Abstract
Background: The Consortium on the Genetics of Schizophrenia (COGS) is an ongoing, National Institute of Mental Health–funded, 7-site collaboration investigating the occurrence and genetic architecture of quantitative endophenotypes related to schizophrenia. The purpose of this article is to provide a description of the COGS structure and methods, including participant recruitment and assessment. Methods: The hypothesis-driven recruitment strategy ascertains families that include a proband with a Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition diagnosis of schizophrenia, and at least one unaffected full sibling available for genotyping and endophenotyping, along with parents available for genotyping and (optional depending on age) endophenotyping. The family structure is selected to provide contrast in quantitative endophenotypic traits and thus to maximize the power of the planned genetic analyses. Probands are recruited from many sources including clinician referrals, local National Alliance for the Mentally Ill chapters, and advertising via the media. All participants undergo a standardized protocol that includes clinical characterization, a blood draw for genotyping, and endophenotype assessments (P50 suppression, prepulse inhibition, antisaccade performance, continuous performance tasks, letter-number span, verbal memory, and a computerized neurocognitive battery). Investigators participate in weekly teleconferences to coordinate and evaluate recruitment, clinical assessment, endophenotyping, and continuous quality control of data gathering and analyses. Data integrity is maintained through use of a highly quality-assured, centralized web-based database. Results: As of February 2006, 355 families have been enrolled and 688 participants have been endophenotyped, including schizophrenia probands (n = 154, M:F = 110:44), first-degree biological relatives (n = 343, M:F = 151:192), and community comparison subjects (n = 191, M:F = 81:110). Discussion: Successful multisite genetics collaborations must institute standardized methodological criteria for assessment and recruitment that are clearly defined, well communicated, and uniformly applied. In parallel, studies utilizing endophenotypes require strict adherence to criteria for cross-site data acquisition, equipment calibration and testing and software equivalence, and continuous quality assurance for many measures obtained across sites. This report describes methods and presents the structure of the COGS as a model of multisite endophenotype genetic studies. It also provides demographic information after the first 2 years of data collection on a sample for whom the behavioral data and genetics of endophenotype performance will be fully characterized in future articles. Some issues discussed in the reviews that follow reflect the challenges of evaluating endophenotypes in studies of the genetic architecture of endophenotypes in schizophrenia.
Keywords: neurophysiology, neurocognitive, genes
Introduction
Decades of research have shown that genetic factors play a key role in the pathogenesis of schizophrenia (eg, Faraone et al,1 Gottesman and Shields,2 Braff and Freedman3). Indeed, replicable linkage for schizophrenia has been reported at a number of chromosomal locations (eg, Badner and Gershon,4 Baron5). However, identification of a specific schizophrenia susceptibility gene, or a set of genes, remains elusive. One strategy that holds promise for understanding the genetic architecture of this heterogeneous disorder is the analysis of discrete and neurobiologically relevant “endophenotypic” abnormalities rather than relying on the noisy and heterogeneous clinical phenotype of schizophrenia.3,6–10
The Consortium on the Genetics of Schizophrenia (COGS) is a 7-site National Institute of Mental Health (NIMH) sponsored collaboration seeking to understand the genetic basis of 6 primary and multiple secondary candidate endophenotypes, including a neurocognitive battery,11,12 in schizophrenia patients and their relatives. The study is unusual in both its national scope and focus on the collection of extensive endophenotype measures in families. The 7 participating sites are Harvard University, Mount Sinai School of Medicine, University of California Los Angeles, University of California San Diego (central administrative site), University of Colorado, University of Pennsylvania, and University of Washington. The COGS was designed to enable the reliable collection of endophenotype measures across all 7 sites so as to maximize the power of genetic linkage and association studies. This article outlines the rigorous selection and quality assurance (QA) procedures that were considered essential for a successful multisite, endophenotype-based neuropsychiatric genetic study.
The COGS endophenotype battery includes 6 primary neurophysiologic and neurocognitive measures that have been reported to exhibit quantitative neurobiological deficits in schizophrenia patients and their first-degree biological relatives. The neurophysiological measures include P50 event-related potential suppression,13–15 prepulse inhibition (PPI) of the startle response,16–18 and the antisaccade oculomotor task.19,20 Neurocognitive measures include the Continuous Performance Test (CPT21–23), tests of verbal memory (cf, Braff et al,17Cirillo et al,24 Faraone et al25), and working memory (cf, Perry et al,26 Conklin et al,27 Gold et al,28 Park and Holzman29). In addition, a computerized neurocognitive battery (CNB), developed at the University of Pennsylvania, was included to characterize the participants and to provide additional endophenotypes.11,12
The 6 candidate endophenotype measures have been selected based on empirical evidence that they fulfill 6 criteria: (1) they show moderate to large effect sizes between schizophrenia patients and control participants, (2) clinically unaffected relatives also have deficits compared with normal controls, (3) these endophenotypes have adequate psychometric properties of heritability, stability, and between-site reliability, (4) in the extant literature, the measures have a known or partially explicated neurobiological substrate (usually assessed via animal model and functional brain imaging studies) that is relevant to schizophrenia, (5) the range of scores reported on these measures is sufficiently broad for control participants, clinically unaffected family members, and schizophrenia patients to support using them to test genetic hypotheses, and (6) at the time of the study's design, medication effects were not known to irretrievably obscure schizophrenia-normal differences and effect sizes. Other considerations include the ease with which uniformity of training and administration (eg, manuals, videotapes, and hands-on training in groups) of these endophenotypes can be achieved in a large, multisite study. The COGS will evaluate the unique and shared genetic vs environmental determinants of these endophenotypes. Recruitment and research design goals are to assess at least 420 clearly defined schizophrenia pedigrees (1680 participants) and 525 community comparison subjects (CCSs) over 5 years. Since the start of the COGS project, additional more specific measures of verbal memory have been added across all sites. In addition, site-specific pilot data from brain imaging, mismatch negativity, and functional outcome measures have been obtained for evaluation as new candidate endophenotypes.
Successful multisite collaborations, especially those that involve complex endophenotyping, require careful standardization and continuous quality-assured administration of measures across sites. The COGS involves standardized clinical characterization, collection of neurophysiological and neurocognitive data, and genotyping from a sample of probands, their first-degree relatives, and CCSs collected across 7 sites. Before initiating subject testing and data collection, 6 months of consortium-wide standardization of equipment and test protocols, training and QA efforts, and database development took place. The purpose of the current report is to (1) provide a detailed description of COGS recruitment, clinical and endophenotype assessment, and continuous QA methods and (2) present the preliminary sample characteristics of participants from whom endophenotype data and DNA have been collected in order to guide other, similar efforts and to serve as a definitive point of reference for COGS publications. Specific data and analytic issues, such as cross-site reliability of endophenotypes and statistical analysis of family data, are addressed in several COGS empirical articles that are in various stages of submission or publication (eg, Greenwood et al,30 Horan et al,31 Radant et al32). A separate article that details the statistical analytic approach for DNA analysis is included in the current issue of Schizophrenia Bulletin.33 Finally, because the COGS family DNA will eventually be available to other investigators through the NIMH data-sharing process, we aimed to provide a description that can be used as a resource for those using the COGS database in the future.
METHOD
The COGS infrastructure, governance interaction among sites, and data flow were planned in detail over a 2-year period commencing before the essentially identical 7 individual COGS R01′s were submitted to the NIMH and reviewed by the Genomics Review Committee. Moreover, recruitment, diagnostic assessment, and endophenotype methods were standardized using in-person meetings, written manuals, videotapes, and regularly scheduled teleconferences.
Overview of COGS Structure and Data Flow
The COGS is organized into 7 core units that work interactively to collect, process, and analyze data.
COGS Structure
The Director's Unit at University of California San Diego (UCSD) (David L. Braff, Director) provides scientific leadership, administrative support, and coordination of information across all sites. The Statistical Genetics Core (Nicholas J. Schork, Director), also located at UCSD, provides specialized expertise and data analysis for the statistical genetics component of the consortium. The Bioinformatics and Data Core at Univeristy of California Los Angeles (UCLA) (Jim Mintz, Director) is responsible for constructing and maintaining the secure web-based COGS data warehouse and for providing methodological and statistical consultation to COGS investigators. The Clinical Core Committee (CCC), comprised of one faculty-level representative from each site, oversees the recruitment and clinical assessment of research participants and assists in the development of the clinical components of the COGS database. The Endophenotype Committee (EPC), comprised of the senior QA scientists at each site, oversees procedures for accurate collection, data transmission, and analysis of the 6 primary endophenotypes, secondary phenotypes, and the Penn Neurocognitive Battery measures. The chairperson of the Clinical and Endophenotype Cores rotates annually among investigators at the 7 sites. The General Investigators Meeting allows for the integration and discussion of all issues, especially those raised by the CCC and the EPC. The Principal Investigators Committee (PIC) is the final decision-making body regarding policies, procedures, and publications.
