Abstract
When the supply of environmental nutrients is limited, multicellular animals can make both physiological and behavioral changes so as to cope with nutrient starvation. Although physiological and behavioral effects of starvation are well known, the mechanisms by which animals sense starvation systemically remain elusive. Furthermore, what constituent of food is sensed and how it modulates starvation response is still poorly understood. In this study, we use a starvation-hypersensitive mutant to identify molecules and mechanisms that modulate starvation signaling. We found that specific amino acids could suppress the starvation-induced death of gpb-2 mutants, and that MGL-1 and MGL-2, Caenorhabditis elegans homologs of metabotropic glutamate receptors, were involved. MGL-1 and MGL-2 acted in AIY and AIB neurons, respectively. Treatment with leucine suppressed starvation-induced stress resistance and life span extension in wild-type worms, and mutation of mgl-1 and mgl-2 abolished these effects of leucine. Taken together, our results suggest that metabotropic glutamate receptor homologs in AIY and AIB neuron may modulate a systemic starvation response, and that C. elegans senses specific amino acids as an anti-hunger signal.
Keywords: Starvation, amino acid response, autophagy, hormesis
During nutritional deprivation, individual cells can respond to starvation by modulating intracellular signaling, which in turn induces a starvation response and thereby enhances their survival (Levine and Yuan 2005; Lum et al. 2005; Levine and Kroemer 2008). An important starvation response of individual cells is a change of metabolism (inhibiting anabolic pathways and activating catabolic pathways), so as to generate metabolic substrates to maintain basal cellular activities (Lum et al. 2005). In multicellular organisms, however, the situation is complicated by the need to induce behavioral changes in addition to physiological changes, and more importantly by the danger that uncoordinated starvation responses in individual cells could be harmful to the organism. In Caenorhabditis elegans, for example, we reported previously that excessive autophagy in pharyngeal muscle causes its malfunction, which eventually prevents the recovery of worms from starvation (Kang et al. 2007). Thus, it is important that multicellular organisms ensure their starvation response is coordinated between individual cells, and therefore it is plausible to assume that there are mechanisms by which animals sense starvation systemically.
Since animals cannot synthesize several amino acids, so-called “essential acids,” they must ingest these amino acids from external food sources to maintain homeostasis (Gietzen and Rogers 2006). C. elegans also requires certain amino acids in the diet (Szewczyk et al. 2003). This fact leads to the intriguing possibility that amino acids act as a food or anti-hunger signal. Indeed, treatment with amino acids can inhibit starvation-induced autophagy in cultured mammalian cells (Codogno and Meijer 2005). Furthermore, at the organismal level, intracerebroventricular administration of leucine causes a decrease in food intake in the rat (Cota et al. 2006), suggesting that the possibility could be true. But it is not known whether amino acids modulate a starvation response in multicellular organisms.
Results and Discussion
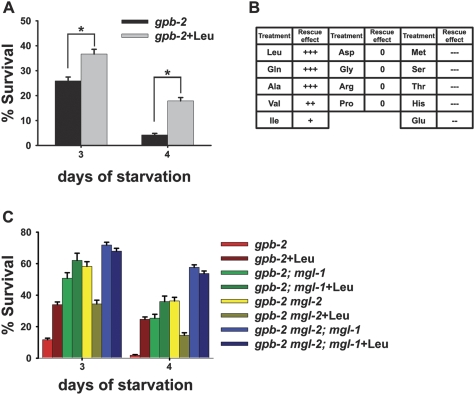
To examine the effect of amino acids on starvation signaling in Caenorhabditis elegans, we screened several amino acids for suppression of the death of the starvation-hypersensitive gpb-2 mutant. We showed previously that starvation signaling is overactivated in this background (You et al. 2006; Kang et al. 2007; Kang and Avery 2008). We found that treatment with a subset of amino acids including leucine, glutamine, alanine, valine, and isoleucine can rescue the death of gpb-2 mutants during starvation, whereas other amino acids did not rescue or worsened survival (Fig. 1A,B; Supplemental Fig. 1). Given the specificity of the rescue effect, it is unlikely that amino acids act solely as a carbon source to maintain nutrient homeostasis; rather, it suggests that amino acids might act as signaling molecules that modulate a starvation response.
Figure 1.
