Abstract
The regulator of G-protein signaling 4 (RGS4, chromosome 1q23.3) plays a critical role in G-protein function. Four common single-nucleotide polymorphisms (SNPs) localized between the 5′ upstream sequence and the first intron, as well as 2 haplotypes derived from these SNPs may confer liability to schizophrenia (SZ). However, the pattern of associations varies among samples. To help clarify the putative associations, we report the following analyses: (1) a comprehensive catalog of common polymorphisms, (2) linkage disequilibrium (LD) and association analyses using these SNPs, and (3) functional analysis based on dual-luciferase promoter assays. We identified 62 SNPs from a 20-kb genomic region spanning RGS4, of which 26 are common polymorphisms with a minor allele frequency (MAF) of >5%. LD analysis suggested 5 clusters of SNPs (r2 > .8). Association analyses using the novel SNPs were consistent with the prior reports, but further localization was constrained by significant LD across the region. The 2 haplotypes reported to confer liability to SZ had significant promoter activity compared with promoterless constructs, suggesting a functional role for both haplotypes. Further analyses of promoter sequences are warranted to understand transcriptional regulation at RGS4. This information will be useful for further analysis of samples in which genetic association of RGS4 polymorphisms with SZ has been reported.
Keywords: RGS4, association, linkage disequilibrium, promoter, schizophrenia
Regulators of G-protein signaling (RGS) function as GTPase-activator proteins for heterotrimeric G-protein alpha (Gα) subunits and accelerate the hydrolysis of Gα-bound GTP.1 Thus, they shorten the duration of intracellular signaling of many G-protein–coupled receptors. To date, over 28 RGS proteins have been identified and most appear to have selective receptor targets.1,2 RGS proteins may therefore modulate intracellular effects of G-protein–coupled neurotransmitters. RGS proteins also appear to be responsive to stress, eg, regulator of G-protein signaling 4 (RGS4) expression is altered following chronic stress in rodents or following dexamethasone treatment of cells in vitro.3 Thus, a regulatory role for RGS4 in maintaining the milieu interior is plausible. RGS genes and their protein products are highly conserved, supporting critical roles in cellular function.4
The role of RGS proteins in several human diseases has been investigated. Drug addiction and brain injury may be associated with altered levels of some RGS mRNA species.5,6 Perhaps, the most extensive evidence relates to RGS4 and schizophrenia (SZ).7 It was initially suggested that RGS4 mRNA levels were significantly lower in postmortem samples from the dorsolateral prefrontal cortex of subjects with SZ, compared with matched controls.8 These studies suggest reduced expression of RGS4 in selected regions of the brain, and replicate studies have been published.9,10 In contrast, others did not detect significant case-control differences in postmortem brain samples.11
Because RGS4 maps to chromosome 1q23.3, a region implicated in prior SZ linkage studies,12 we conducted association studies using RGS4 genetic polymorphisms in family-based samples. These samples evaluate transmission of polymorphisms from heterozygous parents to affected offspring; significant transmission distortion is indicative of genetic association. Among case-parent “trio” families, we detected transmission distortions at 4 individual single-nucleotide polymorphisms (SNPs), and at haplotypes (combinations of these SNPs indicating chromosomal segments), in 2 independent samples of Caucasian ancestry.13 These samples were recruited separately by our group (Pittsburgh [PITT]) and by the National Institute of Mental Health (NIMH) Collaborative Genetics Initiative. The associated SNPs were denoted SNPs 1, 4, 7, and 18 (rs10917670, rs951436, rs951439, and rs2661319, respectively). However, the alleles associated in the PITT sample differed from those in the NIMH sample, leading to associations with 2 different haplotypes in these samples (haplotypes G-G-G-G, and A-T-A-A, respectively). The haplotypes together account for over 80% of the variation in this genomic region in US Caucasian samples. Suggestive transmission distortion with the A-T-A-A haplotype was also detected in an independent Indian sample gathered by our group, prompting us to propose RGS4 as a putative susceptibility gene for SZ.13 The associated region spans the 5′ upstream sequence and the first exon of the gene (6.09 kb). Since the initial report, 3 independent case-control comparisons and 2 family-based analysis have reported significant associations at the same SNPs, but the associated alleles have also differed between studies.14–18 Significant association with any of these SNPs was not detected in a Brazilian,19 a Chinese,20,21 a US sample,22 a UK sample,23,24 or a Japanese sample.25 In view of the discrepancies, we conducted meta-anlaysis of all available genotypes. Our published meta-analyses suggest risk due to at least 2 common haplotypes in the presence of heterogeneity.26 Two other meta-analyses that evaluated smaller subsets of these samples did not find significant association.27,28
There are several explanations for these divergent results, apart from stochastic variation. If an association exists, there could be one unidentified risk allele against the background of these 2 common haplotypes. Alternatively, 2 different (unidentified) risk variants could exist on these haplotypes. Risk may also be conferred by these haplotypes for different subgroups of SZ, and differences in ascertainment criteria could thus lead to divergent associations. Because of linkage disequilibrium (LD), polymorphisms in close proximity at a genomic segment can be correlated at the population level. Thus, the SNPs analyzed to date may indicate primary association with another polymorphism. Hence, the putative primary association, if present, could be in the intronic regions, the promoter sequences, or the upstream or downstream regions. To help discriminate among these possibilities, we identified all common RGS4 polymorphisms and evaluated patterns of LD. These SNPs were genotyped in our family-based and case-control samples.
No exonic variations were detected in the initial study, including the postmortem samples used for the expression analyses (n = 36). Thus, the functional impacts of the associations are uncertain. Because some of the associated SNPs and haplotypes are located in the upstream region of RGS4, we evaluated the possibility that they may alter transcription. Using in vitro dual-luciferase reporter systems, the promoter activities of selected SNPs and haplotypes were analyzed.