Thus, investigators and research staff participate in 4 separate teleconferences, each held twice per month to (1) coordinate and evaluate recruitment and assessment progress and procedures (CCC), (2) discuss and provide continuous QA of the 7-site endophenotyping endeavor (EPC), (3) integrate the CCC and EPC recommendations and discuss COGS-wide issues with faculty and staff (Investigators Committee), and (4) review overall site performance, set policies and publication plans, and resolve administrative issues (PIC). Training and development is an important mission of the COGS. In general, the COGS structure and practice is designed to enhance the training and facilitate authorship contributions of junior faculty, who often serve as site coordinators and/or fill other crucial roles. All procedures are approved by the local institutional review boards (IRBs) at each institution.
Data Flow
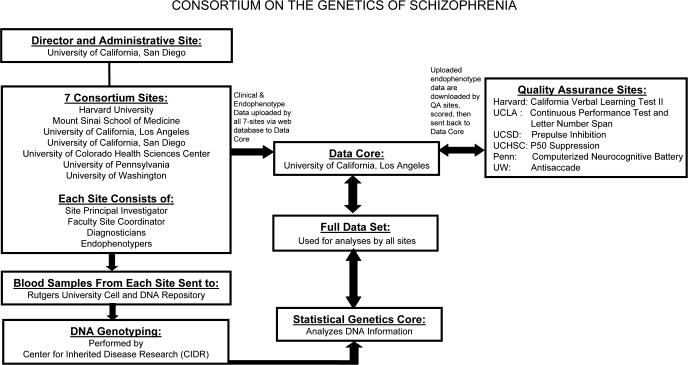
Figure 1 provides an overview of the flow of COGS data. Participants at each COGS site undergo detailed clinical assessment and collection of all 7 endophenotypes. Blood samples for DNA analysis are sent from the local site to the Rutgers University Cell and DNA Repository using standardized collection and shipping procedures.
Fig. 1.
Overview of COGS Structure, Key Interactions, and Data flow.
Data management depends on a constant iterative relationship between the 7 sites and the UCLA Data Core. All data are uploaded to a rigorously “firewalled,” password-protected, web-based data warehouse developed collaboratively by the COGS investigators and maintained by the Central Data Core at UCLA (http://www.npistat.com/cogs/). Backup copies of these data are stored locally at all 7 sites, and all data are deidentified before electronic transmission to the Central Data Core. Clinical assessment data and endophenotype data are uploaded separately. Once the information has been transmitted to the Central Data Core, the QA scientist responsible for each endophenotype regularly downloads all new endophenotype data collected on that measure across all sites. The QA scientists review and score the endophenotype data according to procedures established for each particular measure. Any data questions that arise during this QA process are reviewed with the personnel at the site at which the data were collected. Separation between clinical and endophenotype information ensures that QA scientists remain blinded to diagnosis and other clinical information at the time of data scoring.
Data from the Penn CNB are transmitted electronically from the administrator's laptop to a central repository at the University of Pennsylvania, with immediate e-mail confirmation of receipt to the local administrator, site coordinator, and key PENN personnel. In addition to error messages generated by the computerized scoring program, a PENN research team member assesses the validity of each submitted battery and addresses any questions to the administrator and site coordinator. A weekly tabulation of completed batteries, including weekly and cumulative totals, is e-mailed to each site and to the Central Data Core at UCLA.
After the QA review and scoring are completed, the scored data from each endophenotype and from the CNB are again uploaded to the central data warehouse. Finally, the scored endophenotype data are merged with the clinical data to create a composite COGS data set. The composite data set is updated and distributed to principal investigators (PIs) and site coordinators at each site on a monthly basis. Primary data analyses are conducted using this composite data set.
Cross-Site Training and QA Procedures
From the initial (prefunding) meetings, the COGS Investigators were focused on the absolute need for uniformity in equipment, experimental procedures, physical environments of the testing rooms, experimenter behavior, and an array of other factors (see below). To further ensure data QA, all equipment is calibrated monthly in a standardized fashion, and backup equipment is available to each site in case a malfunction is detected.
Clinical Assessment Training
Diagnostic interviewers learn to competently administer the clinical assessment tools using a standardized training procedure developed by a clinical psychologist (Monica E. Calkins) with extensive training and experience in semistructured interviewing. Before the start of the study, diagnostic interviewers met for a 4-day training workshop at the Central Site at UCSD. When new interviewers are hired, training is supervised by senior clinical site coordinators who implement the training protocol. The fundamental goals of the protocol are to enhance and facilitate the clinical skill and knowledge of psychopathology required to expertly administer semistructured interviews. Accordingly, the protocol consists of didactic sessions, observation, and supervised practice. Ten videotaped didactic sessions cover major psychopathology and differential diagnosis, particulars of semistructured interviewing, interview documentation practices, and item-by-item review of assessment instruments, rating scales, and COGS-specific operational definitions. Contemporaneously, each trainee practices the interview modules with mock participants and observes interviews conducted by senior interviewers. Finally, trainees engage in supervised practice with at least 3 participants and, subsequently, obtain ongoing regular supervision by a senior clinician.
Endophenotype Assessment Training
All 6 endophenotypes have an assigned QA site that reviews and scores data coming from all 7 COGS sites. To ensure that each site is testing according to protocol, UCSD hosted an initial 4-day training workshop attended by testers from all 7 sites along with all 6 endophenotype faculty QA experts prior to initiation of the study. Training materials for each endophenotype include written procedure manuals and instructional videos demonstrating step-by-step administration of the protocol generated at the yearly COGS-wide training sessions. The procedure manuals include scripts for task instructions, which must be stated verbatim to each participant by each tester, and equipment settings. Each site maintains a library of these materials for training of current and newly hired staff. Furthermore, “retraining” is held at UCSD on an annual basis to minimize experimenter drift in methods and train newly hired testers. During these extensive training workshops, each QA expert reviews the endophenotyping methodology and observes all testers as they setup and administer each test, thereby approving these individuals and their respective sites as “COGS certified.”
In addition to the training workshops, the COGS Project Coordinator (Andras Kovach) visits all 7 COGS sites every year in order to ascertain testing procedures across sites. His role is to ensure the standard and consistent practice of administering the endophenotypes according to the original protocol and to inspect the endophenotype equipment. During each 2-day visit, he spends 1 day acting as if he is a naïve CCS and then observes an actual CCS being tested. He compares tester performance during both days to QA-approved checklists on proper protocol and reports any deviations from the protocol of testing or calibration to the QA site for their advice and action. He works with the site PIs and with the faculty QA experts for each endophenotype to ensure that all sites strictly follow COGS testing guidelines.
It is thus a complex and challenging endeavor (for this and other, similar studies) to uniformly collect and ensure the quality of endophenotypes. At the outset, some sites had little or no experience with some endophenotypes. Consequently, the group training, video tutorials, and manuals had to be precisely followed. The yearly retraining and on-site visits yielded very high uniformity and some surprises that cost us lost data for some endophenotypes over short time periods. Slight deviations in instructions and procedures had to be uniformly corrected. For example, deviations in the placement of visual fixation points can cause oculomotor discharge that can alter blink startle measures. Fortunately, with the combination of rigorous video and manualized retraining, onsite inspections, and yearly retraining workshops, projects of this kind can be “quality assured.” The initial “data-based” COGS articles include analyses of site differences to assess the comparability of measurement across sites.
Recruitment
Strategies
Schizophrenia probands have been recruited from clinics/clinician referrals (37%), local chapters of the National Alliance for the Mentally Ill (16%), Web sites (http://www.schizophrenia.com, http://www.SchizophreniaResearch.net, local sites; 9%), and various other sources (38%) including the media, local flyers, and referral from other studies. CCS are recruited primarily through newspaper advertisements (32%), Web sites (25%), local flyers (24%), and various other sources (19%), including referrals from previous participants and other studies. Once identified, potential participants undergo semistructured screening, either in person or on the telephone, the purpose of which is to provide an initial informed determination regarding subject eligibility for study participation. Information obtained in the screening assessment includes referral source, demographic data, current clinical symptoms, brief medical history, family structure, and family history of mental illness. Permission to review relevant medical records is also obtained at this time. Persons identified as potentially eligible are invited to participate in the more detailed, standardized clinical assessment as described below.
