Abstract
It has been conventional for psychiatric research, including the search for predisposing genes, to proceed under the assumption that schizophrenia and bipolar disorder are separate disease entities with different underlying etiologies. These represent Emil Kraepelin's traditional dichotomous classification of the so-called “functional” psychoses and form the basis of modern diagnostic practice. However, findings emerging from many fields of psychiatric research do not fit well with this model. In particular, the pattern of findings emerging from genetic studies shows increasing evidence for an overlap in genetic susceptibility across the traditional classification categories—including association findings at DAOA(G72), DTNBP1 (dysbindin), COMT, BDNF, DISC1, and NRG1. The emerging evidence suggests the possibility of relatively specific relationships between genotype and psychopathology. For example, DISC1 and NRG1 may confer susceptibility to a form of illness with mixed features of schizophrenia and mania. The elucidation of genotype-phenotype relationships is at an early stage, but current findings highlight the need to consider alternative approaches to classification and conceptualization for psychiatric research rather than continuing to rely heavily on the traditional Kraepelinian dichotomy. As psychosis susceptibility genes are identified and characterized over the next few years, this will have a major impact on our understanding of disease pathophysiology and will lead to changes in classification and the clinical practice of psychiatry.
Keywords: schizophrenia, bipolar disorder, psychosis, nosology, diagnosis, classification, genetics
Background
It has been conventional for psychiatric research, including the search for predisposing genes, to proceed under the assumption that schizophrenia and bipolar disorder are separate disease entities with different underlying etiologies. These represent Emil Kraepelin's traditional dichotomous classification of the so-called “functional” psychoses1,2 and form the basis of modern diagnostic practice as defined operationally in DSMIV3 and ICD10.4 However, there has been a long tradition of dissent against the validity of this view,5–7 and the veracity and utility of the so-called Kraepelinian dichotomy have been increasingly challenged by emerging data from many fields of psychiatric research.8–10 The most convincing challenges have come in recent years from genetic studies. These findings are of great potential for informing our understanding of nosology as well as delineating the biological mechanisms that contribute to the pathogenesis of the psychoses.
In this article we will first briefly review the key pieces of genetic evidence that support the existence of a genetic relationship between bipolar disorder and schizophrenia. We will then consider the current data for genes that have, to date, been implicated in both disorders. We will end by considering the implications.
Genetic Evidence Supporting Non-Independence of Schizophrenia and Bipolar Disorder
Most current genetic studies of schizophrenia and bipolar disorder have been based on the assumption of independence, with individual studies typically focusing on only one or the other disorder. Cases with a mix of mood and psychotic features, while common, have been ignored or subsumed into some broader category of either schizophrenia or bipolar disorder. Although there is a substantial body of family, twin, and adoption data that has been interpreted to support the dichotomous view,11,12 some earlier studies and several more recent studies are inconsistent with this view, as are the data emerging from molecular genetic investigations. Although it is beyond the scope of this article to review the data in detail, key pieces of evidence include the following.
Family Studies
Although many family studies have demonstrated that schizophrenia and bipolar disorder tend to “breed true,”13–15 families are known in which there are multiple cases of schizophrenia, bipolar disorder, and cases in which both psychosis and mood disorder occur.16 Further, some studies have shown statistically significant evidence that bipolar disorder occurs at increased rates in relatives of probands with schizophrenia,17 and that schizophrenia occurs at increased frequency in relatives of probands with bipolar disorder.18 Moreover, schizoaffective disorder has been shown to occur at increased rates both in families of probands with schizophrenia19 and in those of probands with bipolar disorder,20 and both schizophrenia and bipolar disorder have been shown to occur at increased rates in families of probands with schizoaffective disorder.20
Twin Studies
Only one twin study has used an analysis that was unconstrained by the diagnostic hierarchy inherent in current classification systems (i.e., the principle that schizophrenia “trumps” mood disorder in diagnosis). This study demonstrated a clear overlap in genetic susceptibility to syndromically defined mania and schizophrenia.21 In addition to supporting the existence of some susceptibility genes that are specific to schizophrenia and others that are specific to bipolar disorder, it suggested that there are others that influence susceptibility to schizoaffective disorder, schizophrenia, and bipolar disorder. A graphic illustration of the varied expression of the same set of susceptibility genes is provided by the Maudsley triplets—a set of genetically identical triplets, two of whom had a lifetime diagnosis of schizophrenia and the third a lifetime diagnosis of bipolar disorder.22
Linkage Studies
Genetic linkage studies have identified some chromosome regions that show convergent or overlapping regions of interest in both schizophrenia and bipolar disorder—including regions of 13q, 22q, 6q, and 18.23,24 There are, however, difficulties in interpreting overlaps in linkage findings from different phenotypes because of the poor localization of linkage signals for complex disorders and difficulties assessing statistical evidence for significant co-occurrence. However, the hypothesis that loci exist that influence susceptibility across the schizophrenia-bipolar divide receives further support from the observation that a genome scan using families selected on the basis of a member with DSMIV schizoaffective disorder, bipolar type, demonstrated genome-wide significance at 1q42 and suggestive linkage at 22q11, with linkage evidence being contributed equally by “schizophrenia” families (i.e., where other members had predominantly schizophrenia) and “bipolar families” (i.e., where other members had predominantly bipolar disorder).25
Studies of Individual Genes
Linkage studies can only provide at best indirect evidence for shared genetic effects. More direct evidence has come from reports implicating variation in the same genes as influencing susceptibility to both schizophrenia and bipolar disorder (see Table 1). In most cases the gene was first implicated in studies of schizophrenia, and the evidence in most cases is strongest for this phenotype. This could reflect the true contribution to the phenotype or may simply reflect the fact that substantially greater resources and samples have been used to date on studies of schizophrenia. We will consider the evidence for each gene in turn.
Table 1.
