Abstract
Eukaryotes possess mechanisms to limit crossing over during homologous recombination, thus avoiding possible chromosomal rearrangements. We show here that budding yeast Mph1, an ortholog of human FancM helicase, utilizes its helicase activity to suppress spontaneous unequal sister chromatid exchanges and DNA double-strand break-induced chromosome crossovers. Since the efficiency and kinetics of break repair are unaffected, Mph1 appears to channel repair intermediates into a noncrossover pathway. Importantly, Mph1 works independently of two other helicases—Srs2 and Sgs1—that also attenuate crossing over. By chromatin immunoprecipitation, we find targeting of Mph1 to double-strand breaks in cells. Purified Mph1 binds D-loop structures and is particularly adept at unwinding these structures. Importantly, Mph1, but not a helicase-defective variant, dissociates Rad51-made D-loops. Overall, the results from our analyses suggest a new role of Mph1 in promoting the noncrossover repair of DNA double-strand breaks.
Keywords: Genome instability, recombination, DNA helicase, crossing over, Fanconi anemia
DNA double-strand breaks (DSBs) that arise during DNA replication or are induced by DNA damaging agents, such as ionizing radiation, are frequently repaired by homologous recombination (HR). In yeast and other eukaryotes, the RAD52 epistasis group of proteins mediate homologous recombination (for reviews, see Paques and Haber 1999; Krogh and Symington 2004). In this process, DSB ends are resected by nucleases to create 3′ ssDNA that becomes coated with the recombinase protein Rad51. The Rad51–ssDNA nucleoprotein filament then carries out a search for a homologous donor DNA sequence and promotes strand invasion of the donor molecule to form a D-loop (Sung and Klein 2006). After D-loop formation, there appear to be two alternative pathways that result in DSB repair. One pathway involves the formation of a double Holliday junction (dHJ) that can be resolved by symmetrical strand cleavage into either a crossover or noncrossover gene conversion (Szostak et al. 1983). An alternative mechanism, called synthesis-dependent strand annealing (SDSA), posits formation mostly of noncrossovers, and in most cases does not involve a dHJ intermediate (for review, see Paques and Haber 1999). In support of the SDSA model of gene conversion, we showed recently that both newly synthesized strands are inherited by the broken recipient DNA molecule (Ira et al. 2006). The choice between crossover and noncrossover is tightly regulated in both mitotic and meiotic cells. In meiotic S. cerevisiae cells the correct number of crossovers are required for proper chromosome segregation; the proportion of gene conversion accompanied by crossing over varies between 25% and 50% depending on the locus (Roeder 1995). In mitotic cells, the proportion of crossovers is much lower, ranging between <1% and 15%, depending on the recombination assay and organism (Esposito 1978; Nassif et al. 1994; Johnson and Jasin 2000; Virgin et al. 2001; Stark and Jasin 2003). These differences are best explained if recombination proceeds most often according to the dHJ model in meiotic cells and via SDSA and other noncrossover means in mitotic cells. However, even in meiotic cells SDSA model explains best at least a part of noncrossover products (McMahill et al. 2007). The suppression of mitotic crossovers, which can lead to gross chromosome aberrations and rearrangements, is thought to be critical for the preservation of genome integrity (Richardson et al. 1998; Shaw and Lupski 2004).
In both mitotic and meiotic Saccharomyces cells, crossover and noncrossover products are generated by genetically and kinetically distinct pathways (Allers and Lichten 2001; Hunter and Kleckner 2001; Ira et al. 2003; Barbera and Petes 2006; McMahill et al. 2007). Crossover and noncrossover pathways have different genetic requirements in mammalian cells as well (Guillon et al. 2005). In yeast mitotic cells, we and others demonstrated that two DNA helicases, Sgs1 and Srs2, suppress crossovers in DSB-induced and spontaneous recombination, but in different ways (Ira et al. 2003; Lo et al. 2006; Robert et al. 2006). Sgs1 was also shown to suppress crossing over in the meiotic cycle, particularly when three or four chromatids are involved in recombination (Jessop et al. 2006; Oh et al. 2007, 2008; Jessop and Lichten 2008). Srs2 appears to promote steps in the SDSA pathway, whereas Sgs1 and its associated topoisomerase Top3 reduce crossovers apparently by resolving the dHJ in a noncrossover manner. The postulated role of Sgs1–Top3 is strongly supported by biochemical data showing that the human ortholog of Sgs1/Top3, BLM/Topo IIIα, is able to dissolve dHJs to yield noncrossovers (Wu and Hickson 2003). RECQ5, a homolog of the BLM helicase, is involved in the suppression of crossing over (Wang et al. 2003; Hu et al. 2005).
By exploiting published information regarding the synthetic lethal growth phenotypes of srs2∆ with other mutations (Tong et al. 2004), we sought to identify novel yeast genes that specifically affect the proportion of crossover outcome. Among synthetic interactors with srs2Δ in yeast and its Escherichia coli homolog uvrD there are a number of genes encoding proteins working in later steps of recombination including Sgs1–Top3 and Rad54 in yeast and the ruvABC resolvase in E. coli (Palladino and Klein 1992; Stewart et al. 1997; Gangloff et al. 2000; Solinger and Heyer 2001; Solinger et al. 2002; Bugreev et al. 2006; Magner et al. 2007). It has been hypothesized that in the absence of the anti-recombinase Srs2/uvrD (Krejci et al. 2003; Veaute et al. 2003) and enzymes that resolve recombination intermediates, toxic intermediates accumulate to cause cell death (Gangloff et al. 2000; Fabre et al. 2002; Magner et al. 2007). Therefore, we presumed that it would be possible to identify novel proteins involved in late stages of recombination that direct DSB repair toward crossover or noncrossover by screening all SRS2 synthetic interactors.
We deleted the yeast genes previously demonstrated to have synthetic interaction with srs2Δ (Tong et al. 2004) in a strain with which we could follow the frequency of crossovers and noncrossovers accompanying the repair of an HO endonuclease-induced DSB. We found that only the deletion of MPH1 or RMI1 leads to an increase in the frequency of crossing over. Deletion of RMI1 causes a twofold increase in crossover frequency, and this phenotype is epistatic to sgs1Δ, consistent with the premise that Rmi1 influences the dHJ dissolution activity of the Sgs1–Topo IIIα complex (Wu and Hickson 2003; Chang et al. 2005; Mullen et al. 2005). The absence of the Mph1 helicase, yeast's ortholog of the Fanconi anemia (FA) protein FANCM, elevates crossover frequency by more than threefold. Importantly, this crossover regulatory role of Mph1 is independent of Sgs1 and Srs2. Mph1, like Sgs1, suppresses spontaneous unequal sister chromatid exchange as well. We present chromatin immunoprecipitation (ChIP) and biochemical data to implicate Mph1 in the dissociation of the Rad51-made D-loop intermediate. These results implicate Mph1 in channeling recombination intermediates into the noncrossover pathway(s), and have implications as to an analogous role of the human FANCM protein.
Results
Mph1 and Rmi1/Nce4 promote noncrossover outcomes in mitotic cells
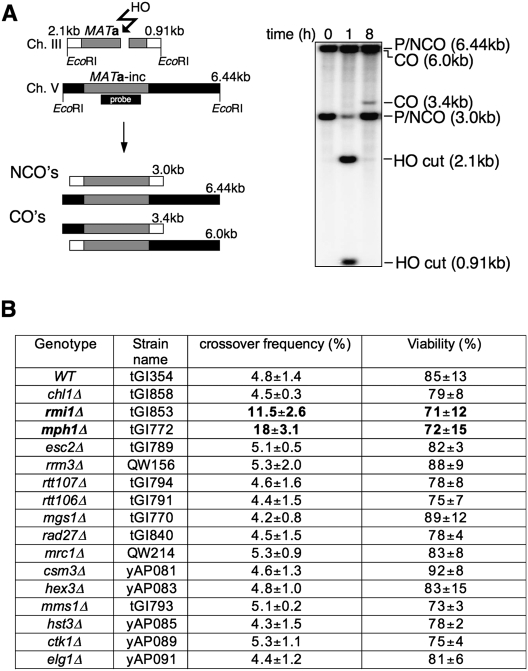
To identify proteins involved in post-synaptic stages of recombination we examined genes (CHL1, RMI1/NCE4, MPH1, ESC2, RRM3, RTT107, RTT106, MMS1, MGS1, RAD27, MRC1, CSM3, HEX3, HST3, CTK1, and ELG1) whose deletion shows a negative synthetic interaction with srs2Δ (Tong et al. 2004) for their possible role in the regulation of crossover formation in mitotic cells. SGS1–TOP3 and MUS81–MMS4 have been tested previously (Ira et al. 2003). RAD54 was not tested here because it is required for all gene conversion (Sugawara et al. 2003). We used an ectopic recombination system where the DSB created by HO endonuclease within a MAT a sequence on chromosome V is repaired by HR with MAT a-inc sequences on chromosome III (Fig. 1A). In wild-type yeast cells (tGI354), the HO break is repaired efficiently by Rad51-dependent gene conversion, with ∼5% of the repair events being accompanied by crossing over (Ira et al. 2003).
