 427, 439, 446, 456, 464, 465, 469, 474, 484, 611, 872, 915, 916, CAM19, CAM63, CAM72, CAM07, MI05/03721-2 | Guadarrama goat | Spain (central) | II |
| 51, 53, 55, 56 | Holstein cattle | Spain (Balearic Islands) | II |
| 682 | Holstein cattle | Spain (central) | II |
| 25, 27, 33, 34, 35 | Holstein cattle | Spain (north) | II |
| 10 | Limousine cattle | Spain (north central) | II |
| 1 | Mouflon | Spain (north central) | II |
| 46, 72, 78, 83, 87, 90, 94, 106, 45b, N10, N11, N21, N24, N29, N42, N64, N65, N90, N105, N109, N124 | Murciano-Granadina goat | Spain (south central) | II |
| 733 | Bullfighting cattle | Spain (north central) | III |
| CAM40, CAM42 | Guadarrama goat | Spain (central) | III |
DNA extraction from cultures.
Cultures were grown on solid Middlebrook 7H11 medium for up to 12 weeks. For MAPAC array analysis, 109 cells were scraped and emulsified by passage 10 times through a 25-gauge needle into 650 μl mycobacterial lysis buffer (8.6 ml H2O, 0.5 M EDTA [pH 8.0], 5 M NaCl, 1 M Tris-HCl, 10% sodium dodecyl sulfate [SDS], 1 mg/ml lysozyme [catalog no. L-6876; Sigma, United Kingdom]), 0.15 mg/ml proteinase K (catalog no. P-2308; Sigma, United Kingdom), and 0.5 mg/ml lipase (catalog no. L8525-1MU; Sigma, United Kingdom) and incubated at 37°C in a rotator for 1 h. Samples were added to lysing matrix B (catalog no. 6911-100; Qbiogene, United Kingdom) in 1.9-ml ribolyser reaction tubes, mechanically disrupted in a ribolysing machine (Hybaid, United Kingdom) at 6,500 rpm for 45 s, and iced for 10 min. Lysate (220 μl) was then added to 200 μl of Qiagen DNAeasy AL lysis buffer, mixed, and applied to a DNAeasy column. Ethanol (100%; 200 μl) was then added and the tube sealed and mixed. Columns were washed in 500 μl Qiagen lysis buffers 1 and 2, with centrifugation at 8,000 × g for 1 min, and then eluted in 90 μl DNA/RNase-free H2O overnight on the column at 4°C. DNA from single colonies with a large or small morphological appearance was prepared for PCR analysis of variable genomic islands (vGIs) by being lysed in mycobacterial lysis buffer and then extracted after ribosylation using standard phenol, phenol-chloroform, and ethanol precipitations into 50 μl DNA/RNase-free H2O overnight. DNA for PCR screening of specific genes within LSPs was prepared from a resuspension of a loopful of colonies growing on solid media into 200 μl of sterile deionized water and heat inactivation at 100°C for 10 min and then cleared by centrifugation at 8,000 × g for 1 min.
Microarray design and optimization.
The design strategy undertaken to generate a microarray with 60-mer oligonucleotide reporters that represented all annotated genes in M. avium subsp. paratuberculosis K-10 (GenBank accession no. NC_002944) and M. avium subsp. hominissuis 104 (GenBank accession no. NC_008595) followed the general design principles for multistrain arrays described previously (20). However, the approach taken for the MAPAC array involved an initial optimization phase to empirically select an optimal set of oligonucleotide reporters to subsequently progress to oligonucleotide synthesis and spotting using more standard robotic arraying technology (19).
For the optimization phase, multiple oligonucleotide reporters were designed in silico to represent each of the annotated genes in M. avium subsp. paratuberculosis K-10 and M. avium subsp. hominissuis 104, ensuring standard oligonucleotide design criteria of matched melting temperature and lack of secondary structures or polymeric repeats (Oxford Gene Technology [OGT], United Kingdom). Furthermore, design criteria aimed to minimize potential cross-hybridization by intrastrain paralogues while maintaining identity to interstrain orthologues. This set of 15,000 oligonucleotides, plus their associated mismatched control oligonucleotides (∼15,000), were then arrayed at high density using inkjet in situ synthesis technology by OGT and hybridized with DNA from the two reference strains, namely, M. avium subsp. paratuberculosis K-10 and M. avium subsp. hominissuis 104. Based on the hybridization performance of each oligonucleotide in the inkjet in situ synthesis arrays, in terms of both relation to the mismatched control and intensity in each channel, a subset of 5,744 optimally performing oligonucleotides were selected as the final oligoset for the spotted array.
The optimal set of 60-mer oligonucleotides were synthesized (Operon Biotechnologies, Germany), supplied in 384-well plates, and resuspended at 50 mM in 50% dimethyl sulfoxide. These oligonucleotide reporters were then arrayed at high density on aminosilane-coated UltraGaps slides (Corning) by use of a MicroGrid II (BioRobotics) arraying robot. Microarrays were postprint processed according to the slide manufacturer's instructions to rehydrate, fix, and UV cross-link the oligonucleotides.
DNA labeling and microarray hybridization.
DNA from the test strain and the M. avium subsp. paratuberculosis K-10 reference strain was fluorescently labeled and hybridized to the microarray using protocols described previously (12). Briefly, 1 μg of DNA was labeled by random priming with Klenow polymerase to incorporate either Cy3 or Cy5 dCTP (GE Healthcare) for the test strain or the reference strain, respectively. Equal amounts of the Cy3- and Cy5-labeled samples were copurified through a Qiagen MinElute column (Qiagen), mixed with a formamide-based hybridization solution (1× MES [morpholineethanesulfonic acid], 1 M NaCl, 20% formamide, 0.02 M EDTA, 1% Triton X-100), and denatured at 95°C for 2 min. The labeled sample was loaded on to a prehybridized (3.5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% SDS, 10 mg/ml bovine serum albumin) microarray under two 22- by 22-mm LifterSlips (Erie Scientific), sealed in a humidified hybridization cassette (Corning), and hybridized overnight by immersion in a water bath at 55°C for 16 to 20 h. Slides were washed once in 400 ml 1× SSC, 0.06% SDS at 55°C for 2 min and twice in 400 ml 0.06× SSC for 2 min.
Microarray data analysis.
Microarrays were scanned using an Affymetrix 428 scanner, and signal intensity data were extracted using BlueFuse for Microarrays v3.5 (BlueGnome, Cambridge, United Kingdom). The intensity data were further postprocessed using BlueFuse to exclude both controls and low-confidence data (P < 0.1) prior to normalization by two-dimensional Lowess (window size of 20) and median centering. Further analysis of the normalized data was undertaken using BlueFuse, GeneSpring 7.3.1 (Agilent Technologies), and Eisen Cluster (13).
Analysis methods for CGH calling to determine the genes that were present in the test strain only, present in the reference strain only, or present in both strains were undertaken as described previously for twofold and 3-standard deviation (SD) approaches (41), using a hidden Markov model for CGH calling (29), or utilizing the BlueFuse CGH calling algorithm with parameters of >2 SD for the genome, a log2 threshold of >1 or ≤1, and a minimum region size of 1.