Summary of current weight of available evidence supporting several of the more promising genes implicated in the pathogenesis of categorically defined schizophrenia, bipolar disorder, and mixed bipolar-psychosis phenotypes*
| Gene/Locus | Chromosomal Location | Evidence in Schizophrenia | Evidence in Cases with Mixed Bipolar-Psychosis Features | Evidence in Bipolar Disorder |
| Dysbindin | 6p22 | + + + + + | + | |
| Neuregulin 1 | 8p12 | + + + + | + | + |
| DISC 1 | 1q42 | + + + | ++ | + |
| COMT | 22q11 | + | + | |
| DAOA(G72)/G30 | 13q33 | + + | + + | |
| BDNF | 11p13 | + | + + |
More + symbols indicate greater evidence. The scale is relative. Note that very few studies have been undertaken explicitly using cases with mixed phenotype (although many “schizophrenia” and bipolar disorder” samples will include a substantial proportion of cases with a mix of mood and psychotic features). Note that the limitations of the current approach of examining diagnostic categories makes it difficult to determine whether overlaps represent a non-specific contribution to susceptibility across traditional categories or a relatively specific contribution to susceptibility to one or more domains of the phenotype (such as depression or paranoid ideation).
NRG1
NRG1 was first implicated in schizophrenia in the Icelandic population after a systematic study of 8p21-22 revealed association between schizophrenia and a multi-marker haplotype at the 5′ end of NRG1.26 Strong evidence for association with the same haplotype was subsequently found in a large sample from Scotland,27 with considerably weaker support in our own UK sample.28 These and subsequent studies of NRG1 in schizophrenia have been reviewed recently in Schizophrenia Bulletin.29 Overall, there is strong evidence from several studies that genetic variation in NRG1 confers risk to schizophrenia, but as yet, and in spite of extensive re-sequencing, specific susceptibility and protective variants have not been identified. NRG1 has not yet been extensively studied in bipolar disorder. However, the one study published to date found significant evidence for association of the core Icelandic risk haplotype with susceptibility to bipolar disorder with a similar effect size to that seen by the same group in schizophrenia.30 In the bipolar cases with predominantly mood-incongruent psychotic features, the effect was greater, as was the case in the subset of schizophrenia cases (n = 27) who had experienced mania. These findings should be treated with caution pending replication, but they suggest that NRG1 plays a role in influencing susceptibility to both bipolar disorder and schizophrenia, and that it may exert a specific effect in the subset of functional psychosis that has manic and mood-incongruent psychotic features.
Dysbindin (DTNBP1)
Evidence implicating dystrobrevin binding protein 1 (DTNBP1), also known as dysbindin, in schizophrenia was first reported by Straub et al.,31 and there is now quite impressive support from a number of studies reviewed recently in Schizophrenia Bulletin.32 However, once again various markers and haplotypes have been associated, and the actual susceptibility variants have yet to be identified. Only one published study has so far examined SNPs from dysbindin in bipolar disorder. Raybould and colleagues33 found no significant associations in bipolar disorder as a whole but found modestly significant evidence for association in the subset of bipolar cases with predominantly psychotic episodes of mood disturbance with a similar pattern of findings to that seen by this group in schizophrenia. This finding suggests that variation in dysbindin confers risk to psychosis rather than to mood disorders per se, although replication is required. Recent work in the Irish Study of High-Density Schizophrenia Families has shown that schizophrenic patients with negative symptoms were more likely to inherit the dysbindin risk haplotype,34 raising the possibility that negative symptoms might also define the sub-group of psychotic bipolars that are particularly likely to carry the dysbindin risk haplotype.
G72(DAOA)/G30
This locus was first implicated in studies of schizophrenia by Chumakov and colleagues,35 who undertook association mapping in the linkage region on chromosome 13q22-34. They found associations in French Canadian and Russian populations in markers around two novel genes, G72 and G30, which are overlapping but transcribed in opposite directions. G72 is a primate-specific gene expressed in the caudate and amygdala. Using yeast two-hybrid analysis, evidence for physical interaction was found between G72 and D-amino-acid oxidase (DAO). DAO is expressed in the human brain, where it oxidizes D-serine, a potent activator of NMDA glutamate receptor. Co-incubation of G72 and DAO in vitro revealed a functional interaction with G72 enhancing the activity of DAO. Consequently, G72 has now been named D-amino-acid oxidase activator (DAOA).
Associations between DAOA and schizophrenia have subsequently been reported in samples from Germany,36 China,37 Ashkenazi Jews,38 and both the United States and South Africa,39 as well as a small sample of very early onset psychosis subjects from the United States.40 There is no consensus concerning the specific risk alleles or haplotypes across studies.
Currently this is the best-supported locus for bipolar disorder. At least five independent datasets contribute evidence that variation at the DAOA/G30 locus on chromosome 13q influences susceptibility to bipolar disorder, including three US family samples,41,42 a German case-control sample,36 and a large UK case-control sample.43 In all studies, evidence for association came from both individual SNPs as well as multilocus haplotypes, although there is variation between studies in the associated SNPs and haplotypes. No pathologically relevant variant has yet been identified, and the biological mechanism remains to be elucidated. The largest study to date was of 2831 individuals, of whom 709 had DSMIV schizophrenia, 706 had bipolar-I disorder, and 1416 were ethnically matched controls.43 The authors identified significant association with bipolar disorder but failed to find association with schizophrenia. Analyses across the traditional diagnostic categories revealed significant evidence for association in the subset of cases (n = 818) in which episodes of major mood disorder had occurred. A similar pattern of association was observed in both bipolar cases and schizophrenia cases in which individuals had experienced major mood disorder. In contrast, there was no evidence for association in the subset of cases (n = 1153) in which psychotic features occurred. This finding requires replication, but the data as they stand suggest that, despite being originally reported as a schizophrenia susceptibility locus, variation in DAOA/G30 does not primarily increase susceptibility for prototypical schizophrenia or psychosis. Instead, it appears that variation in DAOA/G30 influences susceptibility to episodes of mood disorder across the traditional bipolar and schizophrenia categories. Importantly, these findings also imply that whether or not significant associations are seen in schizophrenia will depend upon the proportion of cases that have suffered from episodes of mood disorder, and remind us of the importance of sample differences in the replicability of genetic association studies.