Figure 1.
Screen for genes regulating the crossover frequency identifies Mph1. (A) Ectopic gene conversion is induced by DSBs generated by the HO endonuclease within a 1.9-kb MAT a sequence (gray rectangle) that replaced the ARG5,6 gene on chromosome V. The MAT a-inc sequence on chromosome III is a donor for recombination, and shares 1403-bp and 530-bp homology on opposite sides of the DSB. EcoRI noncrossover fragments (NCOs) of 6.4 kb and 3 kb can be distinguished from crossover fragments (COs) of 6 kb and 3.4 kb, and quantified on Southern blots. The probe used to detect crossovers and noncrossovers was a MAT a fragment overlapping the first 200 bp on each side of the HO break. (B) Viability resulting from DSB repaired by ectopic recombination was measured by dividing colony-forming units on YEP-galactose over those on YEP-dextrose. Crossover frequency among the product in SRS2 synthetic interactors was determined 8 h after break induction as the intensity ratio of the Southern blot signal corresponding to gene conversion with crossover and that corresponding to gene conversion both with and without crossover.
We measured the frequency of crossovers among the recombination products 8 h after HO induction, the time when most cells have already completed repair (Ira et al. 2003). The crossover frequency was determined in Southern blots as a ratio of the intensity of the band corresponding to gene conversion with crossing over to the intensity of bands corresponding to gene conversions both with and without a crossover. Viability was determined as a ratio of the number of cells on YEP medium with galactose (HO break induction) to the number of cells on YEP with dextrose (no DSB). Among all tested genes only the deletion of MPH1 (tGI772) or RMI1/NCE4 (tGI853) showed a marked change in crossover frequency: mph1Δ cells had 18% crossovers (a more than threefold increase compared with wild type), while the rmi1Δ/nce4Δ deletion doubled the level of crossovers (Fig. 1B). Deletion of MPH1 did not seem to affect the kinetics of DSB repair. The efficiency of repair in mph1Δ mutant cells as measured by cell viability, although very slightly decreased, is not statistically distinguishable from wild-type cells (Figs. 1B, 2A). Similarly, rmi1Δ did not change the kinetics or efficiency of repair (Fig. 1B; data not shown). These results suggest that Mph1 and Rmi1 proteins actively regulate some key aspect of the DSB repair to favor noncrossover products. The synthetic interaction of MPH1 and SRS2 was suppressed by the deletion of the RAD51 gene (Fig. 2B), as in the case with SGS1 and SRS2 (Gangloff et al. 2000). Therefore, initiation of recombination is responsible for slow growth of mph1Δ srs2Δ mutant cells, and Mph1 probably plays an important role in later stages of recombination. Rmi1/Nce4 forms a complex with Sgs1 and Top3 (Chang et al. 2005; Mullen et al. 2005). Therefore, we tested whether the sgs1Δ and rmi1Δ/nce4Δ mutations are epistatic in crossover suppression. Indeed, rmi1Δ (tGI853) single and rmi1Δ sgs1Δ (tGI880) double mutants show the same frequency of crossovers (11%) (Supplemental Fig. S1). In conclusion, in the suppression of crossover formation, Rmi1 and Sgs1 appear to act in the same pathway, whereas, as will be demonstrated below, Mph1 functions in a novel pathway.
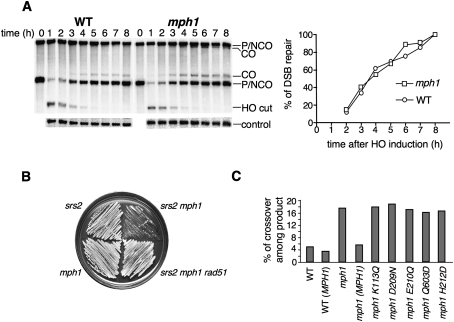
Figure 2.
MPH1 helicase channels DSB repair to noncrossovers. (A) Kinetics of DSB repair in wild type and mph1Δ mutant, determined by dividing the normalized Southern blot signals corresponding to product at different times by the signal corresponding to the maximal product at 8 h after break induction in wild-type cells. (B) Comparison of growth rate between srs2Δ and mph1Δ. Slow growth rate of double mutant srs2Δ mph1Δ is suppressed by elimination of RAD51. (C) Crossover level among products in mph1Δ and helicase point mutants is shown. The mph1Δ strain was complemented with plasmids carrying either wild-type or mutant MPH1 sequences.
Mph1 regulates crossovers in allelic and sister chromatid recombination
To ask whether the effect of mph1Δ on crossovers is specific to ectopic recombination, where homologous sequences are limited to about 2 kb, we tested the impact of Mph1 on crossovers in allelic recombination in a diploid strain where there is essentially unlimited homology on both sides of the DSB (Supplemental Fig. S1). The HO break was induced on one chromosome III, while the donor chromosome III carried a single base pair mutation within the HO recognition site. The frequency of crossovers was scored as the number of reciprocal sectors involving the distal markers THR4 and thr4ΔrURA3 (16 kb away from HO break). Wild-type cells showed 13% ± 2% crossovers, while mph1/mph1 cells exhibited 29% ± 4% crossing over; the overall repair efficiency was not changed (Supplemental Fig. S1). Therefore, the effect of Mph1 helicase on gene conversion is not limited to ectopic recombination. In the same assay, srs2Δ and sgs1Δ diploids increase the frequency of crossovers by twofold to threefold with no change in the efficiency of DSB repair (Ira et al. 2003; Lo et al. 2006).
A high level of intersister chromatid exchanges is a hallmark of Bloom syndrome cells (Chaganti et al. 1974) and of mutants, such as sgs1∆, deficient in the BLM ortholog (Onoda et al. 2004). We examined the frequency of sister chromatid exchanges in mph1Δ cells in a system where recombination between sister chromatids carrying direct truncated his3 repeats, his3-Δ5′ and his3-Δ3′, gives His+ colonies (Fasullo and Davis 1987). The results showed that mph1Δ cells (yGI012) have a 2.5-fold increase (3.0 ± 0.3 × 10−6) of unequal intersister exchanges per cell division when compared with wild-type cells (yNN301; 1.2 ± 0.5 × 10−6). In conclusion, Mph1 appears to be a general crossing-over suppressor in mitotic recombination.
Helicase activity of Mph1 is required for crossover suppression
Mph1 is a member of the DEAH family of helicases (Scheller et al. 2000). To test whether the function of Mph1 in HR depends on its helicase domains, we quantified crossovers in mph1Δ mutant cells transformed with multicopy 2-μm plasmids (pYES2) carrying either the wild-type MPH1 gene (yDS024) or five different MPH1 alleles with mutations in the conserved helicase motifs—I (K113Q), II (D209N, E210Q, and H212D), and IV (Q603D)—predicted to impair the helicase activity (strains yDS018–22). Consistent with this premise, one of these mutants, D209N, was expressed in yeast cells, purified, and shown to lack both ATPase and helicase activities (Supplemental Fig. 4). Suppression of the high-crossover phenotype was observed for the wild-type MPH1 gene only (Fig. 2C). Importantly, overexpression of MPH1 in mph1Δ or wild-type cells did not affect the efficiency of DSB repair or viability (data not shown). These results implicate the helicase activity of Mph1 in crossover suppression.
Mph1 suppresses crossovers independently of Sgs1 and Srs2
The single sgs1Δ and srs2Δ mutants elevate the frequency of crossovers from 5% to 12% and 17%, respectively. The double mutant mph1Δ sgs1Δ (tGI787), which grows well, shows 32% crossing over among the products with no decrease in DSB repair compared with wild type (Fig. 3A). Therefore, Sgs1 and Mph1 work in distinct crossover-suppressing pathways. Although the srs2Δ mph1Δ cells (yDS65) are growth-=impaired, they still grow well enough to test the frequency of crossovers. The double mutant shows 33% of crossover product demonstrating that Mph1 functions independently of Srs2 (Fig. 3A). As we showed previously (Ira et al. 2003), overexpression of Rad51 in srs2Δ (tGI548) but not in sgs1Δ (tGI542) mutants reduces the proportion of cells that can repair a DSB from 30% to 12%, and increases the frequency of crossovers among the remaining products from 17% to 28%. Overexpression of Rad51 in the mph1Δ mutant (tGI835) has no effect on DSB repair efficiency or crossover level, suggesting that the Mph1 function is distinct from that of Srs2 (Fig. 3A). Although Mph1, Sgs1, and Srs2 define three different pathways of noncrossover formation, the fact that the crossover increase in double mutants of mph1∆ with either sgs1∆ or srs2∆ is slightly more than additive could mean that these helicases also fulfill partially overlapping roles.