Inclusion/Exclusion Criteria
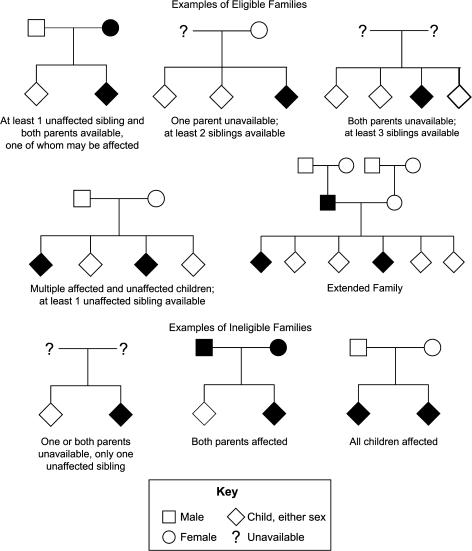
All participants who are endophenotyped are between the ages of 18–65 and able to understand and provide informed consent. Parents older than 65 have their blood drawn for genotyping purposes but do not routinely undergo endophenotyping. Eligible families fulfill the following criteria: (1) a proband meets Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV) criteria for schizophrenia as determined by the Best Estimate Final Diagnosis (BEFD) procedures described below, (2) both biological parents are available for genotyping, and (3) at least one full sibling who is unaffected with schizophrenia is available for endophenotyping and genotyping. First-degree biological relatives include parents and siblings of the proband. Examples of eligible and ineligible family pedigrees are depicted in figure 2. This family configuration was selected to optimize the study of quantitative phenotypic variation, in which contrasts between individuals with high and low phenotype values generate greater power.34–37 Probands having only one available parent but 2 or more available siblings (with at least one unaffected by schizophrenia) are also included, as are probands having no available parents but 3 or more available siblings (with at least one unaffected by schizophrenia), though these configurations are less powerful for the proposed analyses (cf, Ott38). Multigenerational (extended) families are included when an additional relative (eg, nephew or child of the proband) meets criteria for schizophrenia, and the proband has living parents and at least one unaffected sibling available (see figure 2).
Fig. 2.
Examples of Eligible and Ineligible Family Pedigrees. Shaded symbols represent individuals affected with schizophrenia.
Having met these basic eligibility requirements, participants are excluded if they fulfill any of the exclusion criteria presented in table 1. To minimize genetic heterogeneity, families that include only a proband with schizoaffective disorder (either bipolar or depressed subtype) are not included. Additionally, families with evidence of “parental bilineality” (ie, both parents with schizophrenia) or no “contrast” in sibships (all sibs have a diagnosis of schizophrenia) are not included (see figure 2). Again, the family structure criteria were developed to maximize power for quantitative genetic analyses of the endophenotypic measures.
Table 1.
Exclusion Criteria for Endophenotype Assessment
| Exclusion Criterion | All Participants | Proband | CCS |
|---|---|---|---|
| Does not meet basic family inclusion criteria (see text: “Methods—Recruitment”) | X | ||
| Adopted or family history unknown | X | ||
| Outside study age range of 18–65a | X | ||
| Unable to understand consent due to language or competency | X | ||
| Physically unable to participate in testing of at least one endophenotype. | X | ||
| Previous endophenotype testing in the last 1 mob | X | ||
| Previous neuropsychological testing in the last 3 mob | X | ||
| Positive illicit drug or alcohol screen at the time of testing | X | ||
| Severe systemic illness (e.g., congestive heart disease, brittle insulin dependent diabetes mellitus, Lupus) that interferes with ability to be endophenotyped | X | ||
| Parents not living (unless sibship is large) or unavailable for genotyping | X | ||
| Both parents diagnosed with schizophrenia | X | ||
| Siblings unavailable for endophenotyping and genotyping | X | ||
| All siblings diagnosed with schizophrenia | X | ||
| Electroconvulsive treatment in the last 6 mo | X | X | |
| Alcohol or substance abuse in the past 1 mo | X | X | |
| Alcohol or substance dependence not in remission for 6 mo | X | X | |
| Vision and hearing problems | X | X | |
| Significant head injury (loss of consciousness > 15 min and/or neurological sequelae) | X | X | |
| Neurological illness (e.g., seizures, stroke, Parkinson disease) | X | X | |
| Less than one 1 month psychiatrically stable | X | X | |
| Estimated premorbid IQ < 70 per Wide Range Achievement Test-Third Edition (WRAT-3) | X | X | |
| History of psychosis in themselves or a family member (1st or 2nd degree) | X | ||
| Cluster A personality disorder | X | ||
| Current treatment with antipsychotic agents | X |
Note: CCS, community comparison subject.
Parents older than 65 have their blood drawn for genotyping purposes and undergo diagnostic assessment if permissible but do not routinely undergo endophenotyping.
If otherwise eligible for participation, she/he will typically be recontacted after the specified time has elapsed for possible participation.
If subjects fulfill the basic eligibility requirements described above, all available first-degree family members complete diagnostic and endophenotype testing regardless of comorbid medical and psychiatric conditions. CCS, who serve as a reference group against which to gauge performance of probands and family members, participate in all components of testing, including diagnostic and endophenotype assessment, and blood draw. To parallel psychiatric comorbidity in relatives of probands, nonpsychotic axis I psychopathology is accepted in approximately 30% of CCS, but clinical stability and/or remission is required. Although relatives and CCS with medical and psychiatric conditions are included in the sample, data are coded to allow subset analyses restricted to medically and psychiatrically healthy participants (eg, Calkins et al19). Data on these subjects provide the opportunity to test the influence of clinical conditions that may affect endophenotype performance and provide a comparison sample for relatives who are not affected by schizophrenia but are afflicted with other psychiatric conditions.
Assessment Batteries
The assessment protocol is typically completed over the course of 2 days, which are scheduled in as close temporal proximity as possible. The first session consists of the clinical assessment battery and blood draw, and the second includes the endophenotype assessment battery.
Clinical Assessment Battery
All participants undergo an extensive, standardized assessment protocol that was developed to provide a comprehensive evaluation of psychotic, mood, and substance-related disorders. The assessment procedure includes the following components.
Diagnostic Interview for Genetics Studies.
The Diagnostic Interview for Genetics Studies (DIGS), which was developed by the NIMH Genetics Initiative for genetic studies of schizophrenia and mood disorders, provides for a detailed assessment of psychotic, mood, and substance-related symptoms, which in turn allows for the reliable differential diagnosis of related disorders.39 In addition, it provides self-reported demographic and medical history data. Several instruments embedded within the DIGS allow for Mini-Mental Status Examination, functional assessment (Global Assessment Scale, with standardized anchors40), clinical symptom ratings via the Scale for the Assessment of Positive Symptoms (SAPSs41) and the Scale for the Assessment of Negative Symptoms (SANSs42), and assessment of schizotypal and other axis II cluster A personality features via a modified version of the Structured Interview for Schizotypy.43
Family Interview for Genetic Studies.
The Family Interview for Genetic Studies (FIGS), also developed by the NIMH Human Genetics Initiative,44 is a method of systematically gathering information about psychiatric disorders in the family from an informant. Although this method has low sensitivity compared with direct assessment of each family member, its specificity is high.1 In the COGS, FIGS information is gathered independently from all available family members, resulting in the creation of a composite pedigree that is informed by multiple participants. FIGS administration involves drawing a family pedigree—which is sent to the Statistical Genetics Core—obtaining psychiatric information about relatives in the pedigree, and following up on endorsed symptom categories, including depression, mania, substance-related disorders, psychosis, and cluster A personality disorders.
Brief Psychiatric Rating Scale—Expanded Version, Anxiety Scale.45.
This scale from a version of the Brief Psychiatric Rating Scale developed by the UCLA Clinical Research Center's Diagnosis and Psychopathology Unit is administered to assess symptoms of anxiety spectrum disorders. It provides a standardized method of eliciting descriptions about and rating the severity of anxiety symptoms, including apprehension, tension, fear, panic, or worry.
Narrative Summary.
After gathering all sources of information, including available medical records, the interviewer prepares a comprehensive narrative summary of the results of the diagnostic assessment. The narrative provides behavioral observations along with an integrated timeline of the onset, duration, description, and temporal relationship of symptoms.
Best-Estimate Final Diagnoses.
For each participant, the BEFD process begins at the point of initial contact, where screening information and informal observations are recorded by recruiters. Following the diagnostic assessment battery for all participants, all available sources of information, including DIGS (and its embedded instruments), FIGS, rating scales, narrative summary, and medical records are reviewed by at least 2 faculty-level clinicians from the site assessing the participant. The clinicians then assign DSM-IV lifetime BEFD based on consensus review of these materials. Prior to the beginning of data collection, a subcommittee of clinicians reviewed and operationalized standardized diagnostic criteria for major disorders. The criteria were then discussed and approved by the PI's and are taught to new staff during their training. In addition to routine communications among the CCC at biweekly meetings, these DSM-IV based criteria ensure that perceived ambiguities in DSM-IV criteria are handled in a uniform manner across sites. A pilot study of intersite reliability of BEFD for 5 cases across all 7 sites, in which each site reviewed identical assessment materials, yielded 100% agreement for the BEFD of schizophrenia.