A subset of amino acids rescues the starvation hypersensitivity of gpb-2 mutants, and mgl-1 and mgl-2 are involved in the process. Starvation survival analyses were performed as described in the Materials and Methods. (A) Treatment with leucine partially rescues the starvation hypersensitivity of gpb-2 mutants. Percent of worms surviving to adulthood on NGM plates with HB101 bacteria after incubation in M9 buffer in the absence of food with or without leucine for the indicated time. Error bars are standard errors estimated assuming a Poisson distribution, and similar results were obtained in three independent experiments. (*) P < 0.001 (χ2 test of independence). (B) Summary of the rescue effect of amino acids on the survival of gpb-2 mutants during starvation. (+) Positive effects on survival; (0) no effects on survival; (−) negative effects on survival. For details, see Supplemental Figure 1. (C) Mutation of either mgl-1 or mgl-2 can rescue death of gpb-2 mutants during starvation, and double mutation causes an enhanced rescue effect. Percent of worms surviving to adulthood on NGM plates with HB101 bacteria after incubation in M9 buffer in the absence of food with or without leucine for the indicated time. Error bars are standard errors estimated assuming a Poisson distribution, and similar results were obtained in three independent experiments.
Since the amino acids observed to have a rescue effect do not fall into specific subgroups (essential amino acids/nonessential amino acids or polar/nonpolar), we reasoned that there must be a mechanism(s) to sense a broad spectrum of amino acids. It has recently been shown that a broad spectrum of amino acids can be sensed by class 3 G-protein-coupled receptors, which include the extracellular calcium sensing receptor, heterodimeric taste receptors and GPRC6A (Conigrave and Hampson 2006). MGL-1 and MGL-2 metabotropic G-protein-coupled glutamate receptors are the closest homologs of CaR in the C. elegans genome (Dillon et al. 2006). Thus, we hypothesized that MGL-1 and MGL-2 were involved in the modulation of amino acid response and starvation response. To test this, we made gpb-2; mgl-1, gpb-2 mgl-2 and gpb-2 mgl-2; mgl-1 mutants. Mutation of either mgl-1 or mgl-2 partially rescued death of gpb-2 mutants during starvation, and double mutation caused an additive effect, suggesting that mgl-1 and mgl-2 modulate a starvation response in a parallel manner (Fig. 1C). Mutation of either mgl-1 or mgl-2 did not abolish the response to leucine, while mutations of both mgl-1 and mgl-2 made gpb-2 mutants resistant to leucine treatment, suggesting that both mgl-1 and mgl-2 are involved. Until 5 d of starvation, there is no decrease in the survival of wild-type or mgl-1, mgl-2, and mgl-2; mgl-1 mutants, suggesting that the resistance to leucine treatment observed in gpb-2 mgl-2; mgl-1 mutants does not result from full rescue of gpb-2 mutants by mutations of mgl-1 and mgl-2 (Kang et al. 2007; data not shown). We observed similar results with alanine and glutamine (Supplemental Fig. 2A).
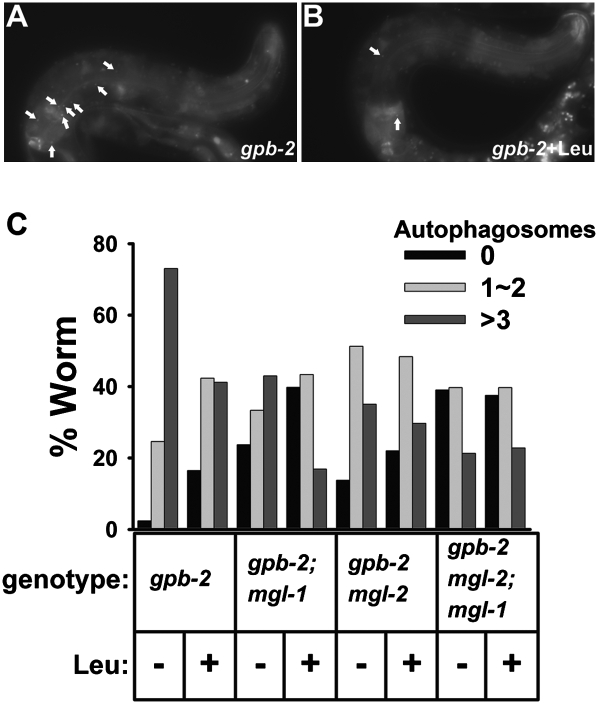
One cause of death in gpb-2 mutants during starvation is excessive autophagy in pharyngeal muscle, which causes malfunction of the pharynx, the C. elegans feeding organ (Kang et al. 2007). To test whether mgl-1 and mgl-2 are involved in the modulation of autophagy in pharyngeal muscle of gpb-2 mutants during starvation, we generated mutant strains carrying an integrated transgene that expressed a GFP-tagged version of LGG-1, a specific marker for autophagy (Melendez et al. 2003; Kang et al. 2007; Klionsky et al. 2008). We found that mutation of either mgl-1 or mgl-2 decreased the excessive levels of autophagy in pharyngeal muscle, and double mutation showed an additive effect on decreasing autophagy, suggesting that mgl-1 and mgl-2 can modulate autophagy in a parallel manner (Fig. 2). Treatment with leucine also decreased excessive levels of autophagy during starvation, consistent with the hypothesis that amino acids act as anti-hunger signals. However, leucine had no effect on levels of autophagy when mgl-1 and mgl-2 were both mutated, suggesting that they are necessary for the modulation of autophagy by amino acids. We also observed similar results with alanine and glutamine (Supplemental Fig. 2C). Interestingly, treatment with glutamate slightly increased levels of autophagy in gpb-2 mutants, yet glutamate had no effect on levels of autophagy in gpb-2 mgl-2; mgl-1 mutants, consistent with survival data (Supplemental Fig. 2B).
