Abstract
Signaling through mitogen-activated protein kinases (MPKs) cascades is a complex and fundamental process in eukaryotes, requiring MPK-activating kinases (MKKs) and MKK-activating kinases (MKKKs). However, to date only a limited number of MKK–MPK interactions and MPK phosphorylation substrates have been revealed. We determined which Arabidopsis thaliana MKKs preferentially activate 10 different MPKs in vivo and used the activated MPKs to probe high-density protein microarrays to determine their phosphorylation targets. Our analyses revealed known and novel signaling modules encompassing 570 MPK phosphorylation substrates; these substrates were enriched in transcription factors involved in the regulation of development, defense, and stress responses. Selected MPK substrates were validated by in planta reconstitution experiments. A subset of activated and wild-type MKKs induced cell death, indicating a possible role for these MKKs in the regulation of cell death. Interestingly, MKK7- and MKK9-induced death requires Sgt1, a known regulator of cell death induced during plant innate immunity. Our predicted MKK–MPK phosphorylation network constitutes a valuable resource to understand the function and specificity of MPK signaling systems.
Keywords: MAPK, Arabidopsis, phosphorylation network, protein microarrays, cell death
Signaling through mitogen-activated protein kinase (MPK) cascades is a fundamental and conserved process in eukaryotes. MPK signaling cascades are composed of three main signaling elements—MKK-activating kinase (MKKK), MPK-activating kinase (MKK), and MPK—activated through consecutive phosphorylation events in response to extracellular or intracellular signals. The activated MPK phosphorylates a variety of cytoplasmic and nuclear substrates, with profound effects on their localization, activity state, stability, and transcript levels (Whitmarsh 2007). MPK substrates regulate many essential cellular processes in response to a stimulus.
MPK signaling cascades form complex interconnected networks within cells (Pedley and Martin 2005). Traditional genetic and biochemical methods have identified MKKK/MKK/MPK signaling modules with overlapping roles in controlling cell division, development, hormone signaling and synthesis, and response to abiotic stress and pathogens. For example, for Arabidopsis, the MEKK1–MKK4/5–MPK3/6 module was found to participate in flagellin-mediated innate immune signaling (Asai et al. 2002), the MEKK1–MKK1/2–MPK4/6 module was activated by various stress treatments (Ichimura et al. 2000; Teige et al. 2004; Meszaros et al. 2006; Brader et al. 2007), MKK3–MPK6 module was activated by jasmonic acid (JA) (Takahashi et al. 2007), and the YODA–MKK4/5–MPK3/6 cascade was established as a key regulator of stomatal development and patterning (Wang et al. 2007).
Eukaryotes have multiple MKKKs, MKKs, and MPKs (humans have 16 MKKKs, seven MKKs, and nearly 20 MPKs, and Arabidopsis has 60–80 predicted MKKKs, 20 MKKs, and at least 20 MPKs) and two fundamental problems are (1) understanding which upstream kinases regulate particular downstream kinases and (2) identification of the downstream substrates that are targeted by these pathways. These problems have been difficult to resolve because of cellular interactions, redundancy, and functional pleiotropy (Friedman and Perrimon 2006). Unbiased high-throughput studies that explore interactions between signaling molecules and their targets are essential for a systematic analysis of the signaling networks. In this regard high-density protein microarrays offer an effective approach to identify potential kinase substrates (Feilner et al. 2005; Ptacek et al. 2005).
In order to identify novel MPK targets and to study MPK signaling cascades in an unbiased and high-throughput manner, we employed high-density Arabidopsis protein microarrays containing 2158 proteins. The protein microarrays were screened with MPK probes phosphorylated and activated in planta by specific MKKs. Functional MKK/MPK modules were selected by combinatorial pairing of 10 MPKs and nine wild-type or constitutively active MKKs. We identified 570 putative MPK phosphorylation targets, with an average of 128 targets per activated MPK. Several WRKY and TGA transcription factors were shown to be phosphorylated in vivo when coexpressed with specific MKK/MPK modules. Furthermore, coexpression studies of several MKK/MPK/Substrate modules implicate MKK7, MKK9, and MPK10 in the regulation of programmed cell death. The cell death phenotype of MKK7- and MKK9-expressing plants was dependent on Sgt1, a known regulator of innate immunity induced cell death. This result indicates that MKK7- and MKK9-activated signals functions upstream of Sgt1 and also suggest that MKK7 and MKK9 modules may function during plant innate immunity. Based on our MPK target list, we constructed a phosphorylation network of MKK/MPK/Substrate pathways. Analysis of phosphorylation network suggests an important role for MPK signaling in the transcriptional control of the stress response and development.
Results
Identification of functional MKK–MPK modules
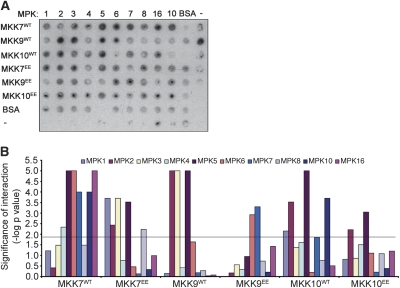
To identify novel MPK effectors we employed high-density Arabidopsis protein microarrays containing 2158 proteins. The protein microarrays were screened with in planta phosphorylated and activated MPKs by specific MKKs. We first carried out a comprehensive analysis of enzymatic activity and specificity of MKKs, by evaluating the interactions of 180 binary combinations of nine wild-type MKK (MKKwt) and constitutively active (MKKEE) proteins (MKK1 to MKK7, MKK9, and MKK10), and 10 wild-type MPK proteins (MPK1 to MPK8, MPK10, and MPK16). For the MKKEE proteins two Ser/Thr (S/T) residues in the predicted consensus sequence S/TXXXXXS/T of the activation loop, were replaced with glutamic acid (Glu, E) (Supplemental Fig. 1). The predicted activation motif is conserved in all Arabidopsis MKKs with the exception of MKK10, and is very similar to the one present in animal MKKs (Matsuoka et al. 2002). For MKK10, only the S197 residue located between the conserved kinase domains VII and VIII was mutated to Glu (Supplemental Fig. 1).
MKKwt, MKKEE, and MPK were transiently expressed in Nicotiana benthamiana and recombinant proteins were affinity purified from tissue harvested at 2–3 d post-infiltration (dpi) or 4–5 dpi as described previously (Popescu et al. 2007). To identify all possible functional MKK-MPK modules, in vitro phosphorylation assays with purified protein and myelin basic protein (MBP) were performed with all binary combinations of MKKs and MPKs. In agreement with previous observations (Yang et al. 2001), MKKEE mutants showed increased in vitro kinase activity compared with the wild-type MKKs. (Supplemental Table 1). However, in some cases—for example, MKK7 and MKK9—the wild-type kinases were more active than the MKKEE proteins (Fig. 1A; Supplemental Table 1). It is possible that these plant MKKs are also regulated through the phosphorylation of other residues located within or outside the activation loop. Indeed, phosphorylation events within the activation loop of several plant, animal, and yeast MKKs were reported to completely inactivate their kinase activity (Xing et al. 2001; Yang et al. 2001; Matsuoka et al. 2002; Gopalbhai et al. 2003). Furthermore, spatial and temporal regulators of MKK phosphorylation are probably needed to ensure specific downstream activation of MPKs. Future identification of all MKK phosphorylation sites and their effects on enzymatic activity will be necessary for a detailed understanding of plant MKK regulation.
Figure 1.
Comprehensive analysis of in vitro MKK substrate specificity. (A) Combinations of MKK/MPK purified fusion proteins comprising nine wild-type MKK (MKKWT) and activation loop mutants (MKKEE) and 10 MPKs, were mixed in 96-well plates with MBP and kinase buffer in the presence of [γ-32P]-ATP. The proteins were transferred to PVDF membranes and phosphorylated MBP was detected on film. A representative blot is shown. (B) The kinase activity, indicated by phosphorylated MBP, was estimated by measuring the amount of incorporated [γ-32P]-ATP in all reactions in two or three technical replicates. Spot intensities were obtained from the PhosphorImager exposure. Graphical representation of the likelihood of interaction (−log [P-value]) for the blot in A is shown. The level of significance (0.0143) was represented as the black threshold line at 1.8 U on the Y-axis.