Endophenotype Assessment Battery
The endophenotype and neuropsychological battery is administered in one of 2 fixed orders (1 through 6 from the numbered tests below or 6 through 1) that take a total testing time of 4–6 hours. A break is typically scheduled between the neurophysiologic and the neurocognitive testing batteries to help minimize subject fatigue. For each of the 6 endophenotypes, a single primary outcome measure is used for subsequent analyses, based on prior evidence of maximal group separation and relationship to specific clinical features. In addition, secondary measures are obtained from the test session itself (eg, blink latency in the PPI session) or via “extension” of the tests (eg, smooth pursuit eye movement, following the antisaccade testing). Finally, each site can “recall” subjects for additional testing, such as neuroimaging or other EEG testing, per their locally approved IRB consents. The endophenotype assessment battery includes the following measures:
P 50 Suppression: Evoked Potential Recording. Participants lay supine in a recliner chair with their eyes open while listening to paired auditory click stimuli (intrapair interval of 0.5 seconds and an interpair interval of 10 seconds), delivered 50 dB over the participant's auditory threshold.46 Electroencephalographic activity is recorded at Cz, and artifact free trials are averaged and digitally bandpass filtered (10–100 Hz). The conditioning P50 wave is identified as the most positive peak 40–80 ms after the conditioning stimulus, measured relative to the preceding negative trough. The test wave is identified as the most positive peak occurring after the test stimulus, within ±10 ms of the latency of the conditioning response. The ability to filter out extraneous stimuli is measured by the ratio of the test amplitude to the conditioning amplitude, T/C ratio. This is alternatively expressed as the P50 percent suppression = 1−T/C ratio. The duration of testing is approximately 45 minutes (QA site: Univeristy of Colorado).
Antisaccade. Ocular motor recordings are obtained in a quiet, darkened room. Participants are seated in front of a flat screen monitor with head stabilized using a chin rest or bite bar. Horizontal eye movements are measured using an Eye Trac Model 310 eye movement monitor and infrared spectacle (4-ms time constant) mounted on eyeglass frames (Applied Science Laboratories, Waltham, MA). Recordings of the eye movements are digitized using an A–D board connected to an IBM-compatible computer so that they can be scored off-line for direction of response, saccadic reaction time, and saccadic accuracy. Stimuli consist of filled circles subtending about 1 degree of visual angle. A stimulus is presented at central fixation for 2400–3600 ms. During the last 200 ms of this period, an antisaccade cue stimulus appears (for 800 ms) 10° or 15° to the left or right of the central fixation dot. Participants are instructed to move their eyes as quickly and accurately as possible to the cue's mirror image (same amplitude, opposite direction). Proportion of correct responses is the primary dependent measure. The task takes approximately 30 minutes (QA Site: Univesity of Washington).
The PPI. Subjects sit in a recliner chair with eyes open. Two small cup electrodes are placed over the orbicularis oculi muscles bilaterally. Hearing thresholds are determined. Startle stimuli are presented binaurally through headphones. Each test session includes 70-dB[A] broadband background noise, prepulse stimuli, and acoustic startle stimuli that are generated by the SR-LAB system. Startle is elicited by a 115-dB[A] 40-ms burst of broadband noise. PPI is induced by 20-ms bursts of broadband noise occurring 30, 60, and 120 ms before the startling stimulus. Prepulse intensities are 16 dB[A] above the background. PPI is defined as the percent reduction in startle magnitude in the presence of the prepulse compared with the magnitude in the absence of the prepulse. PPI testing takes approximately 40 minutes (QA site: UCSD).
Attention: Degraded Stimulus CPT. The degraded stimulus CPT (DS-CPT)22 requires sustained focused attention in a situation that includes a substantial perceptual discrimination burden but not working memory demands. The UCLA computer program allows this task to be administered in a convenient and highly reliable way using a PC, 15-inch NEC AccuSync 50 Monitor, and Microsoft Basic Mouse.47 Single digits 0 through 9 are presented one at a time in quasi-random order at a rate of one per second, with individual exposure times of 29 ms. A random 40% of the pixels in each digit and in the background are changed from black to white, or vice versa, to create a highly blurred image. The participant's task is to monitor the rapid series of digits and to respond as quickly as possible with a button press to each blurred 0 that appears. Targets appear in 25% of 480 digit presentations; nontarget digits that are perceptually similar to targets are presented in another 25%, while the remainder of the presentations (50%) involve other single digits. Participants are told that they will never be sure that a given stimulus is a 0 but that they should respond to the digits that look most like a 0. Participants are introduced to the stimuli and practice the task before administration. Hit rate and false alarm rate are determined and are used to calculate signal detection theory indices48 for signal/noise discrimination (d') and response criterion (natural log of beta), with d′ being the primary summary measure. The DS-CPT is administered during the break within the California Verbal Learning Test, Second Edition (CVLT-II, described below), and takes approximately 15 minutes to administer. The Continuous Performance Task-Identical Pairs version21 is also administered as a secondary measure (QA site: UCLA).
Working Memory: Letter-Number Span Test. In this test of working memory, participants are presented with clusters of intermixed numbers and letters (eg, G4K2).28,49 The test is administered under 2 conditions: (1) forward condition, in which participants repeat each series back in the presented order, without reordering (ie, transient online storage and retrieval) and (2) reordered condition, in which the participants mentally rearrange the order of the presented letters and numbers (ie, executive-functioning working memory) so that numbers are recalled first in ascending order and letters are then recalled in alphabetical order. Raw scores are calculated, with the score for the executive-functioning working memory condition being used as the primary dependent variable. Both conditions of the letter-number span can be administered in 15 minutes (QA Site: UCLA).
Verbal Memory: CVLT-II. Verbal learning and memory are assessed using the CVLT-II, an established test for assessing declarative memory, a learning curve, strategies of learning (ie, serial vs semantic processing), and interference effects that may impair recall.50,51 Administration of the CVLT-II involves 5 trials of a 16-word list. These words can be grouped into 4 semantic clusters (embedded in the test construction) for more efficient recall. Recall is assessed immediately after each learning trial, after one administration of a second list of 16 words (an interference condition), and after a 20-minute delay. Recall trials include free and then cued (in which participants recall the words after being cued with semantic category names) recall conditions. Recognition is also tested. The primary dependent measure is the CVLT-II Total Recall score (sum of 5 trials). Administration time is about 20 minutes. Other tests/measures (the DS-CPT in this protocol) may be administered during the 20-minute delay between recall conditions (QA site: Harvard).
University of Pennsylvania CNB. The CNB12,52–54 is administered on portable Macintosh computers in a fixed order using clickable icons. It was abbreviated for the COGS to reduce redundancy with core endophenotypes and includes brief standardized rest periods. The COGS CNB uses 8 tasks to assess 6 neurocognitive domains, described in detail elsewhere: abstraction and mental flexibility (Penn Conditional Exclusion Test55), attention and working memory (letter-n-back, 1-back, and 2-back conditions56), face memory (Penn Face Memory Test57), spatial memory (Visual Object Learning Test58), spatial processing (Computerized Judgment of Line Orientation12), sensorimotor dexterity (Computerized Finger-Tapping Task and Motor Praxis test), and emotion processing (Penn Emotion Recognition Test-4059). For each domain, 3 summary functions are calculated: (1) Accuracy, which reflects the number of correct responses; (2) Speed, the median response time for correct answers; and (3) Efficiency, which reflects both accuracy and speed (accuracy/log [speed]). Scoring of the battery is automated and produces scaled scores for each neurocognitive domain. The total administration time is approximately 60 minutes (QA site: PENN).
Results
From the start of testing in the fall of 2003 until February 2006, 355 families were enrolled, 792 participants were interviewed, and 688 participants were endophenotyped. Endophenotyped participants included 154 probands, 343 first-degree relatives (n of siblings = 209, n of parents = 134), and 191 CCS. Each of these individuals completed at least one endophenotype, with the majority completing all endophenotypes. Reasons for missing data and criteria for removing data from any particular endophenotype are described in individual endophenotype articles. This report presents demographic information on the preliminary sample because recruitment and testing of participants are ongoing.
Family Structure
Table 2 presents a summary of families according to the number of first-degree relatives who were endophenotyped as of February 2006. In 159 families, ≥1 member completed the endophenotype assessment. Unaffected sibling pairs will be critical to COGS genetics analyses. In 5 families, the proband had a sibling who was also affected with schizophrenia. These families, however, all included at least one other unaffected sibling who had completed endophenotype assessment, so the “contrast outcome” requirement (at least one sibling with and one sibling without schizophrenia) was met. Of the families with endophenotypes, 21 did not yet have a sibling who had completed the battery (see table 2, Row 1). Thus, at the time these data were downloaded, there were a total of 138 families with ≥1 unaffected sibling who had been endophenotyped (see table 2, Rows 2–7).