Measures of specificity and sensitivity for each of these approaches were determined by comparison with the expected results as predicted by BLAST analysis of the oligonucleotide reporter sequences against the reference genome sequences, as described previously (41). Further cluster analysis to investigate the relatedness of strains and highlight the genomic regions of interest was undertaken using only genes called by the BlueFuse CGH algorithm either test strain specific or reference strain specific in any one of the 12 M. avium subsp. paratuberculosis strains analyzed. Clustering of log ratio data was performed in Eisen Cluster to cluster arrays using only a standard, uncentered Pearson correlation by average linkage, as the genes were maintained in genome order.
Genes that fell within a >1.5 or <2 log2 threshold increase or decrease relative to the M. avium subsp. paratuberculosis K-10 reference normalized standard and were contiguous in the genome were flagged as belonging to a vGI.
PCR.
PCR primers (AltaBioscience, United Kingdom) were designed using Primer 3 software (http://biotools.umassmed.edu/bioapps/primer3_www.cgi) to specifically amplify open reading frames (ORFs) in LSP and vGI gene loci (Table 2) . PCR was carried out using an Expand high-fidelity PCR system (Roche Diagnostics, Germany). Each reaction was performed with a 50-μl volume, containing 5 μl of DNA sample and 1× Expand HiFiPLUS reaction buffer containing 1.5 mM MgCl2, 0.200 mM PCR nucleotide mix PCR grade, upstream and downstream primers (2 μM each), 5 μl of dimethyl sulfoxide (Sigma, United Kingdom), and 2.5 U of Expand HiFiPLUS enzyme blend, under the following conditions: denaturation at 94°C for 3 min followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 58°C for 45 s, and extension at 72°C for 1 min 30 s, with a final cycle of extension at 72°C for 5 min.
TABLE 2.
PCR primer pairs and amplicon sizes of M. avium subsp. paratuberculosis ORFs used in this work
| Region | ORFa | Primers (5′-3′)b | Size (bp) |
|---|---|---|---|
| vGI-10 | Pre-16S rRNA gene | (F) TTGGCCATACCTAGCACTCC; (R) GCGCAGCGAGGTGAATTT | 97 |
| vGI-1 | MAP0101 | (F) GGTTACCGACTTGGTCCAGA; (R) CCCGTCAGATCCATTACGAC | 238 |
| —c | MAP0160 | (F) ATGCTTCGCGATACTTCCAA; (R) TGAGCACCTTGTTCAAATCG | 178 |
| vGI-4 | MAP0859c | (F) CCGGCGTACCTACAGACATT; (R) GAGCGATACAGGCGAAAGAC | 255 |
| vGI-4 | MAP0865c | (F) CCCGATAGCTTTCCTCTCCT; (R) GATCTCAGACAGTGGCAGGTG | 609 |
| INDEL4 | MAP1435 | (F) TGATTGCGTTCACGTCGTC; (R) AACAGCGCATCGATCACATA | 265 |
| — | MAP2729 | (F) GTGGCGGACAACGACTTC; (R) GATCTGCTCTCGCAGTTCG | 216 |
| INDEL15 | MAP3584 | (F) GCGTTGGATCCTTTCGTG; (R) GTCCAGGCCGTCGAGATAG | 633 |
| vGI-13 | MAP3746 | (F) ATGACAAGGACACCCGAAAG; (R) AGTGCAGAACTCACGCAATG | 239 |
| INDEL12 | MAV_4125 | (F) TCACCTGTCCAGATCAACGA; (R) CGGGATCAGCTTGAGATACC | 303 |
| INDEL12 | MAV_4126 | (F) GAACATGAACACCGAGGTCAC; (R) CACACGTACTCGTTGGCGTA | 306 |
Annotations are from GenBank.
F, forward; R, reverse.
—, not associated with vGI or LSP.
Quantitative PCR (qPCR) against vGI-associated genes and gene controls for normalization was performed in duplicate in two separate experiments (see Table S1 in the supplemental material) using a Power SYBR green qPCR kit (ABgene, United Kingdom) according to the manufacturer's specifications with a Stratagene MX3000P instrument (Stratagene, United Kingdom). Estimates of the copy number for each gene were made using calibration curves obtained for each PCR primer pair against a dilution curve of M. avium subsp. paratuberculosis K-10 reference DNA using MxPro software (Stratagene, United Kingdom). The estimated total sample copy numbers determined for each sample were initially normalized against the estimated total copy numbers of MAP0160. qPCR from MAP0160 was not available on all colonies from one experiment; therefore, MAP0101 was used to normalize samples in this case. To decrease any bias introduced by variations in amplification efficiencies between these normalizing genes, the final increases/decreases were calculated using the averages of MAP0101 and MAP0160 results from both experiments.
Microarray data accession numbers.
Fully annotated microarray data have been deposited in BμG@Sbase (accession no. A-BUGS-35 and E-BUGS-69) (http://bugs.sgul.ac.uk/E-BUGS-35 and http://bugs.sgul.ac.uk/E-BUGS-69, respectively) and also ArrayExpress (accession no. A-BUGS-35 and E-BUGS-69).
RESULTS
MAPAC microarray validation.
An evaluation of the MAPAC microarray performance was made by hybridizing labeled DNA from the sequenced reference strains M. avium subsp. paratuberculosis K-10 and M. avium subsp. hominissuis 104 (Fig. 1). Comparison of these hybridization data with BLAST predictions indicated that the array was correctly identifying genes specific to M. avium subsp. paratuberculosis K-10 or M. avium subsp. hominissuis 104 and also genes shared between M. avium subsp. paratuberculosis K-10 and M. avium subsp. hominissuis 104. Measures of sensitivity and specificity, using the various analysis approaches to identify genes present or absent/highly divergent in M. avium subsp. hominissuis 104 compared to M. avium subsp. paratuberculosis K-10, supported the validation of the array using the twofold (sensitivity of 98%, specificity of 98%), 3-SD (sensitivity of 98%, specificity of 98%), hidden Markov model (sensitivity of 91%, specificity of 96%), or BlueFuse CGH (sensitivity of 98%, specificity of 99%) approach.
FIG. 1.
Microarray data for validation hybridization comparing the two sequenced reference strains, using M. avium subsp. hominissuis 104 as the test strain and M. avium subsp. paratuberculosis K-10 as the reference strain per other strain comparisons. Scatter plots show the signal intensities for the test (y axis) versus the reference (x axis) strain channels. Data points are colored according to the BLAST prediction based on the sequence: black, predicted to be present in both the test and reference strains; light gray, predicted to be present in the test strain only; dark gray, predicted to be present in the reference strain only. Diagonal lines represent twofold cutoffs applied to each strain for each of the analysis methods used. MAH, M. avium subsp. hominissuis; MAP, M. avium subsp. paratuberculosis.
The BlueFuse CGH calling protocol was established as providing a reliable and automated call on strain-specific genes for the two reference strains, with the best balance of false positives (2%) to false negatives (1%), and therefore this method was chosen for the analysis of all other strains. A subset of genes that were called either test strain specific or reference strain specific in any one of the test strains was selected. This subset of genes represented the genes of interest that would include genomic differences between strains and so was subjected to clustering. A summary of the clustering results and genomic loci of interest are presented in Table 3, with a complete clustering tree provided in Fig. S1 in the supplemental material. A full set of hybridization ratios with predicted gene functions for each gene are supplied in Table S2 in the supplemental material.