MATERIALS AND METHODS
Sample Collection
The recruitment of NIMH and PITT families as well as the controls for the PITT families has been described.13 Briefly, the NIMH sample consisted of families ascertained on the basis of 2 affected first-degree relatives with schizophrenia/schizoaffective disorder (Diagnostic and Statistical Manual for Mental Disorders, Revised Third Edition criteria) (http://zork.wustl.edu/nimh). From the entire sample, we identified 39 case-parent trios. Only one affected person was selected from each family. The PITT families were recruited without regard to familiality and consisted of case-parent trios (n = 154, including earlier reported 93 trios). The probands were diagnosed with SZ or schizoaffective disorder (Diagnostic and Statistical Manual for Mental Disorders, Fourth Edition criteria). In addition, neonatal cord blood samples were obtained from live births at Magee-Women's Hospital, Pittsburgh, and served as unscreened, population-based controls (n = 92). All participants reported Caucasian ancestry. Ethnicity was based on self-report (maternal report for neonatal samples). Written informed consent was obtained from all participants. The University of Pittsburgh Institutional Review Board did not require informed consent from neonatal control individuals.
SNP Identification and Genotyping
SNPs were identified initially by sequencing separate pools of DNA from cases and controls (n = 200 samples, each group).29,30 DNA pools were prepared by mixing equal concentrations of DNA, after quantifying individual samples with the Pico Green fluorescent method, using the supplier's protocol (http://probes.invitrogen.com). We can thus detect SNPs with frequency greater than 5%–10%.30 All SNPs were genotyped using single base extension SnaPshot assays (Applied Biosystems, Foster City, CA).31 Eight DNA samples were resequenced at each SNP to verify genotypes from SnaPshot assays. These samples were used as positive controls. Details of the polymerase chain reaction (PCR) primers, multiplex PCR, and SnaPshot reaction conditions, and extension primers information are available at our Web site (http://www.pitt.edu/∼nimga/research/rgs4). There was 100% concordance between the SNaPshot assays and genotypes obtained by sequencing.26 We also designed primers flanking the 500-bp complex repeat (L1-like family) in the RGS4 upstream region and the PCR-amplified products were analyzed on 2% agarose gels and stained with ethidium bromide.
Reporter Constructs and Dual-Luciferase Assay for Promoter activity
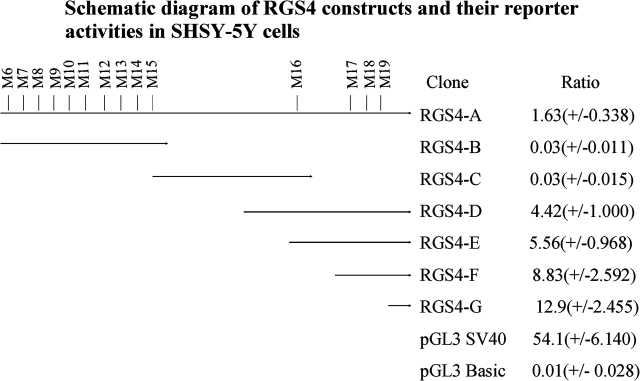
A 6.7-kb upstream region of RGS4 gene was examined (RGS4-A). In order to localize promoter activity, this 6.7-kb region was amplified in the form of 3 overlapping fragments (denoted RGS4-B, RGS4-C, and RGS4-D, figure 3) using Kpn1-tailed PCR-specific sense and Nhe1-tailed antisense primers (RGS4-B: −6735 to −4446 bp, RGS4-C: −4992 to −1583 bp, and RGS4-D: −3045 to −1 bp). We also generated 5′ RGS4 promoter deletion constructs from RGS4-D using Kpn1 and Nhe1 restriction enzymes. These fragments were denoted RGS4-E (−2364 to −1 bp), RGS4-F (−1078 to −1 bp), RGS4-G (−311 to −1 bp). For constructs RGS4-A, D to G, a common 3′ anchor Nhe1-tailed antisense primer (RGS4-5UTR_R-Nhe1) was used starting at −1 from the translation start site. Fragments thus generated were inserted into pGL3 basic promoterless vector at Kpn1 and Nhe1 sites, upstream of the luciferase gene. The integrity of each construct was checked by restriction enzyme digestion and sequencing. The following forward and reverse primers were used in the current study (restriction enzyme recognition sites are indicated with italics): RGS4-A_F, ggggtacccc gtctggctcaaacaccatac; RGS4-B_R, ctagctag aagcatagaggacttaagtact; RGS4-C_F, ggggtacccc tgtctattcagattcttcttg; RGS4-C_R, ctagctag aagtctctagccgcccataa; RGS4-D_F, ggggtacccc atcaaatctcattttagataccacct; RGS4-E_F, ggggtacccc aagtgaacactccttgaataaaatgtgtaaaatt; RGS4-F_F, ggggtacccc acctatagggcttaatattcttacaa; RGS4-G_F, ggggtacccc tacttttcagaaggattttctctgc; RGS4-5UTR_R-Nhe1, ctagctag ctagcttatttaacagcttggaattcgc.
Fig. 3.
Schematic diagram of RGS4 constructs and their reporter activities in SHSY-5Y cells. RGS4 constructs of different lengths encompassing different regions of RGS4 5′ region-RGS4-A (−6735 to −1 bp), RGS4-B (−6735 to −4446 bp), RGS4-C (−4992 to −1583), RGS4-D (−3045 to −1), RGS4-E (−2364 to −1 bp), RGS4-F (−1078 to −1 bp) and RGS4-G (−311 to −1 bp); pGL3 SV40 promoter positive control; and pGL3 basic promoterless controls were cotransfected with pRL-TK. The luciferase activities for each construct were evaluated as ratio relative to the activity of pRL-TK vector cotransfected with test vector constructs. The experiments were independently repeated 3 times or more, with replicable results. The data are presented as mean values with SEs.