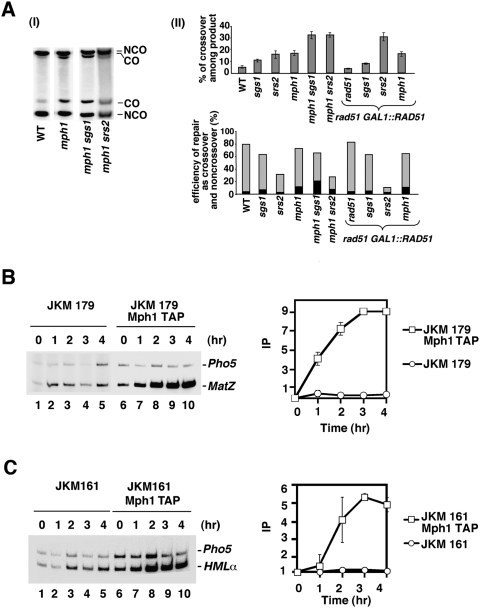
Figure 3.
Mph1 is recruited to DSBs, and works independently of Sgs1 and Srs2. (A, panel I) Southern blot analysis of gene conversion with and without crossovers in strains lacking the indicated helicases. (Panel II, top) Percentage of crossovers in cells that repaired the DSB. (Bottom) Percentage of crossovers (black) and noncrossovers (gray) among all cells with the HO cut. These percentages were determined by dividing the normalized intensities of the crossover or noncrossover band on Southern blot by the intensity of parental uncut MATa band before break induction (time, 0 h) (B) Time-course ChIP experiments showing recruitment of TAP-tagged Mph1 to nonrepairable HO break (strain JKM179) or to the HMLα donor sequence in strain (JKM 161) that can repair the break by gene conversion.
Mph1 is recruited to DSBs
We used ChIP to examine whether Mph1 is recruited to the HO break. For this purpose, we constructed TAP-tagged Mph1 strains in backgrounds that harbor or lack the HML donor sequence. The tagged strains are just as resistant to the genotoxin MMS as the untagged counterpart (data not shown), thus verifying that affinity tagging has no adverse effect on the biological activity of Mph1. At various times after HO induction, an aliquot of the yeast cultures was processed for ChIP, using IgG Sepharose beads to precipitate TAP-tagged Mph1 and associated DNA. Radioactive PCR with the appropriate primer sets was then used to amplify the target sequences: MAT Z (the invading strand) and HMLα (the recombination donor) (Wolner et al. 2003; Kwon et al. 2008). As shown in Figure 3B, Mph1 is recruited to the MAT Z region in the donorless strain (JKM179) after DSB induction. The maximal enrichment of ∼10-fold occurs 3 h after DSB induction. We also found an enhanced association of Mph1 with the HMLα donor in the switching strain (JKM161), with a fivefold enrichment of Mph1 occurring 3h after DSB induction (Fig. 3C).
Mph1 binds and unwinds D-loops
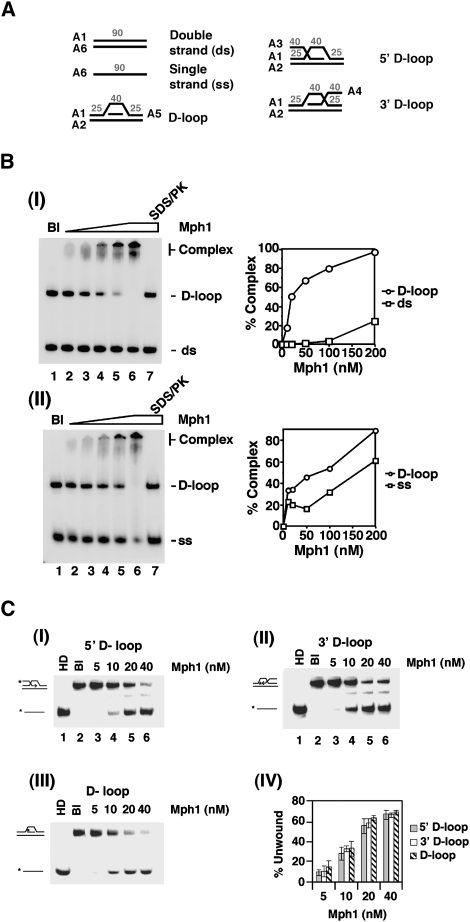
We used a DNA mobility shift assay with radiolabeled DNA substrates to investigate whether purified Mph1 protein (Prakash et al. 2005) has affinity for various DNA structures, including the D-loop and the HJ that are generated during recombination. In these experiments, we incubated Mph1 with both the D-loop substrate containing a 5′ tail and one of the remaining DNA substrates, dsDNA, ssDNA, DNA bubble, and the HJ. At low stringency (50 mM KCl), Mph1 shows a significant preference for D-loop over HJ (Supplemental Fig. 2B), ssDNA or dsDNA (Fig. 4B), but seems to have the same affinity for DNA bubble (Supplemental Fig. 2B). The specificity of Mph1 for D-loop over DNA bubble was revealed under conditions of increasing stringency (75 to 200 mM KCl) (Supplemental Fig. 3A). As shown in Supplemental Figure 3B, Mph1 apparently has the same affinity for D-loops that bear no overhang, a 3′ overhang, or a 5′ overhang.
Figure 4.
DNA binding and D-loop unwinding by Mph1. (A) DNA substrates used for DNA-binding and DNA-unwinding assays. The oligonucleotides used for constructing the substrates and the sizes of the DNA regions in the substrates are indicated and their sequences are given in Supplemental Table 2. In the central portion of the D-loop substrates, the unpaired strand bears no homology with the paired duplex region. (B) Mph1 (10–200 nM) was incubated with the 5′ D-loop substrate (50 nM) and dsDNA (50 nM) (panel I) or ssDNA (50 nM) (panel II). The results from these DNA mobility shift experiments were plotted. (C) Mph1 (5 to 40 nM) was incubated with D-loop substrates (50 nM each) that harbored a 5′ tail (panel I), a 3′ tail (panel II), or no tail (panel III). The heat-denatured substrate (HD) was run in lane 1 and the reaction blank was run in lane 2. The results from these DNA unwinding experiments were plotted in panel IV.
We next asked whether Mph1 can dissociate D-loops (with no overhang, a 3′ overhang, or a 5′ overhang), a DNA bubble, and a HJ. The results showed that Mph1 unwinds the three D-loop substrates with the same efficiency (Fig. 4C); as will be shown later, D-loop unwinding requires ATP hydrolysis by Mph1. In contrast, Mph1 is much less capable of dissociating the DNA bubble or the HJ under conditions where the majority of the three D-loop substrates are unwound (Fig. 4C; Supplemental Fig. 2C,D).
Effect of Mph1 on the Rad51-catalyzed D-loop reaction
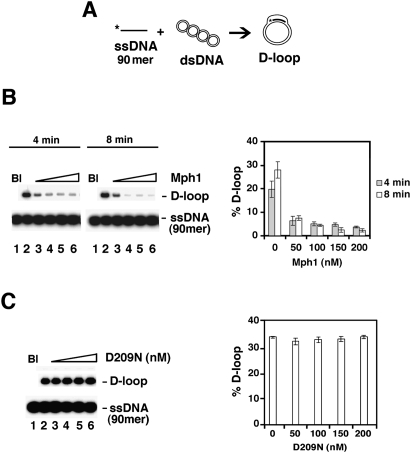
During recombination, a nucleoprotein complex of Rad51 and ssDNA invades a homologous duplex molecule to produce a D-loop intermediate (San Filippo et al. 2008). Since Mph1 binds and unwinds D-loop structures (Fig. 4), we asked whether it affects the Rad51-mediated D-loop reaction. In this reaction, a radiolabeled oligonucleotide is paired with a supercoiled target duplex by Rad51 in conjunction with its accessory factor Rad54 and the ssDNA-binding protein RPA to yield the D-loop product (Fig. 5A). We saw a significant inhibition of product formation by Mph1 after 4 min of incubation (Fig. 5B). The same results were obtained when we used ssDNA substrates that bear a region of nonhomology at the 3′ or 5′ end (see below).
Figure 5.
Effect of Mph1 on the Rad51-mediated D-loop reaction. (A) Schematic of the D-loop reaction. The ssDNA substrate was the 90-mer Oligonucleotide D1, homologous to positions 1932–2022 of pBluescript SK DNA (see Supplemental Table 2 for detailed sequence). (B) D-loop reactions without (lane 2) or with (lanes 3–6) Mph1 (50, 100, 150, and 200 nM) were analyzed after 4 or 8 min of incubation. The reaction blank (Bl) was run in lane 1. The results were plotted. (C) D-loop reactions without (lane 2) or with (lanes 3–6) mph1 D209N (50, 100, 150, and 200 nM) were analyzed after 8 min of incubation. The reaction blank (Bl) was run in lane 1. The results were plotted. The concentration of Rad51 was 0.8 μM and of Oligonucleotide D1 was 2.4 μM (nucleotides).