Disrupted in Schizophrenia 1 (DISC1)
This gene was implicated through studies of an extended pedigree in which a balanced chromosomal translocation (1;11)(q42;q14.3) showed strong evidence for linkage to a fairly broad phenotype comprising schizophrenia, bipolar disorder, and recurrent depression.44 The translocation was found to disrupt two genes on chromosome 1: DISC1 and DISC2.44,45 DISC2 contains no open reading frame and may regulate DISC1 expression via anti-sense RNA.45 A small pedigree has recently been reported in which a 4bp deletion in exon 12 of DISC1 cosegregates with schizophrenia and schizoaffective disorder.46 Interestingly, DISC 1 and 2 are located close to the chromosome 1 markers implicated in two Finnish linkage studies of schizophrenia.47,48 The Edinburgh group that identified DISC1 found no linkage evidence in their own schizophrenia sample but did find suggestive evidence for linkage in bipolar disorder.49 More recently, Hamshere and colleagues reported genome-wide significant evidence for linkage at this locus in a linkage study of schizoaffective disorder, bipolar type.50 DISC1 is certainly an interesting candidate gene for mental disorder, but it is important to remember that translocations exert effects on genes other than those directly disrupted. For example, there are several mechanisms by which a translocation can influence the expression of neighboring genes. In order to unequivocally implicate DISC1 and/or 2 in the pathogenesis of schizophrenia, it is necessary to identify mutations or polymorphisms that are associated with schizophrenia in non-deleted cases, and are not in linkage disequilibrium with neighboring genes. Negative studies were initially reported by the Edinburgh group with a small number of markers,51 and by a group that focused on the 5' end of the gene in a large Japanese sample.52 More recently, several groups have reported positive findings,53–56 although in no case are the results compelling, and there is little agreement as to the specific markers or haplotypes showing association. Interestingly, in three of these studies, associations were observed with bipolar disorder as well as schizophrenia,53–55 and in one, the strongest association was observed with schizoaffective disorder.54
While no consistent pattern of association has yet emerged and no pathogenically relevant variants have been established, the convergence of the linkage data are strongly suggestive that variation in DISC1 or another gene in this region influences susceptibility to mood-psychosis phenotypes that cut across the traditional Kraepelinian divide.
COMT
COMT lies in the chromosome 22q11 region implicated in schizophrenia, bipolar disorder, and schizoaffective disorder bipolar type by linkage studies23,28,50,57,58 and by its deletion in Velo-cardio-facial syndrome, in which adult sufferers frequently develop affective and psychotic disorders. COMT has been intensively studied because of its key role in dopamine catabolism. Most studies have focused upon a valine to methionine change at codon 158 of the brain-predominant membrane-bound form of COMT (MB-COMT) and codon 108 of the soluble form (S-COMT). The valine allele confers higher activity and thermal stability to both forms of COMT59 and has been fairly consistently associated with reduced performance in tests of frontal lobe function.60,61 The results in schizophrenia have been mixed, with recent meta-analyses62–64 reporting no overall evidence for association with the valine allele. Some studies have found stronger evidence for association between haplotypes at COMT than for the Val/Met locus alone,59,65–67 although a recent study found no evidence for association in a study of more than 2800 individuals, including almost 1200 schizophrenics.68 Moreover, it is difficult to reconcile the stronger evidence for association with haplotypes at COMT with the observation that COMT activity, at least in peripheral tissues,69 is largely dictated by the Val/Met locus, a finding that may also be true in the brain.59 The evidence does not support a simple role for Val/Met 158 in susceptibility to schizophrenia, although a small effect on susceptibility cannot be excluded, nor can a role in phenotype modification. It also remains possible that variation elsewhere in COMT, or in a neighboring gene such as ARVCF66,70 confers susceptibility.
COMT has been studied less in bipolar disorder, although some evidence for association at the Val/Met polymorphism was found in one meta-analysis71 and modest haplotypic evidence in one Israeli study.72 It is extremely likely that genetic variation in this region influences susceptibility across the psychosis spectrum, although it is not yet clear that COMT itself is the only (or the major) susceptibility gene at this locus. If it is, the mechanism is complex and the phenotype not yet adequately defined.
Brain Derived Neurotrophic Factor (BDNF)
A functional candidate gene that has attracted a great deal of recent interest in mood disorder is Brain Derived Neurotrophic Factor (BDNF).73 BDNF plays an important role in promoting and modifying growth, development, and survival of neuronal populations and, in the mature nervous system, is involved in activity-dependent neuronal plasticity,74 processes that are prominent in the synaptic plasticity hypothesis of mood disorder, which focuses on the functional and structural changes induced by stress and antidepressants at the synaptic level. The BDNF gene lies on chromosome 11p13 and encodes a precursor peptide (proBDNF), which is cleaved proteolytically to form the mature protein.75 The 11p13 chromosomal location of BDNF has been implicated in some linkage studies of bipolar disorder but not in meta-analyses of linkage studies. Only one frequent, non-conservative polymorphism in the human BDNF gene has been identified, a single nucleotide polymorphism (SNP) at nucleotide 196 within the 5′ pro-BDNF sequence that causes an amino acid substitution of valine to methionine at codon 66 (Val66Met) and may have a functionally relevant effect by modifying the processing and trafficking of BDNF.76
There have been three positive reports using family-based association studies of Caucasian bipolar disorder samples of European-American origin and the Val66Met SNP: two are in adult bipolar samples,77,78 and one is with a small childhood onset sample.79 All have shown over-transmission of the common Val allele. Evidence with multilocus haplotypes was stronger in one study.78 There have been four case-control association studies (of European,80,81 Chinese,82 and Japanese origin83) to date, in which there is no evidence for an allelic or genotypic association. In our own UK case-control study of over 1000 bipolar cases, we found no significant evidence for association of the Val allele (or multilocus haplotypes) with bipolar disorder but some evidence for association within the subset of cases in which rapid cycling of mood had occurred at some during illness.84 This finding has been replicated in a re-analysis of one of the original family samples showing evidence for association.85 BDNF has attracted less interest in schizophrenia, although there has been a positive report of association at Val66Met and a 2 locus haplotype including this polymorphism.86 Of substantial interest is the recent finding of association with depression in schizophrenia as well as in unipolar disorder.87 Such a finding has some consistency with the rapid cycling finding in bipolar disorder because depression is a prominent feature of such cases.