Figure 2.
mgl-1 and mgl-2 modulate autophagy in the pharyngeal muscles of gpb-2 mutants during starvation. (A) Representative images of gpb-2 mutants with or without leucine after 3 d of starvation. The arrows show representative GFP∷LGG-1-positive punctate structures that label preautophagosomal and autophagosomal structures. (B) Quantification of autophagy in the pharyngeal muscles of worms of the indicated genotype after 3 d of starvation with or without leucine. Data come from three independent experiments (n = 80–141).
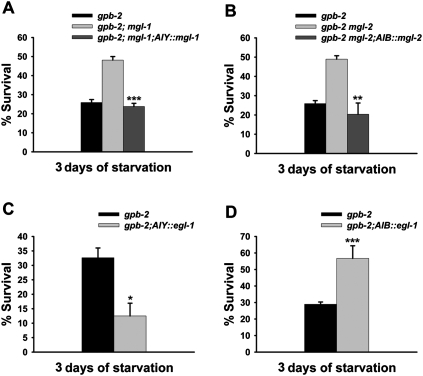
It has been reported that mgl-1 and mgl-2 are mainly expressed in a limited number of neurons (http://www.wormbase.org). These expression patterns, together with the results showing that mgl-1 and mgl-2 can modulate levels of autophagy in pharyngeal muscles (Fig. 2), lead to the intriguing hypothesis that mgl-1 and mgl-2 can modulate a starvation response in a cell-nonautonomous manner. To test this, we examined whether expression of mgl-1 or mgl-2 in specific neurons could restore the starvation-sensitivity of gpb-2; mgl-1 and gpb-2 mgl-2 mutants. Among the neurons where mgl-1 and mgl-2 are expressed, AIY and AIB are particularly interesting because it has been reported recently that AIY and AIB neurons control food- and odor-evoked behaviors (Chalasani et al. 2007). We found that AIY-specific expression of mgl-1 and AIB-specific expression of mgl-2 could restore the starvation-sensitivity of gpb-2; mgl-1 and gpb-2 mgl-2 mutants, respectively (Fig. 3A,B). In contrast, expressing either mgl-1 or mgl-2 in a subset of MGL-1- or MGL-2-expressing neurons using either the glr-4 promoter (to express in RMD, RIB, and 17 additional neurons) or nmr-2 promoter (to express in AVE and five additional neurons) did not restore the starvation-sensitivity (Supplemental Fig. 3A–C). These data support our hypothesis that mgl-1 and mgl-2 mainly act in AIY and AIB neurons, respectively, to modulate a starvation response.
Figure 3.
mgl-1 and mgl-2 mainly act in AIY and AIB neurons, respectively. (A–D) Percent of worms surviving to adulthood on NGM plates with HB101 bacteria after incubation in M9 buffer in the absence of food for the indicated time. Error bars are standard errors estimated assuming a Poisson distribution, and similar results were obtained in three independent experiments. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001 (χ2 test of independence). (A,B) AIY-specific expression of mgl-1 and AIB-specific expression of mgl-2 restore the starvation-sensitivity of gpb-2; mgl-1, and gpb-2 mgl-2, respectively. Data for gpb-2 single mutants in A and B are from Figure 1A. Similar results were obtained in two independent transgenic lines. (C,D) AIY-specific expression of egl-1 exacerbates death of gpb-2 mutants during starvation, whereas AIB-specific expression of egl-1 rescues it.
Based on homology, mgl-1 encodes a group II glutamate receptor, expected to inhibit adenylyl cyclase activity, which is likely to decrease neuronal activity, whereas mgl-2 encodes a group I glutamate receptor, expected to stimulate phospholipase C and likely increase neuronal activity (Dillon et al. 2006). To test whether mgl-1 and mgl-2 modulate starvation response by regulating the activity of AIY and AIB neurons, respectively, we killed AIY and AIB neurons by AIY- and AIB-specific expression of egl-1, the BH3-only protein that promotes cell death (Conradt and Horvitz 1998; Chang et al. 2006). We found that AIY-specific expression of egl-1 exacerbates the starvation sensitivity of gpb-2 mutants and that AIB-specific expression of egl-1 rescues gpb-2 mutants (Fig. 3C,D), suggesting that changes in the neuronal activities of AIY and AIB are sufficient to modulate the starvation response. We confirmed the AIY killing results using a ttx-3 mutation (Supplemental Fig. 3D), which is known to disrupt the function of AIY (Altun-Gultekin et al. 2001). These results, together with recent findings showing that AIY is activated by food or odor presentation and AIB is activated by food or odor removal (Chalasani et al. 2007), suggest that AIY inhibits the starvation response, whereas AIB activates it.