To ascertain the relevance of the Mph1 helicase activity in the attenuation of D-loop formation, we expressed the biologically inactive mph1 D209N mutant protein (Supplemental Fig. 4A), predicted to be deficient in ATPase and helicase activities, in yeast cells. Using the procedure that we originally devised for wild-type Mph1 (Prakash et al. 2005), the mph1 D209N protein was purified to near homogeneity (Supplemental Fig. 4B). During purification, the mph1 D209N mutant protein behaved exactly like the wild-type protein and a yield of the mutant similar to that of wild-type protein was obtained (data not shown). As expected, the mph1 D209N protein is devoid of the ability to hydrolyze ATP or to unwind DNA (Supplemental Fig. 4C,D) but proficient in DNA binding (Supplemental Fig. 5E). Importantly, the inclusion of as much as 200 nM of the mutant mph1 D209N protein has little or no effect on the Rad51-mediated D-loop reaction (Fig. 5C). Taken together, the results presented above demonstrate that Mph1 attenuates the Rad51-mediated D-loop reaction, in a manner that requires its helicase activity.
Mechanism of Mph1 action
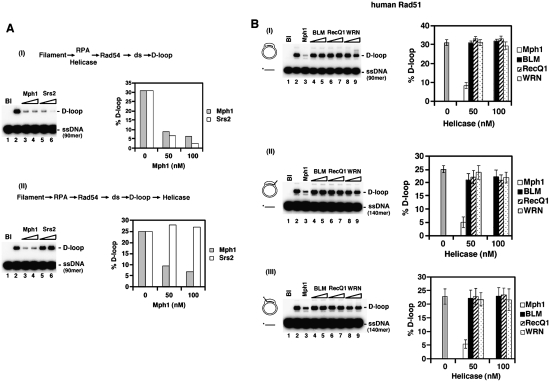
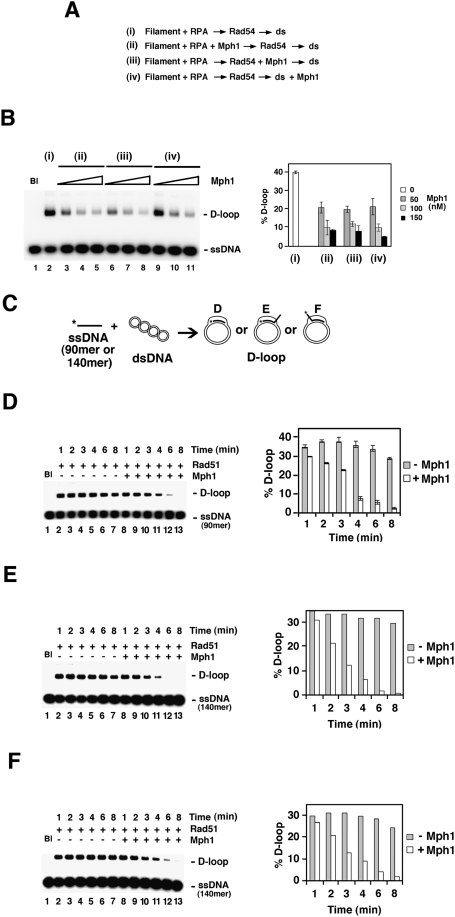
The reduction of D-loop formation in prior experiments could have arisen because of Mph1’s ability to dissociate the D-loop or an ability of Mph1 to disrupt the Rad51 presynaptic filament. A variety of biochemical and electron microscopic analyses were performed to help distinguish between these possibilities. We first carried out order of addition of experiments in which Mph1 or the Srs2 helicase was incorporated either at the beginning of the D-loop reaction or after the D-loop had already been made. Consistent with its Rad51 filament disruptive function (Krejci et al. 2003; Veaute et al. 2003), Srs2 strongly attenuates the D-loop reaction when added with Rad51 to the ssDNA but seems incapable of dissociating the D-loop product (Fig. 7A, below). In contrast, Mph1 efficiently reduces the level of D-loop product regardless of its order of addition (Figs. 6A,B, 7A). Thus, Mph1 likely acts differently than Srs2 in the attenuation of Rad51-mediated D-loop formation.
Figure 7.
Mechanism and specificity of Mph1 action. (A) In I, Mph1 or Srs2 (50 or 100 nM) was added with Rad51 at the beginning of the D-loop reaction, which, following the incorporation of pBluescript SK DNA, was incubated for 8 min. The reaction blank and the reaction without any helicase were run in lanes 1 and 2, respectively. The results were plotted. In II, Mph1 or Srs2 (50 or 100 nM) was added to the D-loop reaction 1 min after product synthesis had commenced, followed by an additional 8-min incubation. The reaction blank and the reaction without any helicase were run in lanes 1 and 2, respectively. The results were plotted. Oligonucleotide D1 was used at 2.4 μM (nucleotides) and Rad51 was at 0.8 μM, as in Figure 6. (B) Mph1, BLM, RecQ1, and WRN (50 or 100 nM) were tested for their ability to dissociate human Rad51-made D-loops that harbored no tail (I), a 3′ tail (II), or a 5′ tail (III). The D-loops were generated with Oligonucleotide D1 (2.4 μM nucleotides with 0.8 μM Rad51), D2 (3.72 μM nucleotides with 1.24 μM Rad51), or D3 (3.73 μM nucleotides with 1.24 μM Rad51) and pBluescript SK duplex DNA, as in Figure 6. The helicases were added with Rad51 at the beginning of the reaction and the incubation time was 8 min. The reaction blank and the reaction without any helicase were run in lanes 1 and 2, respectively. The results were plotted.
Figure 6.
Mph1 acts by dissociating preformed D-loops. (A) Summary of D-loop reactions that did not contain Mph1 (i) or with Mph1 (50, 100, 200 nM) added at different stages (ii–iv). The reactions used Oligonucleotide D1 as the ssDNA substrate. (B) Results from the reactions summarized in A. The reaction blank was run in lane 1. The results were plotted. (C) Schematic of D-loop reactions utilizing ssDNA substrates that gave no tail (90-mer Oligonucleotide D1), a 50-nucleotide 3′ tail (the 140-mer Oligonucleotide D2), or a 50-nucleotide 5′ tail (the 140-mer Oligonucleotide D3). All three oligonucleotides bear homology with positions 1932 to 2022 of pBluescript SK DNA and their sequence is given in Supplemental Table 2. (D–F) Time course experiments that examined formation the D-loops with no tail (D), a 3′ 50-nucleotide tail (E), or a 5′ 50-nucleotide tail (F). These reactions either contained Mph1 (150 nM; lanes 8–13) or not (lanes 2–7). In D, 2.4 μM (nucleotides) of Oligonucleotide D1 and 0.8 μM of Rad51 were used; in E and F, 3.72 μM of Oligonucleotide D2 or D3 and 1.24 μM of Rad51 were used. The reaction blank (Bl) was run in lane 1. The results were plotted.
Further details regarding Mph1's action mechanism were revealed by conducting detailed time course experiments with substrates that yield D-loops with no overhang, a 3′ overhang, or a 5′ overhang. In the absence of Mph1, the D-loop product reaches its maximal level after 1 min of incubation. The inclusion of Mph1 does not seem to prevent D-loop formation, but rather leads to a gradual disappearance of the D-loop product, such that slightly less D-loop product is seen at the earlier time points (e.g., 1 min) and little D-loop is left after 8 min (Fig. 6C–F).
The conclusion that Mph1 does not significantly affect the integrity of the Rad51–ssDNA nucleoprotein filament was validated biochemically and also by electron microscopy. In the biochemical experiment, preassembled Rad51–ssDNA nucleoprotein filaments were incubated with Mph1 or Srs2 in the presence of RPA, followed by the addition of topologically relaxed DNA to trap the displaced Rad51 molecules. The binding of Rad51 to the relaxed DNA trap induced positive DNA supercoiling, which was removed by calf thymus topoisomerase I to result in the generation of an unwound DNA species, called Form U, upon deproteinization of the reaction mixture (see Supplemental Fig. 5A for schematic). The results showed that, unlike Srs2, Mph1 does not appear to be capable of dismantling the Rad51–ssDNA nucleoprotein filament (Supplemental Fig. 5A). Finally, electron microscopy was used to quantify the dissociation of Rad51–ssDNA nucleoprotein filaments. As reported previously (Krejci et al. 2003; Veaute et al. 2003) and confirmed here, the incubation of preformed Rad51–ssDNA nucleoprotein filaments with Srs2 and RPA led to an efficient exchange of these filaments by RPA–ssDNA complexes (Supplemental Fig. 5B). In contrast, Mph1 had little or no impact on the level of Rad51–ssDNA nucleoprotein filaments (Supplemental Fig. 5B).
Overall, the results support the idea that Mph1’s regulatory action in recombination is related to its ability to dissociate the D-loop intermediate made by Rad51 protein. We propose that this Mph1 activity is important for promoting the noncrossover pathway of recombination.
Comparison of D-loop disrupting activity of Mph1 and RecQ-like helicases
The RecQ-like helicases BLM and WRN can unwind the D-loop structure (van Brabant and Holloman 2000; Orren et al. 2002). Moreover, RECQ1 and BLM have been found to dissociate D-loops made by the human Rad51 protein under specific reaction conditions (Bugreev et al. 2007, 2008). However, within the protein concentration range where Mph1 exerts a strong negative effect on the yeast Rad51-mediated D-loop reaction, none of RECQ1, BLM, and WRN noticeably influences the reaction efficiency (data not shown). As shown in Figure 7B, while Mph1 is equally adept at reducing the level of D-loop product generated by human Rad51, little or no inhibitory effect has been noted for RECQ1, BLM, or WRN. As expected, the RECQ1, BLM, and WRN proteins could unwind a test HJ substrate (Supplemental Fig. 6) and hydrolyze ATP in a ssDNA-dependent manner (data not shown).