Substantial additional genetic and biological work will be required to confirm (or refute) the role of BDNF in influencing susceptibility to mood and psychotic illness. Systematic study of variation across the whole gene is required with study in further independent samples.
Conclusions
Current genetic findings are beginning to provide suggestive evidence that, as implied by the family and twin data, there are genetic loci that contribute susceptibility across the Kraepelinian divide to schizophrenia, bipolar disorder, and schizoaffective disorders (see Table 1). This work is in its early stages, and the findings should be treated with caution, especially given that for none of the genes implicated have specific risk variants been identified. Indeed, it may turn out that some, or all, of the genes discussed contain multiple risk (and protective) variants with effects on different aspects of psychopathology. However, the simplest interpretation of the current published data is that variation in DISC1/2 and NRG1 appears to predispose to both prototypical Kraepelinian illnesses, but there is suggestive evidence that the effects of both genes might be felt most strongly in disorders with features of both schizophrenia and bipolar disorder. Variation in DTNBP1 predominantly predisposes to schizophrenia and possibly a form of illness characterized by prominent negative symptoms, with an effect on bipolar disorder confined to those cases with prominent psychotic features. In contrast, DAOA/G30 and BDNF appear to be most strongly associated with mood disorder, and the extent to which associations with schizophrenia are seen is likely to depend upon the proportion of cases that have experienced mood disorder syndromes.
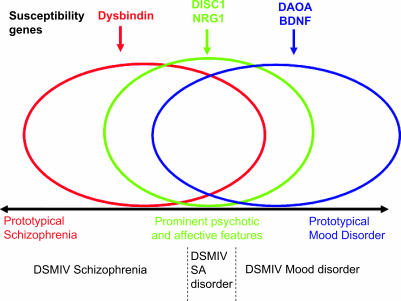
These findings have important implications for classification of the major psychiatric disorders because they suggest an overlap in the biological basis of disorders that have, over the last 100 years, been classified as distinct entities.8 Thus, we can expect that over the coming years molecular genetics will catalyze a re-appraisal of psychiatric nosology as well as providing a path to understanding the pathophysiology that will facilitate development of improved treatments. For example, current genetic findings suggest that rather than classifying psychosis as a dichotomy, a more useful formulation may be to conceptualize alternative categories or a spectrum of clinical phenotypes with susceptibility conferred by overlapping sets of genes8 (see Figure 1). It is important that researchers are willing to embrace and explore such alternative approaches to the clinical phenotype in order to interpret the accumulating data. This will be an iterative process with identified genetic signals allowing refinement of the phenotype and the refined phenotype allowing increased power to detect further genetic signals. To facilitate this approach, it will be important to use large samples that have a full representation of phenotypes across the mood-psychosis spectrum, and detailed, high-quality phenotypic assessments, preferably including dimensional measures88,89. Already it is possible to recognize some common biological features among the genes implicated by current studies, and tentative models have been advanced that postulate the key role of synaptic function.90
Fig. 1.
Simplified hypothesized relationship between specific susceptibility genes (above the black line) and clinical phenotype (below the line) using the model outlined in Craddock and Owen8 The overlapping ellipses represent overlapping sets of genes: red influencing susceptibility to phenotypes with prominent schizophrenia-like features, blue to prominent mood features, and green to phenotypes with a prominent mix of both types of feature. These assignments are based on current data and are likely to require revision as more data accumulate.
In conclusion, accumulating evidence supports the existence of an overlap in genetic susceptibility across the traditional Kraepelinian divide with studies of several genes providing the most compelling such evidence. This work is at an early stage but has the potential to change our conception of psychiatric nosology as well as our understanding of the pathogenesis of psychopathology.
Acknowledgments
The authors' work on the genetics of psychosis and mood disorders is funded through grants from the Wellcome Trust and the MRC. We are indebted to all the participants in our studies.