For transfection, cells from a neuroblastoma cell line (SHSY-5Y, ATCC-CRL-2266) were seeded into 24-well plates at a density of 1.2 × 105 cells/well 1 day prior to transfection, using the manufacturer's protocol (Invitrogen Inc., Carlsbad, CA). Briefly, 1.0 μg of the experimental construct or promoterless basic control was cotransfected with 50 ng of pRL-TK (Renilla Luciferase vector, Promega, Madison, WI) as internal control using Lipofectamine (Invitrogen Inc.). Cells were harvested 24 h later. Transfection was done with passive lysis buffer (Promega). Firefly and Renilla luciferase activity were analyzed using a Dual-Luciferase reporter system (Promega) according to the manufacturer's protocol. Luminescence was measured using a Monolight 2010 luminometer. The promoter activity of each construct was calculated as the ratio of firefly luciferase activity to Renilla Luciferase activity in each experiment. For a positive control of promoter activity, the pGL3-control vector containing SV-40 promoter plus enhancer sequences was used in transfection. Reporter gene assays were performed in 4 independent experiments in triplicates, and data were expressed as mean values with SEs.
Bioinformatic Analysis
Public databases were used to identify additional polymorphisms (http://www.ncbi.nlm.nih.gov; http://genome.ucsc.edu). The GEMS launcher software from the Genomatix package was used to identify potential transcription factor binding sites at the SNPs of interest (http://www.genomatix.de)
Statistical Analysis
PEDCHECK software was used to check for Mendelian inconsistencies.32 LD was estimated using D′ and r2 using HAPLOVIEW and H-Clust software, respectively.33,34 GENEHUNTER software was used for Transmission Disequilibrium Test (TDT) analysis of individual SNPs and haplotypes.35 The Armitage trends test and chi-square tests were employed for comparisons between cases and unrelated controls.
Results
Identification of Additional RGS4 Polymorphisms
We previously sequenced all exons and introns, as well as a 10-kb 5′ upstream region using individual DNA samples from a panel of cases and controls. To enable more extensive coverage, we resequenced a 20-kb genomic region spanning SNPs 1, 4, 7, and 18 and extending into the 3′ region. This region extends from approximately 10 kb upstream to the transcription initiation site, across the coding sequences of RGS4 and into the 3′ untranslated region (figure 1). Using pooled DNA samples from 200 cases and 200 controls, we detected 26 SNPs with MAFs of 5% or more. Polymorphisms were numbered in sequential order from the 5′ to 3′ regions of the gene (M1–M24, see table 1). Previously reported SNPs (SNPs 1, 4, 7, and 18) are denoted. We also identified several rare variants by sequencing different sets of individual samples (Supplementary Table S1). We did not identify any coding SNPs in an additional panel of 48 patients sequenced individually. In addition, we detected a 500-bp repeat showing partial similarity to the L1 repeat family, 10 kb upstream to the RGS4 gene. We did not observe any variations for this repetitive sequence.
Fig. 1.
RGS4 single-nucleotide polymorphisms (SNPs) analyzed in the current report. The numbers M1–M24 refer to the serial numbers used for SNPs in table 1.
Table 1.
RGS4 SNPs Used for Linkage Disequilibrium Analysis
| TDT (T/NT) |
||||||||||
| SNPs | dbSNP ID | Aliasb | Intermarker Distance (bp) | Allele 1 | Allele 2 | Allele 1 Frequency | PITT | P | NIMH | P |
| M1 | rs2842016 | G | C | 0.959 | 27/33 | 11/6 | ||||
| M2a | rs2661352 | 7 | G | C | 0.279 | 27/28 | 29/30 | |||
| M3a | rs2842017 | 18 | C | T | 0.929 | 8/9 | 14/12 | |||
| M4 | rs2842018 | 66 | C | T | 0.929 | 9/8 | 14/12 | |||
| M5a | rs2842019 | 155 | T | A | 0.959 | 7/4 | 11/6 | |||
| M6 | rs10917670 | SNP1 | 5970 | G | A | 0.530 | 64/60 | 13/23 | ||
| M7 | rs2661347 | 2 | A | T | 0.550 | 60/73 | 22/6 | .002 | ||
| M8 | rs2842026 | 245 | G | T | 0.455 | 69/60 | 6/22 | .002 | ||
| M9a | rs951436 | SNP4 | 253 | T | G | 0.558 | 60/73 | 22/6 | .002 | |
| M10 | rs951437 | 169 | A | G | 0.455 | 69/60 | 6/22 | .002 | ||
| M11 | rs951438 | 26 | A | C | 0.455 | 70/61 | 6/22 | .002 | ||
| M12a | rs951439 | SNP7 | 154 | G | A | 0.536 | 69/64 | 13/23 | ||
| M13a | ss35522247 | 85 | C | G | 0.097 | 24/18 | 5/9 | |||
| M14a | ss35522248 | 240 | C | T | 0.966 | 15/10 | 2/8 | |||
| M15a | rs6427711 | 402 | C | T | 0.717 | 63/44 | .06 | 12/11 | ||
| M16 | rs6678136 | 2899 | A | G | 0.470 | 62/73 | 23/13 | |||
| M17 | rs7515900 | 1296 | T | G | 0.454 | 66/72 | 23/13 | |||
| M18 | rs12402634 | 71 | A | C | 0.036 | 2/7 | ||||
| M19 | rs10917671 | 169 | G | A | 0.537 | 72/65 | 13/23 | |||
| M20 | rs2661319 | SNP18 | 924 | A | G | 0.520 | 62/71 | 24/8 | .004 | |
| M21 | rs10917672 | 35 | C | T | 0.958 | 15/10 | 2/7 | |||
| M22 | rs2661317 | 15 | T | G | 0.467 | 70/61 | 9/24 | .009 | ||
| M23a | rs10799897 | 3261 | A | G | 0.523 | 70/62 | 9/23 | .01 | ||
| M24a | rs10759 | 3263 | G | A | 0.321 | 63/69 | 9/17 | |||
Note: NIMH, National Institute of Mental health; PITT, Pittsburgh; SNP, single-nucleotide polymorphism; T/NT, allele 1 transmitted/not transmitted; allele frequency estimates are based on 308 individual parental DNA samples. Only P values of .05 or lower are shown for the TDT analysis.