TABLE 3.
Summary of significant signal divergences from multiple probes in LSPs and vGIs in M. avium subsp. paratuberculosis types I, II, and III and M. avium subsp. hominissuis 104 compared with M. avium subsp. paratuberculosis K-10
Nomenclature of previously annotated LSPs associated with INDELs is given in parentheses.
−, signal >2- and <5-fold less than that for K-10; −−, signal >5-fold less than that for K-10; ., signal >1.5-fold less than and <1.5-fold greater than that for K-10; V−, signal >1.5- and <2-fold less than that for K-10; +, signal >2-fold and <5-fold greater than that for K-10; ++, signal >5-fold greater than that for K-10; V+, signal >1.5- and <2-fold greater than that for K-10.
M. avium subsp. hominissuis 104 genomic loci present in M. avium subsp. paratuberculosis type I and type III strains.
All of the M. avium subsp. paratuberculosis type I and type III strains but none of the M. avium subsp. paratuberculosis type II strains, including the reference strain M. avium subsp. paratuberculosis K-10, showed significant differences in hybridization when analyzed by microarray, suggesting the presence of 89 ORFs also present in the M. avium subsp. hominissuis 104 genome (Table 3). These included three single ORFs (insertion/deletion 1 [INDEL1], or MAV_0339 [tetR regulator]; INDEL8, or MAV_2254 [function unknown]; and INDEL9, or MAV_2223 [IS6120]) and six LSPs (INDEL3, or MAV_3258 to MAV_3270 [previously described as MAV17 {42}]; INDEL5, or MAV_2978 to MAV_2998 [previously described as MAV14 {42}]; INDEL10, or MAV_1975 to MAV_2008 [previously described as MAV7 {42}]; INDEL12, or MAV_4125 to MAV_4130 [previously described as MAV21 {42}]; INDEL14, or MAV_4351 to MAV_4353 [dioxygenase]; and INDEL16, or MAV_5225 to MAV_5243 [previously described as MAV24 {42}]) containing prevalent predicted functions involving lipid metabolism (31). Full putative-function lists are supplied in Table S2 in the supplemental material.
Deletion of M. avium subsp. paratuberculosis genomic loci associated with M. avium subsp. paratuberculosis type I and type III strains.
All M. avium subsp. paratuberculosis type I and type III strains arrayed by MAPAC showed a significant decrease in hybridization relative to that for M. avium subsp. paratuberculosis K-10 in a total of 26 ORFs (Table 3). These included INDEL2, or MAV_0775 (an ORF not called in the M. avium subsp. paratuberculosis annotation but positioned between MAP0660 and MAP0661); INDEL6, or MAP1484 to MAP1491; and INDEL7, or MAP1728c to MAP1744 (previously described as LSP locus S2 [24]). All M. avium subsp. paratuberculosis type I strains had additional deletions of INDEL11, or MAP2704 (hemolysin III like), and decreased signals to INDEL13, or MAP3460c (a transposase with eight similar copies in M. avium subsp. paratuberculosis K-10). All M. avium subsp. paratuberculosis type III strains had additional deletions of INDEL4, or MAP1433c to MAP1438c (lipid metabolism), and INDEL15, or MAP3584 (alkanesulfonate monooxygenase). Each of these deleted ORFs was present in M. avium subsp. hominissuis 104. All of the strains tested contained MAP2325, which has previously been reported as deleted from some M. avium subsp. paratuberculosis type I sheep strains isolated in Australia (24).
Screening of M. avium subsp. paratuberculosis strain panel for INDELs by PCR.
To confirm INDELs described by the microarray, we used specific PCRs for INDEL4, INDEL12, and INDEL15 (Table 2) to screen 66 M. avium subsp. paratuberculosis strains recovered from different geographic areas and hosts in Spain, Scotland, and Denmark that had previously been typed by PFGE (Table 1) and confirmed those previously submitted for microarray analysis (n = 12). PCRs for INDEL15 (MAP3584) were positive for all M. avium subsp. paratuberculosis type I strains (n = 6) and M. avium subsp. paratuberculosis type II strains (n = 64) but negative for all M. avium subsp. paratuberculosis type III strains (n = 8). INDEL12 (MAV_4125) PCRs were positive for all M. avium subsp. paratuberculosis type I and type III strains but negative for all M. avium subsp. paratuberculosis type II strains. INDEL12 (MAV_4126) PCRs were positive for all M. avium subsp. paratuberculosis type III strains and type I pigmented strains but negative for all other strains. PCRs for INDEL4 (MAP1435) were positive for all M. avium subsp. paratuberculosis type I and type II strains but negative for all M. avium subsp. paratuberculosis type III strains (Table 4).
TABLE 4.
Summary of specific M. avium subsp. paratuberculosis gene PCRs performed with DNA extracted from M. avium subsp. paratuberculosis strains isolated from various hosts and locations
| Type | Host(s) (no. of strains) | Presence (+) or absence (−) of locus tag:
|
||||
|---|---|---|---|---|---|---|
| MAP3584 | MAP1435 | MAV_4125 | MAV_4126 | MAP865 | ||
| I | Sheep (5) | + | + | + | + | + |
| Sheep (1) | + | − | + | − | + | |
| II | Cattle (15), goat (47), fallow deer (1), mouflon (1) | + | + | − | − | + |
| III | Cattle (3), goat (5) | − | − | + | + | + |
LSP INDEL12 sequence analysis.
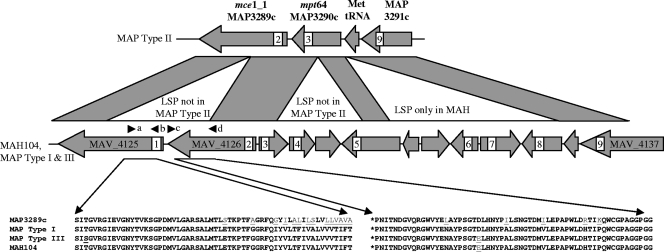
MAPAC results for INDEL12 were not fully consistent with previously described array data covering this region (32). We therefore performed sequencing on PCR products amplified from M. avium subsp. paratuberculosis type I strain M189 (EMBL accession no. FM199950) and M. avium subsp. paratuberculosis type III strain CAM86 (EMBL accession no. FM199949) with primers specific to MAV_4125 and MAV_4126 located within INDEL12 and compared these with the M. avium subsp. paratuberculosis K-10 and M. avium subsp. hominissuis 104 reference genomes (Fig. 2). This showed that in M. avium subsp. paratuberculosis type II strains, as a consequence of a small internal deletion, a fusion of the MAV_4125 C terminus and the MAV_4126 N terminus resulted in a new ORF (MAP3289c), while M. avium subsp. paratuberculosis type I strain M189 and M. avium subsp. paratuberculosis type III strain CAM86 retained both genes with 99% homology to M. avium subsp. hominissuis 104 gene sequences but each containing nonsynonymous sequence divergences specific to each M. avium subsp. paratuberculosis type. MAV_4125 and MAV_4126 are both homologues of M. avium subsp. paratuberculosis mycobacterial cell entry genes, which demonstrates that M. avium subsp. paratuberculosis type II strains contain a decreased complement of these important virulence determinants. In addition, the immediately adjacent sequence alignments show a similar small deletion that has resulted in the replacement of 30% of the C terminus of the M. avium subsp. paratuberculosis type II mpt64 gene (MAP3290c) compared to its homologue MAV_4130 present in both M. avium subsp. paratuberculosis type I and type III strains and M. avium subsp. hominissuis 104.