References
- 1.Kraepelin E. Manic-Depressive Insanity and Paranoia. Edinburgh: Livingstone; 1919. [Google Scholar]
- 2.Kendell RE. Diagnosis and classification of functional psychoses. Br Med Bull. 1987;43:499–513. doi: 10.1093/oxfordjournals.bmb.a072198. [DOI] [PubMed] [Google Scholar]
- 3.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 4th ed. Washington, DC: American Psychiatric Press; 1994. [Google Scholar]
- 4.WHO. The ICD10 Classification of Mental and Behavioural Disorders. Geneva, World Health Organization; 1993. Diagnostic criteria for research. [Google Scholar]
- 5.Brockington IF, Kendell RE, Wainwright S, Hillier VF, Walker J. The distinction between the affective psychoses and schizophrenia. Br J Psychiatry. 1979;135:243–248. doi: 10.1192/bjp.135.3.243. [DOI] [PubMed] [Google Scholar]
- 6.Crow TJ. The continuum of psychosis and its genetic origins. The sixty-fifth Maudsley lecture. Br J Psychiatry. 1990;156:788–797. doi: 10.1192/bjp.156.6.788. [DOI] [PubMed] [Google Scholar]
- 7.Taylor MA. Are schizophrenia and affective disorder related? A selective literature review. Am J Psychiatry. 1992;149:22–32. doi: 10.1176/ajp.149.1.22. [DOI] [PubMed] [Google Scholar]
- 8.Craddock N, Owen MJ. The beginning of the end for the Kraepelinian dichotomy. Br J Psychiatry. 2005;186:364–366. doi: 10.1192/bjp.186.5.364. [DOI] [PubMed] [Google Scholar]
- 9.Van Os J, Gilvarry C, Bale R, Van Horn E, Tattan T, White I, Murray R. A comparison of the utility of dimensional and categorical representations of psychosis. UK700 Group. Psychol Med. 1999;29:595–606. doi: 10.1017/s0033291798008162. [DOI] [PubMed] [Google Scholar]
- 10.Murray RM, Sham P, Van Os J, Zanelli J, Cannon M, McDonald C. A developmental model for similarities and dissimilarities between schizophrenia and bipolar disorder. Schizophr Res. 2004;71:405–416. doi: 10.1016/j.schres.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Gottesman II. Schizophrenia Genesis: The Origins of Madness. New York: Freeman; 1991. [Google Scholar]
- 12.Tsuang MT, Faraone SV. The Genetics of Mood Disorders. Baltimore: The Johns Hopkins University Press; 1990. [Google Scholar]
- 13.Gershon ES, Hamovit J, Guroff JJ, et al. A family study of schizoaffective, bipolar I, bipolar II, unipolar, and normal control probands. Arch Gen Psychiatry. 1982;39:1157–1167. doi: 10.1001/archpsyc.1982.04290100031006. [DOI] [PubMed] [Google Scholar]
- 14.Frangos E, Athanassenas G, Tsitourides S, et al. Prevalence of DSM III schizophrenia among the first-degree relatives of schizophrenic probands. Acta Psychiatr Scand. 1985;72:382–386. doi: 10.1111/j.1600-0447.1985.tb02625.x. [DOI] [PubMed] [Google Scholar]
- 15.Baron M, Gruen R, Asnis L, et al. Schizoaffective illness, schizophrenia and affective disorders: morbidity risk and genetic transmission. Acta Psychiatr Scand. 1982;65:253–262. doi: 10.1111/j.1600-0447.1982.tb00845.x. [DOI] [PubMed] [Google Scholar]
- 16.Pope HG, Jr, Yurgelun-Todd D. Schizophrenic individuals with bipolar first-degree relatives: analysis of two pedigrees. J Clin Psychiatry. 1990;51:97–101. [PubMed] [Google Scholar]
- 17.Tsuang MT, Winokur G, Crowe RR. Morbidity risks of schizophrenia and affective disorders among first degree relatives of patients with schizophrenia, mania, depression and surgical conditions. Br J Psychiatry. 1980;137:497–504. doi: 10.1192/bjp.137.6.497. [DOI] [PubMed] [Google Scholar]
- 18.Valles V, Van Os J, Guillamat R, et al. Increased morbid risk for schizophrenia in families of in-patients with bipolar illness. Schizophr Res. 2000;42:83–90. doi: 10.1016/s0920-9964(99)00117-6. [DOI] [PubMed] [Google Scholar]
- 19.Kendler KS, Karkowski LM, Walsh D. The structure of psychosis: Latent class analysis of probands from the Roscommon Family Study. Arch Gen Psychiatry. 1998;55:492–499. doi: 10.1001/archpsyc.55.6.492. [DOI] [PubMed] [Google Scholar]
- 20.Rice J, Reich T, Andreasen NC, et al. The familial transmission of bipolar illness. Arch Gen Psychiatry. 1987;44:441–447. doi: 10.1001/archpsyc.1987.01800170063009. [DOI] [PubMed] [Google Scholar]
- 21.Cardno AG, Rijsdijk FV, Sham PC, et al. A twin study of genetic relationships between psychotic symptoms. Am J Psychiatry. 2002;159:539–545. doi: 10.1176/appi.ajp.159.4.539. [DOI] [PubMed] [Google Scholar]
- 22.McGuffin P, Reveley A, Holland A. Identical triplets: non-identical psychosis? Br J Psychiatry. 1982;140:1–6. doi: 10.1192/bjp.140.1.1. [DOI] [PubMed] [Google Scholar]
- 23.Badner JA, Gershon ES. Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry. 2002;7:405–411. doi: 10.1038/sj.mp.4001012. [DOI] [PubMed] [Google Scholar]
- 24.Berrettini W. Evidence for shared susceptibility in bipolar disorder and schizophrenia. Am J Med Genet. 2003;123C:59–64. doi: 10.1002/ajmg.c.20014. [DOI] [PubMed] [Google Scholar]
- 25.Hamshere ML, Williams NM, Segurado R, et al. Evidence for linkage (LOD=3.54) at IQ42 close to DISC1in a genome scan of functional psychosis pedigrees including at least one member with schizoaffective disorder of bipolar type [abstract] American Journal of Medical Genetics. 2004;130B(1):127. [Google Scholar]
