Abstract
Pancreatic ductal adenocarcinoma (PDAC) is characterized by the deregulation of the hedgehog signaling pathway. The Sonic Hedgehog ligand (Shh), absent in the normal pancreas, is highly expressed in pancreatic tumors and is sufficient to induce neoplastic precursor lesions in mouse models. We investigated the mechanism of Shh signaling in PDAC carcinogenesis by genetically ablating the canonical bottleneck of hedgehog signaling, the transmembrane protein Smoothened (Smo), in the pancreatic epithelium of PDAC-susceptible mice. We report that multistage development of PDAC tumors is not affected by the deletion of Smo in the pancreas, demonstrating that autocrine Shh–Ptch–Smo signaling is not required in pancreatic ductal cells for PDAC progression. However, the expression of Gli target genes is maintained in Smo-negative ducts, implicating alternative means of regulating Gli transcription in the neoplastic ductal epithelium. In PDAC tumor cells, we find that Gli transcription is decoupled from upstream Shh–Ptch–Smo signaling and is regulated by TGF-β and KRAS, and we show that Gli1 is required both for survival and for the KRAS-mediated transformed phenotype of cultured PDAC cancer cells.
Keywords: Pancreatic ductal adenocarcinoma, PDAC, hedgehog, Gli, Smoothened, pancreatic cancer
Pancreatic ductal adenocarcinoma (PDAC) carries one of the worst prognoses in oncology, with a 5-yr survival rate of <5%, and is in acute need of new therapeutic options (Bardeesy and DePinho 2002). The early stage of the disease is characterized by pancreatic intraepithelial neoplasia lesions (PanIN) bearing mutations in the Kras proto-oncogene, which progress to malignant PDAC by accumulating mutations in other pathways, most frequently in the tumor suppressor genes p16-Ink4A, Trp53, and Smad4 (Tuveson and Hingorani 2005; Bardeesy et al. 2006a; Hezel et al. 2006). Knowledge of the mutations that occur frequently in human patients has guided the engineering of a new generation of mouse models of PDAC that recapitulate more faithfully the pathology of the human disease (Aguirre et al. 2003; Hingorani et al. 2003), and in which the contribution of other signaling pathways to PDAC tumorigenesis can be assessed.
The Hedgehog pathway has been recently implicated in PDAC formation, following the initial observation that the Sonic Hedgehog ligand (Shh), undetectable in the normal pancreas, is highly expressed in PDAC samples (Berman et al. 2003; Thayer et al. 2003). Canonical hedgehog signaling requires the binding of a hedgehog family ligand such as Shh to the Patched (Ptch) 12 trans-membrane domain receptor, resulting in the activation of the Smoothened (Smo) seven trans-membrane domain protein. Activated Smo induces the nuclear translocation of transcriptionally active members of the Gli transcription factor family and the consequent elevated transcription of Gli target genes, which include Ptch1 and Gli1 (Pasca di Magliano and Hebrok 2003; Jacob and Lum 2007). Constitutive activation of this signaling pathway, through inactivating mutations of the Ptch receptor, activating mutations of Smo, or elevated expression of Gli1 are key tumor-promoting features of basal cell carcinoma, medulloblastoma, and several other cancers (Taipale and Beachy 2001; Rubin and de Sauvage 2006).
Several lines of evidence support the notion that hedgehog signaling plays a functionally important role in the genesis of pancreatic cancer. First, forced expression of Shh during mouse development is sufficient to induce lesions resembling PanINs (Thayer et al. 2003; Morton et al. 2007). Second, a Gli-driven transcriptional program characterized by foregut developmental markers and elevated expression of canonical Gli target genes is evident in PanINs (Thayer et al. 2003; Prasad et al. 2005). Third, activated Kras cooperates with activated Gli2 to induce undifferentiated pancreatic tumors (Pasca di Magliano et al. 2006). Finally, cyclopamine, an inhibitor of Smo activity, is reported to induce apoptosis and to block primary tumor formation (Thayer et al. 2003) or metastatic dissemination (Feldmann et al. 2007) of several PDAC cell lines upon transplantation. These results support the hypothesis that autocrine Shh signaling stimulates PDAC formation (Rustgi 2006; Jacob and Lum 2007). A recent study identifying mutations of downstream hedgehog signaling components, including GLI1 and GLI3, in all human pancreatic cancer cell lines scrutinized, supports the notion that this pathway is important for PDAC formation in a cell-autonomous way (Jones et al. 2008).
While clearly implicating Gli transcription and Shh signaling in PDAC formation, the aforementioned studies did not establish a requirement per se for autocrine Shh signaling nor for Gli transcription in pancreatic ductal tumorigenesis. Studies in which Shh and Gli2 contributions have been interpreted to be, respectively, autocrine and cell-autonomous in nature do not, in fact, exclude a potentially key stromal contribution of hedgehog signaling to PDAC formation (Thayer et al. 2003; Pasca di Magliano et al. 2006; Morton et al. 2007). Indeed, a recent study demonstrates a clear role for Smo-mediated hedgehog signaling in the stroma, but not in the epithelial compartment of transplanted xenografts of hedgehog ligand-expressing tumor cells (Yauch et al. 2008).
To clarify the importance of hedgehog signaling in tumor cells for PDAC development and to better map the key nodes of hedgehog signaling susceptible to potential therapeutic intervention in this disease, we sought to determine if the singular transducer of hedgehog signaling, the Smo coreceptor, is required in the ductal epithelium during PDAC carcinogenesis in a model of oncogenic Kras-driven carcinognenesis. We establish that Smo is dispensable in the pancreatic epithelium during PDAC tumorigenesis and that expression of Gli transcriptional targets is nevertheless maintained in the absence of Smo-mediated hedgehog signaling. We then describe initial investigations into the mechanisms of Smo-independent Gli transcriptional modulation and the requirement of this transcription factor for PDAC cell survival and transformation.
Results
Ablating Smo expression in the pancreas of PDAC-bearing mice
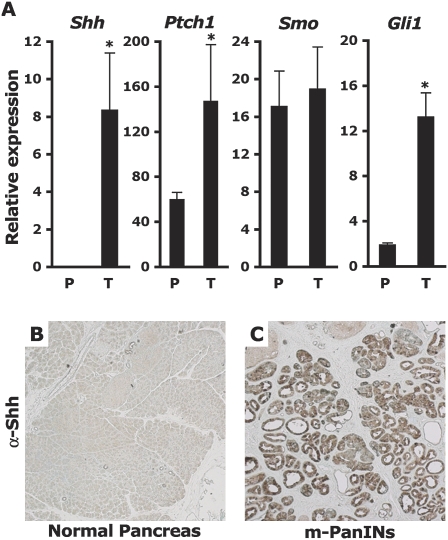
A variation of the genetically engineered PDAC model used in this study was previously described (Bardeesy et al. 2006a): The model consists of an activated KrasG12D conditional allele (LSL-Kras) (Tuveson et al. 2004), a heterozygous conditional Trp53 loss-of-function allele (Trp53F) (Jonkers et al. 2001), and the Cre recombinase targeted to the endogenous Ptf1-p48 locus (p48-Cre) (Kawaguchi et al. 2002). Compound p48-Cre/+; LSL-KrasG12D/+; Trp53F/+ mice present with low-grade PanIN lesions within 4 wk of birth, progress to invasive PDAC between 9 and 13 wk of age, and succumb to the disease 10–29 wk after birth, with 50% of the animals dying within 16 wk of birth. PanIN-like and PDAC lesions from p48-Cre/+; LSL-KrasG12D/+; Trp53F/+ mice express high levels of the Shh mRNA (Fig. 1A). Shh protein expression is not detectable in normal pancreatic tissue (Fig. 1B) but is readily detectable and restricted to the ductal cells in PDAC sections (Fig. 1C). PDAC tumor samples from this model also express appreciable levels of the Smo mRNA, as well as elevated levels of the Ptch1 and Gli1 mRNAs (Fig. 1A).
Figure 1.