FIG. 2.
Diagrammatic genomic alignment of the M. avium subsp. paratuberculosis type II (MAP3289c to MAP3291c) region against M. avium subsp. paratuberculosis type I and III strains and M. avium subsp. hominissuis 104 (MAV_4125 to MAV_4137), showing homologies (gray) and locations of LSPs that have generated the mce1_1 gene by fusing sections of MAV_4125 and MAV_4126 and the mpt64 gene by fusing sections of MAV_4130 and the complement of MAV_4127. The approximate positions of gene probes 1 to 9 used in the MAPAC array are indicated in white boxes. The positions of PCR primer pairs used for detection of MAV_4125 (a and b) and MAV_4126 (c and d) are also shown. Sequence differences (underlined) observed in the MAV_4126 and MAV_4127 genes of M. avium subsp. paratuberculosis and M. avium subsp. hominissuis 104 obtained from PCR amplicons generated using primers a and d are indicated and aligned against MAP3289c obtained from the M. avium subsp. paratuberculosis K-10 genome sequence. MAH, M. avium subsp. hominissuis; MAP, M. avium subsp. paratuberculosis.
vGI analysis.
MAPAC array data revealed 16 vGIs totaling 138 kb (see Table S2 in the supplemental material) that were present and variable in type I and type III isolates but present but not variable in type II isolates. Ten vGIs were present within the M. avium subsp. paratuberculosis K-10 reference genome and absent from the M. avium subsp. hominissuis 104 genome, thus being partially or completely inclusive of M. avium subsp. paratuberculosis LSPs. A comparison of GC% contents of vGIs against those of LSPs not associated with vGIs showed that the vGI group had a GC% significantly (P = 0.0005; Mann-Whitney) lower than that of the LSP group. In addition, 80% of vGIs were immediately bounded by short inverted repeats and 40% were immediately bounded by or contained integrase proteins and transpositional elements, including IS900, IS1311, IS1610, and IS1110 (Table 5).
TABLE 5.
Positions, nomenclature, and GC% of M. avium subsp. paratuberculosis K-10 LSPs and vGIs with associated transposable elements not present in M. avium subsp. hominissuis 104
| LSP namea | LSP locus tag(s) | vGI name | vGI locus tag(s) | GC% | Associated transposition element(s) | Associated short inverted repeat sequence | Associated short inverted repeat location(s) |
|---|---|---|---|---|---|---|---|
| vGI-1a | MAP0071 to MAP0093 | 65.84 | |||||
| MAP1, or LSPp1 | MAP0092 to MAP0108 | vGI-1b | MAP0094 to MAP0103c | 63.89 | MAP0104 (IS1311) | CGGTGATCCGCCG | MAP092 to MAP0103c/MAP0104 |
| MAP2, or LSPp2 | MAP0282c to MAP0284c | vGI-2 | MAP0281 to MAP0283c | 60.57 | CACGCCGACGCC | MAP0280 to MAP0284c | |
| MAP3, or LSPp3 | MAP0387 to MAP0389 | 65.73 | |||||
| vGI-3 | MAP0758 to MAP0774c | 65.81 | CGAGGTCGTCCGCT | MAP0758 to MAP0774c | |||
| MAP4, or LSPp4 | MAP0850c to MAP0866 | vGI-4 | MAP0852 to MAP0866 | 59.91 | MAP0849 (IS1311), MAP0850 (ISMav2), MAP0866 (integrase) | CGGACGGGCGG | MAP0850 to MAP0866 |
| MAP5, or LSPp5 | MAP0956 to MAP0967 | 69.61 | GCGCAGCGCGTCG | MAP0957 to MAP0967c | |||
| MAP6, or LSPp6 | MAP1231 to MAP1237c | vGI-5 | MAP1231 to MAP1236c | 58.60 | TGGGGCTACGC | MAP1230 to MAP1236 | |
| MAP7, or LSPp7 | MAP1344 to MAP1349c | 67.32 | GGCGCTGACGCTG | MAP1344 to MAP1349 | |||
| MAP8, or LSPp8 | MAP1631c to MAP1638c | vGI-6 | MAP1631c to MAP1637c | 61.63 | GCGGCGGGACGAA | MAP1630 to MAP1637c | |
| MAP9, or LSPp9 | MAP1718c to MAP1727 | vGI-7 | MAP1720 to MAP1727 | 65.34 | MAP1722 (IS900) | ||
| MAP11, or LSPp10 | MAP2026 to MAP2029c | vGI-8 | MAP2025c to MAP2031c | 65.80 | MAP2034c (IS900) | GCCGCGCGGGCG | MAP2025c to MAP2031c |
| GACCAAGGCGGC | MAP2026 to MAP2032c | ||||||
| MAP12, or LSPp11 | MAP2148 to MAP2158 | vGI-9 | MAP2151 to MAP2157 | 58.99 | MAP2150 (IS1311), MAP2155 (IS1610), MAP2157 (IS900) | GCGGCGCCGCCGG | MAP2150 to MAP2157 |
| MAP13, or LSPp12 | MAP2178 to MAP2196 | 66.93 | GTCCTCGACGG | MAP2178 to MAP2196 | |||
| MAP14, or LSPp13 | MAP2751 to MAP2769c | 67.46 | TGGGCGGCCTGG | MAP2752 to MAP2769/MAP2770 | |||
| vGI-10 | MAP2443 to MAP2457c | 64.17 | MAP2444c (IS900) | CCGGGATCGCCG | MAP2443 to MAP2457c | ||
| vGI-11 | MAP2523c to MAP2529 | 64.43 | |||||
| vGI-12 | MAP2767c to MAP2769c | 62.02 | MAP2769 (integrase) | CGCGGCAACCG | MAP2767c to MAP2769c | ||
| MAP16, or LSPp14 | MAP3721 to MAP3764 | vGI-13 | MAP3730 to MAP3747c | 64.91 | MAP3748 (IS1110) | CGATGTGCTGCT | MAP3730 to MAP3747c |
| vGI-14 | MAP3749 to MAP3770c | 61.15 | MAP3748 (IS1110), MAP3759c (IS1311) | TTTTCAATAAGCGT | MAP3747c/MAP3748 to MAP3969/MAP3770c | ||
| MAP16, or LSPp15 | MAP3770 to MAP3776c | 65.00 | |||||
| LSPp16 | MAP3814 to MAP3818 | vGI-15 | MAP3815 to MAP3818 | 60.74 | MAP3814c (IS900 like) | CAGGAAGCGGG | MAP3814 to MAP3819 |
| MAP17, or LSPp17 | MAP4266 to MAP4270 | vGI-16 | MAP4266 to MAP4267 | 62.20 | AGACGCAAAAGCCCCCG | MAP4265 to MAP4266 | |
| TGCGGCAGGCG | MAP4266 to MAP4271 | ||||||
| MAP18 | MAP4326c to MAP4328c | 66.06 |
qPCR amplifications designed to amplify MAP0101 within vGI-1, MAP0859c within vGI-4, MAP3746 within vGI-13, and a pre-16S rRNA ribosomal gene sequence within vGI-10 were performed on DNA extractions from single colonies of M. avium subsp. paratuberculosis type II and type III strains to measure variations in gene copy number. qPCR threshold cycle signal values were converted to copy number estimates using calibration curves against DNA from reference strain M. avium subsp. paratuberculosis K-10 for each gene tested. The copy number estimate for each of the tested genes was then normalized against average copy number estimates obtained for MAP0101 and MAP0160 (assumed to be present in single copies in each genome) to provide ratios between copy number estimates (Table 6). Ratios of MAP2729 in vaccine strain II (a known duplication in this strain) illustrated that a fivefold increase could be indicative of duplication using this analysis. All other genes in each of the M. avium subsp. paratuberculosis type II colonies tested showed no significant change in copy number above that of the M. avium subsp. paratuberculosis K-10 reference strain. MAP2729 and MAP0859c showed a trend toward an increase in the ratio of M. avium subsp. paratuberculosis type III isolates but this was not statistically significant from that of M. avium subsp. paratuberculosis type II isolates. However, both pre-16S rRNA gene and MAP3746 qPCRs showed modest increases (P = 0.037) between M. avium subsp. paratuberculosis type III and type II strains with pre-16S rRNA from a large colony of the M. avium subsp. paratuberculosis type III strain CAM86, exhibiting a fourfold increase which was not significantly sufficient to be called a duplication.