- 26.Stefansson H, Sigurdsson E, Steinthorsdottir V, et al. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71:877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stefansson H, Sarginson J, Kong A, et al. Association of neuregulin 1 with schizophrenia confirmed in a Scottish population. Am J Hum Genet. 2003;72:83–87. doi: 10.1086/345442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams NM, Norton N, Williams H, et al. A systematic genomewide linkage study in 353 sib pairs with schizophrenia. Am J Hum Genet. 2003;73:1355–1367. doi: 10.1086/380206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tosato S, Dazzan P, Collier D. Association between the neuregulin 1 gene and schizophrenia: a systematic review. Schizophr Bull. 2005;31:613–617. doi: 10.1093/schbul/sbi043. [DOI] [PubMed] [Google Scholar]
- 30.Green E, Raybould R, McGregor S, Gordon-Smith K, Heron J, Hyde S, et al. The operation of the schizophrenia susceptibility gene, Neuregulin 1 (NRG1) across traditional diagnostic boundaries to increase risk for bipolar disorder. Archives of General Psychiatry. 2005;62:642–648. doi: 10.1001/archpsyc.62.6.642. [DOI] [PubMed] [Google Scholar]
- 31.Straub RE, Jiang Y, MacLean CJ, et al. Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. Am J Hum Genet. 2002 Aug;71(2):337–348. doi: 10.1086/341750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams NM, O'Donovan MC, Owen MJ. Is the dysbindin gene (DTNBP1) a susceptibility gene for schizophrenia? Schizophr Bull. 2005;31:800–805. doi: 10.1093/schbul/sbi061. [DOI] [PubMed] [Google Scholar]
- 33.Raybould R, Green EK, MacGregor S, Gordon-Smith K, Heron J, Hyde S, et al. Bipolar disorder and polymorphisms in the dysbindin (dystrobrevin binding protein 1) gene (DTNBP1) Biological Psychiatry. 2005;57:696–701. doi: 10.1016/j.biopsych.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 34.Fanous AH, van den Oord EJ, Riley BP, Aggen SH, Neale MC, O'Neill FA, Walsh D, Kendler KS. Relationship between a high-risk haplotype in the DTNBP1 (dysbindin) gene and clinical features of schizophrenia. Am J Psychiatry. 2005;162:1824–1832. doi: 10.1176/appi.ajp.162.10.1824. [DOI] [PubMed] [Google Scholar]
- 35.Chumakov I, Blumenfeld M, Guerassimenko O, et al. Genetic and physiological data implicating the new human gene G72 and the gene for D-amino acid oxidase in schizophrenia. Proc Natl Acad Sci USA. 2002;99:13675–13680. doi: 10.1073/pnas.182412499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schumacher J, Jamra RA, Freudenberg J, et al. Examination of G72 and D-amino-acid oxidase as genetic risk factors for schizophrenia and bipolar affective disorder. Mol Psychiatry. 2004;9:203–207. doi: 10.1038/sj.mp.4001421. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, He G, Gu N, et al. Association of G72/G30 with schizophrenia in the Chinese population. Biochem Biophys Res Commun. 2004;319:1281–1286. doi: 10.1016/j.bbrc.2004.05.119. [DOI] [PubMed] [Google Scholar]
- 38.Korostishevsky M, Kaganovich M, Cholostoy A, et al. Is the G72/G30 locus associated with schizophrenia? Single nucleotide polymorphisms, haplotypes, and gene expression analysis. Biol Psychiatry. 2004;56(3):169–176. doi: 10.1016/j.biopsych.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 39.Hall D, Gogos JA, Karayiorgou M. The contribution of three strong candidate schizophrenia susceptibility genes in demographically distinct populations. Genes Brain Behav. 2004;3:240–248. doi: 10.1111/j.1601-183X.2004.00078.x. [DOI] [PubMed] [Google Scholar]
- 40.Addington AM, Gornick M, Sporn AL, et al. Polymorphisms in the 13q33.2 gene G72/G30 are associated with childhood-onset schizophrenia and psychosis not otherwise specified. Biol Psychiatry. 2004;55:976–980. doi: 10.1016/j.biopsych.2004.01.024. [DOI] [PubMed] [Google Scholar]
- 41.Hattori E, Liu C, Badner JA, et al. Polymorphisms at the G72/G30 gene locus, on 13q33, are associated with bipolar disorder in two independent pedigree series. Am J Hum Genet. 2003;72:1131–1140. doi: 10.1086/374822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen YS, Akula N, Detera-Wadleigh SD, et al. Findings in an independent sample support an association between bipolar affective disorder and the G72/G30 locus on chromosome 13q33. Mol Psychiatry. 2004;9:87–92. doi: 10.1038/sj.mp.4001453. [DOI] [PubMed] [Google Scholar]
- 43.Williams NM, Green EK, Macgregor S, et al. Variation at the DAOA/G30 locus influences susceptibility to major mood episodes but not psychosis in schizophrenia and bipolar disorder. Archives of General Psychiatry. doi: 10.1001/archpsyc.63.4.366. In press. [DOI] [PubMed] [Google Scholar]
- 44.Blackwood DH, Fordyce A, Walker MT, et al. Schizophrenia and affective disorders-cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet. 2001;69:428–433. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Millar JK, Wilson-Annan JC, Anderson S, et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- 46.Sachs NA, Sawa A, Holmes SE, Ross CA, DeLisi LE, Margolis RL. A frameshift mutation in Disrupted in Schizophrenia 1 in an American family with schizophrenia and schizoaffective disorder. Mol Psychiatry. 2005;10:758–764. doi: 10.1038/sj.mp.4001667. [DOI] [PubMed] [Google Scholar]
- 47.Ekelund J, Hovatta I, Parker A, et al. Chromosome 1 loci in Finnish schizophrenia families. Hum Mol Genet. 2001;10:1611–1617. doi: 10.1093/hmg/10.15.1611. [DOI] [PubMed] [Google Scholar]
- 48.Ekelund J, Hennah W, Hiekkalinna T, et al. Replication of 1q42 linkage in Finnish schizophrenia pedigrees. Mol Psychiatry. doi: 10.1038/sj.mp.4001536. 2004 Jun 15 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 49.Macgregor S, Visscher PM, Knott SA, et al. A genome scan and follow-up study identify a bipolar disorder susceptibility locus on chromosome 1q42. Mol Psychiatry. 2004;9(12):1083–1090. doi: 10.1038/sj.mp.4001544. [DOI] [PubMed] [Google Scholar]