MKK–MPK pairs with maximum MPK activity were identified by analyzing all significant MBP phosphorylation events after correction for MPK and MKK autophosphorylation (Fig. 1B; Supplemental Table 1). From this analysis, 49 statistically significant MKK–MPK interactions were identified including all 10 previously reported interactions (Supplemental Table 2). Hierarchical clustering analysis was performed to analyze MKK target similarity. Hierarchical clustering analysis revealed that several MKKs clustered along structural similarity lines. For example, MKK1 and MKK2 from Group A and MKK4, MKK5 from Group C clustered together, indicating that they recognize very similar MPK targets (Supplemental Fig. 2). Likewise, Group D kinases MKK7 and MKK9 belong to a separate cluster group (Supplemental Fig. 2). These results suggest a common mechanism of target recognition for structurally similar MPKs.
Detection of MPK phosphorylation substrates using Arabidopsis protein microarrays
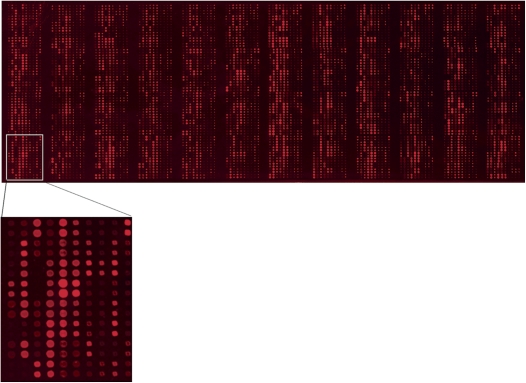
Recently, we described the construction of Arabidopsis protein microarrays containing 1133 unique proteins (Popescu et al. 2007). Using the procedures described in Popescu et al. (2007), we generated a second version of the Arabidopsis protein microarray containing 2158 unique proteins (Fig. 2; Supplemental Table 3). Gene ontology (GO) molecular function categories analyses indicate that 50.4% of proteins are putative transcription factors (30% relative to total TAIR annotations in this GO slim category), 34% are putative nucleic acid-binding proteins (20% relative to TAIR classification), and 16% are protein kinases (13% relative to TAIR classification) (Supplemental Fig. 3).
Figure 2.
Generation of Arabidopsis protein microarrays. A representative protein microarray containing 3840 protein preparations printed in duplicates, representing 2158 unique Arabidopsis proteins. The microarray was probed with anti-cMyc monoclonal primary antibody and Cy5-labeled secondary antibody to assess the amount of protein on the slide. For clarity, an enlarged image of one block is shown below the protein microarray.
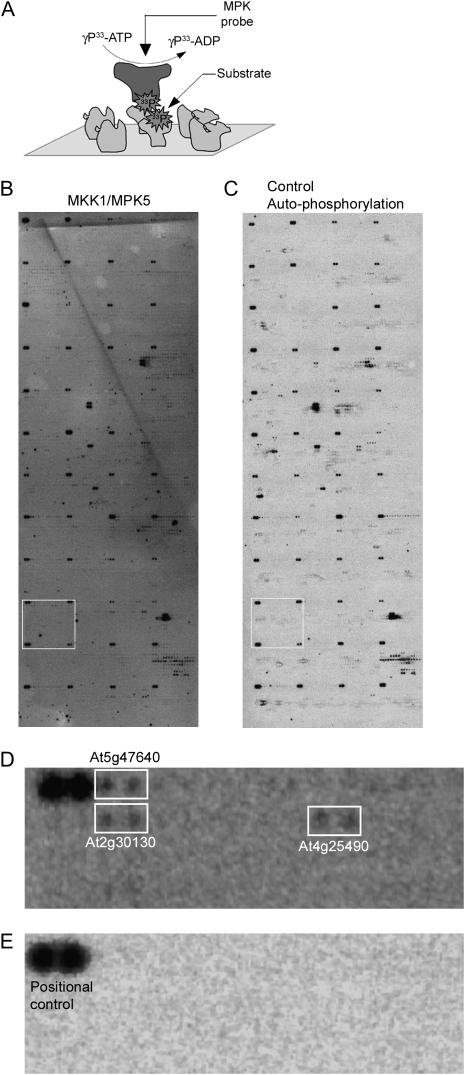
To identify phosphorylation substrates of MPKs the Arabidopsis protein microarrays were incubated with 33P-γ-ATP and 10 different MPK recombinant proteins that were activated in planta by two different MKKs (Supplemental Table 2). A total of 75 protein microarrays were analyzed with in planta activated MPK proteins. For each MPK probe under study, two to five microarrays were assayed (Supplemental Table 4). An additional seven microarrays were incubated with only kinase buffer and 33P-γ-ATP; these served as negative controls. Several control spots were included on the slides: buffer-only, albumin, and autophosphorylating proteins as positional controls. The latter included AT3g20530 and AT4G23650, which were identified in preliminary experiments as the two most active autophosphorylating kinases; these were spotted in duplicate in the first position of each array block as positive controls. A representative protein microarray probed with MPK5 activated by MKK1 is shown in Figure 3. Three putative MPK5 targets, At5g47640 (HAP3B), At2g30130 (ASL5, LBD12), and At4g25490 (CBF1) are shown in Figure 3D.
Figure 3.
Identification of MPK phosphorylation substrates on protein microarrays. (A) Schematic of kinase assays on protein microarrays. MPK-TAP fusion proteins were purified from N. benthamiana overexpressing 20 selected in vitro functional combinations of MKK/MPK. Purified MPK proteins in kinase buffer were overlaid on protein microarrays in the presence of [γ-33P]-ATP and incubated for 1 h at 30°C. Phosphorylated MPK substrates on the protein microarrays were detected by exposure to Kodak film. (B,C) A representative microarray probed with activated MPK5 (B) and a representative negative control overlaid with kinase buffer and [γ-33P]-ATP to detect autophosphorylating proteins (C) are shown. Bright dark spots at the corner of each block represents positional controls spotted in duplicate to help with subsequent grid alignment and spot identification. (D,E) Zoomed-in area of block 26 marked as white rectangle from microarrays represented in B and C are shown in D and E, respectively. White rectangle regions in D represent putative MPK5 phosphorylation targets for which signal is present in the MPK5 probed array (D) but absent in the autophosphorylation control array (E).
MKK/MPK/substrate phosphorylation network
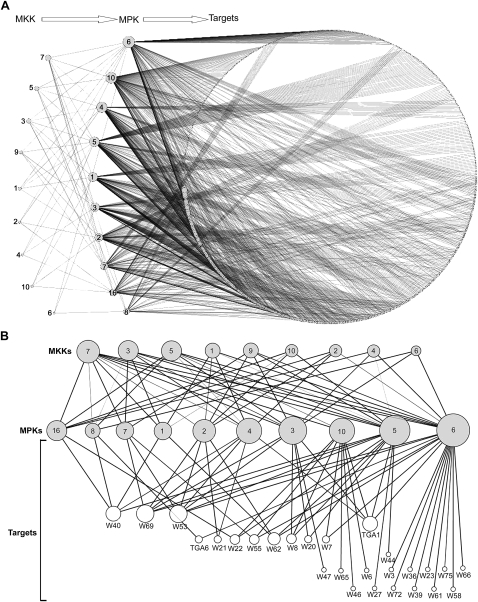
MPK phosphorylation substrates were inferred from the protein microarrays using a Bayesian decision tree method. Predicted preliminary targets were scored according to the number of phosphorylation events detected in all technical replicates and the number of MPK probes that phosphorylated them. Targets identified in less than two replicates for the same MPK probe were eliminated. In addition, all autophosphorylating proteins identified in the control microarrays incubated with only 33P-γ-ATP were removed from the target list. Following this analysis, 570 proteins were selected as high-confidence MPK targets (Supplemental Table 5) and a MKK/MPK/Substrate network was constructed using all predicted phosphorylation events (Fig. 4A). The network contains 589 nodes representing MKKs, MPKs, and predicted targets, and 1331 edges representing predicted phosphorylation events. In this representation, the node size is proportional with its degree of connectivity, and the edge width is proportional with the likelihood of the phosphorylation event it represents, as calculated using the causal network analysis.