TABLE 6.
Ratios of qPCR-derived copy number estimates of genes from M. avium subsp. paratuberculosis type II and type III strains
| Strain | Ratio (n-fold) of copy no.a
|
|||||
|---|---|---|---|---|---|---|
| Pre-16S rRNA gene | MAP3746 | MAP2729 | MAP0859c | MAP0101 | MAP0160 | |
| Type III strains | ||||||
| 841 | 2.1 | 2.52 | 1.87 | 1.88 | 1.09 | 0.92 |
| CAM86 large colony | 4.09 | 1.9 | 1.51 | 1.35 | 0.82 | 1.28 |
| CAM86 small colony | 3.27 | 0.87 | 0.66 | 0.61 | 0.75 | 1.49 |
| CAM87 | 1.65 | 1.41 | 0.52 | 0.86 | 0.78 | 1.39 |
| Type II strains | ||||||
| 456 large colony | 0.93 | 1.06 | 0.71 | 0.74 | 0.88 | 1.15 |
| 456 small colony | 1.32 | 1.12 | 0.67 | 1.01 | 1 | NTb |
| CAM63 large colony | 1 | 1 | 1 | 1 | 1 | 1 |
| CAM63 small colony | 0.91 | 1 | 1.31 | 1.1 | 1 | NT |
| Vaccine II | 1.09 | 1.24 | 5.16 | 1.65 | 1.02 | 0.98 |
| K-10 | 1 | 1 | 1 | 1 | 1 | 1 |
Significance between M. avium subsp. paratuberculosis type II and type III strains for the pre-16S rRNA gene and MAP3746 showed a P value of 0.037; all other values were nonsignificant.
NT, not tested.
A comparison of large and small colonies picked from the same culture slant showed no significant difference between ratios in any of the tested genes in two M. avium subsp. paratuberculosis type II strains. However, there were significant increases (two- to threefold) from genes within vGI-4 (represented by MAP0859c) and vGI-13 (represented by MAP3746) when comparing small and large colonies of M. avium subsp. paratuberculosis type III CAM86 strains (Table 6).
DISCUSSION
This work describes the design, validation, and application of the MAPAC pan-genome microarray comprised of optimized oligonucleotide reporters to generate a specific signal for each of the shared and unique ORFs present in the M. avium subsp. hominissuis 104 and M. avium subsp. paratuberculosis K-10 genomes. Validation performed using reference strain genome preparations demonstrated excellent sensitivity and specificity in determining known genomic differences with low false-positive and false-negative rates.
The MAPAC array was applied in this study to characterize the genomes of a representative panel of 12 M. avium subsp. paratuberculosis strains, including types I, II, and III. A comparison of MAPAC results with other published arrays (24, 32, 35, 42) confirmed previously annotated LSPs within M. avium subsp. paratuberculosis K-10 and M. avium subsp. hominissuis 104 genomes. Minor differences were observed at the very ends of some LSPs, and these could be explained by variations in the locations of reporters within ORFs during array designs. Additional small divergences not reported by other array formats were also detected. Consistent genetic features were found in each group of strains within an M. avium subsp. paratuberculosis type. Comparative array data demonstrated that all M. avium subsp. paratuberculosis type I (n = 6) and type III (n = 8) strains tested contained nine separate genomic loci (62 ORFs) not present or deleted in M. avium subsp. paratuberculosis type II strains but highly homologous in base sequence and gene order to the reference genome of M. avium subsp. hominissuis 104. These included the insertion sequence IS6120 (MAV_2223), a set of mce genes (MAV_4125 to MAV_4130, MAV_4351, and MAV_4353) involved in taurine metabolism, and the previously described LSPs MAV17 (MAV_3258 to MAV_3270), MAV14 (MAV_2978 to MAV_2998), and MAV24 (MAV_5225 to MAV_5243) (31, 37, 42).
All M. avium subsp. paratuberculosis type I pigmented strains had deletions of MAP2704, a hemolysin III homologue (25) associated with virulence and invasion of the MAC in human disease. They also lacked MAP3460c, a transposase, possibly reflecting a variation in copy number of this gene present in nine similar copies in the M. avium subsp. paratuberculosis K-10 genome. All M. avium subsp. paratuberculosis type III strains contained a deletion of the M. avium subsp. paratuberculosis-specific region MAP1433c to MAP1438c, which has putative functions suggesting alterations in lipid and fatty acid metabolism, and also a deletion of MAP3584, an alkanesulfonate monooxygenase putatively involved in sulfur metabolism. Differences in M. avium subsp. paratuberculosis genomotypes within INDEL4, INDEL12, and INDEL15 were confirmed by using PCRs designed to screen for these characteristic deletions and M. avium subsp. hominissuis 104 homologous loci within M. avium subsp. paratuberculosis types I, II, and III. These results were fully consistent with MAPAC data from a panel of 66 M. avium subsp. paratuberculosis strains, including 60 M. avium subsp. paratuberculosis type II cattle isolates, 3 M. avium subsp. paratuberculosis type I isolates from sheep in Scotland and Denmark, and 3 M. avium subsp. paratuberculosis type III intermediate strains from Spanish goats and bullfighting cattle.