Table 2.
Families with Completed Endophenotype Assessments
| Number of Parents with Endophenotypes |
||||
|---|---|---|---|---|
| Number of Siblings with Endophenotypes | 0 | 1 | 2 | Total Number of Families |
| 0 | 14 | 5 | 2 | 21 |
| 1 | 37 | 12 | 46 | 95 |
| 2 | 15 | 5 | 5 | 25 |
| 3 | 9 | 1 | 2 | 12 |
| 4 | 3 | 0 | 0 | 3 |
| 5 | 1 | 1 | 0 | 2 |
| 6 | 1 | 0 | 0 | 1 |
| Total number of families | 80 | 24 | 55 | 159 |
Note: Includes 5 families in which proband had not yet been endophenotyped.
Demographics
Basic demographic characteristics of endophenotyped participants are presented in tables 3 (age, education, parental education, and sex) and 4 (ethnicity). Preliminary analyses indicate that relatives are significantly older than CCS and patients, who do not differ from each other. Importantly, siblings do not differ in age from CCS. Thus, in order to evaluate the potential impact of age on endophenotype performance in relatives, we will be able to compare the subgroup of siblings to the CCS, in addition to the standard approach of using age as a covariate. However, because the parents of the probands tend to be older than the CCS, the COGS is enhancing recruitment of older CCS who will be better matched in age to parents of the probands.
Table 3.
Demographic Characteristics of COGS Participants
| PRO | REL | CCS | Post hoc significant at P < 0.05 (following significant group effect)a | |
|---|---|---|---|---|
| n | 154 | 343 | 191 | |
| Age (years)a | ||||
| Mean | 35.3 | 45.5 | 37.3 | REL > CCS |
| SD | (11.0) | (13.8) | (12.1) | REL > PRO |
| Education (years)a | ||||
| Mean | 13.5 | 15.4 | 15.4 | CCS > PRO |
| SD | (2.8) | (2.8) | (2.8) | REL > PRO |
| Parental education (years)a | ||||
| Mean | 15.7 | 14.7 | 14.9 | PRO > REL |
| SD | (3.4) | (3.6) | (3.0) | PRO > CCS |
| Sex: M:Fa | 110:44 | 151:192 | 81:110 | PRO disproportionately male vs CCS and REL |
Note: aP < 0.05 by univariate ANOVA or chi square. Post hoc analyses were pairwise Tukey least significant difference or pairwise (2 × 2) chi square. Parental education is the level of education achieved by the participant's mother or father, whichever is highest. PRO, proband; REL, relative; CCS, community comparison subject.
As expected, probands have achieved less education than CCS and relatives. Notably, relatives and CCS do not significantly differ in their years of education (see table 3) making this direct relative-CCS comparison straightforward regarding possible education effects. Because participant's educational attainment is associated with illness status (eg, Isohanni et al60), some COGS endophenotype analyses will use education of the most educated parent rather than of the participant as an analytic variable. Relatives and CCS are well matched in both personal and parental education, indicating that educational disparities are unlikely to account for any endophenotype deficits observed in relatives.
As anticipated, the groups are not balanced by sex. The proband group is disproportionately male compared with the relative group and CCS who do not differ from each other. Our recruitment efforts have therefore been altered to recruit more male controls. Sex balance is important so that any observed endophenotype impairments will not be attributable to varying distributions of males and females.
The sample is composed of participants from many ethnicities, with the majority of European descent. see (table 4) Because ethnicity itself is a complex, sometimes subjective, and often misunderstood construct, our statistical genetics strategy will account for any final observed group × ethnicity interactions.61,62 Individual articles reporting analyses of endophenotypes will evaluate the existence of any group differences in these demographic variables at the given point of time. Where relevant for particular endophenotypes, demographic variables will then be entered as factors or covariates in the analyses.
Table 4.
Parental Ethnicity of Probands and Community Comparison Subjects
| Father's Ethnicity |
||||||||
|---|---|---|---|---|---|---|---|---|
| Mother's Ethnicity | European | African American | Asian | Native American | Hispanic | Other | Unknown | Total |
| European | 98/118 | 0/0 | 1/0 | 2/1 | 1/1 | 2/1 | 2/1 | 106/122 |
| African American | 0/0 | 9/30 | 0/0 | 0/0 | 0/1 | 0/0 | 0/0 | 9/31 |
| Asian | 3/1 | 0/0 | 9/8 | 0/0 | 0/0 | 0/0 | 0/0 | 12/9 |
| Native American | 1/0 | 0/2 | 0/0 | 1/1 | 0/2 | 0/0 | 0/0 | 2/5 |
| Hispanic | 1/2 | 0/0 | 0/0 | 1/0 | 9/10 | 0/0 | 0/0 | 11/12 |
| Other | 1/1 | 0/0 | 1/0 | 0/0 | 0/0 | 5/6 | 0/0 | 7/7 |
| Unknown | 3/2 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 4/3 | 7/5 |
| Total | 107/95 | 9/23 | 11/7 | 4/0 | 10/6 | 7/6 | 5/4 | 154/191 |
Note: n of probands/n of community comparison subjects. Ethnicity codes follow Diagnostic Interview for Genetic Studies coding rules. Other = special populations including genetic isolates and outliers (eg, Old Order Amish, Sardinian, Ashkenazi, Sephardic), Pacific Islanders, and indigenous Australians.
Clinical Characteristics
Table 5 presents lifetime BEFD in each participant group. The most common comorbid conditions among probands are substance-related disorders, both abuse and dependence. Approximately 4% of relatives were diagnosed with schizophrenia spectrum disorders (ie, schizophrenia, schizoaffective disorder, psychotic disorder not otherwise specified, and schizotypal and paranoid personality disorders). In both the relative and the CCS groups, substance abuse, substance dependence, and major depressive disorder (MDD)–single episode each affect between 7–10% of participants. Thus, relatives and CCS in this sample exhibit similar patterns of axis I nonschizophrenia spectrum psychopathology, except that more relatives (12%) than CCS (5%) have experienced MDD-recurrent. Note, however, that unlike relatives, potential CCS would have been excluded if they had current substance abuse (past 1 month) or dependence (past 6 months) (see table 5). Again, where relevant for particular endophenotypes, conditions such as mood disorders and substance-related disorders, as well as psychotropic medications, will be handled as factors or covariates in the analyses.
Table 5.
DSM-IV Lifetime Best Estimate Final Diagnoses of COGS Participants
| Participant Group |
||||||
|---|---|---|---|---|---|---|
|
DSM-IV Axis Diagnostic Category |
PRO (n = 154) |
REL (n = 343) |
CCS (n = 191) |
|||
| n | % | n | % | n | % | |
| Axis I | ||||||
| Schizophrenia | 154 | 100 | 6 | 2 | 0 | 0 |
| Schizoaffective disorder | 0 | 0 | 2 | 1 | 0 | 0 |
| Psychotic disorder, NOS | 0 | 0 | 1 | <1 | 0 | 0 |
| Major depressive disorder | 1a | <1 | 34 | 10 | 18 | 9 |
| Single episode | ||||||
| Recurrent | 0 | 0 | 42 | 12 | 9 | 5 |
| Dysthymic disorder | 1a | <1 | 14 | 4 | 3 | 2 |
| Depressive disorder, NOS | 14 | 9 | 4 | 1 | 0 | 0 |
| Bipolar I disorder | 0 | 0 | 3 | 1 | 1 | 1 |
| Bipolar disorder, NOS | 0 | 0 | 1 | <1 | 0 | 0 |
| Mood disorder due to a general medical condition | 0 | 0 | 1 | <1 | 0 | 0 |
| Substance dependence | 34 | 22 | 25 | 7 | 13 | 7 |
| Substance abuse | 28 | 18 | 29 | 8 | 17 | 9 |
| Axis II | ||||||
| Schizotypal personality disorder | — | — | 5 | 1 | 0 | 0 |
| Paranoid personality disorder | — | — | 1 | <1 | 0 | 0 |
Note: DSM-IV, Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition; PRO, proband; REL, relative; CCS, community comparison subject; n, number of participants within the participant group with the diagnosis; %, percentage of participants within the participant group with the diagnosis; NOS, not otherwise specified.
Mood disturbance occurred prior to the onset of schizophrenia.