Hedgehog pathway components are deregulated in PDAC lesions. (A) Expression of Shh, Ptch1, Smo, and Gli1 mRNA in total RNA extracts from ∼12-wk-old LSL-Kras/+; p53F/+ control pancreata (P) (N = 3) or p48-Cre/+; LSL-Kras/+; p53F/+ tumors (T) (N = 3). Levels of mRNAs expressed as a percentage of the mGus control mRNA. Asterisk indicates a P-value <0.01. (B,C) Immuno-histochemical detection of Shh (50×). (B) No Shh detected in normal pancreas. (C) Shh expression detected inside ductal cells in PanIN lesions of 9-wk-old PDAC mice. The panels are representative of multiple fields of pancreatic sections from two control and two PDAC mice.
We compared the formation of PDAC in mice heterozygous and homozygous for a Smoothened conditional loss-of-function allele (SmoF) (Long et al. 2001). The Smo heterozygous mice were used as control since a previously described Smo-null allele did not display haplo-insufficient phenotypes (Zhang et al. 2001). Thus, p48-Cre/+; LSL-KrasG12D/+; Trp53F/+; SmoF/+ mice (PDAC SmoF/+) were compared with p48-Cre/+; LSL-KrasG12D/+; Trp53F/+; SmoF/SmoF mice (PDAC SmoF/F) for their propensity to develop PanIN-like and PDAC lesions and for their overall survival.
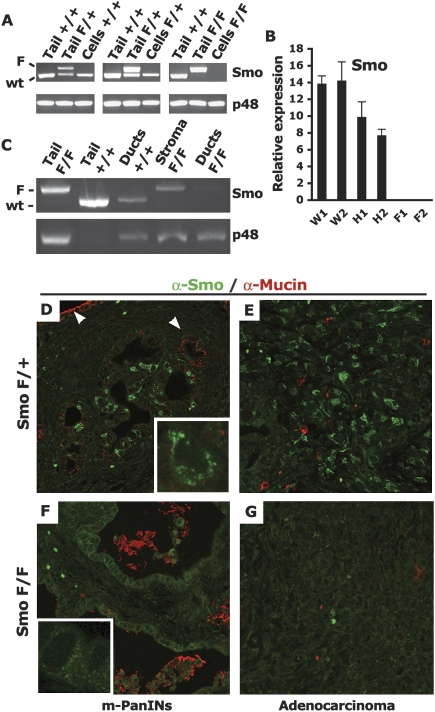
We first established that the Smoothened allele was properly recombined in this model by PCR analysis of PDAC tumor cell lines derived from these mice. Three out of three independently derived PDAC SmoF/+ cell lines carried a recombined SmoF allele, and three out of four independently derived PDAC SmoF/F cell lines had recombined both SmoF alleles (Fig. 2A; data not shown). We confirmed the complete depletion of Smo mRNA in these cells by quantitative real-time RT–PCR (Fig. 2B). Furthermore, to investigate the extent of in vivo recombination of the SmoF allele in pancreatic ducts, we performed a laser capture microdissection of ductal structures from neoplastic pancreas from PDAC SmoF/+ and PDAC SmoF/F mice: Analysis of pools of ductal lesions captured from the two genotypes indicated that the conditional SmoF allele was widely recombined in neoplastic ducts but not in the surrounding stroma (Fig. 2C). Finally, to validate the loss of the Smo protein in PDAC SmoF/F pancreatic ducts, we performed fluorescent immunostaining on sections from PDAC SmoF/+ and PDAC SmoF/F tumors from 9.5-wk-old PDAC-bearing mice. A granular cytoplasmic Smo staining pattern was readily detectable in the PDAC SmoF/+ sections (Fig. 2D,E), similar to what had been described (Incardona et al. 2002). About 30% of the ductal cells in SmoF/+ ductal lesions express high levels of the Smo protein (Fig. 2D). Strong Smo staining was also detected in PDAC SmoF/+ tumors (Fig. 2E). In contrast, Smo staining was depleted in SmoF/F ducts and tumors (Fig. 2F,G). Consecutive sections were stained by H&E to show that the immuno-stained areas depicted in Figure 2, E and G, represent areas of adenocarcinoma (Supplemental Fig. 1). Smo protein depletion in PDAC SmoF/F mice was extensive, ranging from the retention of rare Smo-positive cells in one PDAC SmoF/F mouse to the complete absence of staining in tumor sections from two other PDAC SmoF/F mice. Collectively, these analyses demonstrate that Smo expression is largely ablated in the ductal cell compartment of PDAC SmoF/F mice.
Figure 2.
Smoothened is depleted in ductal cells of PDAC SmoF/F mice. (A) Recombination of the Smo locus in PDAC cell lines. PCR amplification of the Smo locus (Smo) or the p48-Cre transgene (p48). The Smo genotyping procedure amplifies the nonrecombined conditional SmoF allele (upper band, F) and the wild-type (wt) Smo allele (lower band, wild type). The upper PCR band is lost upon Cre recombination of the Smo conditional locus. The input genomic DNA used in each PCR reaction is indicated: (Tail) genomic DNA from mouse tail, 100 ng; (Cells) genomic DNA from PDAC-derived tumor cell lines, 10 ng; (+/+) Smo+/+; (F/+) SmoF/+; (F/F) SmoF/F. (B) Depletion of the Smo mRNA in recombined cell lines. Expression of Smo mRNA in total RNA extracts from Smo+/+ (W), SmoF/+ (H), or SmoF/F (F) PDAC cell lines. Levels of mRNAs expressed as a percentage of the m-Gus control mRNA. Total RNA extracts from two cells lines of each genotype were assayed in triplicate. (C) In vivo recombination of the Smo locus. Genomic DNA from ductal structures or stromal areas isolated by laser-capture microdissection (LCM) from PDAC tumors was subjected to the same PCR amplification as in A. Ducts or stromal areas from two PDAC tumors of each genotype were pooled and subjected to PCR amplification. The input genomic DNA used in each PCR reaction is indicated: (Tail) DNA from mouse tail; (Ducts) LCM-captured ducts from two PDAC tumors; (Stroma) LCM-captured stromal-rich area of two PDAC tumors; (+/+) Smo+/+; (F/+) SmoF/+; (F/F) SmoF/F. (D–G) In vivo depletion of the Smo protein. Immunofluorescent detection of Smo (green) and Muc-1 (red) (630×). The Smo protein is detected in a subset of mucin-negative ducts inside PDAC SmoF/+ PanIN-like lesions (D), but not in mucin-positive ducts (white arrows) as well as in mucin-negative PDAC SmoF/+ adenocarcinoma lesions (E). Smo is undetectable in PDAC SmoF/F PanIN-like lesions (F) orin PDAC SmoF/F adenocarcinoma (G). Granular cytoplasmic Smo staining of an individual PDAC SmoF/+ cell (2520×) (D, insert) absent in individual PDAC SmoF/F cells (F, insert). The panels are representative of multiple fields of pancreatic sections from three PDAC SmoF/+ mice and three PDAC SmoF/F mice.
Smoothened depletion does not impact exocrine pancreatic development
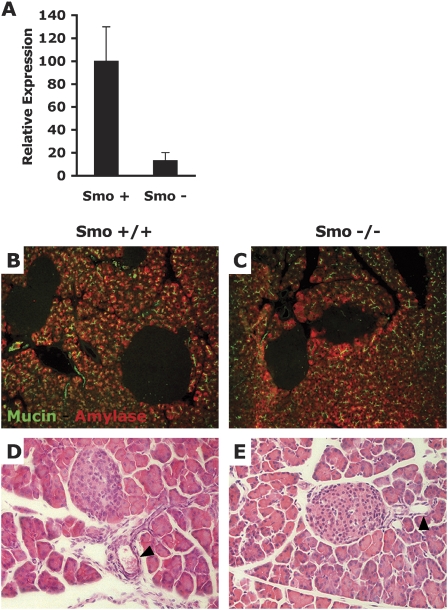
Before characterizing the potential role of Smo during ductal carcinogenesis, we sought to verify that the loss of Smo did not affect normal pancreatic development, since a pre-existing developmental defect in the pancreas could, in principle, complicate the interpretation of any tumor phenotype. For this purpose, we compared pancreas samples from Pdx-Cre; Smo+/+ and Pdx-Cre; SmoF/Null mice, in which slight pancreatic endocrine defects have been observed (J. Lau and M. Hebrok, unpubl.). Quantitative PCR analysis of cDNAs derived from dissected pancreatic buds of 12.5-d-old embryos demonstrates that levels of the endogenous Smo mRNA are decreased by almost 90% in mutant Smo pancreatic bud extracts (Fig. 3A). This result shows that the vast majority of pancreatic progenitor cells of Pdx-Cre; SmoF/Null mice are genetically lacking Smo function. We stained pancreatic sections of wild-type and Smo mutant pancreas with an anti-Muc1 antibody to mark ductal cells, and with an anti-α-amylase antibody to mark acinar cells. No difference could be observed between the two sets of pancreas with either staining (Fig. 3B,C). In addition, we stained wild-type and Smo mutant pancreas with H&E and found no morphological difference (Fig. 3D,E). We conclude that the development of ductal and acinar lineages proceeds normally in the absence of Smoothened and that tumor phenotypes occurring in the absence of Smo will not be impacted by pre-existing ductal or acinar developmental defects.