Tag SNPs defined by the H-Clust method.
SNPs listed as being associated with schizophrenia in our prior publication.13
LD Analysis: 2 SNPs (rs2842030 and rs12753561) Were Not Genotyped Due to Inconsistent Assays
Genotypes for the remaining 24 SNPs were used for LD analysis in a panel of 308 parents from the PITT case-parent trio families (table 2). Using a conservative LD cutoff value (r2 = .8), we observed 5 clusters each with 2 or more SNPs. However, 5 SNPs did not fall into these clusters. Therefore, to genotype “tag” SNPs that represent polymorphisms across this region, it would be necessary to select 10 SNPs (figures 1 and 2). The 4 putatively associated SNPs belonged to 2 clusters, which were in significant LD (r2 > .8).
Table 2.
Marker-to-Marker LD in the RGS4 locus
| SNPa | M1 | M2 | M3 | M4 | M5 | M6 | M7 | M8 | M9 | M10 | M11 | M12 | M13 | M14 | M15 | M16 | M17 | M18 | M19 | M20 | M21 | M22 | M23 | M24 |
| M1 | 0.01 | 0.55 | 0.55 | 1.00 | 0.04 | 0.03 | 0.04 | 0.03 | 0.04 | 0.04 | 0.04 | 0.00 | 0.91 | 0.02 | 0.04 | 0.04 | 0.91 | 0.04 | 0.04 | 0.77 | 0.04 | 0.05 | 0.02 | |
| M2 | 0.95 | 0.03 | 0.03 | 0.02 | 0.21 | 0.14 | 0.15 | 0.14 | 0.15 | 0.15 | 0.23 | 0.04 | 0.01 | 0.65 | 0.23 | 0.21 | 0.01 | 0.21 | 0.12 | 0.00 | 0.11 | 0.01 | 0.01 | |
| M3 | 1.00 | 1.00 | 1.00 | 0.55 | 0.07 | 0.04 | 0.05 | 0.04 | 0.05 | 0.05 | 0.06 | 0.01 | 0.51 | 0.03 | 0.07 | 0.06 | 0.53 | 0.06 | 0.05 | 0.41 | 0.05 | 0.08 | 0.00 | |
| M4 | 1.00 | 1.00 | 1.00 | 0.55 | 0.07 | 0.04 | 0.05 | 0.04 | 0.05 | 0.05 | 0.06 | 0.01 | 0.51 | 0.03 | 0.07 | 0.06 | 0.53 | 0.06 | 0.05 | 0.41 | 0.05 | 0.08 | 0.00 | |
| M5 | 1.00 | 0.98 | 1.00 | 1.00 | 0.04 | 0.03 | 0.04 | 0.03 | 0.04 | 0.04 | 0.04 | 0.00 | 0.91 | 0.02 | 0.04 | 0.04 | 0.91 | 0.04 | 0.04 | 0.77 | 0.04 | 0.05 | 0.02 | |
| M6 | 1.00 | 0.70 | 1.00 | 1.00 | 1.00 | 0.71 | 0.71 | 0.67 | 0.71 | 0.71 | 0.96 | 0.10 | 0.03 | 0.43 | 0.96 | 0.92 | 0.04 | 0.95 | 0.61 | 0.01 | 0.60 | 0.43 | 0.00 | |
| M7 | 1.00 | 0.66 | 0.83 | 0.83 | 1.00 | 0.98 | 0.99 | 1.00 | 0.99 | 0.99 | 0.71 | 0.13 | 0.03 | 0.32 | 0.73 | 0.69 | 0.03 | 0.71 | 0.89 | 0.03 | 0.91 | 0.60 | 0.00 | |
| M8 | 1.00 | 0.68 | 0.83 | 0.83 | 1.00 | 0.98 | 1.00 | 0.95 | 1.00 | 1.00 | 0.71 | 0.13 | 0.03 | 0.32 | 0.73 | 0.68 | 0.03 | 0.71 | 0.86 | 0.02 | 0.90 | 0.60 | 0.00 | |
| M9 | 1.00 | 0.66 | 0.84 | 0.84 | 1.00 | 0.98 | 1.00 | 1.00 | 0.95 | 0.95 | 0.70 | 0.14 | 0.03 | 0.32 | 0.72 | 0.66 | 0.03 | 0.71 | 0.88 | 0.04 | 0.91 | 0.60 | 0.00 | |
| M10 | 1.00 | 0.68 | 0.83 | 0.83 | 1.00 | 0.98 | 1.00 | 1.00 | 1.00 | 1.00 | 0.71 | 0.13 | 0.03 | 0.32 | 0.73 | 0.68 | 0.03 | 0.71 | 0.86 | 0.02 | 0.90 | 0.60 | 0.00 | |
| M11 | 1.00 | 0.68 | 0.83 | 0.83 | 1.00 | 0.98 | 1.00 | 1.00 | 1.00 | 1.00 | 0.71 | 0.13 | 0.03 | 0.32 | 0.73 | 0.68 | 0.03 | 0.71 | 0.86 | 0.02 | 0.90 | 0.60 | 0.00 | |
| M12 | 1.00 | 0.71 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.09 | 0.03 | 0.46 | 0.97 | 0.93 | 0.03 | 0.95 | 0.63 | 0.03 | 0.60 | 0.40 | 0.00 | |
| M13 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.00 | 0.04 | 0.10 | 0.09 | 0.00 | 0.09 | 0.12 | 0.00 | 0.13 | 0.08 | 0.00 | |
| M14 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.01 | 0.03 | 0.03 | 1.00 | 0.03 | 0.03 | 1.00 | 0.03 | 0.04 | 0.02 | |
| M15 | 1.00 | 0.80 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.44 | 0.43 | 0.02 | 0.42 | 0.26 | 0.01 | 0.24 | 0.06 | 0.03 | |
| M16 | 1.00 | 0.71 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.94 | 0.03 | 0.95 | 0.65 | 0.03 | 0.62 | 0.40 | 0.00 | |
| M17 | 1.00 | 0.66 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 1.00 | 0.95 | 1.00 | 0.03 | 0.99 | 0.67 | 0.04 | 0.65 | 0.43 | 0.00 | |
| M18 | 1.00 | 0.71 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.03 | 0.04 | 1.00 | 0.03 | 0.04 | 0.02 | |
| M19 | 1.00 | 0.68 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 1.00 | 0.95 | 1.00 | 1.00 | 1.00 | 0.70 | 0.03 | 0.67 | 0.42 | 0.00 | |
| M20 | 1.00 | 0.58 | 0.84 | 0.84 | 1.00 | 0.86 | 1.00 | 0.99 | 1.00 | 0.99 | 0.99 | 0.89 | 1.00 | 1.00 | 0.84 | 0.89 | 0.93 | 1.00 | 0.94 | 0.03 | 0.96 | 0.59 | 0.00 | |