In this preliminary analysis, probands had a mean (SD) total SANS score of 10.1 (5.9) and a SAPS score of 7.2 (3.9). Global assessment of functioning for the past month among the 3 groups was probands (mean = 44.8, SD = 12.8), relatives (mean = 78.9, SD = 13.5), and CCS (mean = 82.4, SD = 9.8). More detailed clinical data will be presented in future articles.
Discussion
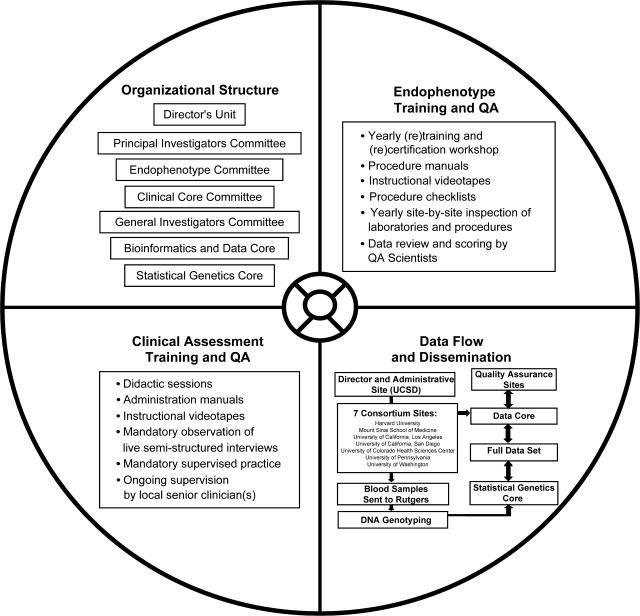
Multisite genetic collaborations, especially these involving multigenerational families, require assessment and recruitment methods that are precisely defined and uniformly applied. These requirements sound straightforward, but in practice entail a great deal of planning and thought, standardization of training and methods, and ongoing retraining to attain cross-site QA. Figure 3 provides a summary of the key components of the multisite collaboration. This article presents preliminary demographic findings on a carefully diagnosed 7-site sample of probands, families, and normal comparison participants for endophenotype performance and genetic substrates, which will both be fully characterized in current and future reports. In the COGS cohort, parental DNA, the ascertainment of a sibship with diagnostic “contrast,” and sibs who are endophenotyped and genotyped are the hallmark minimal requirements for family eligibility. After over 2 years of active recruitment, 688 participants have been successfully evaluated and endophenotyped, including 138 families with at least one sibpair discordant (contrasting) for schizophrenia, and a corresponding sample of 191 CCS. DNA has been collected for genotyping on all participants.
Fig. 3.
Key Components of the COGS Multisite Collaboration.
As emphasized throughout this article, the COGS has been designed and organized in such a way as to facilitate the reliable collection of refined endophenotype measures across all 7-subject ascertainment and clinical centers. The design and goals of the COGS are thus quite different in orientation from other large-scale genetic consortia sponsored by the National Institutes of Health (NIH), which often focus on integrating information provided by participating study sites using very different endpoints and study designs or consider the collection of families with a single disease using common, accepted diagnostic criteria and/or perhaps a few additional clinical or neurocognitive parameters collected in a uniform manner. In this light, the COGS is unique among large-scale NIH-sponsored genetic consortia in that it focuses on collection of multiple subclinical measures—that are difficult enough to assay at any single site—in a uniform manner across all data collection sites so as to maximize the power of genetic linkage and association studies.
Caveats and Challenges
Because the central aim of the COGS is to explore the familiality and genetics of quantitative endophenotypic traits, sufficient contrasts between individuals with high and low phenotypic variation will generate adequate statistical power for informative genetic analyses.34,37,63–66 This approach is distinct from the strategies used for studying a single qualitatively defined disease, where affected sibling pairs and multiplex families provide the most efficient and powerful design for allele-sharing linkage analysis.67 When studying quantitative phenotypes underlying or contributing to qualitative disease outcomes like schizophrenia, it is important to include families that do not exclusively have affected individuals because, if impaired performance on these phenotypes truly associate with schizophrenia, one would actually reduce the variation exhibited by this trait in the samples and thereby reduce power for studying the determinants of the quantitative phenotypes.
However, the requirement of relatively intact families could lead to the ascertainment of a group of families that is perhaps “healthier,” or at least better connected, than those participating in other studies. It is possible that intact families (especially probands and their siblings) are not representative of “all” schizophrenia kindred (eg,Horan et al31). For example, perhaps these volunteer families carry less of a “paranoid” or “suspicious” genetic load, which could make positive findings less likely and therefore reduce false positive (or increase false negative) results. Similarly, families willing to participate in a lengthy diagnostic and testing protocol, which can be tedious, may be different from families in other schizophrenia family studies that are not as demanding of time and effort. To the extent that such differences are associated with endophenotype performance, participants from these families may not show the typical expected pattern of endophenotype impairments observed in families recruited with other strategies. Future analyses will address the impact of this ascertainment strategy on clinical features and endophenotype performance of COGS participants in comparison with those from other family genetic studies.
The inclusion of CCS in the COGS provides the additional comparative data necessary for evaluating the grouped results of neurophysiological and neurocognitive tests. CCS both with and without a lifetime history of nonpsychotic mental disorders are included in the COGS. While our field continues to struggle with the ideal criteria for selecting comparison subjects (eg, Kendler68), we believe that the COGS criteria provides a sufficient number of CCS diagnosed with psychiatric conditions to allow both (1) meaningful subanalyses of the impact of comorbid diagnoses on endophenotype measurements and (2) direct comparisons of the CCS group to the relatives, a portion of whom also have psychiatric conditions.
There are several other challenges and unique opportunities available to the COGS. First, while the minimum COGS family structure requires participation of a discordant sibpair and 2 living parents, additional siblings are not always available or willing to participate. If these nonparticipating siblings share relevant clinical characteristics that cluster with endophenotype abnormalities (eg, schizophrenia and schizotypy), differences between relatives and CCS may be more difficult to detect. Fortunately, the availability of FIGS data on nonparticipating relatives will enable an estimate of the magnitude of these potential biases. Second, the cross-sectional design may lead to diagnostic misclassification, especially among young adult siblings of affected probands who are still at high risk for developing schizophrenia. This concern is addressed in part by the comprehensive assessment of subsyndromal and schizotypal symptoms in subsections of the DIGS, and by the strategy of using neurobiological endophenotypes, rather than clinical phenotypes, as the trait of interest in genetic analyses. Finally, in the interest of efficiency, the structured portion of the DIGS interview has been modified slightly to emphasize the major diagnoses that are relevant to schizophrenia. Thus, some diagnoses (eg, phobias) may be less thoroughly assessed than others (eg, major depression, schizophrenia, bipolar disorder, and substance-use disorders).
In summary, the COGS has established a rigorous infrastructure for participant recruitment, clinical characterization, measurement of multiple endophenotypes, data management, and ultimately genotyping. The structure and function of this endeavor has and will serve as a model for other endophenotyping consortia for medical and neuropsychiatric disorders. This multisite study of patients with schizophrenia, their family members, and CCS will help to clarify the genetic architecture of quantitative endophenotypes underlying schizophrenia susceptibility, as described in other articles of this Special Issue of Schizophrenia Bulletin and in the first generation of COGS original data papers cited in those reviews.
Acknowledgments
This work was supported by the NIMH. We are grateful for the generosity of time and effort by the families who make this COGS research possible. We thank the following key personnel for their dedicated efforts to the COGS: Harvard University (RO1-MH065562; MH43518; Commonwealth Research Center of the Massachusetts, Department of Mental Health): Lynda Jacobs, Monica Landi, Erica Lee, Andrea Roe, Frances Schopick, and Alison Thomas; Mount Sinai School of Medicine (RO1-MH065554): Leigh Baitler, Rui Ferreira, Robert Fieo, Frances Schopick, Christopher Smith, and Rebecca West; University of California Los Angeles (RO1-MH65707): William Horan and Mark Sergi; University of California, San Diego (R01-MH065571): Andras Kovach, Joyce Sprock, Katrin Meyer-Gomes, Barbara Haugeland, Kari Tweedale, Sheldrick Holmes, and Emmeline Crowley; University of Colorado (RO1-MH65588): Jamey Ellis, Jeff Hollis, Vicki Pender, Bernadette Sullivan, Bettye Clement, Christopher Cason, and Alexis Ritvo; University of Pennsylvania (RO1-MH65578): Felipe Da Silva, Alexandra Duncan Ramos, Jarrod Gutman, CarlaAnn Henry, Paul Hughett, Jennifer Jackson Greene, Adrienne Mishkin, J. Daniel Ragland, Leslie Ramsey, David Rice, Jan Richard, Devon Seward, and Robert Witalec; University of Washington (R01-MH65558): Kate B. Alvey, Andrew C. David, Sean P. Meichle, and Denise O. Pritzl. Portions of this article were presented at the American College of Neuropsychopharmacology Meeting, December 2005, Waikoloa, Hawaii.