Figure 3.
Ductal and acinar pancreatic development is normal in the absence of Smo function. (A) Quantitative RT–PCR comparison of Smo mRNA in total RNA extracts from dissected pancreatic buds of 12.5-d Pdx-Cre; Smo+/+ (Smo+) embryos and Pdx-Cre; SmoF/Null (Smo−) embryos. (B,C) Anti-Muc1 and anti-α-amylase staining of pancreatic sections from Pdx-Cre; Smo+/+ mice (B) and Pdx-Cre; SmoF/Null mice (C). (D) H&E staining of pancreatic sections from Pdx-Cre; Smo+/+ mice and Pdx-Cre; SmoF/Null mice. Arrows indicate pancreatic ducts. The panels are representative of multiple fields of pancreatic sections from two Pdx-Cre; Smo+/+ and two Pdx-Cre; SmoF/Null mice.
Effect of Smoothened depletion on PDAC formation
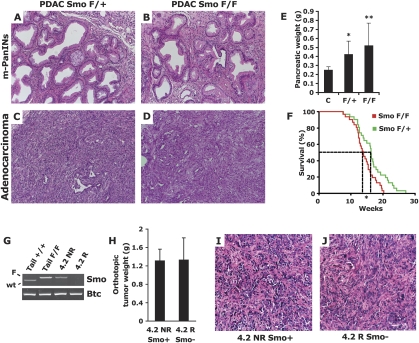
In the transgenic model of PDAC used in our study, the pancreas undergoes profound phenotypic changes following the activation of mutant KrasG12D and the heterozygous deletion of Trp53 throughout the developing pancreas. Following a process of acinar-ductal metaplasia, virtually all the exocrine pancreas is replaced with neoplastic ductal structures by 9.5 wk of age. At this stage, adenocarcinoma lesions are commingled with numerous ductal foci containing PanIN-like lesions, separated from each other by a thick activated stroma composed of activated fibroblasts and immune cells (Fig. 4A,C). When we compared the histopathology of PDAC SmoF/+ and PDAC SmoF/F pancreas, we found no appreciable difference between the pathology of SmoF/+ and SmoF/F mice. The analysis of pancreata from five mice in each cohort revealed similar foci of PanIN-like lesions surrounded by an activated stroma (Fig. 4A,B) along with scattered invasive adenocarcinoma lesions characterized by a prominent desmoplastic component (Fig. 4C,D). Thus, the acinar-ductal metaplasia and the formation of PanIN and adenocarcinoma lesions were not qualitatively affected by the deletion of Smo in PDAC SmoF/F mice. We also noted a complete absence of Smo staining in the ducts of mucin-negative PanIN-like lesions (Fig. 2F) and in PDAC tumor areas (Fig. 2E), highlighting that these advanced neoplastic lesions can develop in the absence of Smo.
Figure 4.
Genetic depletion of Smo in the pancreatic epithelium does not affect PDAC tumorigenesis. (A–D) H&E analysis of the histopathology of PDAC SmoF/+ (A,C) and PDAC SmoF/F (B,D) lesions reveals no overt difference in the presentation of PanIN-like lesions (A,B) or PDAC (C,D). (E) No significant difference in the pancreatic weight of PDAC SmoF/+ mice (F/+; N = 10) and PDAC SmoF/F mice (F/F; N = 11) could be detected [(**) P = 0.378)], but a significant difference is observed between the pancreatic weight of tumor-bearing mice and control non-tumor-bearing mice [N = 13; (*) P < 0.01]. (F) The mean survival of a cohort of PDAC SmoF/+ mice (green line; N = 31) was significantly greater [17 d; (*) P < 0.05] than that of a PDAC SmoF/F mice (red line; N = 31). The panels are representative of multiple fields of pancreatic sections from eight PDAC SmoF/+ mice and eight PDAC SmoF/F mice. (G) In vitro recombination of the Smo locus in the PDAC 4.2 R cell line derived from the nonrecombined PDAC 4.2 NR cell line. PCR amplification of the Smo locus (Smo) or the Betacellulin control locus (Btc). Smo genotyping: unrecombioned SmoF allele (upper band, F) and wild-type Smo allele (lower band, wild type). The upper PCR band is lost upon Cre recombination of the Smo conditional locus. The input genomic DNA is indicated: (Tail) genomic DNA from mouse tail; (4.2 NR) genomic DNA from nonrecombined PDAC cell line; (4.2 R) genomic DNA from in vitro recombined PDAC cell line. (H) Average pancreatic tumor weight from nude mice orthotopically injected with 4.2 NR (N = 8, Smo+) or 4.2 R cells (N = 8, Smo−). No significant difference was detected. (I,J) H&E staining of 4.2 NR (Smo+) and 4.2 R(Smo−) xenograft tumor sections. The panels are representative of multiple fields of pancreatic sections from four 4.2 NR and four 4.2 R pancreatic tumors.
Given that no normal pancreas remained at 9.5 wk of age, we used whole pancreatic weight as a proxy measurement for tumor burden. Using this metric, we found an approximate doubling of the pancreatic weight of PDAC mice compared with Cre-negative control animals. Notably, we found no statistical difference in pancreatic tumor weight between the SmoF/+ and the SmoF/F animals (Fig. 4E). Moreover, we aged two cohorts of 31 mice each to evaluate a possible survival difference between PDAC SmoF/+ and PDAC SmoF/F mice. If Shh produced by PDAC cancer cells promoted tumor growth in an autocrine manner, we expected the PDAC SmoF/F mice, in which ductal Shh signal transduction is lost, to live longer than their PDAC SmoF/+ littermates. However, to our surprise, PDAC SmoF/F animals died on average 17 d earlier than their PDAC SmoF/+ counterparts (Fig. 4F). This statistically significant difference may be explained by slight genetic variations among the different mouse strains that were bred to generate the PDAC SmoF/F and PDAC SmoF/+ compound mice. The lack of a survival advantage in PDAC SmoF/F mice, however, clearly demonstrates that Smo deletion does not alleviate PDAC-induced morbidity in these mice.
Since a few Smo-positive cells persisted in one of three PDAC SmoF/F mice analyzed, a possibility arose that Smo function inside PDAC tumors may be conveyed by a few isolated cells that possess stem cell-like properties. In such a scenario, even a few remaining Smo-positive cells might be sufficient to carry out the tumor cell-intrinsic function required for PDAC tumor growth, and thus ablating Smo in >90% of the cancer cells may not be sufficient to produce an observable defect in PDAC tumorigenesis. To address this possibility, we conducted a second experiment in which Smo function was entirely ablated. One of the pancreatic tumor-derived cell lines we obtained from PDAC SmoF/F mice had undergone transformation without recombining its Smo conditional alleles (Fig. 4G, lane 3). We proceeded to delete Smo in these cells by infecting them in vitro with an adenovirus expressing the Cre recombinase. Through this process, we obtained genetically matched PDAC cell lines that differed only by the recombination status of their Smo conditional allele (Fig. 4G), which we named 4.2 NR (nonrecombined) and 4.2 R (recombined). We orthotopically injected 10,000 cells from each cell line into the pancreases of two cohorts of eight nude mice to assess their propensity to develop PDAC tumors in vivo. After 2.5 wk, the two cohorts of mice were sacrificed, and the weight of their pancreatic tumors was measured. No difference was found between the tumor weight of nude mice injected with 4.2 NR or with 4.2 R cells (Fig. 4H). In addition, no morphological difference was evident in H&E-stained sections of these tumors (Fig. 4I,J). To ensure that the nonrecombined Smo allele had not spontaneously recombined in vivo in the 4.2 NR cells, we genotyped the Smo locus in the derivative tumors after sacrifice, and detected the nonrecombined allele in 4.2 NR, but not the 4.2 R tumor genomic DNA (Supplemental Fig. 2), as well as the wild-type Smo allele present in whole-tumor DNA because of Smo wild-type host mesenchymal cells present in the tumor.