- 50.Hamshere ML, Bennett P, Williams N, Segurado R, Cardno A, Norton N, et al. Genome-wide linkage scan in schizoaffective disorder: Significant evidence for linkage (LOD=3.54) at 1q42 close to DISC1, and suggestive evidence at 22q11 and 19q13. Archives of General Psychiatry. 2005;62:1081–1088. doi: 10.1001/archpsyc.62.10.1081. [DOI] [PubMed] [Google Scholar]
- 51.Devon RS, Anderson S, Teague PW, et al. Identification of polymorphisms within Disrupted in Schizophrenia 1 and Disrupted in Schizophrenia 2, and an investigation of their association with schizophrenia and bipolar disorder. Psychiatr Genet. 2002;11:71–78. doi: 10.1097/00041444-200106000-00003. [DOI] [PubMed] [Google Scholar]
- 52.Kockelkorn TT, Arai M, Matsumoto H, et al. Association study of polymorphisms in the 5′ upstream region of human DISC1 gene with schizophrenia. Neurosci Lett. 2004;368(1):41–45. doi: 10.1016/j.neulet.2004.06.048. [DOI] [PubMed] [Google Scholar]
- 53.Hennah W, Varilo T, Kestila M, et al. Haplotype transmission analysis provides evidence of association for DISC1 to schizophrenia and suggests sex-dependent effects. Hum Mol Genet. 2003;12:3151–3159. doi: 10.1093/hmg/ddg341. [DOI] [PubMed] [Google Scholar]
- 54.Hodgkinson CA, Goldman D, Jaeger J, et al. Disrupted in schizophrenia 1 (DISC1): association with schizophrenia, schizoaffective disorder, and bipolar disorder. Am J Hum Genet. 2004 Nov;75(5):862–872. doi: 10.1086/425586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomson PA, Wray NR, Millar JK, et al. Association between the TRAX/DISC locus and both bipolar disorder and schizophrenia in the Scottish population. Mol Psychiatry. 2005;10:657–668. doi: 10.1038/sj.mp.4001669. 616. [DOI] [PubMed] [Google Scholar]
- 56.Callicott JH, Straub RE, Pezawas L, et al. Variation in DISC1 affects hippocampal structure and function and increases risk for schizophrenia. Proc Natl Acad Sci USA. 2005;102:8627–8632. doi: 10.1073/pnas.0500515102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lewis CM, Levinson DF, Wise LH, et al. Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: Schizophrenia. Am J Hum Genet. 2003 Jul;73(1):34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Potash JB, Zandi PP, Willour VL, et al. Suggestive linkage to chromosomal regions 13q31 and 22q12 in families with psychotic bipolar disorder. Am J Psychiatry. 2003;160(4):680–686. doi: 10.1176/appi.ajp.160.4.680. [DOI] [PubMed] [Google Scholar]
- 59.Chen X, Wang X, O'Neill AF, et al. Variants in the catechol-o-methyltransferase (COMT) gene are associated with schizophrenia in Irish high-density families. Mol Psychiatry. 2004;9:962–967. doi: 10.1038/sj.mp.4001519. [DOI] [PubMed] [Google Scholar]
- 60.Egan MF, Goldberg TE, Kolachana BS, et al. Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci USA. 2001;98:6917–6922. doi: 10.1073/pnas.111134598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malhotra AK, Kestler LJ, Mazzanti C, et al. A functional polymorphism in the COMT gene and performance on a test of prefrontal cognition. Am J Psychiatry. 2002;159:652–654. doi: 10.1176/appi.ajp.159.4.652. [DOI] [PubMed] [Google Scholar]
- 62.Glatt SJ, Faraone SV, Tsuang MT. Association between a functional catechol O-methyltransferase gene polymorphism and schizophrenia: meta-analysis of case-control and family-based studies. Am J Psychiatry. 2003;160:469–476. doi: 10.1176/appi.ajp.160.3.469. [DOI] [PubMed] [Google Scholar]
- 63.Fan JB, Zhang CS, Gu NF, Li XW, Sun WW, Wang HY, et al. Catechol-O-methyltransferase gene Val/Met functional polymorphism and risk of schizophrenia: a large-scale association study plus meta-analysis. Biol Psychiatry. 2005;57:139–144. doi: 10.1016/j.biopsych.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 64.Munafo MR, Bowes L, Clark TG, Flint J. Lack of association of the COMT (Val158/108 Met) gene and schizophrenia: a meta-analysis of case-control studies. Mol Psychiatry. 2005;10:765–770. doi: 10.1038/sj.mp.4001664. [DOI] [PubMed] [Google Scholar]
- 65.Shifman S, Bronstein M, Sternfeld M, et al. A highly significant association between a comt haplotype and schizophrenia. Am J Hum Genet. 2002;71:1296–1302. doi: 10.1086/344514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sanders AR, Rusu I, Duan J, et al. Haplotypic association spanning the 22q11.21 genes COMT and ARVCF with schizophrenia. Mol Psychiatry. doi: 10.1038/sj.mp.4001586. 2004 Aug 31 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 67.Handoko HY, Nyholt DR, Hayward NK, et al. Separate and interacting effects within the catechol-O-methyltransferase (COMT) are associated with schizophrenia. Mol Psychiatry. doi: 10.1038/sj.mp.4001606. 2004 Oct 26 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 68.Williams HJ, Glaser B, Williams NM, Norton N, Zammit S, Macgregor S, et al. No association between polymorphisms in COMT and schizophrenia in two large samples. Am J Psychiatry. 2005;162:1736–1738. doi: 10.1176/appi.ajp.162.9.1736. [DOI] [PubMed] [Google Scholar]
- 69.Lachman HM, Papolos DF, Saito T, Yu YM, Szumlanski CL, Weinshilboum RM. Human catechol-O-methyltransferase pharmacogenetics: description of a functional polymorphism and its potential application to neuropsychiatric disorders. Pharmacogenetics. 1996;6(3):243–250. doi: 10.1097/00008571-199606000-00007. [DOI] [PubMed] [Google Scholar]