Figure 4.
Reconstituted weighted MKK/MPK/Substrate phosphorylation networks. (A) The reconstituted network contains nine MKKs, 10 MPKs, and 570 MPK substrates represented as nodes. The size of the nodes is proportional with their degree of connectivity. The network edges indicate identified phosphorylation events. The width of each edge is proportional with the likelihood of the phosphorylation event it represents. The network was generated using Cytoscape version 2.5.1. (B) MKK/MPK/transcription factor hierarchical phosphorylation subnetwork including all predicted WRKY (W) and TGA (T) transcription factors. The nodes represent proteins and the edges represent phosphorylation events. The node size is equal with its degree of connectivity, and edge width is proportional with the likelihood of the phosphorylation event. The network was generated using Cytoscape 2.5.1.
The majority of Arabidopsis MPKs phosphorylated a similar number of substrates, ranging from 40 to 184, with an average of 128 targets per kinase (Supplemental Fig. 4A). MPK6, MPK8, and MPK16 deviated significantly from this average; MPK6 phosphorylated 184 targets and MPK8 and MPK16, structurally similar kinases, were the least active, phosphorylating only 40 and 69 substrates respectively (Supplemental Fig. 4A).
Interestingly, a large proportion of the MPK substrate phosphorylation events detected on the protein microarrays were specific. Our analysis identified 290 MPK phosphorylation targets (51% of total) that were phosphorylated by only one MPK (Supplemental Fig. 4B). Only 94 targets (16%) were phosphorylated by two or more MPKs. A smaller number of substrates were phosphorylated by more than three MPKs, and no substrates were found to be phosphorylated by all MAPKs (Supplemental Fig. 4B; Supplemental Table 5). These results indicate that although MPKs are related in sequence they exhibit unique substrate specificities in vitro.
We analyzed the percentage of shared MPK phosphorylation substrates by comparing each MPK with all other MPKs. We observed that MPK1, MPK2, MPK7, MPK8, and MPK16 share a large number of phosphorylation targets. Between 46% and 67% of MPK7, MPK8, and MPK16 substrates are shared with MPK1 and MPK2. Likewise, MPK1 and MPK2 phosphorylate over 50% identical substrates. MPK6 shares the largest percentage of substrates with MPK3 (40%), and MPK3 also shares >50% of its targets with MPK4 (Supplemental Table 6).
Arabidopsis MPKs phosphorylate mostly transcriptional regulators that are associated with developmental and stress responses
To further characterize the predicted MPK targets, we determined the overrepresentation of GO functional categories among MPK phosphorylation substrates relative to the total number of proteins in each respective category present on the protein microarray. Interestingly, we found significant enrichment in substrates with transcription factor activity (33%), response to stress (32%), response to abiotic and biotic stimulus (29%), and development (29%) (Supplemental Fig. 5A). Further subcategorization of the MPK substrates with transcription factor activity revealed that 11.5% have known or predicted roles in development and 14% of them have roles in or response to abiotic and biotic stimuli (Supplemental Fig. 5B).
Transcription factors represent the largest group of MPK targets identified in our screen. 33% of all proteins annotated as having transcription factor activity were phosphorylated by at least one MPK on the microarray. Taking into account that our protein microarray contains over 65% of all predicted transcription factors, we estimate that a large fraction of the Arabidopsis transcription factors are regulated by MPK phosphorylation.
Several transcription factors families, such as MYB and MYB-related proteins, bZIPs, AP2/EREBP, homeobox, and WRKY, had a large proportion of phosphorylated members on protein microarrays (Supplemental Fig. 5C). Of the 15 ethylene response factor (ERF) subfamily members identified as MPK phosphorylation substrates in our analyses, four of them (AT2G44840, AT5G61600, AT4G34410, AT4G17490) are also induced at the transcriptional level by treatments with chitin and methyl jasmonate (Libault et al. 2007). Moreover, 19 other phosphoproteins annotated as transcription factors or transcriptional regulators, and identified in the same study to be induced by chitin or other abiotic and biotic stress treatments, were also identified in our screen.
MPKs phosphorylate WRKY and TGA transcription factors
WRKY and TGA transcription factors perform central functions in the regulation of developmental processes, senescence, and defense responses against pathogens (Eulgem and Somssich 2007). Previous studies suggested that WRKY proteins constitute an important link between pathogen-activated MPK signaling pathways and downstream transcriptional reprogramming. WRKY25 and WRKY33 interact with the MPK4 substrate MKS1 (Andreasson et al. 2005). Arabidopsis WRKY53 is phosphorylated in vitro by MKK1 (Miao and Zentgraf 2007) and tobacco NtWRKY1 is phosphorylated in vitro by SIPK (Menke et al. 2005). To explore this relationship further, we focused our attention on the WRKY and TGA transcription factors phosphorylated by MPKs on protein microarrays. We identified a significant number of WRKYs as in vitro MPK targets (25 out of 55) (Fig. 4B). The largest number of WRKYs and TGAs were phosphorylated by MPK5, MPK6, and MPK10 (Supplemental Fig. 6). Our analyses indicate that 19 WRKY/TGAs (70%) were phosphorylated by one or two MPKs, demonstrating a high specificity of signal activation and output through the WRKY/TGA effectors.
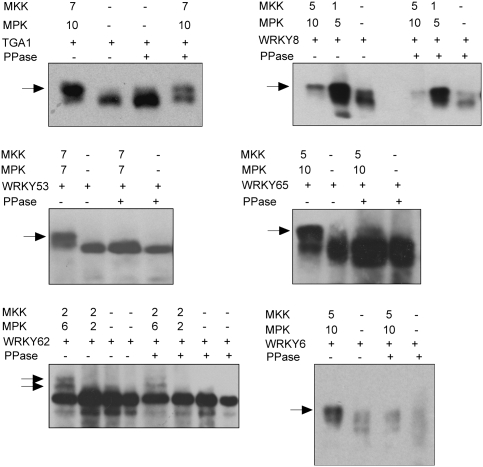
To validate our protein microarray results, a subset of WRKY and TGA effectors were expressed in the presence or absence of activating MKK/MPK modules, and their phosphorylation status was assessed by mobility shifts in SDS-PAGE (Fig. 5). As negative controls, all tissue extracts were incubated with phosphatases. WRKY53, WRKY62, and WRKY6 were phosphorylated only when they were coexpressed with MKK7/MPK7, MKK2/MPK6, and MKK5/MPK10 modules, respectively. WRKY65 and TGA1 were phosphorylated in the presence of MKK5/MPK10 and the MKK7/MPK10 modules, respectively.
Figure 5.
In planta validation of MPK targets. MKK-HA, MPK-CFP, and WRKY/TGA-TAP proteins were expressed together in N. benthamiana. As negative controls, WRKY/TGA-TAP proteins were expressed alone. Total protein extracts were separated on SDS-PAGE, and blots were probed with anti-cMyc monoclonal antibody. Phosphorylated and nonphosphorylated isoforms of WRKY/TGA factors were detected by mass shifts. Total protein extracts subjected to protein phosphatase 1 or λ phosphatase treatment were run alongside nontreated extracts as controls. Arrows represent phosphorylated protein isoforms.
Several intriguing observation were made during the course of these experiments. First, WRKY6 and WRKY8 proteins expressed alone in vivo showed a low level of phosphorylation, possibly due to endogenous N. benthamiana active MPKs. However, increased phosphorylation of these two factors was observed in the presence of MKK/MPK-activating modules. Second, phosphatase treatment of the tissue expressing WRKY6 by itself resulted in rapid degradation of the WRKY6 protein, suggesting a phosphorylation-dependent regulation of protein stability (Fig. 5).