Thus, targeted ablation of Smo in pancreatic epithelial cells does not affect the tumor grade or tumor burden of mice engineered to develop PDAC, nor does it confer a survival advantage. We conclude that autocrine Shh signaling mediated by Smo is not required in pancreatic ductal cells for the onset and progression of PDAC.
Smo-independent mechanisms of Gli target genes maintenance
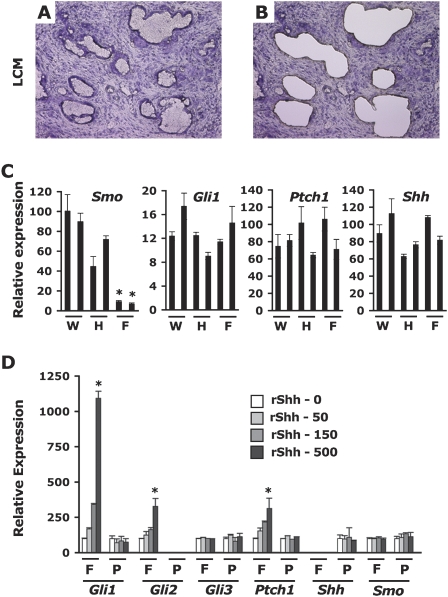
Our experiments demonstrate that expression of Smo in the pancreatic ductal epithelium is dispensable for the initiation and progression of PDAC. One anticipates that the loss of Shh signaling via Smo would result in the down-regulation of hedgehog target genes such as Gli1 and Ptch1. This would in turn suggest that the Gli transcriptional program could be down-regulated without adverse effect to pancreatic tumor cells, a somewhat unexpected result in light of studies that implicate Gli transcription as a salient characteristic of this disease (Thayer et al. 2003; Prasad et al. 2005). To explore this matter in vivo, we performed laser capture microdissections of ductal structures from frozen tumor sections obtained from PDAC Smo+/+, PDAC SmoF/+, or PDAC SmoF/F mice (Fig. 5A,B). We extracted total RNA from these microdissected ducts to evaluate the level of expression of the Ptch1 and Gli1 transcripts, two known Gli target genes. We also assayed the expression of the Shh transcript. We found that, whereas Smo expression is greatly reduced in microdissected neoplastic SmoF/F ducts compared with analogous SmoF/+ and Smo+/+ ducts, the expression levels of Ptch1 and Gli1 are not significantly affected by the genetic ablation of Smo (Fig. 5C). This unexpected observation reveals that the transcription of Gli1 and Ptch1 is maintained in neoplastic ductal cells through Smo-independent mechanisms.
Figure 5.
Ptch1 and Gli1 expression are maintained in vivo in Smo-depleted neoplastic ductal cells. (A,B) Laser microdissection of pancreatic ducts before (A) and after (B) capture on H&E-stained frozen tumor sections (50×). (C) Expression of Smo, Ptch1, Shh, and Gli1 mRNA in total RNA extracts from pools of laser capture microdissected pancreatic ducts from two PDAC Smo+/+ (W), two PDAC SmoF/+ (H), and two PDAC SmoF/F (F) tumors. Levels of mRNAs expressed as a percentage of the m-Gus control mRNA. (D) Relative expression (% mGus) of Hedgehog/Gli signaling components following stimulation with increasing concentrations of recombinant Shh (0–50–150–500 ng/mL) in primary pancreatic fibroblasts (F) or in PDAC cell line 3.3 (P). Asterisk indicates a P value <0.01 (C) or <0.001 (D).
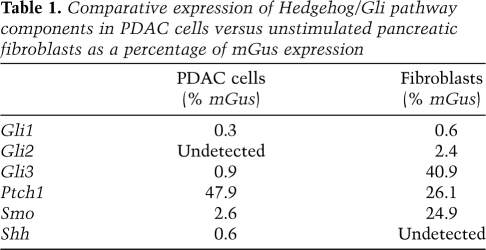
Given the lack of effect on Gli1 and Ptch1 transcription upon Smo abrogation in neoplastic pancreatic ducts, we asked if PDAC cancer cells were capable of transducing hedgehog signals. We investigated this question by comparing Smo-positive neoplastic PDAC cells with Smo-positive primary pancreatic fibroblasts for their in vitro ability to induce Gli target genes upon exposure to exogenous recombinant Shh (rShh). Once grown to confluence, both cell types were incubated in low serum conditions for 16 h, then stimulated with increasing amounts of rShh for 8 h (Fig. 5D). In response to rShh, the transcription of Gli target genes was markedly induced in primary pancreatic fibroblasts, but not in PDAC cells. Admittedly, absolute levels of the various components of the Hedgehog/Gli machinery varied significantly between these two cell types (Table 1), but both cell types express readily detectable levels of Smo and Ptch1, suggesting that their lack of responsiveness is not due to a lack of Shh receptors. We conclude that both PDAC cells and pancreatic fibroblasts express Gli1 and Ptch1 transcripts. However, Smo-positive PDAC cells do not respond to direct hedgehog ligand stimulation, indicating that their expression of Gli1 and Ptch1 is decoupled from upstream Shh/Ptch/Smo signaling.
Table 1.
Comparative expression of Hedgehog/Gli pathway components in PDAC cells versus unstimulated pancreatic fibroblasts as a percentage of mGus expression
TGF-β and Kras signaling impact Gli target genes independently of Smo
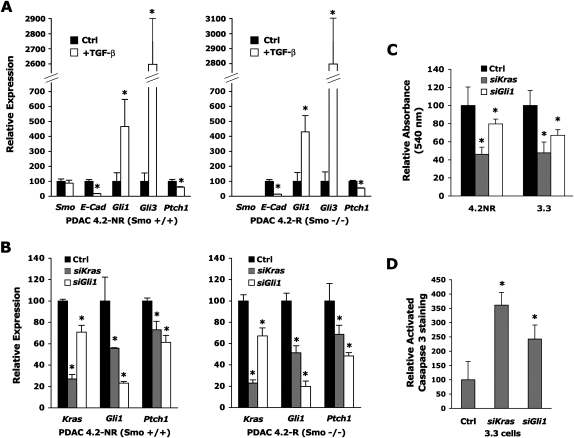
We next sought to identify pathways that could be regulating the observed Gli1 and Ptch1 transcription in PDAC cells independently of Smo-mediated signaling. TGF-β has been reported to induce Gli1 and Gli2 mRNA levels in several cell lines (Dennler et al. 2007). Given that TGF-β1 is highly expressed by PDAC tumor cells in vivo in a closely related murine PDAC genetic model (Bardeesy et al. 2006b), as well as in the model used in this study (C. Chaudury and D. Hanahan, unpubl.), we assessed the impact of TGF-β1 signaling on Gli1 transcription in the genetically matched pair of Smo wild-type (4.2 NR) and Smo mutant (4.2 R) PDAC cell lines introduced above. We cultured these cells with or without recombinant TGF-β1 and measured the effect on the expression of Gli1, Gli2, Gli3, Ptch1, and E-cadherin, the latter being a transcript known to be strongly down-regulated by the TGF-β pathway (Fig. 6A). We found that regardless of the Smo status of the PDAC cells, incubation with TGF-β resulted in a marked 4.5-fold up-regulation of Gli1 and a >25-fold elevation of Gli3; Gli2 remained undetectable in these cells (data not shown), but Ptch1 was decreased by 40%, while E-cadherin, as expected, decreased by 90% upon TGF-β1 exposure. These marked effects on E-cad, Gli1, Gli3, and Ptch1 were observed in two other independently derived PDAC cell lines (data not shown).
Figure 6.