Since mgl-1 and mgl-2 are similar to metabotropic glutamate receptors, it is possible that glutamate neurotransmission is involved in the modulation of mgl-1 and mgl-2. To test this possibility, we looked at the effect of mgl-1 and mgl-2 on starvation survival in an eat-4 mutant background. eat-4 encodes a vesicular glutamate transporter, and glutamate neurotransmission is impaired in eat-4 mutants (Lee et al. 1999), including transmission from the known presynaptic partners of AIY and AIB (Chalasani et al. 2007). If amino acids modulate the activity of mgl-1 and mgl-2 through glutamate neurotransmission, one would expect mutation of eat-4 to abolish the effects of mgl-1, mgl-2, and leucine. However, this is not the case. mgl-1, mgl-2, and leucine have pronounced effects on starvation survival in the absence of eat-4. This suggests that glutamatergic neurotransmission is not necessary for the activation of MGL-1 and MGL-2. We suggest, rather, that amino acids can directly modulate the activity of mgl-1 and mgl-2 (Supplemental Fig. 4). An alternative possibility, however, is that other vesicular glutamate transporters function redundantly with eat-4, and therefore they modulate the activity of mgl-1 and mgl-2.
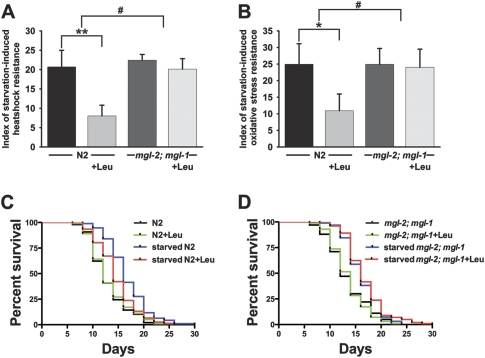
Our data in gpb-2 mutants indicate that amino acids act as an anti-hunger signal and that mgl-1 and mgl-2 are involved in the systemic starvation response. To evaluate whether treatment with amino acids can also suppress starvation response in wild-type worms and whether mgl-1 and mgl-2 are involved in that process, we took advantage of the concept of starvation-induced hormesis. Hormesis indicates beneficial effects of low doses of treatments known to be harmful at higher doses (Gems and Partridge 2008). It has been reported that short-term starvation (1–2 d) increases oxidative-stress resistance and extends life span in C. elegans (Cypser et al. 2006; Weinkove et al. 2006). We confirmed that starvation induces heat-shock resistance, oxidative stress resistance, and life span extension in wild-type worms, and found that treatment with leucine partially suppressed these effects of starvation, suggesting that amino acids act as an anti-hunger signal in wild-type worms (Fig. 4A–C). More interestingly, we saw no effect of leucine on the starvation-induced heat-shock resistance, oxidative stress resistance, and life span extension in mgl-2; mgl-1 mutants, suggesting that mgl-1 and mgl-2 are likely involved in wild-type amino acid responses, as they are in the gpb-2 starvation response (Fig. 4A,B,D). AIY-specific expression of mgl-1 and AIB-specific expression of mgl-2 restored the effect of leucine on the starvation-induced heat-shock and oxidative stress resistance in mgl-2; mgl-1 mutants, further supporting our hypothesis that mgl-1 and mgl-2 acts in AIY and AIB neurons (Supplemental Fig. 5).
Figure 4.
Starvation induces hormesis, treatment with leucine partially suppresses it in wild-type worms, and the effect of leucine is abolished in mgl-2; mgl-1 mutants. (A,B) Indexes of starvation-induced heat-shock resistance and oxidative stress resistance were calculated as described in the Materials and Methods. Starvation induces heat-shock resistance and oxidative stress resistance, treatment with leucine partially suppresses it in wild-type worms, and the effect of leucine is not observed in mgl-2; mgl-1 mutants. Data were combined from at least four independent experiments (n = 389∼2458 for each heat-shock experiment and n = 213∼1181 for each oxidative stress resistance). (*) P < 0.05; (**) P < 0.01 (Student's t-test, paired); (#) P < 0.05 (Student's t-test, unpaired). (C,D) Life span curves represent combined data from two independent experiments (n = 76∼101). Starvation extends life span, and treatment with leucine partially suppresses the life span extension in wild-type worms (starved N2, 16.74 ± 0.04 vs. starved N2 with leucine, 14.61 ± 0.05, P < 0.005, Mantel-Cox Log-rank test). Effect of leucine is not observed in mgl-2; mgl-1 mutants (starved mgl-2; mgl-1, 16.23 ± 0.03 vs. starved mgl-2; mgl-1 with leucine, 16.73 ± 0.04).