In other mycobacterial species, such as the Mycobacterium tuberculosis group, phylogenetic diversity and variability of host specificity or pathogenesis can be attributed mostly to gene deletions or the creation of pseudogenes via mutations. Comparison of our results with previous studies confirmed that all M. avium subsp. paratuberculosis types contain the Mycobacterium avium subsp. avium serotype 2 gene cassette but have diverged into two major phylogenetic branches originating from an IS900-positive progenitor. The M. avium subsp. paratuberculosis type II genomotype has undergone a series of major genomic deletion events which at some point in the fairly recent past has had a rapid worldwide distribution and then diverged further through more limited deletions of single genes, transposition events, and genomic transformations that may have been fixed as a result of geographical enclosures. M. avium subsp. paratuberculosis type I and type III strains form separate phylogenies that appear to have retained much of the M. avium subsp. avium serotype 2 progenitor genome but similarly are becoming more diverse as a result of rearrangements and separate single gene deletions. The design of the MAPAC array was such that it incorporated both M. avium subsp. hominissuis 104 and M. avium subsp. paratuberculosis K-10 genomes and by definition therefore could look for deletions only within this combined complement. Additional unknown genomic regions (not in the reference genomes) could therefore theoretically exist in some of these tested strains and would not be detected by our array. Further full-genome sequencing of more M. avium subsp. paratuberculosis strains is required to resolve this issue.
There appears to be a trend, but not an exclusivity of host preference, between some M. avium subsp. paratuberculosis types in particular areas. In this study, we report on M. avium subsp. paratuberculosis type III isolates in Spain that have frequently been isolated from goats and are the most predominant cause of M. avium subsp. paratuberculosis infection in cattle bred specifically for bullfighting. M. avium subsp. paratuberculosis type I pigmented strains are isolated predominantly from sheep and appear to be geographically restricted, while M. avium subsp. paratuberculosis type II strains are predominant in cattle and deer and present in many other animal species. Host-pathogen interplay could also contribute to conditions likely to promote host-specific adaptations through gene redundancies, which, while not creating host exclusivity, may positively select for genomotypes and thus phenotypes associated with discrete animal groups. This is reflected in the unique pigmentation or very slowly growing phenotypes characteristic of particular M. avium subsp. paratuberculosis type I strains and the different pathways of host intracellular signaling induced between M. avium subsp. paratuberculosis type I and type II strains during macrophage processing (26).
Of particular interest was the presence in M. avium subsp. paratuberculosis type I and type III strains of a pair of mycobacterial cell entry genes (mce, MAV_4125 and MAV_4126). In M. avium subsp. paratuberculosis type II strains, as a consequence of a small internal deletion, these appear to have gone through a process that has resulted in combining the C terminus of MAV_4125 and the N terminus of MAV_4126 to form a new mce gene (MAP3289c) with 87 to 89% identity. Using specific PCR, we have demonstrated that M. avium subsp. paratuberculosis type I pigmented and M. avium subsp. paratuberculosis type III strains contain both mce gene homologues but that M. avium subsp. paratuberculosis type II strains have only MAP3289c. The precise mechanism of the mce gene function is not fully established; however, the loss of mce regions in other pathogenic mycobacteria can profoundly affect virulence by decreasing the initiation of infection through cell entry (38) and important pathogenic mechanisms, such as granuloma formation (15). In the M. avium subsp. paratuberculosis type II K-10 reference genome, mce genes occur in seven separate clusters containing 6 to 10 ORFs (5). MAP3289c has 94% identity to mce1A (MAP3604) from the mce1 cluster. MAP3604 is unique as it is the only mce gene homologue that is not associated with an mce gene cluster located immediately downstream, suggesting that this may be an ancillary gene copy. Differences in mce gene complement have previously been demonstrated for other members of the MAC (32) and have been proposed as important factors in defining variations in host pathogenesis. This work now shows that M. avium subsp. paratuberculosis types can also be defined through their mce complement and suggests that mce gene redundancy may be an important process in defining phenotypic differences between M. avium subsp. paratuberculosis types.
In addition, it is shown that an immediately adjacent deletion event in this genomic region has removed a series of small ORFs (MAV_4127 to MAV_4129) in M. avium subsp. paratuberculosis type II strains but not M. avium subsp. paratuberculosis type I or type III strains. This has resulted in a fusion of the N terminus of MAV_4130 with the complementary sequence of MAV_4126 and created an M. avium subsp. paratuberculosis type II-specific gene, MPT64 (MAP3290). The homologue of MAV_4130 in M. tuberculosis (Rv1980c) is a major immunogenic protein associated with mycobacterial persistence through the inhibition of apoptosis in multinucleated giant cells during granuloma formation (27). The replacement of 30% of MAV_4130, represented as MAP3290 in M. avium subsp. paratuberculosis type II strains, could therefore be a significant determinant in diverse mechanisms of pathogenesis and host persistence associated with infections involving this genomotype.
In some of the strains tested in this study, the MAPAC array was sufficiently sensitive to detect clusters of genes with significantly altered ratios which were consistently close to the duplication or deletion threshold as determined by analysis based on the distribution of the complete genome data set. These could be grouped into 16 regions of consecutive genes, described here as vGIs. Bioinformatic analysis revealed that vGIs corresponded closely to M. avium subsp. paratuberculosis LSPs previously described as conserved in all M. avium subsp. paratuberculosis isolates and contained a significantly lower GC% content than LSPs not associated with vGIs (P = 0.0005). vGIs were not detected within the M. avium subsp. paratuberculosis K-10 reference strain and were observed only rarely in the M. avium subsp. paratuberculosis type II strains tested but were frequently partially elevated (suggesting duplication) in the majority of M. avium subsp. paratuberculosis type III strains and decreased (suggesting depletion) in type I strains. vGI-5 (MAP1231 to MAP1237) forms part of a previously described genomic region associated with the emergence of rough- and smooth-colony forms of M. avium subsp. avium generated through homologous recombinatory events (3). Other vGI regions comprise a diverse group of genes associated with virulence and survival, including a set of mce genes (vGI-3, or MAP0758 to MAP0774c); part of the 38-kDa-siderophore, low-GC% island (vGI-13, or MAP3730 to MAP3747c; vGI-14, or MAP3749 to MAP3758c) (40); the virulence regulator oxyR (vGI-6, or MAP1631 to MAP1637) (30); and a region containing the F57 diagnostic gene (vGI-4, or MAP0851 to MAP0866) (34). Differences in ratios obtained from vGIs in M. avium subsp. paratuberculosis type I and type III isolates were significantly less than those observed with LSPs between M. avium subsp. paratuberculosis K-10 and M. avium subsp. hominissuis 104 and were often close to threshold values derived during normalization against the complete genome data set. However, the varied morphological appearances observed with M. avium subsp. paratuberculosis type III strains grown on solid media suggested that these cultures contained a mixture of variant forms, mostly of the rough type, exhibiting low and high rates of growth. We hypothesized that the total genomic DNA extracted for array analysis from a mixture of colonies with variations in duplications or deletions would contain a heterogeneous population of genomes that could generate the types of fluctuations in signals we have observed. Therefore, to confirm that these subtle, potentially mixed signal differences were significant and not a hybridization phenomenon perhaps associated with low GC%, we designed gene-specific qPCRs directed at ORFs inside vGI regions and showed that three of the four vGI regions tested had significant increases in M. avium subsp. paratuberculosis type III strains in comparison with M. avium subsp. paratuberculosis type II strains. It was of particular interest that one vGI region (MAP2443 to MAP2457) included the ribosomal (rrn) operon and that qPCR directed against a 16S pre-rrn region of the genome showed significant increases (P = 0.037) in overall copy number from M. avium subsp. paratuberculosis type III strains over that from M. avium subsp. paratuberculosis type II strains. Importantly, the differences in signals observed from vGI-associated genes were significantly less than the increases observed from a known gene duplication included as a control (MAP2729 in vaccine strain II). This suggested that the variation of signal in vGI regions occurs in only a proportion of the bacterial population within each culture and represents either a mixed population of fixed deletions and duplications or possibly a combination of transient variations induced during particular growth phases. We thus compared vGI qPCR signals from single large and small colonies picked from the same culture slant. The results obtained from this analysis indicated significant increases in vGI copy number in large colonies over that in small colonies in M. avium subsp. paratuberculosis type III strain CAM86 but not in two M. avium subsp. paratuberculosis type II strains, suggesting an association with growth phenotype and a potential for variability between M. avium subsp. paratuberculosis types.