| M21 | 0.91 | 0.08 | 0.83 | 0.83 | 0.91 | 0.54 | 1.00 | 0.77 | 1.00 | 0.77 | 0.77 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.03 | 0.04 | 0.02 | |
| M22 | 1.00 | 0.56 | 0.85 | 0.85 | 1.00 | 0.87 | 1.00 | 0.99 | 1.00 | 0.99 | 0.99 | 0.88 | 1.00 | 1.00 | 0.83 | 0.89 | 0.93 | 1.00 | 0.93 | 1.00 | 1.00 | 0.63 | 0.00 | |
| M23 | 1.00 | 0.12 | 1.00 | 1.00 | 1.00 | 0.66 | 0.90 | 0.90 | 0.90 | 0.90 | 0.90 | 0.64 | 0.90 | 1.00 | 0.38 | 0.63 | 0.68 | 1.00 | 0.67 | 0.83 | 1.00 | 0.87 | 0.02 | |
| M24 | 1.00 | 0.11 | 0.36 | 0.36 | 1.00 | 0.01 | 0.02 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.04 | 1.00 | 0.20 | 0.04 | 0.05 | 1.00 | 0.04 | 0.09 | 1.00 | 0.09 | 0.19 |
Note: SNP, single-nucleotide polymorphism; LD, linkage disequilibrium; SNP numbers M6, M9, M12, and M20 are SNP1, SNP4, SNP7, and SNP18, respectively, as previously reported13; number of Caucasian samples used in the LD analysis = 308 parents of schizophrenia probands; correlation coefficient r2 above diagonal, Lewontin D′ below.
Numbers are same as indicated in table 1.
Fig. 2.

Cluster analysis of 24 single-nucleotide polymorphisms (SNPs) from the parental samples (n = 308). The vertical broken bar represents the cutoff value used to designate the tag SNPs, which are indicated with an asterisk. SNPs 1, 4, 7, and 18 (our previous report13) are represented by the numbers M6, M9, M12, and M20, respectively) (arrow indicated). The numbers M1–M5 and M21–M24 represent SNPs outside the 20-kb “critical region” as reported in our previous report.13
TDT and Case-Control Analysis
We genotyped the SNPs listed in table 1 using samples from the NIMH case-parent trios (n = 39 families), the PITT case-parent trios (n = 154 families), and the community controls (n = 92). Analysis of SNPs 1, 4, 7, and 18 has been reported on earlier in these samples.13 Consistent with our published result, transmission distortion was confined to the larger of the 2 clusters observed in the 20-kb region of interest in the NIMH sample (table 1, figure 2). Note that in our earlier report, significant transmission distortion for SNP1 was also reported in the NIMH sample. This analysis relied on restriction enzyme-based assays. Using the more accurate SNaPshot assay, only trends for overtransmission were noted. In contrast, analysis of the PITT trio sample did not reveal any significant transmission distortion, though a trend was observed for rs6427711 (M15; 63 transmissions vs 44 nontransmissions, P = .06). This sample was enlarged from the previous report.13 Consistent with the initial report, no significant differences were observed for any of these SNPs when PITT cases were compared with the community controls.
5′ Upstream Functional Analysis
The clone RGS4-A, spanning a large genomic fragment appeared to increase expression in comparison with the promoterless construct. Clones containing partial sequences RGS4-B and RGS4-C did not drive reporter expression. However, clone RGS4-D did increase expression of the reporter, suggesting that the SNPs upstream to RGS4-D construct may function as repressors. RGS-D includes the region immediately 5′ to translation start point (∼3.0 kb), suggesting the presence of a core promoter element in this region. Further deletion mutants showed that the promoter activity could be restricted to successively smaller clones (RGS4-E, RGS4-F, and RGS4-G), all of which harbor a 311-bp region, upstream from the translation start point. This 311-bp region may encompass the core promoter sequence for RGS4. To evaluate relative promoter activity in the 2 haplotypes putatively associated with SZ, we identified 2 individuals homozygous for the respective haplotypes (G-G-G-G and A-T-A-A, denoted the PITT and NIMH haplotypes, respectively). We constructed clones corresponding to clone RGS4-F and evaluated their promoter activities. These clones encompass SNPs M17 and M19, as listed in table 1. No significant differences were noted (PITT: 8.83 ± 2.59; NIMH: 8.95 ± 0.51).