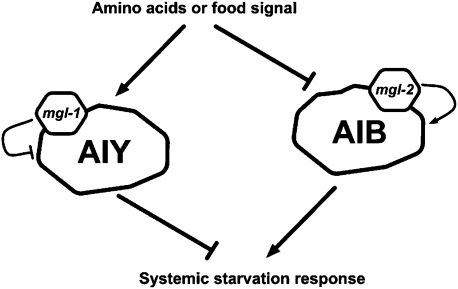
It is reasonable to suppose that the stress response should be coordinated in multicellular organisms, because uncoordinated stress responses in individual cells within a multicellular organism could impair the function of tissues, and eventually be harmful for the survival of the organism. However, little is known about whether the stress response previously thought to be cell-autonomous can be systemically regulated. A recent study elegantly showed that, in C. elegans, the heat-shock response is systemically regulated through the activity of AFD (Prahlad et al. 2008), which was known previously to sense ambient temperature (Mori 1999). By analogy, our study suggests that the starvation response can be systemically regulated through mgl-1 and mgl-2, presumably regulating the activity of AIY and AIB interneurons (Fig. 5), which were known previously to modulate behaviors in response to food or odor presentation or removal (Chalasani et al. 2007).
Figure 5.
Model of the systemic regulation of starvation response in C. elegans. Amino acids or food signal modulates the activity of AIY and AIB neurons, probably through MGL-1 and MGL-2, respectively, which in turn regulate the systemic starvation response.
Since both AIY and AIB are amphid interneurons, one might expect that that they do not directly sense environmental amino acids. It is possible that AIY and AIB receive synapses from certain chemosensory neurons (for example, ASE, AWC, or ASI, based on known neural connectivity) (White et al. 1986) that directly sense environmental amino acids, so as to modulate a systemic starvation response depending on that information. However, it is also possible that AIY and AIB directly respond to internal amino acids, previously absorbed from the environment. The latter possibility is plausible based on the fact that, in rodents, a sensory role for either taste or odor is not involved in the sensing of essential amino acids, and it is suggested that a chemosensor for essential amino acids is in the brain (Gietzen and Rogers 2006). More importantly, our eat-4 mutation data suggest that glutamate neurotransmission is not involved in the regulation of mgl-1 (AIY) and mgl-2 (AIB) in response to amino acids, suggesting the possibility that mgl-1 and mgl-2 may directly sense internal amino acids as in the case of extracellular calcium sensing receptor. It would be interesting to test if AIY and AIB could directly sense amino acids by calcium imaging.
Surprisingly, treatment with leucine decreased survival of gpb-2 mgl-2 mutants during starvation. One possible explanation is that leucine has an additional negative effect on starvation survival, which is downstream from mgl-1, and is masked by the positive effect of leucine through mgl-2. Indeed, in our preliminary experiments, we observed that treatment with leucine decreased mgl-2 mutants survival at later times (9 d of starvation), compared with wild-type or mgl-1 and mgl-2; mgl-1 mutants (data not shown), supporting the possibility.
In summary our data suggest that mgl-1 and mgl-2 are involved in the two related systemic processes: amino acid response and starvation response. Our current model (Fig. 5) suggests that the amino acid signal, which may be a component of the food signal, activates AIY neurons and inhibits AIB neurons by modulating the activities of MGL-1 and MGL-2, respectively. AIY then inhibits the starvation response, whereas AIB activates it. Given that there is no direct connection between AIY/AIB neurons and pharyngeal muscle, it is reasonable to think that specific hormones, perhaps neuropeptides, act downstream from AIY and AIB neurons to modulate the systemic starvation response in several tissues. Further experiments are needed to elucidate whether the systemic signal is indeed one or more neuropeptides and, if so, which peptides regulate the systemic starvation response downstream from AIY and AIB. Our model system, the starvation-hypersensitive gpb-2 mutants, might be helpful to find such peptide signals.
While this paper was under review, Greer et al. (2008) published a study describing the role of metabotropic glutamate receptors in the fat accumulation of C. elegans. They found that daf-7 inactivation (which mimics the status under unfavorable environmental conditions such as reduced food availability) causes fat accumulation via metabotropic glutamate receptors, supporting our model that metabotropic glutamate receptors are involved in the modulation of amino acid response and starvation response.