Overexpression of MKK7 and MKK9 promotes cell death in N. benthamiana
The relationship between cell death and the activation of MPK cascades has been previously documented in plants (Pitzschke and Hirt 2006). During our MKK–MPK module expression studies in N. benthamiana, we noticed the induction of cell death following infiltrations with certain MKKs in the presence or absence of cognate MPKs. Consequently, we initiated further experiments to investigate the potential connection between the activation of MKKs and cell death signaling. In this regard, wild-type and activated MKKs were expressed in N. benthamiana, alone or in combinations with their in vitro identified MPK partners, to observe the timing and the spread of the cell death phenotype (Supplemental Table 7).
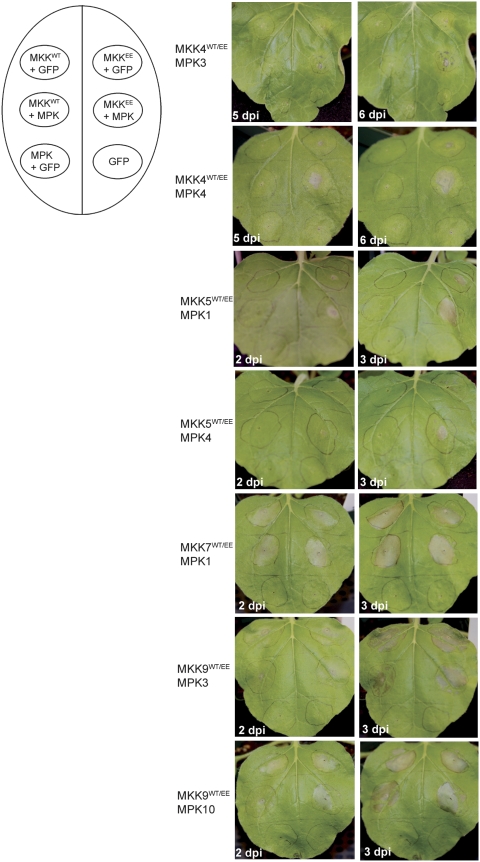
Overexpression of wild-type or activated MKK7 and MKK9 resulted in extensive cell death in N. benthamiana plants at 2–3 dpi (Fig. 6). Subtle differences of the timing and severity of the cell death exists between the wild-type and activated proteins, with MKK7EE and MKK9EE showing a slightly faster and more extensive cell death in the infiltrated area compared with wild type (Fig. 6). Overexpression of MPK with GFP and GFP alone controls did not cause cell death during this timeframe.
Figure 6.
Cell death phenotype induced by overexpression of several MKK/MPK modules in N. benthamiana. The diagram on the left represents the position on the leaf and the description of the infiltrated samples. The phenotype of infiltrated leaves shown on the right was monitored for several days post infiltration (dpi). Representative leaf pictures from two consecutive days for each experiment are shown.
In the case of MKK4 and MKK5, only overexpression of activated MKK4EE with MPK3 or MPK4, and of activated MKK5EE with MPK1 or MPK4 resulted in cell death. Expression of MKK4EE and MKK5EE in the presence of GFP caused limited cell death. Infiltrations of MKKwt proteins with cognate MPKs or GFP did not cause any death (Fig. 6).
In our experiments, overexpression of MPK10, MPK3, and MPK6 alone also triggered cell death at 4–5 dpi (data not shown). Previously, cell death has been observed following overexpression of the tobacco homologues of MPK3 and MPK6 (Ren et al. 2006). Taken together, our results indicate that MKK4-, MKK5-, MKK7-, and MKK9-activated signaling cascades may be involved in controlling cell death pathways in plants.
MKK7 and MKK9-induced cell death requires function of Sgt1
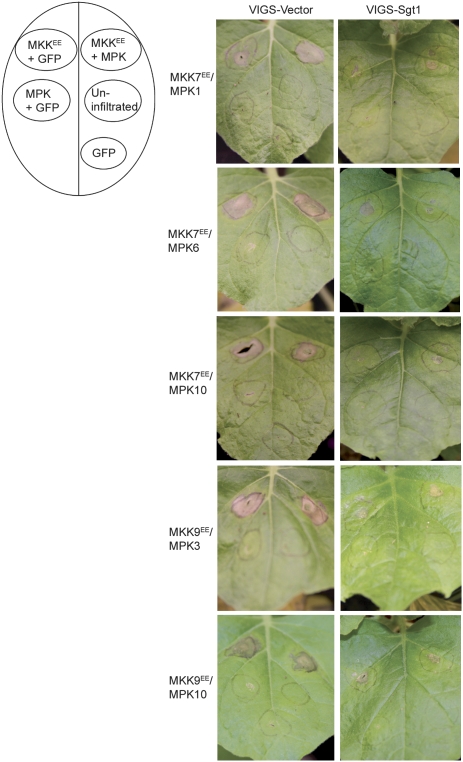
Certain forms of plant cell death are associated with the resistance pathways during pathogen infection (Greenberg and Yao 2004). Thus, we tested whether the cell death triggered by activated MKKs is related to the cell death activated during innate immune response in plants. For these experiments, we analyzed the expression of MKKEE–MPK, and individual MKKEE and MPK proteins in plants silenced for immune response modulators, Sgt1, RAR1, and EDS1 (Shirasu and Schulze-Lefert 2003; Wiermer et al. 2005).
We silenced N. benthamiana homolog of Sgt1 (NbSgt1) using our well-established virus-induced gene silencing (VIGS) system (Liu et al. 2002a; Dinesh-Kumar et al. 2003). In NbSgt1-silenced plants, MKK7EE- and MKK9EE-induced cell death was significantly reduced compared with the control VIGS vector-alone-silenced plants (Fig. 7). However, there was no difference in the induction of cell death by MKK4EE and MKK5EE in NbSgt1-silenced and control-silenced plants (data not shown). In addition, there was no significant difference in the MKK7EE, MKK9EE, MKK4EE/MPK3, or MPK4- and MKK5EE/MPK1- or MPK4-induced cell death in NbRAR1- and NbEDS1-silenced plants compared with the control-silenced plants (data not shown).
Figure 7.
MKK7- and MKK9-induced cell death requires Sgt1. The diagram on the left represents the position on the leaf and the description of the infiltrated samples. MKK7EE and MKK9EE were expressed, individually or in combination with cognate MPKs in Sgt1-silenced or VIGS vector-silenced N. benthamiana plants. Representative leaf pictures from two consecutive days for each experiment are shown.
Thus, the cell death triggered by MKK7- and MKK9-activated signaling cascades requires Sgt1, and suggests a potential role for MKK7 and MKK9 modules during plant innate immunity.
Discussion
We took a high-throughput approach by employing Arabidopsis protein microarrays to understand the MKK/MPK/Substrate signaling pathways. Specifically, we identified in vitro partners of nine MKK and 10 MPK proteins, and generated a phosphorylation network including 570 new and previously known signaling molecules and pathways. Moreover, our study uncovers a range of putative phosphorylation targets of MPKs, thus inferring and predicting in an unbiased manner their role in various cellular pathways. Results described here constitute a starting point for future, in depth in vivo studies of MPKs and their substrates.
A previous high-throughput search for MPK3 and MPK6 substrates using protein microarrays containing ∼1600 denatured Escherichia coli-expressed Arabidopsis proteins yielded 48 and 39 substrates, respectively (Feilner et al. 2005). Although, a direct comparison between the two studies is not possible due to the use of E. coli vs. plant expressed proteins, our analyses identified several common phosphorylation targets. The At2g02820, At3g60390, and At1g77450 transcription factors as well as representatives of several other protein families including several casein kinase isoforms, phospholipases, SNF1-related protein kinases and ribosomal proteins were identified in both studies. However, our study identified a much wider variety of putative MPK3 and MPK6 targets, including 38 new targets. In addition, we identified substrates of an additional eight MPKs from Arabidopsis.