Discussion
Mph1 is an ATP-dependent helicase with 3′–5′ polarity (Prakash et al. 2005), which was initially identified in a screen for mutants with increased spontaneous mutation rate as measured by the formation of canavanine resistant colonies (Entian et al. 1999). Based on the epistatic relationship of MPH1 and RAD51 with respect to canavanine reversion rate and MMS damage sensitivity, Schürer et al. (2004) proposed that Mph1 functions in one of several HR pathways. To understand the action mechanism of Mph1, we carried out genetic studies to unveil a major regulatory role of this helicase in DSB repair by promoting noncrossover formation.
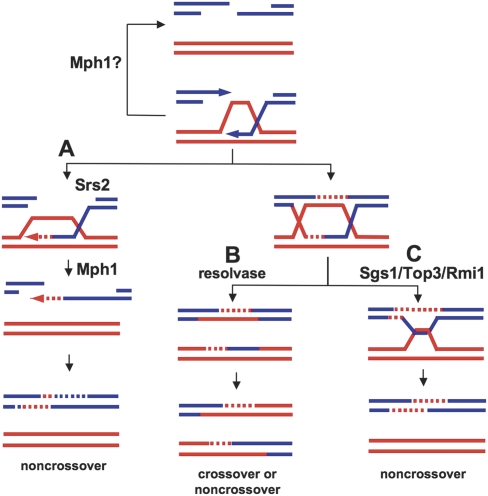
Other helicases, RecQ homologs and Srs2, also suppress crossovers associated with gene conversion. Mechanistically, the human BLM helicase works together with topoisomerase IIIα and the BLAP75/Rmi1 protein to branch migrate and decatenate dHJs into noncrossovers (Wu and Hickson 2003; Raynard et al. 2006; Wu et al. 2006). It is expected that Sgs1, the likely BLM ortholog in Saccharomyces cerevisiae, also functions in this manner to resolve dHJs into noncrossover during DSB repair. Thus, BLM and Sgs1 act late in the regulation of recombination outcome, to direct the resolution of the dHJ structure formed via second DNA end capture to yield noncrossovers (Fig. 8). On the other hand, the S. cerevisiae Srs2 helicase, which facilitates the SDSA mode of recombinational repair in lieu of the crossover pathway, appears to act early in the DSB repair reaction (Ira et al. 2003; Robert et al. 2006; Sung and Klein 2006). Purified Srs2 utilizes the free energy from ATP hydrolysis to disrupt the Rad51–ssDNA nucleoprotein filament (Krejci et al. 2003; Veaute et al. 2003), and Srs2’s function in promoting noncrossovers may be related to its ability to remove Rad51 from ssDNA. One possible scenario is that Srs2 acts early by dissociating Rad51 from the second, unpaired ssDNA tail to prevent second end capture, whereas Mph1 unwinds the D-loop structure without Rad51 removal. These Mph1 and Srs2 activities would act independently to attenuate formation of the dHJ, the likely precursor to crossovers (Fig. 8).
Figure 8.
Model depicting regulation of exchange frequency by DNA helicases during DSB-induced recombination. (A) Both Mph1 and Srs2 promote the noncrossover SDSA pathway by minimizing the possibility of creating a dHJ. Mph1 displaces the invading strand after DNA synthesis has commenced, thus preventing dHJ formation and promoting SDSA. Mph1 may also inhibit recombination if it dissociates D-loop before invading strand-initiated DNA synthesis. Srs2 possibly removes Rad51 from the unpaired 3′ DNA tail to prevent second end capture and dHJ formation. (B) When the D-loop becomes extended and more stable, dHJs are formed and can be resolved into crossovers and noncrossovers by a yet-unknown resolvase. (C) Sgs1 helicase in complex with Rmi1 and Top3 resolves these mitotic dHJs into noncrossovers.
Since Mph1 influences the outcome rather than the efficiency of recombinational repair events, it very likely acts by shunting a DNA intermediate into the SDSA pathway. As revealed in ChIP experiments, Mph1 is targeted to DSBs in cells, suggesting that its action in recombination regulation is direct. Our biochemical results provided evidence that Mph1 regulates recombination pathway choice by dissociating the invading DNA strand from the Rad51-made D-loop (Fig. 7). Consistent with this deduction, overexpression of Rad51 in mph1Δ cells affects neither the DSB repair efficiency nor crossover level. Should Mph1-mediated D-loop dissociation occur prior to the commencement of DNA synthesis primed from the invading strand (Fig. 8), then Mph1 would fulfill an anti-recombination function instead. However, since deletion of MPH1 has little effect on the repair efficiency (this study) and at most a modest effect on the frequency of spontaneous recombination (Schürer et al. 2004), the postulated anti-recombination function may constitute only a minor component of Mph1’s biological role. We note that, even though Srs2 can unwind model DNA substrates that resemble the D-loop and can transverse dsDNA coated with Rad51 (Dupaigne et al. 2008), under our experimental conditions, Mph1 appears to be much more adept at dissociating Rad51-made D-loops. Additional studies will be needed to assess the relative efficiency of the D-loop dissociative function of Mph1 and Srs2, and also to ascertain whether Mph1 is capable of removing a limited number of Rad51 molecules from ssDNA.
It is not clear whether the hypermutable phenotype of mph1 mutants (Entian et al. 1999; Schürer et al. 2004) is related to the ability of the Mph1 helicase to dissociate Rad51-made D-loops. The Sgs1 helicase plays distinct roles in DSB end resection (Gravel et al. 2008; Mimitou and Symington 2008; Zhu et al. 2008), the DNA replication checkpoint (Frei and Gasser 2000), the control of homeologous recombination (Myung et al. 1999), and in recombination regulation via DNA branch migration (Karow et al. 2000) and dHJ dissolution (Wu and Hickson 2003). Given this, it seems reasonable to consider the possibility that Mph1’s involvement in mutation avoidance is unrelated to its D-loop dissociative function.
Likely orthologs of Mph1 have been identified across many species including humans. The best characterized is archaeal Hef helicase from Pyrococcus furiosus. Hef protein, in addition to its helicase domain, also carries an endonuclease domain similar to Mus81 and Rad1/XPF nucleases (Komori et al. 2002, 2004). Recently, a human homolog of Mph1, FANCM, was identified as a part of the FA core complex 1 (Meetei et al. 2005; Mosedale et al. 2005). Initial studies suggested that FANCM protein does not have DNA unwinding activity; however, its helicase domains are required for DNA damage resistance (Xue et al. 2008).
The function of the FA pathway, comprising at least 12 proteins, remains unclear. Based on the finding that BRCA2, a key protein in recombination, and FANCJ (also known as BACH or BRIP), a DNA helicase that interacts with BRCA1, are components of the FA pathway, a role of this pathway in HR has been proposed (Howlett et al. 2002; Bridge et al. 2005; Litman et al. 2005). Whether the FA pathway plays a regulatory role in HR or constitutes a subpathway of HR is not known. Here we demonstrate a very specific role for yeast's FANCM homolog in suppressing crossovers associated with DSB-induced gene conversion. The possibility that FA proteins are involved in post-synaptic stages of recombination like Mph1 is supported by the interaction of Fanconi proteins and proteins involved in the resolution of HJs, BLM and XRCC3 (Wu and Hickson 2003; Liu et al. 2004; Pichierri et al. 2004; Hussain et al. 2006). Finally, similar to the yeast mph1Δ mutant, chicken DT40 cells deficient in FANCC, FANCD2, or FANCJ also show increased SCEs (Niedzwiedz et al. 2004; Bridge et al. 2005; Hirano et al. 2005; Yamamoto et al. 2005).
Crossovers are potentially dangerous, and occur much less frequently in mitotic than meiotic cells (Esposito 1978; Nassif et al. 1994; Johnson and Jasin 2000; Virgin et al. 2001; Stark and Jasin 2003). Specifically, crossovers may lead to loss of heterozygosity (Beumer et al. 1998) and can result in chromosomal aberrations such as translocations, inversions, or deletions. Indeed, crossovers between low-copy repeats may account for many human disorders (for review, see Shaw and Lupski 2004). Now the means by which the low proportion of exchanges is achieved becomes evident with the demonstration that three different helicases, Sgs1/Rmi1/Top3, Srs2, and Mph1, act in apparently unique and independent ways to suppress crossover formation.
Materials and methods
Yeast strains and plasmids
All yeast strain used in this study are listed in Supplemental Table 1. Yeast strains used in this study for mitotic crossover analysis are derivatives of tGI354 (Δho Δhml∷ADE1 MATa-inc Δhmr∷ADE1 ade1 leu2-3,112 lys5 trp1∷hisG ura3-52 ade3∷GAL∷HO arg5,6∷GAL∷MATa). All Mph1 overexpression plasmids were constructed in 2 μ vector pYES2, where MPH1 is under the control of the GAL1 promoter.