Materials and methods
Strains
Strains were maintained as described (Brenner 1974) at 19°C. All worms were maintained and grown on Escherichia coli HB101 bacteria. The following strains were generated using standard genetic procedures: gpb-2(ad541) I, gpb-2(ad541) I; mgl-1(tm1811) X, gpb-2(ad541) mgl-2(tm355) I, gpb-2(ad541) mgl-2(tm355) I; mgl-1(tm1811) X, gpb-2(ad541) I; adIs2122[GFP∷lgg-1 rol-6(d)], gpb-2(ad541) I; mgl-1(tm1811) X; adIs2122, gpb-2(ad541) mgl-2(tm355) I; adIs2122, gpb-2(ad541) mgl-2(tm355) I; mgl-1(tm1811) X; adIs2122, gpb-2(ad541) I; mgl-1(tm1811) X; adEx2241[pttx-3∷mgl-1 rol-6(d)], gpb-2(ad541) mgl-2(tm355) I; adEx2242[podr-2b∷mgl-2 rol-6(d)], gpb-2(ad541) I; adEx2243[pttx-3∷egl-1 rol-6(d)], gpb-2(ad541) I; adEx2244[podr-2b∷egl-1 rol-6(d)], gpb-2(ad541) I; eat-4(ky5) III, gpb-2(ad541) I; eat-4(ky5) III; mgl-1(tm1811) X, gpb-2(ad541) mgl-2(tm355) I; eat-4(ky5) III, mgl-2(tm355) I; mgl-1(tm1811) X, mgl-2(tm355) I; mgl-1(tm1811) X;[pttx-3∷mgl-1 podr-2b∷mgl-2 rol-6(d)], gpb-2(ad541) I; ttx-3(ks5) X, gpb-2(ad541) I; mgl-1(tm1811); [pglr-4∷mgl-1 rol-6(d)], gpb-2(ad541) I mgl-2(tm355); [pglr-4∷mgl-2 rol-6(d)], gpb-2(ad541) I mgl-2(tm355); [pnmr-2∷mgl-2 rol-6(d)].
Molecular biology
cDNA corresponding to the entire coding sequence of egl-1 was amplified and cloned under cell-specific promoters as indicated. Primer sequences were as follows: egl-1: 5′-GCTCTAGAATGTCCAACGTTTTTGACGTTCAATCT-3′ and 5′-CGCTCGAGTTAAAAAGCGAAAAAGTCCAGAAGACG-3′; Template, egl-1 DNA in pBluescript from Scott Cameron and Malia Potts (University of Texas Southwestern Medical Center). Expression in AIY and AIB was achieved using the ttx-3 and odr-2b promoters, respectively, from Cornelia Bargmann (The Rockefeller University).
cDNA corresponding to the entire coding sequence of mgl-2 was obtained using SpeI and XhoI restriction enzymes from mgl-2 cDNA in pBluescript from Isao Katsura (National Institute of Genetics) and cloned under the odr-2b promoter.
PCR construction of mgl-1 with ttx-3 promoter was achieved as follows: The ttx-3 promoter region was amplified from a Pttx-3∷mod-1 cDNA∷GFP plasmid (from Cornelia Bargmann, The Rockefeller University) using primers 5′-GTCTCATTTAAATTTTCAGAGCTTAAAAATGG-3′ and 5′-TGTTTGCCTCATATTGACACCGAAGACAATTATT-3′ (PCR#1). cDNA corresponding to the entire coding sequence of mgl-1 was amplified from mgl-1 cDNA in pMW118 (from Isao Katsura, National Institute of Genetics) using primers 5′-CTTCGGTGTCAATATGAGGCAAACATTTCGGAA-3′ and 5′-TCATAAGAAAGTATCGTGAGCAG-3′ (PCR#2). PCR#1 and #2 were fused using primers 5′-GTCTCATTTAAATTTTCAGAGCTTAAAAATGG-3′ and 5′-TCATAAGAAAGTATCGTGAGCAG-3′.
Expression in RMD/RIB and AVE was achieved using the glr-4 (RMD, RIB, AVA, SMD, SAA, SIB, RIM, AVH, FLP, RMG, DVA, AUA, PVD, URY, URA, SAB, RIF, DB, and PVU) and nmr-2 (AVE, AVA, AVD, RIM, AVG, and PVC) promoters, respectively (Greer et al. 2008), from Kaveh Ashrafi (University of California at San Francisco).