MAPKs and transcriptional control of gene expression
A well-known function of eukaryotic MPKs is the control of gene expression at the transcription level (Chen and Thorner 2007). We identified several WRKY/TGA transcription factors to be phosphorylated by active MKK/MPK modules. Among them, WRKY53 was phosphorylated by activated MPK7 both in protein microarray assays and in vivo. WRKY53 is known to function in early stages of senescence (Miao and Zentgraf 2007), and may have a role in disease resistance (Wang et al. 2006). MPK7 gene expression is strongly up-regulatated during senescence (Zimmermann et al. 2005); therefore, it is possible that WRKY53 functions as a physiological MPK7 substrate to regulate gene expression during senescence and disease resistance.
WRKY62, which we found to be activated by several MPKs including MPK5 and MPK6, is known to be induced by SA and JA, and may be involved in the SA-mediated suppression of JA-responsive genes (Mao et al. 2007). Myc2 (JIN1) was recently shown to function downstream from the activated MKK3/MPK6 module (Takahashi et al. 2007) to regulate diverse JA-dependent functions. Our results support previous observations on a role for MPK6 in mediating JA responsive gene expression, and suggest that MPK6 modulates WRKY62 and Myc2 (JIN1) functions via phosphorylation.
WRKY6 and TGA1 were both phosphorylated by activated MPK10. WRKY6 may be a regulator of PCD, and was found to be induced by wounding, infection with bacterial pathogens and pathogen elicitors, and following treatment with SA, JA, and ethylene (Robatzek and Somssich 2001). TGA1 is known for its role in NPR1-dependent pathogen defense and basal and systemic acquired resistance (Despres et al. 2003; Kesarwani et al. 2007). It is possible that MPK10 regulates the activation of cellular pathways involved in disease resistance through phosphorylation of WRKY/TGA transcription factors.
Previous studies have shown that R2R3 MYB transcription factors (FLP and Myb88) and bHLH proteins (SPCH, FAMA, and MUTE) control cell type differentiation during stomata development (Lai et al. 2005; Pillitteri et al. 2007). In our study, Myb88 was phosphorylated by several MPKs including MPK6, and MUTE was phosphorylated by MPK4. These results are in agreement with a recent report that showed stomata patterning and development being regulated by MPK3 and MPK6 (Wang et al. 2007).
Our analyses also identified a group of >20 Agamous-like MADS-box transcription factors (AGLs) as MPK substrates. Half of the AGL substrates were phosphorylated by MPK6 or MPK1, the rest were targeted by MPK3, MPK4, and MPK10. MPK6 was recently demonstrated to play a significant role in flower development (Bush and Krysan 2007). Additionally, the flower repressor FLC (flowering locus C), the FLC paralog MAF5.2 (MADS affecting flowering 5 variant 2), and REF6 (relative of early flowering) (Noh et al. 2004) were also phosphorylated by MPKs. Given the significant number of AGLs we identified as MPK substrates, we hypothesize an extensive role for MPK signaling in flower development and in the control of flowering time.
The role of MPK signaling cascades and protein phosphorylation in auxin signaling has been documented in numerous studies. For example, auxin was shown to activate an unknown MPK in Arabidopsis seedling roots (Mockaitis and Howell 2000). A tobacco MPK cascade activated in reponse to oxidative stress was found to negatively regulate auxin signal transduction pathway (Kovtun et al. 1998). Recently, the Arabidopsis MKK7 was shown to control polar auxin transport (Dai et al. 2006). However, there are no MPK substrates known to affect gene expression in response to auxin. We identified 12 auxin-related transcription factors as MPK substrates: eight IAA proteins (IAA1, IAA5, IAA8, IAA11, IAA13, IAA15, IAA24, and IAA31) and four auxin response factors (ARF2, ARF3, ARF10, and ARF18). These results suggest an extensive role of MPK network in auxin signaling.
Several recent reports describe the identification of new MPK targets in plants: ACS1/2 (Liu and Zhang 2004), EIN3 (Yoo et al. 2008), NtMAP65-1 (Sasabe et al. 2006), MKS1 (Andreasson et al. 2005), and VIP1 (Djamei et al. 2007). All these proteins, with the exception of VIP1, were not present on our protein microarrays used in the kinase experiments. To investigate whether these MPK substrates and/or their homologs can be also detected on protein microarrays, Map65-1, Map65-3, MKS1, ACS1, ACS4, ACS5, ACS7, ACS8, and EIN3 were cloned, expressed in N. benthamiana and printed on a protein chip. The protein chip was probed with purified MPK6 activated with MKK2 or MKK5 and weak phosphorylation signals were detected for Map65-1 and ACS1 proteins (data not shown). Low expression levels in our plant expression system for VIP1, EIN3, and other ACS family proteins (data not shown) most likely precluded the detection of a phosphorylation signal for these targets.
MAPK signaling cascades and regulation of cell death in plants
MPK cascades are functionally involved in regulating programmed cell death in plants. For example, activation of the tobacco homologs of MPK6 and MPK3 by a constitutively active MKK (Romeis et al. 1999; Yang et al. 2001) and ectopic expression of active mutants of SIPK (Zhang and Liu 2001), MKK4 and MKK5 has been previously shown to result in innate immunity related cell death (Ren et al. 2006). Here, we show that overexpression of MKK7- and MKK9-induced cell death in N. benthamiana. MKK7 and MKK9 are very similar proteins from the class D of the Arabidopsis MKKs. MKK7 was proposed to belong to a class of autoactivating MKK proteins based on the presence of an acidic residue (Asp) at the third position in the consensus activation loop S/TXXXXXS/T (Cardinale et al. 2002; Dai et al. 2006). MKK9 also possesses an Asp residue at the same position. Most likely, overexpression of both wild-type MKK7 and MKK9 results in their activation in the absence of an upstream MKKK. It is possible that the additional two acidic residues present in the activation loop of MKK7 EE and MKK9 EE resulted in their increased activation and thus, a more dramatic cell death phenotype. Moreover, our experiments with Sgt1-silenced plants suggest that MKK7 and MKK9 are involved in regulating innate immunity induced cell death response. This hypothesis is supported by previous studies showing that MKK7 regulates plant basal and systemic acquired resistance (Zhang et al. 2007), and that MKK9 activates MPK3/6 in protoplasts (Yoo et al. 2008).
Although it is likely that the accelerated death phenotype observed with the activated kinases in our experiments resulted as a consequence of substrate activation by the functional MKK/MPK combinations, future experiments will determine whether the signaling components function as a module in controlling cell death, or they activate separate pathways through endogenous signaling molecules.
Complex control of cellular processes by MPKs
Our global analysis of MPK signaling network circumvents the inherent bias of previous studies of signaling networks guided by traditional genetic approaches and reveals potential roles of MPK signaling in the control of different cellular processes. A complex image of signaling pathways emerges from our analysis. MPKs share a core set of effectors but also possess sets of highly specific effectors to differentiate the response. Moreover, the reconstructed signaling network supports the hypothesis of a combinatorial control of transcription through selective phosphorylation of a large number of substrates. We hypothesize that signaling cascades control transcription through a set of common response pathways and subsets of highly specific targets activated by combinations of MKKs and MPKs. Our analysis of signaling cascades components, coupled with data on their spatial and temporal gene expression and subcellular localization studies, should provide the basis for further studies of the MAPK signaling components.
Materials and methods
Cloning and preparation of activated MPK proteins
Wild-type Arabidopsis thaliana Col 0 MKK genes were obtained from the ABRC cDNA collection (MKK9, MKK4, and MKK3) or were PCR-amplified from genomic DNA (MKK5, MKK6, MKK7, and MKK10) and cloned into pYY63 (MKK-HA) using a Gateway cloning method. All MPK ORFs were subcloned into pLIC-cTAP (MPK-TAP) using the ligation-independent cloning method (Popescu et al. 2007). The Ser and Thr residues in the putative activation loop sequence S/TxxxxxS/T of all wild-type MKKs were replaced with Glu using a PCR-based mutagenesis scheme. Primer sequences will be made available upon request. All final clones were sequence confirmed.