Analysis of crossover frequency and DSB repair kinetics
The crossover frequency among recombination products in the ectopic recombination assay was determined as a ratio of the intensity of the band corresponding to gene conversion with crossover to the intensity of bands corresponding to gene conversion both with and without crossover. We measured the frequency of crossovers among the recombination products 8 h after HO induction. DNA isolated from cells was digested with EcoRI enzyme and separated on 0.8% agarose gel. DNA was transferred to Nylon+ membrane and hybridized with a MAT a probe corresponding to 200 base pairs (bp) on each side of HO cut site. To calculate kinetics of repair we normalized the DNA amount using DNA probe specific for APA1 gene.
Unequal sister chromatid exchange
Unequal SCE was measured using a system described by Fasullo and Davis (1987). The rates of spontaneous mitotic SCE were determined by the method of median using 11 independent colonies for each rate calculation. At least three independent rate calculations were done for each strain and significance of the differences was calculated by the Mann-Whitney U-test (Zarr 1999).
Viability
The viability of mutants after HO-induced DSB breakage was calculated as a ratio of the number of cells on YEP with galactose (HO break induction) to the number of cells on YEP with dextrose (no DSB break).
ChIP assay
The donorless yeast strain used in this study was derived the strain JKM179. The strain capable of undergoing DSB-induced gene conversion was derived from JKM161 (see Supplemental Table 1). Both strains harbor the HO gene under the control of the galactose inducible GAL10 promoter. A TAP epitope was attached at the C terminus of the chromosomal MPH1 gene to facilitate Mph1 immunoprecipitation. The expression of TAP-tagged Mph1 in these strains was verified by Western blotting with polyclonal antibodies raised against purified Mph1. To perform ChIP assays, cells were grown in YP media containing 3% glycerol until they reached mid-log phase, followed by the addition of galactose (2%) to induce the HO endonuclease. At the designated times, 45-mL cell aliquots were taken, treated with formaldehyde (1%) for 20 min, and then quenched with 125 mM glycine for 5 min. Cell lysates were prepared and incubated with IgG Sepharose beads for 2 h (Amersham Biosciences). The beads were washed extensively, followed by incubation for 6 h at 65°C to reverse protein–DNA cross-links. Radioactive semiquantitative PCR was used to amplify the MAT Z or HMLα locus and the PHO5 sequence, which was included as the internal control (Wolner et al. 2003). The PCR reaction mixtures were resolved in a 6% nondenaturing polyacrylamide gel run in TBE buffer (40 mM Tris borate at pH 7.4, 0.5 mM EDTA) and quantified by phosphorimaging analysis. The MAT Z or HMLα signal at each time point was divided by the corresponding PHO5 signal and normalized to the 0-h time-point signal, as described (Wolner et al. 2003; Kwon et al. 2008).
Expression and purification of the mph1 D209N protein
The D209N mutation was introduced into Mph1 using the QuickChange Site-Directed Mutagenesis kit (Stratagene) and pYES2-Mph1 (2μ, URA3, and GAL1-MPH1) (Prakash et al. 2005) as the template. The protein expression plasmid pYES2-mph1 D209N was maintained in the protease-deficient strain BJ5464 (Supplemental Table 1), and protein expression was induced by galactose addition. The mutant mph1 protein was purified to near homogeneity as described previously for the wild-type counterpart (Prakash et al. 2005).
Purification of other proteins
Yeast Rad51 protein and BLM helicase were purified from yeast cells tailored to express them, as described (Sung and Stratton 1996; Raynard et al. 2006), and WRN helicase was purified from HighFive insect cells infected with a recombinant bacculovirus that expresses it, as described (Brosh et al. 2000; Raynard et al. 2006). Yeast Rad54 protein, Srs2 helicase, RECQ1 helicase, human Hop2–Mnd1 complex, and yeast and human RPA were purified to near homogeneity from E. coli cells tailored to express them, as described (Sigurdsson et al. 2001; Krejci et al. 2003; Cui et al. 2004; Van Komen et al. 2006; Chi et al. 2007). The BLM, RECQ1, and WRN helicase preparations have been used in several of our published studies (e.g., Raynard et al. 2006; Hu et al. 2007) and they possess a level of DNA-dependent ATPase activity as high as or higher than that reported in the literature (Brosh et al. 1999, 2000; Cui et al. 2004; data not shown) and, as expected (Brosh et al. 1999, 2000; Cui et al. 2004), are adept at unwinding a HJ test substrate (Supplemental Fig. 6).
DNA substrates
The oligonucleotides (Integrated DNA Technology) used in this study for DNA-binding and helicase assays are listed in Supplemental Table 2. To construct the DNA substrates, selected gel-purified oligonucleotides were 5′ end-labeled with [γ-32P] ATP using T4 polynucleotide kinase and then annealed to their partner oligonucleotides by heating an equimolar amount of the oligonucleotides at 95°C for 10 min in buffer H (50 mM Tris-HCl at pH 7.5, 10 mM MgCl2, 100 mM NaCl) and slow cooling to room temperature. Hybridized DNA substrates were separated from unannealed oligonucleotides in a 5% nondenaturing polyacrylamide gel run in TBE buffer and recovered from the gel by electroelution in dialysis tubing in TBE buffer at 4°C. The structure of the DNA substrates used in this study can be found in individual figures and their legends, and the sequence of these oligonucleotide is provided in Supplemental Table 2.
DNA helicase assay
The indicated concentration of Mph1 or mph1 D209N protein was incubated at 30°C with the radiolabeled DNA substrate (50 nM) in 15 μL of buffer R (30 mM Tris-HCl at pH 7.5, 1 mM DTT, 100 μg/mL BSA) containing 2.5 mM MgCl2, 2 mM ATP, and 50 mM KCl. The reaction was stopped after 10 min by treatment with SDS (0.2% final) and proteinase K (0.5 mg/mL) for 2 min at 30°C. The reaction mixtures were resolved in a 10% polyacrylamide gel in TAE buffer at 4°C. Gels were dried onto Whatman DE81 paper (Whatman International Limited) and then analyzed in a Personal Molecular Imager FX PhosphorImager (Bio-Rad).
DNA mobility shift assay
The indicated concentration of Mph1 or mph1 D209N was incubated with the radiolabeled DNA substrate (50 nM) at 30°C in 10 μL of buffer R containing 50 mM KCl for 5 min. After the addition of 2 μL of gel loading buffer (50% glycerol, 20 mM Tris-HCl at pH 7.4, 2 mM EDTA, 0.05% orange G), the reaction mixtures were resolved in a 5% polyacrylamide gel in TAE buffer. Gels were dried and subjected to phosphorimaging analysis as above. Where indicated, reaction mixtures were treated with 0.5% SDS and 0.5 mg/mL proteinase K (SDS/PK) at 37°C for 3 min before electrophoresis.
D-loop reaction mediated by yeast or human Rad51 protein
The reaction (final volume of 12.5 μL) was performed in Buffer R containing 2.5 mM MgCl2, 2.0 mM ATP, 50 mM KCl, and an ATP-regenerating system consisting of 20 mM creatine phosphate and 20 μg/mL creatine kinase. The indicated radiolabeled oligonucleotide (2.4 μM nucleotides D1 or 3.72 μM nucleotides D2 or D3) was incubated with yeast or human Rad51 (0.8 μM for Oligonucleotide D1 or 1.24 μM for Oligonucleotide D2 or D3) for 5 min at 37°C to assemble the Rad51–ssDNA nucleoprotein filament, followed by the incorporation of yeast Rad54 (150 nM, for yeast Rad51) or the human Hop2–Mnd1 complex (300 nM, for human Rad51) and yeast RPA (200 nM, for yeast Rad51) or human RPA (200 nM, for human Rad51) and a 2-min incubation at 23°C. The D-loop reaction was initiated by the addition of pBluescript replicative form I DNA (37 μM base pairs). The reaction mixtures were incubated at 30°C for the indicated times, deproteinized by treatment with SDS (0.5%) and proteinase K (0.5 mg/mL) at 37°C for 3 min, and then resolved in a 0.9% agarose gel in TAE buffer. Gels were dried onto Whatman DE81 paper and then analyzed in the phosphorimager. To test the effect of the DNA helicases on the D-loop reaction, they were incorporated into the reaction as indicated in each case.
Acknowledgments
We thank Ketan Patel, Wojciech Niedzwiedz, and Susan Rosenberg for sharing unpublished data; Steven Brill for strains; Marc Pypaert for guidance in the EM work; and Quiqin Wu for technical assistance. This project was supported by NIH grants GM80600, GM20056, ES07061, GM57814, and GM53738; Wellcome Trust grant GR076476; EMBO/HHMI start-up program ME888; and grant Kr 914/6-1 of the Deutsche Forschungsgemeinschaft.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1737809.
Supplemental material is available at http://www.genesdev.org.