Significantly, 11 of 16 vGIs were found to be immediately flanked by insertion sequences, including IS900, IS1311, and integrases associated with phage elements. In addition, both M. avium subsp. paratuberculosis type I and type III strains but none of the type II strains contained the insertion sequence IS6120 (MAV_2223). This element is present in other mycobacteria, including M. avium subsp. hominissuis 104 (seven copies) and Mycobacterium smegmatis (two copies). The potential to control the variation in gene copy number in vGIs through mechanisms involving transpositional elements resulting in genomic duplications and deletions would significantly increase the adaptability of any strain. The presence of multiple vGIs in M. avium subsp. paratuberculosis type I and type III strains indicates that a range of factors could be influenced by these genomic rearrangements and that fluctuations in transposase activity, if acting in this way, may effect a crude form of global gene control.
In conclusion, this study defines the genomic diversity among the three major groups of M. avium subsp. paratuberculosis. It shows that M. avium subsp. paratuberculosis type I and type III strains contain a larger genomic complement than M. avium subsp. paratuberculosis type II strains and these additional regions encode ORFs with the putative functional capacity to promote different phenotypic characteristics that may result in alterations of disease pathogenesis. The increased potential for plasticity provided by vGIs suggests that previously undescribed mechanisms may exist in M. avium subsp. paratuberculosis, increasing adaptability through transient genomic alterations. Further work is necessary to clarify the significance of these comparisons and the influences they may have on mechanisms of strain/host adaptation.
Supplementary Material
Footnotes
Published ahead of print on 1 December 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Becq, J., M. C. Gutierrez, V. Rosas-Magallanes, J. Rauzier, B. Gicquel, O. Neyrolles, and P. Deschavanne. 2007. Contribution of horizontally acquired genomic islands to the evolution of the tubercle bacilli. Mol. Biol. Evol. 24:1861-1871. [DOI] [PubMed] [Google Scholar]
- 2.Behr, M. A., and V. Kapur. 2008. The evidence for Mycobacterium paratuberculosis in Crohn's disease. Curr. Opin. Gastroenterol. 24:17-21. [DOI] [PubMed] [Google Scholar]
- 3.Belisle, J. T., K. Klaczkiewicz, P. J. Brennan, W. R. Jacobs, Jr., and J. M. Inamine. 1993. Rough morphological variants of Mycobacterium avium. Characterization of genomic deletions resulting in the loss of glycopeptidolipid expression. J. Biol. Chem. 268:10517-10523. [PubMed] [Google Scholar]
- 4.Bull, T. J., E. J. McMinn, K. Sidi-Boumedine, A. Skull, D. Durkin, P. Neild, G. Rhodes, R. Pickup, and J. Hermon-Taylor. 2003. Detection and verification of Mycobacterium avium subsp. paratuberculosis in fresh ileocolonic mucosal biopsy specimens from individuals with and without Crohn's disease. J. Clin. Microbiol. 41:2915-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casali, N., and L. W. Riley. 2007. A phylogenomic analysis of the Actinomycetales mce operons. BMC Genomics 8:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castellanos, E., A. Aranaz, B. Romero, L. de Juan, J. Alvarez, J. Bezos, S. Rodríguez, K. Stevenson, A. Mateos, and L. Domínguez. 2007. Polymorphisms in gyrA and gyrB genes among Mycobacterium avium subsp. paratuberculosis type I, II, and III isolates. J. Clin. Microbiol. 45:3439-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins, D. M., D. M. Gabric, and G. W. de Lisle. 1990. Identification of two groups of Mycobacterium paratuberculosis strains by restriction endonuclease analysis and DNA hybridization. J. Clin. Microbiol. 28:1591-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Juan, L., J. Alvarez, A. Aranaz, A. Rodriguez, B. Romero, J. Bezos, A. Mateos, and L. Dominguez. 2006. Molecular epidemiology of types I/III strains of Mycobacterium avium subspecies paratuberculosis isolated from goats and cattle. Vet. Microbiol. 115:102-110. [DOI] [PubMed] [Google Scholar]
- 9.de Juan, L., J. Alvarez, B. Romero, J. Bezos, E. Castellanos, A. Aranaz, A. Mateos, and L. Dominguez. 2006. Comparison of four different culture media for isolation and growth of type II and type I/III Mycobacterium avium subsp. paratuberculosis strains isolated from cattle and goats. Appl. Environ. Microbiol. 72:5927-5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Juan, L., A. Mateos, L. Dominguez, J. M. Sharp, and K. Stevenson. 2005. Genetic diversity of Mycobacterium avium subspecies paratuberculosis isolates from goats detected by pulsed-field gel electrophoresis. Vet. Microbiol. 106:249-257. [DOI] [PubMed] [Google Scholar]
- 11.Dohmann, K., B. Strommenger, K. Stevenson, L. de Juan, J. Stratmann, V. Kapur, T. J. Bull, and G. F. Gerlach. 2003. Characterization of genetic differences between Mycobacterium avium subsp. paratuberculosis type I and type II isolates. J. Clin. Microbiol. 41:5215-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feller, M., K. Huwiler, R. Stephan, E. Altpeter, A. Shang, H. Furrer, G. E. Pfyffer, T. Jemmi, A. Baumgartner, and M. Egger. 2007. Mycobacterium avium subspecies paratuberculosis and Crohn's disease: a systematic review and meta-analysis. Lancet Infect. Dis. 7:607-613. [DOI] [PubMed] [Google Scholar]
- 15.Gioffre, A., E. Infante, D. Aguilar, M. P. Santangelo, L. Klepp, A. Amadio, V. Meikle, I. Etchechoury, M. I. Romano, A. Cataldi, R. P. Hernandez, and F. Bigi. 2005. Mutation in mce operons attenuates Mycobacterium tuberculosis virulence. Microbes Infect. 7:325-334. [DOI] [PubMed] [Google Scholar]
- 16.Griffiths, T. A., K. Rioux, and J. De Buck. 2008. Sequence polymorphisms in a surface PPE protein distinguish types I, II, and III of Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 46:1207-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris, N. B., and R. G. Barletta. 2001. Mycobacterium avium subsp. paratuberculosis in veterinary medicine. Clin. Microbiol. Rev. 14:489-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hermon-Taylor, J., and T. Bull. 2002. Crohn's disease caused by Mycobacterium avium subspecies paratuberculosis: a public health tragedy whose resolution is long overdue. J. Med. Microbiol. 51:3-6. [DOI] [PubMed] [Google Scholar]
- 19.Hinds, J., K. G. Laing, J. A. Mangan, and P. D. Butcher. 2002. Glass slide microarrays for bacterial genomes, p. 83-89. In B. W. Wren and N. Dorrell (ed.), Methods in microbiology: functional microbial genomics. Academic Press, London, United Kingdom.