The MPK-TAP clones were coinfiltrated into N. benthamiana with specific MKK-HA constructs. Only the MPK recombinant proteins were purified from infiltrated tissue as described (Popescu et al. 2007). Purified MPKs were used to evaluate the activity using MBP as a substrate. Briefly, one-fifth of the eluted volume was mixed with kinase buffer (25 mM Tris-HCl at pH 7.5, 1 mM EGTA, 10 mM MnCl2, 1 mM DTT) in the presence of [γ-32P] ATP (25 μCi/mL; Amersham Biosciences) for 30 min at 30°C. The reactions were stopped by adding 4× SDS-sample loading buffer and then run on a SDS-PAGE gel. The gels were dried and exposed to BioMax XAR film.
VIGS and protein overexpression
N. benthamiana plants were grown in pots at 25°C under continuous light. NbSGT1, NbRar1, NbEDS1 VIGS silencing vectors and VIGS vector alone controls used in this study has been described previously (Liu et al. 2002a,b). Agrobacterium strain GV2260 cultures containing TRV2-NbSgt1 or TRV2-NbRAR1 or TRV2-NbEDS1 or TRV2 vector alone were adjusted to an OD600 = 1.0, were mixed with pTRV1 OD600 = 1.0 in a 1:1 ratio, and infiltrated onto N. benthamiana plants. At 8 d post-infiltration leaves of these silenced plants were infiltrated with various combinations of MKKs and/or /MPKs; and monitored for induction of cell death up to 10 d. Leaf images were acquired using a Camedia E20 digital camera (Olympus) fitted with an MCON-35 macro extension lens (Olpymus).
Preparation of protein microarrays
Recombinant Arabidopsis proteins for microarray printing were generated using transient expression by Agoinfiltration and immunoaffinity purification procedures as described in Popescu et al. (2007). Briefly, ATEC ORFs were transformend into Agrobacterium tumefaciens GV2260. Liquid cultures of transformed A. tumefaciens were infiltrated into N. benthamiana 4-wk-old plants. Two leaves were infiltrated per construct and two constructs were infiltrated per plant. Tissue was collected at 4–5 dpi, grinded in liquid nitrogen, and stored at −80°C for protein purification. Eight-hundred microliters of total tissue extract were incubated at 4°C with IgG-covered Sepharose beads as described in Popescu et al. (2007) in the absence of phosphatase inhibitors. Beads were then washed extensively in Wash Buffer and Cleavage Buffer, and incubated overnight at 4°C with Precision Protease (Amersham Biosciences). The following day, the eluate was recovered, mixed with glycerol to a final concentration of 30%, and stored at −80°C. Purified protein preparations (3840), generated as described above, were deposited in duplicates onto amino-silane-coated microscope slides UltraGAPS (Corning Incorporated Corning) using a 48-pin ESI contact printer (Genomic Solutions). The proteins were printed in 48 blocks in a 10 × 16 spotting pattern. Printed slides were incubated overnight at 4°C and then transferred to −80°C for long-term storage. Several control spots were included in the slides: positional autophosphorylation controls (AT3g20530 and AT4G23650) spotted in the first two positions (row 1, column 1) and (row 1, column 2) of each of the 48 blocks, buffer-only and BSA spots.
In vitro kinase assays and kinase assays on protein microarrays
In vitro kinase assay were done in 96-well plates using purified recombinant proteins. 180 MKK and MPK combinations were incubated with 1 mM ATP in kinase buffer for 15 min at 30°C. A mixture of bovine brain dephosphorylated MBP (Upstate Biotechnologies, Cell Signaling Solutions) (0.25 μg/μL per reaction) and [γ-32P] ATP (0.2 mM per milliliter per one reaction) were added and incubated for 15 min at 30°C. Fifty microliters ofPBS were added to each reaction and the mixtures were transferred to nitrocellulose by running through a dot-blot apparatus. The membranes were washed for 2 h in PBS-Tween (0.1%) with three changes of buffer and exposed Kodak, BioMax XAR film for various lengths of time. For kinase assays on protein microarray, ∼500 ng of purified MPK-TAP protein in 200 μL of kinase buffer with [γ-33P] ATP per slide was overlaid and covered with a hybridization slip (Hybrislip, Grace Bio-Labs) and incubated for 30 min at 30°C in a humidified chamber. The slides were washed three times for 30 min in Washing Buffer (50 mM Tris-HCl at pH 7.5, 0.5% SDS) and once in double-distilled water, spun dry, covered in Saran Wrap, and exposed to X-ray Kodak film. Films with different exposure times were scanned at 4800-dot-per-inch resolution and acquired images were analyzed using GenePix software (Axon Instruments 4200A).
Phosphatase assays
For phosphatase assays, ∼40 μg of total protein extract were treated with protein phosphatase 1 and λ protein phosphatase (New England Biolabs), for 30 min to 1 h at room temperature, in the manufacturer supplied buffer. Treated extract were mixed with SDS sample buffer and separated on SDS-PAGE. Blots were probed with anti-cMyc mouse monoclonal antibodies (Santa Cruz Biotechnologies).
Protein immunoblotting
Total purified protein extracts were run on SDS-PAGE gels (in-house prepared or Novex Tris-Glycine gels 8% or 10% from Invitrogen), blotted onto PVDF Immobilon-P Transfer membrane (Millipore) and probed with primary antibodies anti-HA (Roche) or anti-cMyc mouse monoclonal antibody (Santa Cruz Biotechnologies). For the immunoscreening of the protein microarrays, printed protein slides were blocked in Super-Block Buffer (Pierce) for 2 h and then probed with anti-cMyc mouse antibody (Santa Cruz Biotechnologies) and Cy5-labeled anti-mouse secondary antibody as described (Popescu et al. 2007).
Data analysis of MBP phosphorylation assays
The MBP measurements are affected by MPK autophosphorylation and MKK autophosphorylation (phosphorylation in the absence of an active MPK). MPK and MKK autophosphorylation were robustly filtered by repeatedly selecting and changing outlier points such that to be within two standard deviations of the median. Data from each experiment was normalized to the average MBP autophosphorylation level on each plate. Variation of MPK autophosphorylation was estimated from all replicated data sets. Variation of MKK autophosphorylation was also estimated within each data set. Significant MKK–MPK phosphorylation events were identified by testing (one-side t-test) against the null hypothesis that the average signal intensity in repeated MBP assays is not larger than the average of MPK and MKK autophosphorylation level, assuming that the variance is due to both MPK and MKK autophosphorylation. To control the error rate in subsequent network identification steps, the false discovery rate (FDR) was controlled using the method of Benjamini and Hochberg (1995). The FDR rate was controlled at 0.05 level on the entire set of MKK phosphorylation events, with the P-value threshold of 0.0143. The decision was made by selecting all significant MPK phosphorylation events by constitutively activated and wild-type MKKs.