Starvation survival analyses
Starvation survival analyses were performed as described (You et al. 2006) with a few modifications. After collection of L1 worms from synchronization by egg preparation, we incubated them in 3 mL of sterilized M9 buffer with or without leucine for the time indicated in the figures at 19°C. At each time point, an aliquot from each sample tube was placed on a plate seeded with E. coli HB101. The number of worms surviving to L4 or adulthood was determined after 3 d further growth at 19°C. The number from day 1 of starvation was used as control and as the denominator to calculate the percentage of worms recovering after starvation. Since starvation-survival was influenced by assay conditions, all relevant experimental data were examined and compared within the same experiment. For initial screening, we used 10.9 mM amino acids. Since we found in preliminary exploratory experiments that high concentrations of leucine less efficiently rescued survival of gpb-2 mutants, we used 3.6 mM leucine in all subsequent experiments except the hormesis experiments (Fig. 4), in which we used 7.2 mM leucine. Leucine has a rescue effect on gpb-2 mutants in the range of 3.6–10.9 mM.
C. elegans autophagy analysis
Autophagy analysis was performed as described (Kang et al. 2007). For light microscopic analysis of autophagy, starved L1 animals carrying an integrated transgene that expresses a GFP∷LGG-1 fusion were collected at 3 d of starvation. GFP-positive punctate regions were visualized in the pharyngeal muscles of L1 animals using a Zeiss Axioplan 2 compound microscope (Zeiss Corporation).
Starvation-induced heat-shock resistance analyses
After collection of L1 worms after synchronization by egg preparation, we incubated them in 3 mL of M9 buffer with or without leucine for 0 d (nonstarved group) or 2 d (starved group). We equally divided each group into two subgroups and gave a heat shock (90 min, 35°) to one of them (heat-shock subgroup). We calculated the index of starvation-induced heat-shock resistance as follows: index of starvation-induced heat-shock resistance = (number of surviving starved worms with heat shock/number of surviving starved worms without heat shock) – (number of surviving unstarved worms with heat shock/number of surviving unstarved worms without heat shock).
Starvation-induced oxidative stress resistance analyses
After collection of L1 worms after synchronization by egg preparation, we incubated them in 3 mL of M9 buffer with or without leucine for 0 d (nonstarved group) or 2 d (starved group). We equally divided each group into two subgroups and gave an oxidative stress (H2O2 10 mM, 40 min) to one of them (H2O2 group). We calculated the index of starvation-induced oxidative stress resistance as follows: index of starvation-induced oxidative stress resistance = (number of nonarrested starved worms with H2O2/number of nonarrested starved worms without H2O2) – (number of nonarrested unstarved worms with H2O2/number of nonarrested unstarved worms without H2O2).
Starvation-induced life span extension analyses
Life span analyses were performed as described (Tullet et al. 2008) with a few modifications. After collection of L1 worms after synchronization by egg preparation, animals were allowed to develop at 19°C. When animals reached adulthood, they were collected and incubated in sterilized M9 buffer with or without leucine for 0 h (nonstarved group) or 40 h (starved group). They were transferred to a plate seeded with E. coli HB101, kept at 19°C, and score every 2 d as dead or alive. Animals that crawled off the plate, exploded, or died as bags of worms were excluded from analysis at the time of death. Life spans were measured from adulthood. Survival curve P values were calculated by the Mantel-Cox Log Rank test using Prism statistical software (Graphpad).
Acknowledgments
We thank S. Cameron, B. Levine, M. Cobb, and C. Bargmann for helpful discussions; S. Cameron, C. Bargmann, K. Ashrafi, I. Katsura, and T. Ishihara for providing plasmids; the C. elegans Genetics Center and S. Mitani (National Bioresource Project) for providing strains; and Y. Choi for helping statistic analyses. C.K. thanks M.S. Kim and D. Kang for unfailing support and encouragement. This work was supported by research grant HL46154 from the U.S. Public Health Service.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1723409.
Supplemental material is available at http://www.genesdev.org.