TGF-β and activated Kras signaling impact Gli and Ptch1 expression in a Smo-independent manner. (A) Expression of Smo, Ptch1, E-Cad, Gli1, and Gli3 mRNA in total RNA extracts from wild-type Smo (4.2 NR) or Smo mutant (4.2 R) PDAC cell lines 48 h after stimulation with 5 ng/mL recombinant TGF-β1. Levels of mRNAs expressed as a percentage of the mGus control mRNA. (B) Expression of Kras, Gli1, and Ptch1 mRNA in total RNA extracts from wild-type Smo (4.2 NR) or Smo mutant (4.2 R) PDAC cell 48 h after transfection with control siRNA pools or siRNA pools targeting Kras or Gli1. Levels of mRNAs expressed as a percentage of the mGus control mRNA. (C) Absorbance at 540 nm of two PDAC cell lines (4.2 NR and 3.3) incubated with MTT (see the Materials and Methods) 72 h after transfection with control siRNA pools or siRNA pools targeting Kras or Gli1 and after 24 h of serum starvation. (D) Relative change in Activated Caspase 3 immuno-fluorescent staining of mouse PDAC 3.3 cells 60 h after transfection with control siRNA pools (Ctrl) or siRNA pools targeting Kras or Gli1, following 12 h of serum starvation. Staining was evaluated in five fields each containing at least 500 cells for each condition. Act-Casp-3-positive cells expressed as a percentage of DAPI nuclei; the percentage in Ctrl-treated cells was set at 100%. Asterisk indicates a P-value <0.01 (A), <0.05 (B,C), or <0.005 (D).
Given that the mouse PDAC model is engineered to express the activated Kras oncogene, the most prevalent genetic event detected in human PDAC, we investigated if Kras itself might also be involved in regulating Gli1 and Ptch1 mRNA levels. We transfected the genetically matched Smo wild-type and Smo mutant PDAC cells with siRNA constructs targeted at Kras or Gli1 and measured the impact of this treatment on the expression of Kras, Gli1, Gli2, Gli3, and Ptch1 after 48 h. We found that depleting 80% of Kras expression with Kras-targeted siRNAs resulted in a significant down-regulation of the Gli1 and Ptch1 mRNAs in both PDAC lines (Fig. 6B). Interestingly, depleting 80% of Gli1 expression with Gli1-targeted siRNAs not only resulted in the decreased expression of Ptch1, but also of Kras itself (Fig. 6B), suggesting reciprocal feedback regulation. Gli2 remained undetectable, and Gli3 was unaffected in PDAC tumor cells following Kras depletion (data not shown). Thus, we demonstrate that both TGF-β1 and Kras regulate Gli1 and Ptch1 expression independently of Smo-mediated signaling.
GLI1 is required for PDAC cell survival and for KRAS-driven transformation
Next, we asked if the maintenance of Gli1 expression in PDAC cells was functionally important for PDAC cancer cell homeostasis. We first performed cell growth assays on mouse PDAC cells treated with siRNA constructs targeting Kras and Gli1. In two independent mouse PDAC cell lines (4.2 NR and 3.3), we found that both Kras and Gli1 siRNA targeting resulted in significantly decreased cell numbers 72 h after transfection and 24 h after serum deprivation (Fig. 6C). We repeated the siRNA targeting and stained 3.3 cell cultures with an anti-cleaved caspase-3 antibody that marks cells undergoing apoptosis. We find that apoptosis increases markedly in 3.3 cells treated with Gli1 and Kras siRNAs (Fig. 6D).
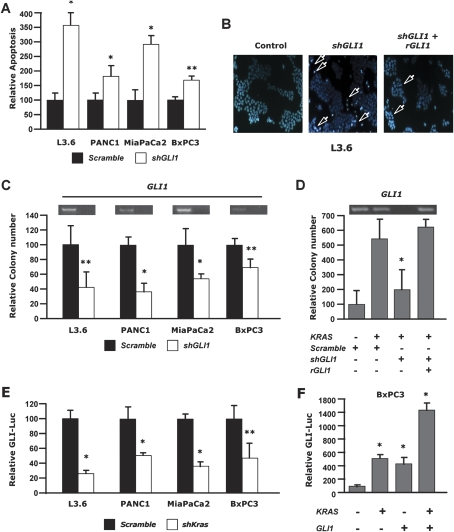
We then asked if our findings in mouse PDAC cells also applied to human PDAC cells. We transfected four human PDAC cell lines with a shRNA targeting GLI1 and compared it with a scrambled shRNA. Three lines (L3.6, PANC1, and MiaPaCa2) contained an activating mutation in KRAS, whereas a fourth, BxPC3, was wild type for KRAS; all four lines express comparable levels of KRAS mRNA (data not shown). We found that upon challenge with cyclohexamide, apoptosis was markedly increased in all four human PDAC cell lines (Fig. 7A,B). We then asked if this decrease in cellular fitness also impacted the propensity to form colonies in soft agar, a transformation assay that measures anchorage-independent cell growth and approximates the malignant potential of tumor cells. Colony formation in soft agar was markedly impaired following GLI1 depletion in all three KRAS mutant cell lines but had a less notable effect on KRAS wild-type BXPC3 cells (Fig. 7C), suggesting that the GLI1 requirement for cellular transformation was more acutely detected in the context of mutant KRAS. To investigate this possibility, we transfected KRAS wild-type BxPC3 cells with an oncogenic KRAS construct, which resulted in a significant increase in colony formation (Fig. 7D). Remarkably, in the context of oncogenic KRAS, BxPC3 colony formation was much more sensitive to GLI1 depletion. This sensitivity was confirmed to be GLI1-specific, as it could be rescued by a cotransfected resistance GLI1 cDNA construct (rGLI1) that is not targeted by the GLI1 shRNA (Fig. 7D, right bar). Colony formation induced by wild-type KRAS overexpression in BxPC3 cells was less pronounced and less sensitive to GLI1 depletion than with mutant oncogenic KRAS (data not shown).
Figure 7.
GLI1 is required for survival and maintenance of the transformed phenotype of human PDAC cancer cells. (A) Relative changes in chromatin condensation/margination and nuclear fragmentation (proxy for apoptosis) in L3.6, PANC1, MiaPaCa2, and BxPC3 pancreatic cancer cells 48 h after transfection with shGLI1 or scramble control shRNA and subsequent treatment with 25 mM cycloheximide. More than 300 cells in four high-power fields were counted; apoptotic cells are expressed as a percentage of total cells. (B) Hoescht 33342 staining of L3.6 PDAC cells transfected with scramble shRNA, shGLI1, or shGLI1 alongside an shGLI1-resistant GLI1 cDNA construct (rGLI1). Arrows indicate nuclear morphological patterns indicative of apoptosis (blue, Hoechst 33342). (C) Relative colony formation (neoplastic anchorage-independent growth) of L3.6, PANC1, MiaPaca2, and BxPC3 pancreatic cancer cells assayed following transfection with shGLI1 or scramble shRNA (control) plasmids. (D) Relative colony formation of BxPC3 pancreatic cancer cells (wild-type KRAS) transfected with oncogenic KRAS and scramble control shRNA, shGli1, or shGli1 alongside an shGLI1-resistant GLI1 cDNA rescue construct (rGli1). (E) Relative changes in Luciferase activity in L3.6, PANC1, MiaPaCa2, and BxPC3 pancreatic cancer cells transfected with a Gli-Luciferase reporter and with shKRAS or scramble control shRNA. (F) Relative changes in Luciferase activity in BXPC3 (wild-type KRAS) pancreatic cancer cells transfected with a GLI-Luciferase reporter with or without KRAS or GLI1 expression constructs. (*) P-value <0.01; (**) P-value <0.05.
We next tested the prediction, based on the results from the mouse model, that human PDAC cell lines in which phenotypic effects of GLI1 depletion were observed would nevertheless be unresponsive to Shh stimulation, in support of our interpretation that endogenous GLI1 regulation is decoupled from upstream Shh signaling in PDAC cells. We exposed L3.6 and PANC1 cells to exogenous recombinant Shh, monitoring the expression of a GLI-luciferase reporter. There was no effect of Shh on the GLI reporter, whereas it was readily induced in a fibroblastic cell line (CH10T1/2) (Supplemental Fig. 3A). In both PDAC cells and fibroblasts, in contrast, transfection of GLI1 markedly increased transcription from the GLI-luciferase reporter, demonstrating that the GLI reporter can respond to elevated GLI transcriptional activity in both cell types (Supplemental Fig. 3B).
Since GLI1 mediates important functions of oncogenic KRAS in human PDAC cells, we investigated the relationship between KRAS and GLI transcription. We found that shRNA-mediated depletion of KRAS in human PDAC cells leads to a marked down-regulation of GLI transcription, as assayed by the activity of a GLI-luciferase reporter (Fig. 7E). Moreover, oncogenic KRAS cooperates with GLI1 to induce elevated GLI transcription when transfected into KRAS wild-type BxPC3 cells (Fig. 7F).