References
- 1.Faraone SV, Tsuang D, Tsuang MT. Genetics of Mental Disorders: A Guide for Students, Clinicians, and Researchers. New York, NY: Guildford; 1999. [Google Scholar]
- 2.Gottesman, Shields J. Schizophrenia and Genetics; a Twin Study Vantage Point. New York, NY: Academic Press Inc.; 1972. [Google Scholar]
- 3.Braff DL, Freedman R. Endophenotypes in studies of the genetics of schizophrenia. In: Davis KL, Charney DS, Coyle JT, Nemeroff C, editors. Neuropsychopharmacology: The fifth generation of progress. Philadelphia, PA: Lippincott Williams and Wilkens; 2002. pp. 703–716. [Google Scholar]
- 4.Badner JA, Gershon ES. Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry. 2002;7:405–411. doi: 10.1038/sj.mp.4001012. [DOI] [PubMed] [Google Scholar]
- 5.Baron M. Genetics of schizophrenia in the new millennium: progress and pitfalls. Am J Hum Genet. 2001;68:299–312. doi: 10.1086/318212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottesman, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 7.Gould TD. Gottesman Psychiatric endophenotypes and the development of valid animal models. Genes Brain Behav. 2006;5:113–119. doi: 10.1111/j.1601-183X.2005.00186.x. [DOI] [PubMed] [Google Scholar]
- 8.Iacono WG. Identifying genetic risk for psychopathology. In: Routh DK, DeRubeis RJ, editors. The Science of Clinical Psychology: Accomplishments and Future Directions. Washington, DC: American Psychological Association; 1998. pp. 3–22. [Google Scholar]
- 9.Seidman LJ. Clinical neuroscience and epidemiology in schizophrenia. Harv Rev Psychiatry. 1997;3:338–342. doi: 10.3109/10673229709030562. [DOI] [PubMed] [Google Scholar]
- 10.Tsuang MT, Faraone SV, Lyons MJ. Identification of the phenotype in psychiatric genetics. Eur Arch Psychiatry Neurol Sci. 1993;243:131–142. doi: 10.1007/BF02190719. [DOI] [PubMed] [Google Scholar]
- 11.Gur RC, Ragland JD, Moberg PJ, et al. Computerized neurocognitive scanning: II. The profile of schizophrenia. Neuropsychopharmacology. 2001;25:777–788. doi: 10.1016/S0893-133X(01)00279-2. [DOI] [PubMed] [Google Scholar]
- 12.Gur RC, Ragland JD, Moberg PJ, et al. Computerized neurocognitive scanning: I. Methodology and validation in healthy people. Neuropsychopharmacology. 2001;25:766–788. doi: 10.1016/S0893-133X(01)00278-0. [DOI] [PubMed] [Google Scholar]
- 13.Adler LE, Freedman R, Ross RG, Olincy A, Waldo MC. Elementary phenotypes in the neurobiological and genetic study of schizophrenia. Biol Psychiatry. 1999:8–18. doi: 10.1016/s0006-3223(99)00085-2. [DOI] [PubMed] [Google Scholar]
- 14.Siegel C, Waldo M, Mizner G, Adler L, Freedman R. Deficits in sensory gating in schizophrenic patients and their relatives. Evidence obtained with auditory evoked responses. Arch Gen Psychiatry. 1984;41:607–612. doi: 10.1001/archpsyc.1984.01790170081009. [DOI] [PubMed] [Google Scholar]
- 15.Adler LE, Pachtman E, Franks RD. Neurophysiological evidence for a deficit in neuronal mechanisms involved in sensory gating in schizophrenia. Biol Psychiatry. 1982;17:639–654. [PubMed] [Google Scholar]
- 16.Braff DL, Stone C, Callaway E, Geyer M, Glick I, Bali L. Prestimulus effects on human startle reflex in normals and schizophrenics. Psychophysiology. 1978;15:339–343. doi: 10.1111/j.1469-8986.1978.tb01390.x. [DOI] [PubMed] [Google Scholar]
- 17.Braff DL, Geyer MA, Light GA, et al. Impact of prepulse characteristics on the detection of sensorimotor gating deficits in schizophrenia. Schizophr Res. 2001;49:171–178. doi: 10.1016/s0920-9964(00)00139-0. [DOI] [PubMed] [Google Scholar]
- 18.Cadenhead KS, Swerdlow NR, Shafer KM, Diaz M, Braff DL. Modulation of the startle response and startle laterality in relatives of schizophrenia patients and schizotypal personality disordered subjects: evidence of inhibitory deficits. Am J Psychiatry. 2000;157:1660–1668. doi: 10.1176/appi.ajp.157.10.1660. [DOI] [PubMed] [Google Scholar]
- 19.Calkins ME, Curtis CE, Iacono WG, Grove WM. Antisaccade performance is impaired in medically and psychiatrically healthy biological relatives of schizophrenia patients. Schizophr Res. 2004;71:167. doi: 10.1016/j.schres.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 20.Fukushima J, Fukushima K, Chiba T, Tanaka S, Yamashita I, Kato M. Disturbances of voluntary control of saccadic eye movements in schizophrenic patients. Biol Psychiatry. 1988;23:670–677. doi: 10.1016/0006-3223(88)90050-9. [DOI] [PubMed] [Google Scholar]
- 21.Cornblatt BA, Risch NJ, Faris G, Friedman D, Erlenmeyer-Kimling L. The continuous performance test, identical pairs version (CPT-IP) I. New findings about sustained attention in normal families. Psychiatry Res. 1988;26:223–338. doi: 10.1016/0165-1781(88)90076-5. [DOI] [PubMed] [Google Scholar]
- 22.Nuechterlein KH, Parasuraman R, Jiang Q. Visual sustained attention: image degradation produces rapid sensitivity decrement over time. Science. 1983;220:327–329. doi: 10.1126/science.6836276. [DOI] [PubMed] [Google Scholar]
- 23.Rosvold HE, Mirsky AF, Sarason I, Bransome ED, Beck LH. A continuous performance test of brain damage. J Consult Psychol. 1956;20:343–350. doi: 10.1037/h0043220. [DOI] [PubMed] [Google Scholar]
- 24.Cirillo M, Seidman LJ. A review of verbal declarative memory function in schizophrenia: from clinical assessment to genetics and brain mechanisms. Neuropsychol Rev. 2003;13:43–77. doi: 10.1023/a:1023870821631. [DOI] [PubMed] [Google Scholar]
- 25.Faraone SV, Seidman LJ, Kremen WS, Pepple JR, Lyons MJ, Tsuang MT. Neuropsychological functioning among the nonpsychotic relatives of schizophrenic patients: a diagnostic efficiency analysis. J Abnorm Psychol. 1995;104:286–304. doi: 10.1037//0021-843x.104.2.286. [DOI] [PubMed] [Google Scholar]
- 26.Perry W, Potterat EG, Braff DL. Self-monitoring enhances Wisconsin Card Sorting Test performance in patients with schizophrenia: performance is improved by simply asking patients to verbalize their sorting strategy. J Int Neuropsychol Soc. 2001;7:344–352. doi: 10.1017/s1355617701733085. [DOI] [PubMed] [Google Scholar]
- 27.Conklin HM, Curtis CE, Calkins ME, Iacono WG. Working memory functioning in schizophrenia patients and their first-degree relatives: cognitive functioning shedding light on etiology. Neuropsychologia. 2005;43:930–942. doi: 10.1016/j.neuropsychologia.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 28.Gold JM, Carpenter C, Randolph C, Goldberg TE, Weinberger DR. Auditory working memory and Wisconsin Card Sorting Test performance in schizophrenia. Arch Gen Psychiatry. 1997;54:159–166. doi: 10.1001/archpsyc.1997.01830140071013. [DOI] [PubMed] [Google Scholar]
- 29.Park S, Holzman PS. Schizophrenics show spatial working memory deficits. Arch Gen Psychiatry. 1992;49:975–982. doi: 10.1001/archpsyc.1992.01820120063009. [DOI] [PubMed] [Google Scholar]
- 30.Greenwood TA, Braff DL, Cadenhead KS, et al. 2006. The Consortium on the Genetics of Schizophrenia (COGS): preliminary heritability analyses of endophenotypic measures for schizophrenia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horan WP, Braff DL, Nuechterlein KH, et al. 2006. Verbal working memory impairments in individuals with schizophrenia and their first-degree relatives: findings from the Consortium on the Genetics of Schizophrenia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radant A, Dobie DJ, Calkins ME, et al. Schizophr Res. Successful multi-site measurement of antisaccade performance deficits in schizophrenia. [DOI] [PubMed] [Google Scholar]
- 33.Schork NJ, et al. Schizophr Bull. 2007. [Google Scholar]