- 70.Li T, Ball D, Zhao J, et al. Family-based linkage disequilibrium mapping using SNP marker haplotypes: application to a potential locus for schizophrenia at chromosome 22q11. Mol Psychiatry. 2000 Jul;5(4):452. doi: 10.1038/sj.mp.4000752. [DOI] [PubMed] [Google Scholar]
- 71.Craddock N, Dave S, Greening J. Association Studies of Bipolar Disorder. Bipolar Disorders. 2001;3:284–298. doi: 10.1034/j.1399-5618.2001.30604.x. [DOI] [PubMed] [Google Scholar]
- 72.Shifman S, Bronstein M, Sternfeld M, et al. COMT: a common susceptibility gene in bipolar disorder and schizophrenia. Am J Med Genet. 2004;128B:61–64. doi: 10.1002/ajmg.b.30032. [DOI] [PubMed] [Google Scholar]
- 73.Green E, Craddock N. Brain-derived neurotrophic factor as a potential risk locus for bipolar disorder: evidence, limitations, and implications. Curr Psychiatry Rep. 2003;5(6):469–476. doi: 10.1007/s11920-003-0086-1. [DOI] [PubMed] [Google Scholar]
- 74.Duman RS. The neurochemistry of mood disorders: preclinical studies. In: Charney DS, Nestler EJ, Bunney BS, editors. The Neurobiology of Mental Illness. New York: Oxford University Press; 1999. pp. 333–347. [Google Scholar]
- 75.Seidah NG, Benjannet S, Pareek S, Chretien M, Murphy RA. Cellular processing of the neurotrophin precursors of NT3 and BDNF by the mammalian proprotein convertases. FEBS Lett. 1996;379:247–250. doi: 10.1016/0014-5793(95)01520-5. [DOI] [PubMed] [Google Scholar]
- 76.Egan MF, Kojima M, Callicott JH, et al. The BDNF Val66Met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 77.Sklar P, Gabriel SB, McInnis MG, et al. Family-based association study of 76 candidate genes in bipolar disorder: BDNF is a potential risk locus. Brain-derived neutrophic factor. Mol Psychiatry. 2002;7:579–593. doi: 10.1038/sj.mp.4001058. [DOI] [PubMed] [Google Scholar]
- 78.Neves-Pereira M, Mundo E, Muglia P, et al. The brain-derived neurotrophic factor gene confers susceptibility to bipolar disorder: evidence from a family-based association study. Am J Hum Genet. 2002 Sep;71:651–655. doi: 10.1086/342288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Geller B, Badner JA, Tillman R, et al. Linkage disequilibrium of the brain-derived neurotrophic factor Val66Met polymorphism in children with a prepubertal and early adolescent bipolar disorder phenotype. Am J Psychiatry. 2004 Sep;161(9):1698–1700. doi: 10.1176/appi.ajp.161.9.1698. [DOI] [PubMed] [Google Scholar]
- 80.Oswald P, Del-Favero J, Massat I, Souery D, Claes S, Van Broeckhoven C, Mendlewicz J. Non-replication of the brain-derived neurotrophic factor (BDNF) association in bipolar affective disorder: a Belgian patient-control study. Am J Med Genet. 2004;129B(1):34–35. doi: 10.1002/ajmg.b.30056. [DOI] [PubMed] [Google Scholar]
- 81.Skibinska M, Hauser J, Czerski PM, et al. Association analysis of brain-derived neurotrophic factor (BDNF) gene Val66Met polymorphism in schizophrenia and bipolar affective disorder. World J Biol Psychiatry. 2004 Oct;5(4):215–220. doi: 10.1080/15622970410029936. [DOI] [PubMed] [Google Scholar]
- 82.Hong CJ, Huo SJ, Yen FC, et al. Association study of a brain-derived neurotrophic-factor genetic polymorphism and mood disorders, age of onset and suicidal behavior. Neuropsychobiology. 2003;48(4):186–189. doi: 10.1159/000074636. [DOI] [PubMed] [Google Scholar]
- 83.Nakata K, Ujike H, Sakai A, et al. Association study of brain-derived neurotrophic factor (BDNF) gene with bipolar disorder. Neuroscience Letters. 337:17–20. doi: 10.1016/s0304-3940(02)01292-2. [DOI] [PubMed] [Google Scholar]
- 84.Green EK, Raybould R, Macgregor S, et al. Genetic variation at brain-derived neurotrophic factor (BDNF) is associated with rapid cycling in a UK bipolar disorder case-control sample of over 3000 individuals. Br J Psychiatry. doi: 10.1192/bjp.bp.105.009969. In press. [DOI] [PubMed] [Google Scholar]
- 85.Müller DJ, De Luca V, Sicard TT, et al. The brain derived neurotrophic factor (BDNF) gene is associated with rapid cycling bipolar disorder. Br J Psychiatry. doi: 10.1192/bjp.bp.105.010587. In press. [DOI] [PubMed] [Google Scholar]
- 86.Neves-Pereira M, Cheung JK, Pasdar A, et al. BDNF gene is a risk factor for schizophrenia in a Scottish population. Mol Psychiatry. 2005;10:208–212. doi: 10.1038/sj.mp.4001575. [DOI] [PubMed] [Google Scholar]
- 87.Schumacher J, Jamra RA, Becker T, et al. Evidence for a relationship between genetic variants at the brain-derived neurotrophic factor (BDNF) locus and major depression. Biol Psychiatry. 2005;58:307–314. doi: 10.1016/j.biopsych.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 88.Levinson DF, Mowry BJ, Escamilla MA, Faraone SV. The Lifetime Dimensions of Psychosis Scale (LDPS): description and interrater reliability. Schizophr Bull. 2002;28:683–695. doi: 10.1093/oxfordjournals.schbul.a006972. [DOI] [PubMed] [Google Scholar]
- 89.Craddock N, Jones I, Kirov G, et al. The Bipolar Affective Disorder Dimension Scale (BADDS)—a dimensional scale for rating lifetime psychopathology in bipolar spectrum disorders. BMC Psychiatry. 2004;4:19. doi: 10.1186/1471-244X-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Harrison PJ, Owen MJ. Genes for schizophrenia? Recent findings and their pathophysiological implications. Lancet. 2003;361(9355):417–419. doi: 10.1016/S0140-6736(03)12379-3. [DOI] [PubMed] [Google Scholar]