Thus KRAS is both required and sufficient for the induction of GLI transcriptional activity in PDAC cancer cells that evidence SHH-independent GLI transcription, and GLI1 demonstrably contributes to PDAC cancer cell survival and to malignant cellular phenotypes mediated by KRAS.
Discussion
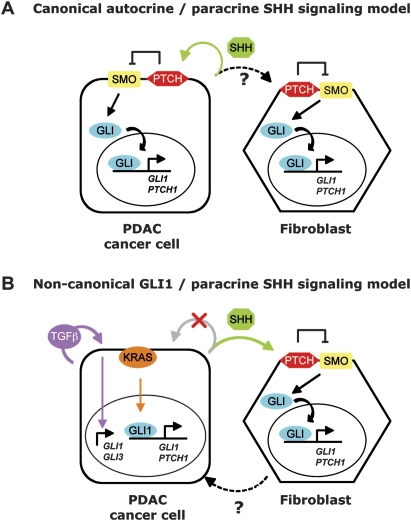
Hedgehog signaling is deregulated in pancreatic adenocarcinoma, and functional studies have implicated this pathway in pancreatic tumorigenesis: Shh overexpression is sufficient to initiate PanIN-like precursor lesions (Berman et al. 2003; Thayer et al. 2003) and to accelerate tumor formation in mouse orthotopic xenotransplants (Morton et al. 2007), while GLI transcription synergizes with activated Kras to induce aggressive undifferentiated pancreatic tumors (Pasca di Magliano et al. 2006). In addition, cyclopamine, a Smo inhibitor, has a clearly deleterious effect on a subset of human PDAC cell lines (Berman et al. 2003; Thayer et al. 2003; Feldmann et al. 2007). Collectively, these studies suggested that autocrine Shh signaling in neoplastic ductal cells was important for PDAC carcinogenesis, alongside a potential but untested interplay with the tumor stroma (Fig. 8A).
Figure 8.
A new conceptualization of functionally significant hedgehog signaling and GLI transcription in pancreatic ductal carcinogenesis. (A) The conventional model of Hh/Gli signaling in PDAC. Earlier studies suggested that autocrine signaling by the SHH ligand was functionally important in PDAC cells (green arrow). Possible in vivo signaling to the stroma was an untested hypothesis (dotted black arrow), and hedgehog signaling in cancer cells and stromal cells such as cancer-associated fibroblasts was thought to follow canonical hedgehog signaling, whereby SHH binds to PTCH and induces SMO signaling resulting in the nuclear translocation of active GLI transcription factors and transcriptional activation of GLI target genes such as GLI1 and PTCH1. (B) A refined model of Hh/Gli signaling in PDAC. Our study demonstrates that PDAC tumor cells (1) require GLI1 for transformation and survival, (2) do not transduce functionally significant SHH ligand signals through the SMO coreceptor (crossed-out gray arrow), and (3) continue to express GLI transcriptional targets independently of SMO via noncanonical regulation of GLI target genes mediated in part by KRAS (orange arrow) and TGFβ (purple arrow). In parallel to noncanonical GLI transcription in PDAC cancer cells, SHH produced by cancer cells signals in a paracrine manner (green arrow) to the surrounding mesenchyme and may play a key paracrine function in PDAC pathogenesis (Yauch et al. 2008). In response to SHH, fibroblasts secrete signaling molecules that may stimulate tumor cell growth (Yauch et al. 2008) (dotted black arrow).
A recent study, however, reports that a potent and specific inhibitor of Smo signaling down-regulates Gli transcription in the stroma of transplanted PDAC tumors, but not in the cancer cells. Furthermore, inhibition of hedgehog signaling in the stroma is functionally important, as it is sufficient to impair tumor growth in subcutaneous xenotransplants of hedgehog ligand-positive cancer cells (Yauch et al. 2008). Other data from the same group also show that pancreatic ductal epithelial cells are not susceptible in vivo to oncogenic Smo signaling, whereas expression of oncogenic Smo in mesenchymal cells induces mesenchymal tumors (Tian and de Sauvage, unpubl.). These new observations, in concert with the data presented herein, support an alternative model for the role of hedgehog signaling in PDAC formation, in which hedgehog ligands secreted by the pancreatic ductal epithelium do not stimulate the cancer cells in an autocrine manner, but rather serve a paracrine signaling function, resulting in the canonical Smo-dependent activation of GLI transcription in adjacent mesenchymal cells. Moreover, our study supports the proposition that neoplastic pancreatic ductal cells do not transduce hedgehog ligand signals, as we show that, unlike fibroblastic cells, PDAC cells do not induce Gli transcription following incubation with Shh. We also demonstrate that autocrine Smo-mediated hedgehog signaling is neither limiting nor functionally required in the ductal epithelium for the development of PDAC, since genetically ablating Smo in the pancreatic epithelium has no effect on PDAC tumorigenesis.
However, while gain or loss of Smo function does not affect pancreatic ductal cells, similarly targeted expression of a downstream effector of hedgehog signaling, the transcription factor Gli2, induces pancreatic neoplasia and accelerates Kras-induced carcinogenesis, albeit of a nondifferentiated kind (Pasca di Magliano et al. 2006). Moreover, a GLI-mediated transcriptional program is clearly induced in PanIN lesions in vivo (Prasad et al. 2005), arguing that GLI transcription inside ductal cells may be important for pancreatic tumorigenesis. Remarkably, we observed no significant decrease in the levels of expression of Gli target genes following the genetic ablation of Smo in neoplastic ductal cells, showing that expression of Gli target genes in PDAC cells is decoupled from upstream Hh/Ptch/Smo signal transduction. We find that two signaling molecules prominently involved in PDAC tumorigenesis, KRAS and TGFβ, regulate the Smo-independent expression of Gli target genes in mouse PDAC cells. Furthermore, we find that GLI1 is required in human PDAC cell lines for survival and for KRAS-mediated cellular transformation. The nonresponsiveness to Shh signaling, along with the demonstrable requirement of Gli1 function in mouse and human PDAC cells, may help explain why genes that are in other circumstances downstream effectors or regulators of hedgehog signaling, including GLI1 and GLI3, were recently reported to be mutated in 100% of 24 human PDAC-derived cell lines whose genome was comprehensively scanned for mutations (Jones et al. 2008). In contrast, the PTCH and SMO coreceptors, which we demonstrate are unimportant in pancreatic cancer cells per se, are not prone to mutational alteration in such cells, consistent with the conceptual refinement of paracrine hedgehog signaling in PDAC (Fig. 8B).
In conclusion, the results of this study, together with data from the de Sauvage group (Yauch et al. 2008), shed new light on the complex circuitry of hedgehog signaling in PDAC pathogenesis, in which canonical paracrine Shh signaling is functionally important in the mesenchymal component of the tumor stroma, whereas SMO-independent, noncanonical, cancer cell-autonomous, KRAS-driven GLI1 transcription is required in the tumor parenchyma (Fig. 8B). The collective knowledge of this and other recent studies suggests a dual strategy for innovative therapeutic targeting of PDAC—that of inhibiting either KRAS itself or Gli transcription in pancreatic cancer cells in conjunction with abrogating SMO-dependent Shh signaling in the tumor stroma. A recent report describing inhibitors of Gli transcription (Lauth et al. 2007) hold in this respect interesting promise worthy of future investigation in concert with the new generation of potent and selective Smo inhibitors (Yauch et al. 2008).
Materials and methods
Mouse strains and genotyping
The LSL-KrasGD12, Trp53F, SmoF, and p48-Cre alleles have been described (Jonkers et al. 2001; Long et al. 2001; Kawaguchi et al. 2002; Tuveson et al. 2004). The Smo allele genotyping was Smo-for (GTTCCCAGGGTTGAAGACAG) used at 0.5 μM and a mix of Smo-rev-wt (ACAGCCAACTCAGCAAAAGC) (wild-type allele, 300-base-pair [bp] band) used at 0.2 μM, and Smo-rev-mut (CTAAAGCGCATGCTCCAGAC) (flox allele, 350-bp band) used at 0.3 μM. The PCR conditions were six cycles (94°C, 1 min; 63°C, 30 sec; 72°C, 45 sec), followed by 32 cycles (94°C, 1 min; 60°C, 30 sec; 72°C, 45 sec), and 5 min at 72°C. PCR bands were separated on 3% agarose gels. All studies were conducted in compliance with University of California Institutional Animal Care and Use Committee guidelines.