- 34.Carey G, Williamson J. Linkage analysis of quantitative traits: increased power by using selected samples. Am J Hum Genet. 1991;49:786–796. [PMC free article] [PubMed] [Google Scholar]
- 35.Gu C, Todorov AA, Rao DC. Genome screening using extremely discordant and extremely concordant sib pairs. Genet Epidemiol. 1997;14:791–796. doi: 10.1002/(SICI)1098-2272(1997)14:6<791::AID-GEPI38>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 36.Risch N, Zhang H. Extreme discordant sib pairs for mapping quantitative trait loci in humans. Science. 1995;268:158–159. doi: 10.1126/science.7777857. [DOI] [PubMed] [Google Scholar]
- 37.Risch N, Zhang H. Mapping quantitative traits loci with extreme discordant sib pairs: sampling considerations. Am J Hum Genet. 1996;58:836–843. [PMC free article] [PubMed] [Google Scholar]
- 38.Ott J. Analysis of Human Genetic Linkage. Baltimore, MD: Johns Hopkins University Press; 1999. [Google Scholar]
- 39.Nurnberger JI, Jr, Blehar MC, Kaufmann CA, et al. Diagnostic interview for genetic studies. Rationale, unique features, and training. NIMH Genetics Initiative. Arch Gen Psychiatry. 1994;51:849–859. doi: 10.1001/archpsyc.1994.03950110009002. [DOI] [PubMed] [Google Scholar]
- 40.Hall RC, Parks J. The modified global assessment of functioning scale: addendum. Psychosomatics. 1995;36:416–417. doi: 10.1016/S0033-3182(95)71656-5. [DOI] [PubMed] [Google Scholar]
- 41.Andreasen NC. The Scale for the Assessment of Positive Symptoms (SAPS) Iowa City, Iowa: The University of Iowa; 1984. [Google Scholar]
- 42.Andreasen NC. The Scale for the Assessment of Negative Symptoms (SANS) Iowa City, Iowa: The University of Iowa; 1983. [PubMed] [Google Scholar]
- 43.Kendler KS, Lieberman JA, Walsh D. The structured interview for schizotypy (SIS): a preliminary report. Schizophr Bull. 1989;15:559–571. doi: 10.1093/schbul/15.4.559. [DOI] [PubMed] [Google Scholar]
- 44.Maxwell ME. FIGS: Clinical Neurogenetics Branch, Intramural Research Program. Betesia, MD: NIMH; 1996. [Google Scholar]
- 45.Ventura J, Lukoff D, Nuechterlein KH, Liberman RP, Green MF, Shaner A. Appendix 1: Brief Psychiatric Rating Scale (BPRS) expanded version (4): scales, anchor points, and administration manual. Int J Methods Psychiatr Res. 1993;3:227–244. [Google Scholar]
- 46.Griffith J, Hoffer LD, Adler LE, Zerbe GO, Freedman R. Effects of sound intensity on a midlatency evoked response to repeated auditory stimuli in schizophrenic and normal subjects. Psychophysiology. 1995;32:460–466. doi: 10.1111/j.1469-8986.1995.tb02097.x. [DOI] [PubMed] [Google Scholar]
- 47.Nuechterlein KH, Asarnow RF. Degraded Stimulus Continuous Performance Test (DS-CPT) Program for IBM-Compatible Microcomputers, Version 8.12. Los Angeles, CA: Nuechterlein and Asarnow; 1999. [Google Scholar]
- 48.Nuechterlein KH. Vigilance in schizophrenia and related disorders. In: Steinhauer SR, Gruzelier JH, Zubin J, editors. Handbook of Schizophrenia, Vol. 5: Neuropsychology, Psychophysiology, and Information processing. Amsterdam, Netherlands: Elsevier; 1991. pp. 397–433. [Google Scholar]
- 49.Wechsler D. Wechsler Adult Intelligence Scale. San Antonio, TX: Psychological Corporation; 1997. [Google Scholar]
- 50.Delis DC, Kramer JH, Kaplan E, Ober BA. California Verbal Learning Test-Adult version. New York, NY: The Psychological Corporation; 1987. [Google Scholar]
- 51.Delis D, Kramer J, Kaplan E, Ober B. California Verbal Learning Test: Second Edition. Adult version. Manual. New York, NY: The Psychological Corporation; 2000. [Google Scholar]
- 52.Calkins ME, Gur RC, Ragland JD, Gur RE. Face recognition memory deficits and visual object memory performance in patients with schizophrenia and their relatives. Am J Psychiatry. 2005;162:1963–1966. doi: 10.1176/appi.ajp.162.10.1963. [DOI] [PubMed] [Google Scholar]
- 53.Gur RE, Nimgaonkar VL, Almasy L, et al. 2006. Neurocognitive endophenotypes in a multiplex multigenerational family study of schizophrenia. under review. [DOI] [PubMed] [Google Scholar]
- 54.Aliyu MH, Calkins ME, Swanson CL, et al. Project among African Americans to explore risks for schizophrenia: assessment and recruitment methods. Schizophr Res. doi: 10.1016/j.schres.2006.06.027. In press. [DOI] [PubMed] [Google Scholar]
- 55.Kurtz M, Ragland J, Moberg P, Gur R. The Penn Conditional Exclusion Test: a new measure of executive-function with alternate forms for repeat administration. Arch Clin Neuropsychol. 2004;19:191–201. doi: 10.1016/S0887-6177(03)00003-9. [DOI] [PubMed] [Google Scholar]
- 56.Ragland JD, Turetsky BI, Gur RC, et al. Working memory for complex figures: an fMRI comparison of letter and fractal N-back tasks. Neuropsychology. 2002;16:370–379. [PMC free article] [PubMed] [Google Scholar]
- 57.Gur RC, Jaggi JL, Ragland JD, et al. Effects of memory processing on regional brain activation: cerebral blood flow in normal subjects. Int J Neurosci. 1993;72:31–44. doi: 10.3109/00207459308991621. [DOI] [PubMed] [Google Scholar]
- 58.Glahn DC, Gur RC, Ragland JD, Gur RE. Reliability, performance characteristics, and construct validity and initial application of the visual object learning test (VOLT) Neuropsychology. 1997;11:602–612. doi: 10.1037//0894-4105.11.4.602. [DOI] [PubMed] [Google Scholar]
- 59.Kohler CG, Turner TH, Bilker WB, et al. Facial emotion recognition in schizophrenia: intensity effects and error pattern. Am J Psychiatry. 2003;160:1768–1774. doi: 10.1176/appi.ajp.160.10.1768. [DOI] [PubMed] [Google Scholar]
- 60.Isohanni I, Jones PB, Jarvelin MR, et al. Educational consequences of mental disorders treated in hospital. A 31-year follow-up of the Northern Finland 1966 birth cohort. Psychol Med. 2001;31:339–349. doi: 10.1017/s003329170100304x. [DOI] [PubMed] [Google Scholar]
- 61.Shriver MD, Kennedy GC, Parra EJ, et al. The genomic distribution of population substructure in four populations using 8,525 autosomal SNPs. Hum Genomics. 2004;1:274–286. doi: 10.1186/1479-7364-1-4-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reiner AP, Ziv E, Lind DL, et al. Population structure, admixture, and aging-related phenotypes in African American adults: the Cardiovascular Health Study. Am J Hum Genet. 2005;76:463–477. doi: 10.1086/428654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abecasis GR, Cookson WO, Cardon LR. The power to detect linkage disequilibrium with quantitative traits in selected samples. Am J Hum Genet. 2001;68:1463–1474. doi: 10.1086/320590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.deAndrade M, Amos CI. Ascertainment issues in variance component models. Genet Epidemiol. 2000;19:333–344. doi: 10.1002/1098-2272(200012)19:4<333::AID-GEPI5>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 65.Iyengar S, Calafell F, Kidd KK. Detection of major genes underlying several quantitative traits associated with a common disease using different ascertainment schemes. Genet Epidemiol. 1997;14:809–814. doi: 10.1002/(SICI)1098-2272(1997)14:6<809::AID-GEPI41>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 66.Todorov AA, Province MA, Borecki IB, Rao DC. Trade-off between sibship size and sampling scheme for detecting quantitative trait loci. Hum Hered. 1997;47:1–5. doi: 10.1159/000154381. [DOI] [PubMed] [Google Scholar]
- 67.Lander ES, Schork NJ. The genetic dissection of complex traits. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 68.Kendler KS. The super-normal control group in psychiatric genetics: possible artifactual evidence for coaggregation. Psychiatr Genet. 1990:45–53. [Google Scholar]