Data analysis of kinase assays
After exposure, the films were scanned and the resulting images were processed with GenePix 2.2. The array list file containing the ID of the printed proteins and their position on the microarray was imported into GenePix. The grid was arranged on the images according to the autophosphorylating positional controls. GPR files were generated for 108 probed microarrays (out of which 10 were controls), and used for data analysis. The average intensity value of duplicate spots was calculated for each printed protein and the resulted values were subjected to the intensity-dependent normalization. Normalized data were filtered by removing positional controls. To identify phosphorylated MPK we used a Bayesian decision method. We modeled the noise on each set of replicated experiments as having a normal distribution ∼N[μBk (i),σBk (i)],i = 1..108; noise parameters, μBk (i) and σBk (i), were estimated independently for each microarray using a robust estimator. The data points whose intensity was greater than two standard deviations above background mean were selected as putative targets (Supplemental Table 8): Protein j is phosphorylated by MPK(i) if s(j) − μBk(i) > 2 * σBk(i), where j is any protein on the microarrays probed with MPK(i), i = 1..10, j = 1..2158. Finally, the initial target predictions TI (i, j) were corrected by removing the autophosphorylating proteins detected in the controls common to each group of MPK phosphorylation assays. The final set of MPK targets TF(i, j) was validated by taking the decision, for each target, over the set of initial target decisions from both combinations of MKK/MPKs. A target was validated if it was present in at least one replicate in both MKK/MPK combinations or if it was present in at least half of the technical replicates: D[S(j),K(i)] = (A1A2 V A1A3 V…A3A4) V (B1B2 V…B3B4) V (A1 V…A4) ^ (B1 V…B4), where Ak = Dk {S(j), K(i) | KK1(i) Bk = Dk [S(j), K(i) | KK2(i)]} i = 1..10, j = 1..2158, k = 1..4. The final predicted MPK substrates were used to construct the MPK/substrates network. To control the error rate in subsequent inferences and analysis of the network, we applied the FDR method (Benjamini and Hochberg 1995). The P-values were computed from a one-side t-test with the null hypothesis that the average signal intensity in replicated microarray experiments is no larger than the background mean (Supplemental Table 9). The mean and variance of background were estimated from all sets corresponding to a MKK/MPK combination. The probability corresponding to MPK/substrate phosphorylation event was conditioned on the probability that the protein was phosphorylated in each of the two MKK/MPK combinations. A causal network was used to compute the weights of MPK -> Effectors phosphorylation network: w(i, j) = −log{P[H(i, j) | KK1 (i)]} − log{P[H(i, j) | KK2 (i)]}, i = 1..10, j = 1..2158, KK1 (i), KK2 (i) the two MKK used for in planta activation of MPK(i). The FDR on the 1331 protein interaction events was controlled at 0.05 with a P-value threshold of 0.0021.
Identification of GO categories for the MPK targets
Overrepresented GO categories in all MPK substrate lists were identified using Cytoscape version 2.2 (Shannon et al. 2003) at http://www.cytoscape.org and the BINGO plug-in software (Maere et al. 2005) at http://www.psb.ugent.be/cbd/papers/BiNGO. Two statistical methods were employed: the Hypergeometric statistical test, and the Benjamini and Hochberg FDR multiple testing correction with P-value of <0.05. Printed proteins were tested against all A. thaliana annotations from TAIR. The MPK targets were subsequently tested against all printed proteins.
Cluster analysis of MPK pathways
Cluster analysis was performed on MPK effector weights (−log [P-values]) using Cluster 2.11 (hierarchical clustering with average linking method) and visualized with TreeView (Eisen Laboratory).
Acknowledgments
This research was supported by NSF grants DBI-0519853 and DBI-0723722.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1740009.
Supplemental material is available at http://www.genesdev.org.
References
- Andreasson E., Jenkins T., Brodersen P., Thorgrimsen S., Petersen N.H., Zhu S., Qiu J.L., Micheelsen P., Rocher A., Petersen M., et al. The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 2005;24:2579–2589. doi: 10.1038/sj.emboj.7600737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.-L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. MAP kinase signaling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- Brader G., Djamei A., Teige M., Palva E.T., Hirt H. The MAP kinase kinase MKK2 affects disease resistance in Arabidopsis . Mol. Plant Microbe Interact. 2007;20:589–596. doi: 10.1094/MPMI-20-5-0589. [DOI] [PubMed] [Google Scholar]
- Bush S.M., Krysan P.J. Mutational evidence that the Arabidopsis MAP kinase MPK6 is involved in anther, inflorescence, and embryo development. J. Exp. Bot. 2007;58:2181–2191. doi: 10.1093/jxb/erm092. [DOI] [PubMed] [Google Scholar]
- Cardinale F., Meskiene I., Ouaked F., Hirt H. Convergence and divergence of stress-induced mitogen-activated protein kinase signaling pathways at the level of two distinct mitogen-activated protein kinase kinases. Plant Cell. 2002;14:703–711. [PMC free article] [PubMed] [Google Scholar]
- Chen R.E., Thorner J. Function and regulation in MAPK signaling pathways: Lessons learned from the yeast Saccharomyces cerevisiae . Biochim. Biophys. Acta. 2007;1773:1311–1340. doi: 10.1016/j.bbamcr.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y., Wang H., Li B., Huang J., Liu X., Zhou Y., Mou Z., Li J. Increased expression of MAP KINASE KINASE7 causes deficiency in polar auxin transport and leads to plant architectural abnormality in Arabidopsis . Plant Cell. 2006;18:308–320. doi: 10.1105/tpc.105.037846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despres C., Chubak C., Rochon A., Clark R., Bethune T., Desveaux D., Fobert P.R. The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell. 2003;15:2181–2191. doi: 10.1105/tpc.012849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh-Kumar S.P., Anandalakshmi R., Marathe R., Schiff M., Liu Y. Virus-induced gene silencing. Methods Mol. Biol. 2003;236:287–293. doi: 10.1385/1-59259-413-1:287. [DOI] [PubMed] [Google Scholar]
- Djamei A., Pitzschke A., Nakagami H., Rajh I., Hirt H. Trojan horse strategy in Agrobacterium transformation: Abusing MAPK defense signaling. Science. 2007;318:453–456. doi: 10.1126/science.1148110. [DOI] [PubMed] [Google Scholar]
- Eulgem T., Somssich I.E. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 2007;10:366–371. doi: 10.1016/j.pbi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Feilner T., Hultschig C., Lee J., Meyer S., Immink R.G.H., Koenig A., Possling A., Seitz H., Beveridge A., Scheel D., et al. High throughput identification of potential Arabidopsis mitogen-activated protein kinases substrates. Mol. Cell. Proteomics. 2005;4:1558–1568. doi: 10.1074/mcp.M500007-MCP200. [DOI] [PubMed] [Google Scholar]
- Friedman A., Perrimon N. High-throughput approaches to dissecting MAPK signaling pathways. Methods. 2006;40:262–271. doi: 10.1016/j.ymeth.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Gopalbhai K., Jansen G., Beauregard G., Whiteway M., Dumas F., Wu C., Meloche S. Negative regulation of MAPKK by phosphorylation of a conserved serine residue equivalent to Ser212 of MEK1. J. Biol. Chem. 2003;278:8118–8125. doi: 10.1074/jbc.M211870200. [DOI] [PubMed] [Google Scholar]
- Greenberg J.T., Yao N. The role and regulation of programmed cell death in plant–pathogen interactions. Cell. Microbiol. 2004;6:201–211. doi: 10.1111/j.1462-5822.2004.00361.x. [DOI] [PubMed] [Google Scholar]
- Ichimura K., Mizoguchi T., Yoshida R., Yuasa T., Shinozaki K. Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J. 2000;24:655–665. doi: 10.1046/j.1365-313x.2000.00913.x. [DOI] [PubMed] [Google Scholar]
- Kesarwani M., Yoo J., Dong X. Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis . Plant Physiol. 2007;144:336–346. doi: 10.1104/pp.106.095299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun Y., Chiu W.-L., Zeng W., Sheen J. Suppression of auxin signal transduction by a MAPK cascade in higher plants. Nature. 1998;395:716–720. doi: 10.1038/27240. [DOI] [PubMed] [Google Scholar]