References
- Altun-Gultekin Z., Andachi Y., Tsalik E.L., Pilgrim D., Kohara Y., Hobert O. A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans . Development. 2001;128:1951–1969. doi: 10.1242/dev.128.11.1951. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans . Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalasani S.H., Chronis N., Tsunozaki M., Gray J.M., Ramot D., Goodman M.B., Bargmann C.I. Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans . Nature. 2007;450:63–70. doi: 10.1038/nature06292. [DOI] [PubMed] [Google Scholar]
- Chang A.J., Chronis N., Karow D.S., Marletta M.A., Bargmann C.I. A distributed chemosensory circuit for oxygen preference in C. elegans . PLoS Biol. 2006;4:e274. doi: 10.1371/journal/pbio.0040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codogno P., Meijer A.J. Autophagy and signaling: Their role in cell survival and cell death. Cell Death Differ. 2005;12:1509–1518. doi: 10.1038/sj.cdd.4401751. [DOI] [PubMed] [Google Scholar]
- Conigrave A.D., Hampson D.R. Broad-spectrum L-amino acid sensing by class 3 G-protein-coupled receptors. Trends Endocrinol Metab. 2006;17:398–407. doi: 10.1016/j.tem.2006.10.012. [DOI] [PubMed] [Google Scholar]
- Conradt B., Horvitz H.R. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell. 1998;93:519–529. doi: 10.1016/s0092-8674(00)81182-4. [DOI] [PubMed] [Google Scholar]
- Cota D., Proulx K., Smith K.A., Kozma S.C., Thomas G., Woods S.C., Seeley R.J. Hypothalamic mTOR signaling regulates food intake. Science. 2006;312:927–930. doi: 10.1126/science.1124147. [DOI] [PubMed] [Google Scholar]
- Cypser J.R., Tedesco P., Johnson T.E. Hormesis and aging in Caenorhabditis elegans . Exp. Gerontol. 2006;41:935–939. doi: 10.1016/j.exger.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon J., Hopper N.A., Holden-Dye L., O'Connor V. Molecular characterization of the metabotropic glutamate receptor family in Caenorhabditis elegans . Biochem. Soc. Trans. 2006;34:942–948. doi: 10.1042/BST0340942. [DOI] [PubMed] [Google Scholar]
- Gems D., Partridge L. Stress-response hormesis and aging: ‘That which does not kill us makes us stronger.’. Cell Metab. 2008;7:200–203. doi: 10.1016/j.cmet.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Gietzen D.W., Rogers Q.R. Nutritional homeostasis and indispensable amino acid sensing: A new solution to an old puzzle. Trends Neurosci. 2006;29:91–99. doi: 10.1016/j.tins.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Greer E.R., Perez C.L., Van Gilst M.R., Lee B.H., Ashrafi K. Neural and molecular dissection of a C. elegans sensory circuit that regulates fat and feeding. Cell Metab. 2008;8:118–131. doi: 10.1016/j.cmet.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C., Avery L. To be or not to be, the level of autophagy is the question: Dual roles of autophagy in the survival response to starvation. Autophagy. 2008;4:82–84. doi: 10.4161/auto.5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C., You Y.J., Avery L. Dual roles of autophagy in the survival of Caenorhabditis elegans during starvation. Genes & Dev. 2007;21:2161–2171. doi: 10.1101/gad.1573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Abeliovich H., Agostinis P., Agrawal D.K., Aliev G., Askew D.S., Baba M., Baehrecke E.H., Bahr B.A., Ballabio A., et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R.Y., Sawin E.R., Chalfie M., Horvitz H.R., Avery L. EAT-4, a homolog of a mammalian sodium-dependent inorganic phosphate cotransporter, is necessary for glutamatergic neurotransmission in Caenorhabditis elegans . J. Neurosci. 1999;19:159–167. doi: 10.1523/JNEUROSCI.19-01-00159.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Yuan J. Autophagy in cell death: An innocent convict? J. Clin. Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum J.J., DeBerardinis R.J., Thompson C.B. Autophagy in metazoans: Cell survival in the land of plenty. Natl. Rev. 2005;6:439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- Melendez A., Talloczy Z., Seaman M., Eskelinen E.L., Hall D.H., Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans . Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- Mori I. Genetics of chemotaxis and thermotaxis in the nematode Caenorhabditis elegans . Annu. Rev. Genet. 1999;33:399–422. doi: 10.1146/annurev.genet.33.1.399. [DOI] [PubMed] [Google Scholar]
- Prahlad V., Cornelius T., Morimoto R.I. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science. 2008;320:811–814. doi: 10.1126/science.1156093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk N.J., Kozak E., Conley C.A. Chemically defined medium and Caenorhabditis elegans . BMC Biotechnol. 2003;3:19. doi: 10.1186/1472-6750-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullet J.M., Hertweck M., An J.H., Baker J., Hwang J.Y., Liu S., Oliveira R.P., Baumeister R., Blackwell T.K. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans . Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinkove D., Halstead J.R., Gems D., Divecha N. Long-term starvation and ageing induce AGE-1/PI 3-kinase-dependent translocation of DAF-16/FOXO to the cytoplasm. BMC Biol. 2006;4:1. doi: 10.1186/1741-7007-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.G., Southgate E., Thomson J.N., Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans . Philos. Trans. R. Soc. Lond. B Biol. Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- You Y.J., Kim J., Cobb M., Avery L. Starvation activates MAP kinase through the muscarinic acetylcholine pathway in Caenorhabditis elegans pharynx. Cell Metab. 2006;3:237–245. doi: 10.1016/j.cmet.2006.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]