- 20.Hinds, J., A. A. Witney, and J. K. Vass. 2002. Microarray design for bacterial genomes, p. 67-82. In B. W. Wren and N. Dorrell (ed.), Methods in microbiology: functional microbial genomics. Academic Press, London, United Kingdom.
- 21.Horan, K. L., R. Freeman, K. Weigel, M. Semret, S. Pfaller, T. C. Covert, D. van Soolingen, S. C. Leão, M. A. Behr, and G. A. Cangelosi. 2006. Isolation of the genome sequence strain Mycobacterium avium 104 from multiple patients over a 17-year period. J. Clin. Microbiol. 44:783-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kennedy, D. J., and G. Benedictus. 2001. Control of Mycobacterium avium subsp. paratuberculosis infection in agricultural species. Rev. Sci. Tech. 20:151-179. [DOI] [PubMed] [Google Scholar]
- 23.Li, L., J. P. Bannantine, Q. Zhang, A. Amonsin, B. J. May, D. Alt, N. Banerji, S. Kanjilal, and V. Kapur. 2005. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc. Natl. Acad. Sci. USA 102:12344-12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marsh, I. B., J. P. Bannantine, M. L. Paustian, M. L. Tizard, V. Kapur, and R. J. Whittington. 2006. Genomic comparison of Mycobacterium avium subsp. paratuberculosis sheep and cattle strains by microarray hybridization. J. Bacteriol. 188:2290-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maslow, J. N., D. Dawson, E. A. Carlin, and S. M. Holland. 1999. Hemolysin as a virulence factor for systemic infection with isolates of Mycobacterium avium complex. J. Clin. Microbiol. 37:445-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Motiwala, A. S., H. K. Janagama, M. L. Paustian, X. Zhu, J. P. Bannantine, V. Kapur, and S. Sreevatsan. 2006. Comparative transcriptional analysis of human macrophages exposed to animal and human isolates of Mycobacterium avium subspecies paratuberculosis with diverse genotypes. Infect. Immun. 74:6046-6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mustafa, T., H. G. Wiker, O. Morkve, and L. Sviland. 2008. Differential expression of mycobacterial antigen MPT64, apoptosis and inflammatory markers in multinucleated giant cells and epithelioid cells in granulomas caused by Mycobacterium tuberculosis. Virchows Arch. 452:449-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naser, S. A., G. Ghobrial, C. Romero, and J. F. Valentine. 2004. Culture of Mycobacterium avium subspecies paratuberculosis from the blood of patients with Crohn's disease. Lancet 364:1039-1044. [DOI] [PubMed] [Google Scholar]
- 29.Newton, R., J. Hinds, and L. Wernisch. 2006. A hidden Markov model web application for analysing bacterial genomotyping DNA microarray experiments. Appl. Bioinformatics 5:211-218. [DOI] [PubMed] [Google Scholar]
- 30.Pagán-Ramos, E., S. S. Master, C. L. Pritchett, R. Reimschuessel, M. Trucksis, G. S. Timmins, and V. Deretic. 2006. Molecular and physiological effects of mycobacterial oxyR inactivation. J. Bacteriol. 188:2674-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paustian, M. L., V. Kapur, and J. P. Bannantine. 2005. Comparative genomic hybridizations reveal genetic regions within the Mycobacterium avium complex that are divergent from Mycobacterium avium subsp. paratuberculosis isolates. J. Bacteriol. 187:2406-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paustian, M. L., X. Zhu, S. Sreevatsan, S. Robbe-Austerman, V. Kapur, and J. P. Bannantine. 2008. Comparative genomic analysis of Mycobacterium avium subspecies obtained from multiple host species. BMC Genomics 9:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pavlik, I., A. Horvathova, L. Dvorska, J. Bartl, P. Svastova, R. du Maine, and I. Rychlik. 1999. Standardisation of restriction fragment length polymorphism analysis for Mycobacterium avium subspecies paratuberculosis. J. Microbiol. Methods 38:155-167. [DOI] [PubMed] [Google Scholar]
- 34.Poupart, P., M. Coene, H. Van Heuverswyn, and C. Cocito. 1993. Preparation of a specific RNA probe for detection of Mycobacterium paratuberculosis and diagnosis of Johne's disease. J. Clin. Microbiol. 31:1601-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Semret, M., D. C. Alexander, C. Y. Turenne, P. de Haas, P. Overduin, D. van Soolingen, D. Cousins, and M. A. Behr. 2005. Genomic polymorphisms for Mycobacterium avium subsp. paratuberculosis diagnostics. J. Clin. Microbiol. 43:3704-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Semret, M., C. Y. Turenne, P. de Haas, D. M. Collins, and M. A. Behr. 2006. Differentiating host-associated variants of Mycobacterium avium by PCR for detection of large sequence polymorphisms. J. Clin. Microbiol. 44:881-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Semret, M., G. Zhai, S. Mostowy, C. Cleto, D. Alexander, G. Cangelosi, D. Cousins, D. M. Collins, D. van Soolingen, and M. A. Behr. 2004. Extensive genomic polymorphism within Mycobacterium avium. J. Bacteriol. 186:6332-6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Senaratne, R. H., B. Sidders, P. Sequeira, G. Saunders, K. Dunphy, O. Marjanovic, J. R. Reader, P. Lima, S. Chan, S. Kendall, J. McFadden, and L. W. Riley. 2008. Mycobacterium tuberculosis strains disrupted in mce3 and mce4 operons are attenuated in mice. J. Med. Microbiol. 57:164-170. [DOI] [PubMed] [Google Scholar]
- 39.Stevenson, K., V. M. Hughes, L. de Juan, N. F. Inglis, F. Wright, and J. M. Sharp. 2002. Molecular characterization of pigmented and nonpigmented isolates of Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 40:1798-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stratmann, J., B. Strommenger, R. Goethe, K. Dohmann, G. F. Gerlach, K. Stevenson, L. L. Li, Q. Zhang, V. Kapur, and T. J. Bull. 2004. A 38-kilobase pathogenicity island specific for Mycobacterium avium subsp. paratuberculosis encodes cell surface proteins expressed in the host. Infect. Immun. 72:1265-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Witney, A. A., G. L. Marsden, M. T. Holden, R. A. Stabler, S. E. Husain, J. K. Vass, P. D. Butcher, J. Hinds, and J. A. Lindsay. 2005. Design, validation, and application of a seven-strain Staphylococcus aureus PCR product microarray for comparative genomics. Appl. Environ. Microbiol. 71:7504-7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu, C. W., J. Glasner, M. Collins, S. Naser, and A. M. Talaat. 2006. Whole-genome plasticity among Mycobacterium avium subspecies: insights from comparative genomic hybridizations. J. Bacteriol. 188:711-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.