- Lai L.B., Nadeau J.A., Lucas J., Lee E.-K., Nakagawa T., Zhao L., Geisler M., Sack F.D. The Arabidopsis R2R3 MYB proteins FOUR LIPS and MYB88 restrict divisions late in the stomatal cell lineage. Plant Cell. 2005;17:2754–2767. doi: 10.1105/tpc.105.034116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M., Wan J., Czechowski T., Udvardi M., Stacey G. Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol. Plant Microbe Interact. 2007;20:900–911. doi: 10.1094/MPMI-20-8-0900. [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhang S. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis . Plant Cell. 2004;16:3386–3399. doi: 10.1105/tpc.104.026609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Marathe R., Dinesh-Kumar S.P. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 2002a;30:415–429. doi: 10.1046/j.1365-313x.2002.01297.x. [DOI] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Serino G., Deng X.-W., Dinesh-Kumar S.P. Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene-mediated resistance response to tobacco mosaic virus. Plant Cell. 2002b;14:1483–1496. doi: 10.1105/tpc.002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S., Heymans K., Kuiper M. BiNGO: A cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- Mao P., Duan M., Wei C., Li Y. WRKY62 transcription factor acts downstream of cytosolic NPR1 and negatively regulates jasmonate-responsive gene expression. Plant Cell Physiol. 2007;48:833–842. doi: 10.1093/pcp/pcm058. [DOI] [PubMed] [Google Scholar]
- Matsuoka D., Nanmori T., Sato K.-i., Fukami Y., Kikkawa U., Yasuda T. Activation of AtMEK1, an Arabidopsis mitogen-activated protein kinase kinase, in vitro and in vivo: Analysis of active mutants expressed in E. coli and generation of the active form in stress response in seedlings. Plant J. 2002;29:637–647. doi: 10.1046/j.0960-7412.2001.01246.x. [DOI] [PubMed] [Google Scholar]
- Menke F., Kang H., Chen Z., Park J., Kumar D., Klessig D. Tobacco transcription factor WRKY1 is phosphorylated by the MAP kinase SIPK and mediates HR-like cell death in tobacco. Mol. Plant Microbe Interact. 2005;18:1027–1034. doi: 10.1094/MPMI-18-1027. [DOI] [PubMed] [Google Scholar]
- Meszaros T., Helfer A., Hatzimasoura E., Magyar Z., Serazetdinova L., Rios G., Bardoczy V., Teige M., Koncz C., Peck S., et al. The Arabidopsis MAP kinase kinase MKK1 participates in defence responses to the bacterial elicitor flagellin. Plant J. 2006;48:485–498. doi: 10.1111/j.1365-313X.2006.02888.x. [DOI] [PubMed] [Google Scholar]
- Miao Y., Zentgraf U. The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell. 2007;19:819–830. doi: 10.1105/tpc.106.042705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockaitis K., Howell S.H. Auxin induces mitogenic activated protein kinase (MAPK) activation in roots of Arabidopsis seedlings. Plant J. 2000;24:785–796. doi: 10.1046/j.1365-313x.2000.00921.x. [DOI] [PubMed] [Google Scholar]
- Noh B., Lee S.H., Kim H.J., Yi G., Shin E.A., Lee M., Jung K.J., Doyle M.R., Amasino R.M., Noh Y.S. Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell. 2004;16:2601–2613. doi: 10.1105/tpc.104.025353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedley K.F., Martin G.B. Role of mitogen-activated protein kinases in plant immunity. Curr. Opin. Plant Biol. 2005;8:541–547. doi: 10.1016/j.pbi.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Pillitteri L.J., Sloan D.B., Bogenschutz N.L., Torii K.U. Termination of asymmetric cell division and differentiation of stomata. Nature. 2007;445:501–505. doi: 10.1038/nature05467. [DOI] [PubMed] [Google Scholar]
- Pitzschke A., Hirt H. Mitogen-activated protein kinases and reactive oxygen species signaling in plants. Plant Physiol. 2006;141:351–356. doi: 10.1104/pp.106.079160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu S.C., Popescu G.V., Bachan S., Zhang Z., Seay M., Gerstein M., Snyder M., Dinesh-Kumar S.P. Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc. Natl. Acad. Sci. 2007;104:4730–4735. doi: 10.1073/pnas.0611615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacek J., Devgan G., Michaud G., Zhu H., Zhu X., Fasolo J., Guo H., Jona G., Breitkreutz A., Sopko R., et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- Ren D., Yang K., Li G., Liu Y., Zhang S. Activation of Ntf4, a tobacco mitogen-activated protein kinase, during plant defense response and its involvement in hypersensitive response-like cell death. Plant Physiol. 2006;141:1482–1493. doi: 10.1104/pp.106.080697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S., Somssich I.E. A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J. 2001;28:123–133. doi: 10.1046/j.1365-313x.2001.01131.x. [DOI] [PubMed] [Google Scholar]
- Romeis T., Piedras P., Zhang S., Klessig D.F., Hirt H., Jones J.D.G. Rapid Avr 9- and Cf-9-dependent activation of MAP kinases in tobacco cell cultures and leaves: Convergence of resistance gene, elicitor, wound, and salicylate responses. Plant Cell. 1999;11:273–288. doi: 10.1105/tpc.11.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasabe M., Soyano T., Takahashi Y., Sonobe S., Igarashi H., Itoh T.J., Hidaka M., Machida Y. Phosphorylation of NtMAP65-1 by a MAP kinase down-regulates its activity of microtubule bundling and stimulates progression of cytokinesis of tobacco cells. Genes & Dev. 2006;20:1004–1014. doi: 10.1101/gad.1408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K., Schulze-Lefert P. Complex formation, promiscuity and multi-functionality: Protein interactions in disease-resistance pathways. Trends Plant Sci. 2003;8:252–258. doi: 10.1016/S1360-1385(03)00104-3. [DOI] [PubMed] [Google Scholar]
- Takahashi F., Yoshida R., Ichimura K., Mizoguchi T., Seo S., Yonezawa M., Maruyama K., Yamaguchi-Shinozaki K., Shinozaki K. The mitogen-activated protein kinase cascade MKK3–MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis . Plant Cell. 2007;19:805–818. doi: 10.1105/tpc.106.046581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teige M., Scheikl E., Eulgem T., Doczi R., Ichimura K., Shinozaki K., Dangl J.L., Hirt H. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis . Mol. Cell. 2004;15:141–152. doi: 10.1016/j.molcel.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Wang D., Amornsiripanitch N., Dong X. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2006;2:e123. doi: 10.1371/journal.ppat.0020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ngwenyama N., Liu Y., Walker J.C., Zhang S. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell. 2007;19:63–73. doi: 10.1105/tpc.106.048298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmarsh A.J. Regulation of gene transcription by mitogen-activated protein kinase signaling pathways. Biochim. Biophys. Acta. 2007;1773:1285–1298. doi: 10.1016/j.bbamcr.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Wiermer M., Feys B.J., Parker J.E. Plant immunity: The EDS1 regulatory node. Curr. Opin. Plant Biol. 2005;8:383–389. doi: 10.1016/j.pbi.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Xing T., Malik K., Martin T., Miki B.L. Activation of tomato PR and wound-related genes by a mutagenized tomato MAP kinase kinase through divergent pathways. Plant Mol. Biol. 2001;46:109–120. doi: 10.1023/a:1010633215445. [DOI] [PubMed] [Google Scholar]
- Yang K., Liu Y., Zhang S. Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc. Natl. Acad. Sci. 2001;98:741–746. doi: 10.1073/pnas.98.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S., Cho Y., Tena G., Xiong Y., Sheen J. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature. 2008;451:789–795. doi: 10.1038/nature06543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Liu Y. Activation of salicylic acid-induced protein kinase, a mitogen-activated protein kinase, induces multiple defense responses in tobacco. Plant Cell. 2001;13:1877–1889. doi: 10.1105/TPC.010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Dai Y., Xiong Y., DeFraia C., Li J., Dong X., Mou Z. Overexpression of Arabidopsis MAP kinase kinase 7 leads to activation of plant basal and systemic acquired resistance. Plant J. 2007;52:1066–1079. doi: 10.1111/j.1365-313X.2007.03294.x. [DOI] [PubMed] [Google Scholar]
- Zimmermann P., Hennig L., Gruissem W. Gene-expression analysis and network discovery using Genevestigator. Trends Plant Sci. 2005;10:407–409. doi: 10.1016/j.tplants.2005.07.003. [DOI] [PubMed] [Google Scholar]