References
- Allers T., Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- Barbera M.A., Petes T.D. Selection and analysis of spontaneous reciprocal mitotic cross-overs in Saccharomyces cerevisiae . Proc. Natl. Acad. Sci. 2006;103:12819–12824. doi: 10.1073/pnas.0605778103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer K.J., Pimpinelli S., Golic K.G. Induced chromosomal exchange directs the segregation of recombinant chromatids in mitosis of Drosophila . Genetics. 1998;150:173–188. doi: 10.1093/genetics/150.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge W.L., Vandenberg C.J., Franklin R.J., Hiom K. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat. Genet. 2005;37:953–957. doi: 10.1038/ng1627. [DOI] [PubMed] [Google Scholar]
- Brosh R.M., Jr, Orren D.K., Nehlin J.O., Ravn P.H., Kenny M.K., Machwe A., Bohr V.A. Functional and physical interaction between WRN helicase and human replication protein A. J. Biol. Chem. 1999;274:18341–18350. doi: 10.1074/jbc.274.26.18341. [DOI] [PubMed] [Google Scholar]
- Brosh R.M., Jr, Li J.L., Kenny M.K., Karow J.K., Cooper M.P., Kureekattil R.P., Hickson I.D., Bohr V.A. Replication protein A physically interacts with the Bloom's syndrome protein and stimulates its helicase activity. J. Biol. Chem. 2000;275:23500–23508. doi: 10.1074/jbc.M001557200. [DOI] [PubMed] [Google Scholar]
- Bugreev D.V., Mazina O.M., Mazin A.V. Rad54 protein promotes branch migration of Holliday junctions. Nature. 2006;442:590–593. doi: 10.1038/nature04889. [DOI] [PubMed] [Google Scholar]
- Bugreev D.V., Yu X., Egelman E.H., Mazin A.V. Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes & Dev. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev D.V., Brosh R.M., Jr, Mazin A.V. RECQ1 possesses DNA branch migration activity. J. Biol. Chem. 2008;283:20231–20242. doi: 10.1074/jbc.M801582200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaganti R.S., Schonberg S., German J. A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc. Natl. Acad. Sci. 1974;71:4508–4512. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M., Bellaoui M., Zhang C., Desai R., Morozov P., Delgado-Cruzata L., Rothstein R., Freyer G.A., Boone C., Brown G.W. RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. EMBO J. 2005;24:2024–2033. doi: 10.1038/sj.emboj.7600684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P., San Filippo J., Sehorn M.G., Petukhova G.V., Sung P. Bipartite stimulatory action of the Hop2-Mnd1 complex on the Rad51 recombinase. Genes & Dev. 2007;21:1747–1757. doi: 10.1101/gad.1563007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S., Arosio D., Doherty K.M., Brosh R.M., Jr, Falaschi A., Vindigni A. Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic Acids Res. 2004;32:2158–2170. doi: 10.1093/nar/gkh540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupaigne P., Le Breton C., Fabre F., Gangloff S., Le Cam E., Veaute X. The Srs2 helicase activity is stimulated by Rad51 filaments on dsDNA: Implications for crossover incidence during mitotic recombination. Mol. Cell. 2008;29:243–254. doi: 10.1016/j.molcel.2007.11.033. [DOI] [PubMed] [Google Scholar]
- Entian K.D., Schuster T., Hegemann J.H., Becher D., Feldmann H., Guldener U., Gotz R., Hansen M., Hollenberg C.P., Jansen G., et al. Functional analysis of 150 deletion mutants in Saccharomyces cerevisiae by a systematic approach. Mol. Gen. Genet. 1999;262:683–702. doi: 10.1007/pl00013817. [DOI] [PubMed] [Google Scholar]
- Esposito M.S. Evidence that spontaneous mitotic recombination occurs at the two-strand stage. Proc. Natl. Acad. Sci. 1978;75:4436–4440. doi: 10.1073/pnas.75.9.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre F., Chan A., Heyer W.D., Gangloff S. Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. 2002;99:16887–16892. doi: 10.1073/pnas.252652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasullo M.T., Davis R.W. Recombinational substrates designed to study recombination between unique and repetitive sequences in vivo. Proc. Natl. Acad. Sci. 1987;84:6215–6219. doi: 10.1073/pnas.84.17.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei C., Gasser S.M. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes & Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- Gangloff S., Soustelle C., Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- Gravel S., Chapman J.R., Magill C., Jackson S.P. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes & Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillon H., Baudat F., Grey C., Liskay R.M., de Massy B. Crossover and noncrossover pathways in mouse meiosis. Mol. Cell. 2005;20:563–573. doi: 10.1016/j.molcel.2005.09.021. [DOI] [PubMed] [Google Scholar]
- Hirano S., Yamamoto K., Ishiai M., Yamazoe M., Seki M., Matsushita N., Ohzeki M., Yamashita Y.M., Arakawa H., Buerstedde J.M., et al. Functional relationships of FANCC to homologous recombination, translesion synthesis, and BLM. EMBO J. 2005;24:418–427. doi: 10.1038/sj.emboj.7600534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett N.G., Taniguchi T., Olson S., Cox B., Waisfisz Q., De Die-Smulders C., Persky N., Grompe M., Joenje H., Pals G., et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- Hu Y., Lu X., Barnes E., Yan M., Lou H., Luo G. Recql5 and Blm RecQ DNA helicases have nonredundant roles in suppressing crossovers. Mol. Cell. Biol. 2005;25:3431–3442. doi: 10.1128/MCB.25.9.3431-3442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Raynard S., Sehorn M.G., Lu X., Bussen W., Zheng L., Stark J.M., Barnes E.L., Chi P., Janscak P., et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes & Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N., Kleckner N. The single-end invasion: An asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- Hussain S., Wilson J.B., Blom E., Thompson L.H., Sung P., Gordon S.M., Kupfer G.M., Joenje H., Mathew C.G., Jones N.J. Tetratricopeptide-motif-mediated interaction of FANCG with recombination proteins XRCC3 and BRCA2. DNA Repair (Amst.) 2006;5:629–640. doi: 10.1016/j.dnarep.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Ira G., Malkova A., Liberi G., Foiani M., Haber J.E. Srs2 and Sgs1–Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G., Satory D., Haber J.E. Conservative inheritance of newly synthesized DNA in double-strand break-induced gene conversion. Mol. Cell. Biol. 2006;26:9424–9429. doi: 10.1128/MCB.01654-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L., Lichten M. Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol. Cell. 2008;31:313–323. doi: 10.1016/j.molcel.2008.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L., Rockmill B., Roeder G.S., Lichten M. Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of sgs1. PLoS Genet. 2006;2:e155. doi: 10.1371/journal.pgen.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.D., Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow J.K., Constantinou A., Li J.L., West S.C., Hickson I.D. The Bloom's syndrome gene product promotes branch migration of Holliday junctions. Proc. Natl. Acad. Sci. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori K., Fujikane R., Shinagawa H., Ishino Y. Novel endonuclease in Archaea cleaving DNA with various branched structure. Genes Genet. Syst. 2002;77:227–241. doi: 10.1266/ggs.77.227. [DOI] [PubMed] [Google Scholar]
- Komori K., Hidaka M., Horiuchi T., Fujikane R., Shinagawa H., Ishino Y. Cooperation of the N-terminal Helicase and C-terminal endonuclease activities of Archaeal Hef protein in processing stalled replication forks. J. Biol. Chem. 2004;279:53175–53185. doi: 10.1074/jbc.M409243200. [DOI] [PubMed] [Google Scholar]
- Krejci L., Van Komen S., Li Y., Villemain J., Reddy M.S., Klein H., Ellenberger T., Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- Krogh B.O., Symington L.S. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Kwon Y., Seong C., Chi P., Greene E.C., Klein H., Sung P. ATP-dependent chromatin remodeling by the Saccharomyces cerevisiae homologous recombination factor Rdh54. J. Biol. Chem. 2008;283:10445–10452. doi: 10.1074/jbc.M800082200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litman R., Peng M., Jin Z., Zhang F., Zhang J., Powell S., Andreassen P.R., Cantor S.B. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. 2005;8:255–265. doi: 10.1016/j.ccr.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Liu Y., Masson J.Y., Shah R., O'Regan P., West S.C. RAD51C is required for Holliday junction processing in mammalian cells. Science. 2004;303:243–246. doi: 10.1126/science.1093037. [DOI] [PubMed] [Google Scholar]
- Lo Y.C., Paffett K.S., Amit O., Clikeman J.A., Sterk R., Brenneman M.A., Nickoloff J.A. Sgs1 regulates gene conversion tract lengths and crossovers independently of its helicase activity. Mol. Cell. Biol. 2006;26:4086–4094. doi: 10.1128/MCB.00136-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magner D.B., Blankschien M.D., Lee J.A., Pennington J.M., Lupski J.R., Rosenberg S.M. RecQ promotes toxic recombination in cells lacking recombination intermediate-removal proteins. Mol. Cell. 2007;26:273–286. doi: 10.1016/j.molcel.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahill M.S., Sham C.W., Bishop D.K. Synthesis-dependent strand annealing in meiosis. PLoS Biol. 2007;5:e299. doi: 10.1371/journal.pbio.0050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meetei A.R., Medhurst A.L., Ling C., Xue Y., Singh T.R., Bier P., Steltenpool J., Stone S., Dokal I., Mathew C.G., et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 2005;37:958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou E.P., Symington L.S. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosedale G., Niedzwiedz W., Alpi A., Perrina F., Pereira-Leal J.B., Johnson M., Langevin F., Pace P., Patel K.J. The vertebrate Hef ortholog is a component of the Fanconi anemia tumor-suppressor pathway. Nat. Struct. Mol. Biol. 2005;12:763–771. doi: 10.1038/nsmb981. [DOI] [PubMed] [Google Scholar]