PDAC cell line isolation and tissue culture of pancreatic fibroblasts
A 5-mm tumor fragment of PDAC was dissected, minced in cold PBS with fine scissors, and transferred in 13.5 mL of PBS in a vessel with a stirring bar. Five-hundred microliters of Clostridium histolyticum Collagenase XI (20 g/mL; Sigma #C7657) were added, and the vessel was stirred at high speed for 20 min at 37°C. The sample was filtered through a 100-μm strainer, and the solid fraction in the strainer was kept; the flow-through was discarded. The solid fraction was transferred to a 50-mL tube, washed three times in PBS, and resuspended in 30 mL of complete DMEM (10% FBS + antibiotics). Large undigested fragments sank, and the supernatant was plated on Collagen-I coated plates. Two weeks later, colonies grew and were cloned into 6-well plates (no collagen). For in vitro recombination of PDAC 4.2 NR, 1 × 109 PFUs of Ad5CMVCre virus (GTVC; http://www.uiowa.edu/∼gene) was added to 5 × 106 cells. After 48 h, cells were trypsinized, resuspended in single cell solution, counted, and serially diluted in 96-well plates (1–10 successive dilutions horizontally, followed by 1–10 successive dilutions vertically); wells containing single colonies of recombined cells were expanded and genotyped (five out of five recovered 4.2R clones were recombined). Serial dilution was also performed to isolate uninfected clones of 4.2 NR. Pancreatic fibroblasts were obtained as follows: One wild-type pancreas was dissected, minced in 2-mm fragments in ice-cold PBS, and transferred to a stirring vessel with a sterile stir bar in 2 mL of PBS. Twenty-five milliliters of Collagenase V (1 mg/mL; Sigma #C9263) were added, and the vessel was stirred at high speed for 15 min at 37°C. Collagenase digestion was stopped with three successive washes in complete DMEM. After resuspension in 10 mL of PBS, the sample was filtered through a 100-μm strainer. Both fractions (flow-through and solid fraction) were kept and spun and resuspended in 2 mL of Trypsin 0.25%. Then they were incubated for 5 min at 37°C. The digestion was stopped with 25 mL of complete DMEM, and the pellets were spun and resuspended in 5 mL of complete DMEM and plated in six-well plates. After ∼2 wk (with regular media changes), cells senesced after five to six passages.
In vitro stimulation of PDAC cells and fibroblasts with rShh and rTGFβ1
Stimulation with 0 to 500 ng/mL recombinant rShh (rmShh-C25II-N; R&D Systems #464SH) was performed in low serum conditions (0.2% FBS) for 8 h, after a 16-h period of low serum exposure, and stimulation with 5 ng/mL recombinant TGFβ1 (R&D Systems #240-B) was performed in 10% FBS for 48 h.
MTT and siRNA assays
Cell growth assays were performed using the Vybrant MTT cell proliferation Assay Kit (Molecular Probes #V-13154), following the manufacturer's recommendations. siRNA assays were performed using Dharmacon siRNA ON-TARGETplus SMART pools (Thermo Scientific; Kras #L-043846-01; Gli1 #L-047917-00; Nontargeting Control #D-001810-10) and DharmaFECT 2 transfection reagents (Thermo Scientific #T-2002).
Quantitative PCR
Total RNA was prepared using the RNeasy Mini (Qiagen) following the manufacturer's recommendations. Total RNA from Laser Microdissected samples was prepared using the RNeasy Micro (Qiagen). DNase treatment and RNA cleanup were performed with the DNA-Free RNA Kit (Zymo Research). cDNA synthesis was performed using iScript (Bio-Rad). PCRs were performed using the following TaqMan assays (Applied Biosystems): Mm00494645_m1 (Gli1), Mm01293117_m1 (Gli2), Mm00492338_m1 (Gli3), Mm00436026_m1 (Ptch1), Mm00436527_m1 (Shh), Mm01162704_m1 (Smo), and Mm03053281_s1 (Kras). The mGus assay was obtained from Integrated DNA Technologies. (F) CTCATCTGGAATTTCGCCGA; (R) GGCGAGTGAAGATCCCCTTC; (Probe) fam-CGAACCAGTCACCGCTGAGAGTAATCG-bhq1. Quantitative PCR reactions were performed on an ABI7900HT Sequence Detection System. Ct values were determined and subtracted to obtain the ΔCt [ΔCt = Ct (test locus) − Ct (control locus)]. Relative fold difference was calculated as 2−ΔCt × 100.
Histological analysis, immunochemistry, and immunofluorescence
Tissues were fixed overnight in zinc-buffered formalin and embedded in paraffin. Sections 7 μm thick were subjected to either H&E staining or an antigen retrieval procedure (Citra; BioGenex). Following inhibiting endogenous peroxydases and blocking the slides, primary antibodies were applied overnight at 4°C. The following antibodies were used: goat anti-Shh (1:100; R&D systems AF445), rabbit anti-Smo (1:100; kind gift from Dr. Pao Tien Chuang) (Gerber et al. 2007), Armenian Hamster anti-Muc1 antibody (1:200; Neomarkers HM-1630-P0), rabbit anti-α-amylase (1:500; Sigma A8273), anti-activated Caspase3 (1:300; Cell Signaling 9661S). Biotinylated or fluorochrome-conjugated antibodies were used as secondary antibodies (1:200; Jackson Immunoresearch). 3-3′-DAB tetrahydrochloride (Sigma D4293) was used as a chromogen.
Laser capture microdissection
Ten micrometers of fresh frozen PDAC sections were applied to membrane slides (Leica 11505151) and subjected to a modified H&E staining protocol (Hematoxylin Solution; Harris modified, Sigma HHS-16; Eosin Y solution with Phloine B, Sigma HT110-3-16; Blueing reagent, 0.1% NH4OH, Sigma 221228). Laser capture microdissection was performed over 3 h following the modified H&E staining with a Leica AS LMD automated microscope.
Soft agar growth assays
All cell lines were electroporated for 10 msec at 360 V using a BTX square-wave electroporator. L3.6, PANC1, MiaPaca2, or BxPC3 pancreatic cancer cells (1 × 107) were transfected with the scramble control or shRNA targeting GLI1. In rescue experiments, a resistant version of the human GLI1 cDNA was cotransfected. Transfected cells were resuspended in serum containing media with 0.33% low-melting-point agarose and plated over a cushion of 0.5% agarose in the same medium in a 60-mm dish. The cells were allowed to grow for 2 wk, after which visible colonies containing >50 cells were counted using an inverted microscope. Three different experiments were performed in triplicate. Control for expression was done by PCR as follows: RNA was prepared from pancreatic tumor cell lines using Trizol reagent as per the manufacturer's instructions (Invitrogen). cDNA was prepared from the RNA using the Superscript kit from Invitrogen. The following oligonucleotide pairs were used in order to amplify specific GLI1 transcripts from the cell line cDNA: 5′-ACTGAAGACCTCTCCAGC-3′ and 5′-GCTGACAGTATAGGCAGA-3′. PCR was carried out using the following conditions: 30 cycles of denaturation for 30 sec at 95°C, annealing for 30 sec 52°C, and extension for 1 min at 72°C. One-tenth of the PCR product was analyzed by separation on a 1% agarose-TAE gel and stained with ethidium bromide.
Apoptosis assays
L3.6, PANC1, MiaPaCa2, and BXPC3 were transfected with the scramble control or shRNA targeting GLI1. Forty-eight hours after transfection, cells were treated with vehicle control (DMSO) or cycloheximide (25 μM). For apoptosis determination, cells undergoing apoptosis were identified by the morphological criteria of nuclear fragmentation, nuclear margination, and organelle disorganization using the DNA-staining Hoescht 33860. Positive cells were counted using a Zeiss confocal microscope. GLI1 expression levels were analyzed by PCR at 48 h post-transfection.
Luciferase assay
All cells were grown and transfected as indicated above. For luciferase reporter assays, 1 × 106 cells were distributed in triplicate in 24-well plates and allowed to recover for 18 h. The cells were subsequently washed, and fresh medium containing 1% FBS was added. Samples were harvested and prepared for luciferase assays following the manufacturer's recommendations (Promega). Total proteins quantitation was used to control for intersample variations in transfection efficiency.