Prediction of Transcription Initiation Sites
We performed software-based analysis with MATINSPECTOR in the GEMS launcher package (http://www.genomatix.de/products/MatInspector). This approach did not reveal any likely transcription initiation sites, though our luciferase assays suggest the presence of a transcription start site in the region 0.311 kb upstream from the translation start site. Many weak matrix similarities for binding sites of interesting transcription factors (PBX1/MEIS, HNF1, PRE, and PAX2) were present (Supplementary Table S2).
Discussion
Recently, extensive genetic association studies of RGS4 polymorphisms have been reported in SZ although, with mixed results. In order to enable further exploration of the associations, we present a comprehensive evaluation of common polymorphisms spanning RGS4. This information will be useful for further analysis of samples in which genetic association of RGS4 polymorphisms with SZ has previously been reported.
By sequencing pooled DNA samples from Caucasian individuals, we identified all SNPs with MAFs exceeding 5%. In addition, we selected informative SNPs from public databases. We did not note any large repeats, insertions, or deletions in the complex, imperfect repeat sequence in the 5′ upstream region of RGS4. Our analyses of 24 SNPs suggest significant LD spanning this region, though less common SNPs were not analyzed (tables 1 and 2). Using a novel clustering algorithm based on r2 analysis, we identified 10 tag SNPs that could reflect common polymorphisms across this region. The limited diversity in this region is similar to that observed in other genomic regions among Caucasians.36,37
The clustering algorithm also identified 2 large clusters comprising 12 SNPs and a number of smaller clusters among the set of 24 SNPs analyzed. These 2 large clusters, consisting of 12 SNPs include all 4 SNPs reported to be associated with SZ in prior reports (figure 2). Remarkably, all but 1 of the 8 SNPs at which significant transmission distortion was noted in the NIMH sample in the present analyses is also included in this cluster. One other associated SNP denoted M23 (rs10799897) is in incomplete LD with this cluster (table 2, figures 1 and 2). Thus, our analyses suggest that the putative association is bounded 5′ by the L1 repeat and extends into the third intron of RGS4 (approximately 6.06 kb). The present analyses, as well as our published report, therefore suggest that the putative association with SZ does not extend beyond RGS4.13
Our initial analyses revealed association in a sample of 93 case-parent trios recruited at PITT.13 The associated alleles and haplotypes in this sample differed from those in the NIMH sample. Intriguingly, these 2 haplotypes comprise the majority of variation among individuals of Caucasian descent. The present analyses in the enlarged sample of 154 trio families from PITT did not reveal significant association at any of the SNPs. The lack of association is not surprising, because our meta-analyses of 2160 trio families revealed only a weak association with both common haplotypes.26 Similar complexity has been reported for other putative liability genes for SZ and, indeed, for other complex diseases.38–40 Evidence for statistical epistasis between RGS4 and polymorphisms in other genes has also increased interest in analyzing these interactions to explain risk for SZ.11,41,42
The initial reports did not suggest any function for the associated polymorphisms. Recently, we have observed that first-episode patients with alleles at the SNPs that may be associated with SZ have differences in the volume of the dorsolateral prefrontal cortex estimated from magnetic resonance imaging scans.43 Consistent with these results, one of these SNPs (rs951436) has also been associated with both differences in brain activation in a working memory task and in gray and white matter volumes in healthy subjects.44 The mechanisms for these associations are uncertain. In the initial steps to understand the function of these SNPs/haplotypes in vitro, we performed reporter-based promoter analysis of selected SNPs/haplotypes from the RGS4 upstream region. Our observations suggested significant promoter activity in a 311-bp fragment, with possible repressor effect due to SNPs M6–M16 (see table 1). There was no significant difference in promoter activity between the haplotype associated in the NIMH sample and another haplotype commonly observed in Caucasian samples that was denoted the “PITT” haplotype (G-G-G-G for SNPs rs10917670, rs951436, rs951439, and rs2661319, respectively). Notably, the PITT haplotype has also been observed to be associated in other samples.16 Thus, both putative associated haplotypes may possess comparable functional activity. These preliminary findings point to the necessity of additional studies aimed at exploring regulatory elements in the putative promoter region. Further exploration to elucidate combinatorial effect of cis-acting factors with different upstream SNPs may help define the transcriptional regulation of RGS4. In this context, our in silico analyses suggest that some of the associated polymorphisms may affect the binding of certain transcription factors. These possibilities need to be evaluated further using in vitro assays.
In conclusion, we have catalogued common RGS4 polymorphisms and observed extensive LD in this region. This information will be useful for ongoing gene mapping studies. Using the novel SNPs, associations consistent with the LD patterns were noted in the NIMH sample. The associated haplotype in the NIMH sample appears to have significant promoter activity. Future studies to further characterize the RGS4 promoter are needed.
Supplementary Materials
Supplementary Tables S1 and S2 are available online at http://schizophreniabulletin.oxfordjournals.org.
Acknowledgments
This work was supported by grants from NIMH (MH56242, MH66263, and MH63480 to Nimgaonkar), the NIMH Conté Center for the Neuroscience of Mental Disorders (MH 45156 to Lewis), and the Mental Health Interventional Research Center (MH 30915). We thank B. Devlin for critical reading of the article. Details of the SNPs analyzed are available on our Web site http://www.pitt.edu/∼nimga/research/rgs4.