- Mullen J.R., Nallaseth F.S., Lan Y.Q., Slagle C.E., Brill S.J. Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1–Top3 complex. Mol. Cell. Biol. 2005;25:4476–4487. doi: 10.1128/MCB.25.11.4476-4487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K., Datta A., Chen C., Kolodner R.D. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat. Genet. 2001;27:113–116. doi: 10.1038/83673. [DOI] [PubMed] [Google Scholar]
- Nassif N., Penney J., Pal S., Engels W.R., Gloor G.B. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 1994;14:1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiedz W., Mosedale G., Johnson M., Ong C.Y., Pace P., Patel K.J. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol. Cell. 2004;15:607–620. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Oh S.D., Lao J.P., Hwang P.Y., Taylor A.F., Smith G.R., Hunter N. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007;130:259–272. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.D., Lao J.P., Taylor A.F., Smith G.R., Hunter N. RecQ helicase, Sgs1, and XPF family endonuclease, Mus81–Mms4, resolve aberrant joint molecules during meiotic recombination. Mol. Cell. 2008;31:324–336. doi: 10.1016/j.molcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onoda F., Seki M., Wang W., Enomoto T. The hyper unequal sister chromatid recombination in an sgs1 mutant of budding yeast requires MSH2. DNA Repair (Amst.) 2004;3:1355–1362. doi: 10.1016/j.dnarep.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Orren D.K., Theodore S., Machwe A. The Werner syndrome helicase/exonuclease (WRN) disrupts and degrades D-loops in vitro. Biochemistry. 2002;41:13483–13488. doi: 10.1021/bi0266986. [DOI] [PubMed] [Google Scholar]
- Palladino F., Klein H.L. Analysis of mitotic and meiotic defects in Saccharomyces cerevisiae SRS2 DNA helicase mutants. Genetics. 1992;132:23–37. doi: 10.1093/genetics/132.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F., Haber J.E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae . Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichierri P., Franchitto A., Rosselli F. BLM and the FANC proteins collaborate in a common pathway in response to stalled replication forks. EMBO J. 2004;23:3154–3163. doi: 10.1038/sj.emboj.7600277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R., Krejci L., Van Komen S., Anke Schürer K., Kramer W., Sung P. Saccharomyces cerevisiae MPH1 gene, required for homologous recombination-mediated mutation avoidance, encodes a 3′ to 5′ DNA helicase. J. Biol. Chem. 2005;280:7854–7860. doi: 10.1074/jbc.M413898200. [DOI] [PubMed] [Google Scholar]
- Raynard S., Bussen W., Sung P. A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIα, and BLAP75. J. Biol. Chem. 2006;281:13861–13864. doi: 10.1074/jbc.C600051200. [DOI] [PubMed] [Google Scholar]
- Richardson C., Moynahan M.E., Jasin M. Double-strand break repair by interchromosomal recombination: Suppression of chromosomal translocations. Genes & Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert T., Dervins D., Fabre F., Gangloff S. Mrc1 and Srs2 are major actors in the regulation of spontaneous crossover. EMBO J. 2006;25:2837–2846. doi: 10.1038/sj.emboj.7601158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder G.S. Sex and the single cell: Meiosis in yeast. Proc. Natl. Acad. Sci. 1995;92:10450–10456. doi: 10.1073/pnas.92.23.10450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J., Sung P., Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Scheller J., Schürer A., Rudolph C., Hettwer S., Kramer W. MPH1, a yeast gene encoding a DEAH protein, plays a role in protection of the genome from spontaneous and chemically induced damage. Genetics. 2000;155:1069–1081. doi: 10.1093/genetics/155.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürer K.A., Rudolph C., Ulrich H.D., Kramer W. Yeast MPH1 gene functions in an error-free DNA damage bypass pathway that requires genes from homologous recombination, but not from postreplicative repair. Genetics. 2004;166:1673–1686. doi: 10.1534/genetics.166.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw C.J., Lupski J.R. Implications of human genome architecture for rearrangement-based disorders: The genomic basis of disease. Hum. Mol. Genet. 2004;13:R57–R64. doi: 10.1093/hmg/ddh073. [DOI] [PubMed] [Google Scholar]
- Sigurdsson S., Trujillo K., Song B., Stratton S., Sung P. Basis for avid homologous DNA strand exchange by human Rad51 and RPA. J. Biol. Chem. 2001;276:8798–8806. doi: 10.1074/jbc.M010011200. [DOI] [PubMed] [Google Scholar]
- Solinger J.A., Heyer W.D. Rad54 protein stimulates the postsynaptic phase of Rad51 protein-mediated DNA strand exchange. Proc. Natl. Acad. Sci. 2001;98:8447–8453. doi: 10.1073/pnas.121009898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinger J.A., Kiianitsa K., Heyer W.D. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol. Cell. 2002;10:1175–1188. doi: 10.1016/s1097-2765(02)00743-8. [DOI] [PubMed] [Google Scholar]
- Stark J.M., Jasin M. Extensive loss of heterozygosity is suppressed during homologous repair of chromosomal breaks. Mol. Cell. Biol. 2003;23:733–743. doi: 10.1128/MCB.23.2.733-743.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart E., Chapman C.R., Al-Khodairy F., Carr A.M., Enoch T. rqh1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S phase arrest. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N., Wang X., Haber J.E. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell. 2003;12:209–219. doi: 10.1016/s1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- Sung P., Klein H. Mechanism of homologous recombination: Mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- Sung P., Stratton S.A. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J. Biol. Chem. 1996;271:27983–27986. doi: 10.1074/jbc.271.45.27983. [DOI] [PubMed] [Google Scholar]
- Szostak J.W., Orr-Weaver T.L., Rothstein R.J., Stahl F.W. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Tong A.H., Lesage G., Bader G.D., Ding H., Xu H., Xin X., Young J., Berriz G.F., Brost R.L., Chang M., et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- van Brabant A.J., Ye T., Sanz M., German J.L., III, Ellis N.A., Holloman W.K. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry. 2000;39:14617–14625. doi: 10.1021/bi0018640. [DOI] [PubMed] [Google Scholar]
- Van Komen S., Macris M., Sehorn M.G., Sung P. Purification and assays of Saccharomyces cerevisiae homologous recombination proteins. Methods Enzymol. 2006;408:445–463. doi: 10.1016/S0076-6879(06)08028-1. [DOI] [PubMed] [Google Scholar]
- Veaute X., Jeusset J., Soustelle C., Kowalczykowski S.C., Le Cam E., Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- Virgin J.B., Bailey J.P., Hasteh F., Neville J., Cole A., Tromp G. Crossing over is rarely associated with mitotic intragenic recombination in Schizosaccharomyces pombe . Genetics. 2001;157:63–77. doi: 10.1093/genetics/157.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Seki M., Narita Y., Nakagawa T., Yoshimura A., Otsuki M., Kawabe Y., Tada S., Yagi H., Ishii Y., et al. Functional relation among RecQ family helicases RecQL1, RecQL5, and BLM in cell growth and sister chromatid exchange formation. Mol. Cell. Biol. 2003;23:3527–3535. doi: 10.1128/MCB.23.10.3527-3535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolner B., van Komen S., Sung P., Peterson C.L. Recruitment of the recombinational repair machinery to a DNA double-strand break in yeast. Mol. Cell. 2003;12:221–232. doi: 10.1016/s1097-2765(03)00242-9. [DOI] [PubMed] [Google Scholar]
- Wu L., Hickson I.D. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Wu L., Bachrati C.Z., Ou J., Xu C., Yin J., Chang M., Wang W., Li L., Brown G.W., Hickson I.D. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc. Natl. Acad. Sci. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Li Y., Guo R., Ling C., Wang W. FANCM of the Fanconi anemia core complex is required for both monoubiquitination and DNA repair. Hum. Mol. Genet. 2008;17:1641–1652. doi: 10.1093/hmg/ddn054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K., Hirano S., Ishiai M., Morishima K., Kitao H., Namikoshi K., Kimura M., Matsushita N., Arakawa H., Buerstedde J.M., et al. Fanconi anemia protein FANCD2 promotes immunoglobulin gene conversion and DNA repair through a mechanism related to homologous recombination. Mol. Cell. Biol. 2005;25:34–43. doi: 10.1128/MCB.25.1.34-43.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarr J.H. Biostatistical analysis. Prentice Hall, Inc; Englewood Cliffs, NJ: 1999. [Google Scholar]
- Zhu Z., Chung W.H., Shim E.Y., Lee S.E., Ira G. Sgs1 helicase and two nucleases dna2 and exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]