Acknowledgments
We are indebted to Drs. Ya-Jun Li and Pao-Tien Chuang for their gift of the anti-Smo antibody. We thank N. Bardeesy and R. DePinho for critical reading of the manuscript; members of the Hanahan and Hebrok laboratories for scientific discussions, in particular, P. Olson, J. Morris, and M. Pasca di Magliano; the Helen Diller Family Comprehensive Cancer Center Genome Analysis Core Facility for TaqMan analysis; and E. Drori, M. Vayner, S. Cacacho, and A. Wang for technical assistance. This work was supported by grants from the NIH to M.H. (DK60533-01A1, CA112537-01), M.E.F.Z. (CA125127, Carole and Bob Daly AACR-PANCAN Career Development Award for Pancreatic Cancer Research, Mayo Clinic Pancreatic SPORE P50 CA102701, University of Iowa/Mayo Clinic Lymphoma SPORE P50 CA097274), and D.H. (program project grant on pancreatic cancer; R. DePinho, P.I.). O.N.S. was a Merck Fellow of the Damon Runyon Cancer Research Foundation (DRG-1831-04).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1753809.
Supplemental material is available at http://www.genesdev.org.
References
- Aguirre A.J., Bardeesy N., Sinha M., Lopez L., Tuveson D.A., Horner J., Redston M.S., DePinho R.A. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes & Dev. 2003;17:3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardeesy N., DePinho R.A. Pancreatic cancer biology and genetics. Nat. Rev. Cancer. 2002;2:897–909. doi: 10.1038/nrc949. [DOI] [PubMed] [Google Scholar]
- Bardeesy N., Aguirre A.J., Chu G.C., Cheng K.H., Lopez L.V., Hezel A.F., Feng B., Brennan C., Weissleder R., Mahmood U., et al. Both p16(Ink4a) and the p19(Arf)–p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc. Natl. Acad. Sci. 2006a;103:5947–5952. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardeesy N., Cheng K.H., Berger J.H., Chu G.C., Pahler J., Olson P., Hezel A.F., Horner J., Lauwers G.Y., Hanahan D., et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes & Dev. 2006b;20:3130–3146. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman D.M., Karhadkar S.S., Maitra A., Montes De Oca R., Gerstenblith M.R., Briggs K., Parker A.R., Shimada Y., Eshleman J.R., Watkins D.N., et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- Dennler S., Andre J., Alexaki I., Li A., Magnaldo T., ten Dijke P., Wang X.J., Verrecchia F., Mauviel A. Induction of Sonic Hedgehog mediators by transforming growth factor-β: Smad3-dependent activation of Gli2 and Gli1 expression in vitro and in vivo. Cancer Res. 2007;67:6981–6986. doi: 10.1158/0008-5472.CAN-07-0491. [DOI] [PubMed] [Google Scholar]
- Feldmann G., Dhara S., Fendrich V., Bedja D., Beaty R., Mullendore M., Karikari C., Alvarez H., Iacobuzio-Donahue C., Jimeno A., et al. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: A new paradigm for combination therapy in solid cancers. Cancer Res. 2007;67:2187–2196. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber A.N., Wilson C.W., Li Y.J., Chuang P.T. The hedgehog regulated oncogenes Gli1 and Gli2 block myoblast differentiation by inhibiting MyoD-mediated transcriptional activation. Oncogene. 2007;26:1122–1136. doi: 10.1038/sj.onc.1209891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezel A.F., Kimmelman A.C., Stanger B.Z., Bardeesy N., Depinho R.A. Genetics and biology of pancreatic ductal adenocarcinoma. Genes & Dev. 2006;20:1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- Hingorani S.R., Petricoin E.F., Maitra A., Rajapakse V., King C., Jacobetz M.A., Ross S., Conrads T.P., Veenstra T.D., Hitt B.A., et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- Incardona J.P., Gruenberg J., Roelink H. Sonic hedgehog induces the segregation of patched and smoothened in endosomes. Curr. Biol. 2002;12:983–995. doi: 10.1016/s0960-9822(02)00895-3. [DOI] [PubMed] [Google Scholar]
- Jacob L., Lum L. Deconstructing the hedgehog pathway in development and disease. Science. 2007;318:66–68. doi: 10.1126/science.1147314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S., Zhang X., Parsons D.W., Lin J.C., Leary R.J., Angenendt P., Mankoo P., Carter H., Kamiyama H., Jimeno A., et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers J., Meuwissen R., van der Gulden H., Peterse H., van der Valk M., Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y., Cooper B., Gannon M., Ray M., MacDonald R.J., Wright C.V. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat. Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- Lauth M., Bergstrom A., Shimokawa T., Toftgard R. Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc. Natl. Acad. Sci. 2007;104:8455–8460. doi: 10.1073/pnas.0609699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F., Zhang X.M., Karp S., Yang Y., McMahon A.P. Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development. 2001;128:5099–5108. doi: 10.1242/dev.128.24.5099. [DOI] [PubMed] [Google Scholar]
- Morton J.P., Mongeau M.E., Klimstra D.S., Morris J.P., Lee Y.C., Kawaguchi Y., Wright C.V., Hebrok M., Lewis B.C. Sonic hedgehog acts at multiple stages during pancreatic tumorigenesis. Proc. Natl. Acad. Sci. 2007;104:5103–5108. doi: 10.1073/pnas.0701158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca di Magliano M., Hebrok M. Hedgehog signalling in cancer formation and maintenance. Nat. Rev. Cancer. 2003;3:903–911. doi: 10.1038/nrc1229. [DOI] [PubMed] [Google Scholar]
- Pasca di Magliano M., Sekine S., Ermilov A., Ferris J., Dlugosz A.A., Hebrok M. Hedgehog/Ras interactions regulate early stages of pancreatic cancer. Genes & Dev. 2006;20:3161–3173. doi: 10.1101/gad.1470806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad N.B., Biankin A.V., Fukushima N., Maitra A., Dhara S., Elkahloun A.G., Hruban R.H., Goggins M., Leach S.D. Gene expression profiles in pancreatic intraepithelial neoplasia reflect the effects of Hedgehog signaling on pancreatic ductal epithelial cells. Cancer Res. 2005;65:1619–1626. doi: 10.1158/0008-5472.CAN-04-1413. [DOI] [PubMed] [Google Scholar]
- Rubin L.L., de Sauvage F.J. Targeting the Hedgehog pathway in cancer. Nat. Rev. Drug Discov. 2006;5:1026–1033. doi: 10.1038/nrd2086. [DOI] [PubMed] [Google Scholar]
- Rustgi A.K. The molecular pathogenesis of pancreatic cancer: Clarifying a complex circuitry. Genes & Dev. 2006;20:3049–3053. doi: 10.1101/gad.1501106. [DOI] [PubMed] [Google Scholar]
- Taipale J., Beachy P.A. The Hedgehog and Wnt signalling pathways in cancer. Nature. 2001;411:349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- Thayer S.P., di Magliano M.P., Heiser P.W., Nielsen C.M., Roberts D.J., Lauwers G.Y., Qi Y.P., Gysin S., Fernandez-del Castillo C., Yajnik V., et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuveson D.A., Hingorani S.R. Ductal pancreatic cancer in humans and mice. Cold Spring Harb. Symp. Quant. Biol. 2005;70:65–72. doi: 10.1101/sqb.2005.70.040. [DOI] [PubMed] [Google Scholar]
- Tuveson D.A., Shaw A.T., Willis N.A., Silver D.P., Jackson E.L., Chang S., Mercer K.L., Grochow R., Hock H., Crowley D., et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- Yauch R.L., Gould S.E., Scales S.J., Tang T., Tian H., Ahn C.P., Marshall D., Fu L., Januario T., Kallop D., et al. A paracrine requirement for hedgehog signalling in cancer. Nature. 2008;455:406–410. doi: 10.1038/nature07275. [DOI] [PubMed] [Google Scholar]
- Zhang X.M., Ramalho-Santos M., McMahon A.P. Smoothened mutants reveal redundant roles for Shh and Ihh signaling including regulation of L/R symmetry by the mouse node. Cell. 2001;106:781–792. [PubMed] [Google Scholar]