References
- 1.De Vries L, Zheng B, Fischer T, Elenko E, Farquhar MG. The regulator of G protein signaling family. Annu Rev Pharmacol Toxicol. 2000;40:235–271. doi: 10.1146/annurev.pharmtox.40.1.235. [DOI] [PubMed] [Google Scholar]
- 2.Riddle EL, Schwartzman RA, Bond M, Insel PA. Multi-tasking RGS proteins in the heart: the next therapeutic target? Circ Res. 2005;96:401–411. doi: 10.1161/01.RES.0000158287.49872.4e. [DOI] [PubMed] [Google Scholar]
- 3.Ni YG, Gold SJ, Iredale PA, Terwilliger RZ, Duman RS, Nestler EJ. Region-specific regulation of RGS4 (regulator of G-protein-signaling protein type 4) in brain by stress and glucocorticoids: in vivo and in vitro studies. J Neurosci. 1999;19:3674–3680. doi: 10.1523/JNEUROSCI.19-10-03674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hepler JR. Emerging roles for RGS proteins in cell signalling. Trends Pharmacol Sci. 1999;20:376–382. doi: 10.1016/s0165-6147(99)01369-3. [DOI] [PubMed] [Google Scholar]
- 5.Kobori N, Clifton GL, Dash P. Altered expression of novel genes in the cerebral cortex following experimental brain injury. Brain Res Mol Brain Res. 2002;104:148–158. doi: 10.1016/s0169-328x(02)00331-5. [DOI] [PubMed] [Google Scholar]
- 6.Zachariou V, Georgescu D, Sanchez N, et al. Essential role for RGS9 in opiate action. Proc Natl Acad Sci USA. 2003;100:13656–13661. doi: 10.1073/pnas.2232594100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levitt P, Ebert P, Mirnics K, Nimgaonkar VL, Lewis DA. Making the case for a candidate vulnerability gene in schizophrenia: convergent evidence for regulator of G-protein signaling 4 (RGS4) Biol Psychiatry. 2006;60:534–537. doi: 10.1016/j.biopsych.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 8.Mirnics K, Middleton FA, Stanwood GD, Lewis DA, Levitt P. Disease-specific changes in regulator of G-protein signaling 4 (RGS4) expression in schizophrenia. Mol Psychiatry. 2001;6:293–301. doi: 10.1038/sj.mp.4000866. [DOI] [PubMed] [Google Scholar]
- 9.Erdely HA, Tamminga CA, Roberts RC, Vogel MW. Regional alterations in RGS4 protein in schizophrenia. Synapse. 2006;59:472–479. doi: 10.1002/syn.20265. [DOI] [PubMed] [Google Scholar]
- 10.Bowden NA, Scott RJ, Tooney PA. Altered expression of regulator of G-protein signalling 4 (RGS4) mRNA in the superior temporal gyrus in schizophrenia. Schizophr Res. 2007;89:165–168. doi: 10.1016/j.schres.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Lipska BK, Mitkus S, Caruso M, et al. RGS4 mRNA expression in postmortem human cortex is associated with COMT Val158Met genotype and COMT enzyme activity. Hum Mol Genet. 2006;15:2804–2812. doi: 10.1093/hmg/ddl222. [DOI] [PubMed] [Google Scholar]
- 12.Brzustowicz LM, Hodgkinson KA, Chow EW, Honer WG, Bassett AS. Location of a major susceptibility locus for familial schizophrenia on chromosome 1q21-q22. Science. 2000;288:678–682. doi: 10.1126/science.288.5466.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chowdari KV, Mirnics K, Semwal P, et al. Association and linkage analyses of RGS4 polymorphisms in schizophrenia. Hum Mol Genet. 2002;11:1373–1380. doi: 10.1093/hmg/11.12.1373. [DOI] [PubMed] [Google Scholar]
- 14.Williams NM, Preece A, Spurlock G, et al. Support for RGS4 as a susceptibility gene for schizophrenia. Biol Psychiatry. 2004;55:192–195. doi: 10.1016/j.biopsych.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 15.Morris DW, Rodgers A, McGhee KA, et al. Confirming RGS4 as a susceptibility gene for schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2004;125:50–53. doi: 10.1002/ajmg.b.20109. [DOI] [PubMed] [Google Scholar]
- 16.Chen X, Dunham C, Kendler S, et al. Regulator of G-protein signaling 4 (RGS4) gene is associated with schizophrenia in Irish high density families. Am J Med Genet B Neuropsychiatr Genet. 2004;129:23–26. doi: 10.1002/ajmg.b.30078. [DOI] [PubMed] [Google Scholar]
- 17.Bakker SC, Hoogendoorn MLC, Hendriks J, et al. The PIP5K2A and RGS4 genes are differentially associated with deficit and non-deficit schizophrenia. Genes Brain Behavior. 2007;6:113–119. doi: 10.1111/j.1601-183x.2006.00234.x. [DOI] [PubMed] [Google Scholar]
- 18.Fallin MD, Lasseter VK, Avramopoulos D, et al. Bipolar I disorder and schizophrenia: a 440-single-nucleotide polymorphism screen of 64 candidate genes among Ashkenazi Jewish case-parent trios. Am J Hum Genet. 2005;77:918–936. doi: 10.1086/497703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cordeiro Q, Talkowski ME, Chowdari KV, Wood J, Nimgaonkar V, Vallada H. Association and linkage analysis of RGS4 polymorphisms with schizophrenia and bipolar disorder in Brazil. Genes Brain Behav. 2005;4:45–50. doi: 10.1111/j.1601-183x.2004.00096.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhang F, St Clair D, Liu X, et al. Association analysis of the RGS4 gene in Han Chinese and Scottish populations with schizophrenia. Genes Brain Behav. 2005;4:444–448. doi: 10.1111/j.1601-183X.2005.00167.x. [DOI] [PubMed] [Google Scholar]
- 21.Liu YL, Shen-Jang Fann C, Liu CM, et al. Evaluation of RGS4 as a candidate gene for schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2006;141:418–420. doi: 10.1002/ajmg.b.30286. [DOI] [PubMed] [Google Scholar]
- 22.Sobell JL, Richard C, Wirshing DA, Heston LL. Failure to confirm association between RGS4 haplotypes and schizophrenia in Caucasians. Am J Med Genet B Neuropsychiatr Genet. 2005;139:23–27. doi: 10.1002/ajmg.b.30221. [DOI] [PubMed] [Google Scholar]
- 23.Rizig MA, McQuillin A, Puri V, et al. Failure to confirm genetic association between schizophrenia and markers on chromosome 1q23.3 in the region of the gene encoding the regulator of G-protein signaling 4 protein (RGS4) Am J Med Genet B Neuropsychiatr Genet. 2006;141:296–300. doi: 10.1002/ajmg.b.30288. [DOI] [PubMed] [Google Scholar]
- 24.Puri V, McQuillin A, Choudhury K, et al. Fine mapping by genetic association implicates the chromosome 1q23.3 gene UHMK1, encoding a serine/threonine protein kinase, as a novel schizophrenia susceptibility gene. Biol Psychiatry. 2007;61:873–879. doi: 10.1016/j.biopsych.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 25.Ishiguro H, Horiuchi Y, Koga M, et al. RGS4 is not a susceptibility gene for schizophrenia in Japanese: association study in a large case-control population. Schizophr Res. 2007;89:161–164. doi: 10.1016/j.schres.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 26.Talkowski ME, Seltman H, Bassett AS, et al. Evaluation of a susceptibility gene for schizophrenia: genotype based meta-analysis of RGS4 polymorphisms from thirteen independent samples. Biol Psychiatry. 2006;60:152–162. doi: 10.1016/j.biopsych.2006.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li D, He L. Association study of the G-protein signaling 4 (RGS4) and proline dehydrogenase (PRODH) genes with schizophrenia: a meta-analysis. Eur J Hum Genet. 2006;14:1130–1135. doi: 10.1038/sj.ejhg.5201680. [DOI] [PubMed] [Google Scholar]
- 28.Guo S, Tang W, Shi Y, et al. RGS4 polymorphisms and risk of schizophrenia: an association study in Han Chinese plus meta-analysis. Neurosci Lett. 2006;406:122–127. doi: 10.1016/j.neulet.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 29.Kwok PY. Approaches to allele frequency determination. Pharmacogenomics. 2000;1:231–235. doi: 10.1517/14622416.1.2.231. [DOI] [PubMed] [Google Scholar]
- 30.Chowdari KV, Northup A, Pless L, et al. DNA pooling: a comprehensive, multi-stage association analysis of ACSL6 and SIRT5 polymorphisms in schizophrenia. Genes Brain Behav. 2007;6:229–239. doi: 10.1111/j.1601-183X.2006.00251.x. [DOI] [PubMed] [Google Scholar]
- 31.Mansour HA, Wood J, Logue T, et al. Association study of eight circadian genes with bipolar I disorder, schizoaffective disorder and schizophrenia. Genes Brain Behav. 2006;5:150–157. doi: 10.1111/j.1601-183X.2005.00147.x. [DOI] [PubMed] [Google Scholar]
- 32.O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 34.Rinaldo A, Bacanu SA, Devlin B, Sonpar V, Wasserman L, Roeder K. Characterization of multilocus linkage disequilibrium. Genet Epidemiol. 2005;28:193–206. doi: 10.1002/gepi.20056. [DOI] [PubMed] [Google Scholar]
- 35.Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 36.Chadha S, Miller K, Farwell L, et al. Haplotype structure of TNFRSF5-TNFSF5 (CD40-CD40L) and association analysis in systemic lupus erythematosus. Eur J Hum Genet. 2005;13:669–676. doi: 10.1038/sj.ejhg.5201367. [DOI] [PubMed] [Google Scholar]
- 37.Costas J, Salas A, Phillips C, Carracedo A. Human genome-wide screen of haplotype-like blocks of reduced diversity. Gene. 2005;349:219–225. doi: 10.1016/j.gene.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 38.Shirts BH, Nimgaonkar V. The genes for schizophrenia: finally a breakthrough? Curr Psychiatry Rep. 2004;6:303–312. doi: 10.1007/s11920-004-0081-1. [DOI] [PubMed] [Google Scholar]
- 39.Owen MJ, Craddock N, O'Donovan MC. Schizophrenia: genes at last? Trends Genet. 2005;21:518–525. doi: 10.1016/j.tig.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 40.Bastian W. Genes with linkage or association with type 2 diabetes mellitus. J Pediatr Endocrinol Metab. 2002;15(Suppl 1):471–484. [PubMed] [Google Scholar]
- 41.Nicodemus KK, Kolachana BS, Vakkalanka R, et al. Evidence for statistical epistasis between catechol-O-methyltransferase (COMT) and polymorphisms in RGS4, G72 (DAOA), GRM3, and DISC1: influence on risk of schizophrenia. Hum Genet. 2007;120:889–906. doi: 10.1007/s00439-006-0257-3. [DOI] [PubMed] [Google Scholar]
- 42.Winantea J, Hoang MN, Ohlraun S, et al. A summary statistic approach to sequence variation in noncoding regions of six schizophrenia-associated gene loci. Eur J Hum Genet. 2006;14:1037–1043. doi: 10.1038/sj.ejhg.5201664. [DOI] [PubMed] [Google Scholar]
- 43.Prasad KM, Chowdari KV, Nimgaonkar VL, Talkowski ME, Lewis DA, Keshavan MS. Genetic polymorphisms of the RGS4 and dorsolateral prefrontal cortex morphometry among first episode schizophrenia patients. Mol Psychiatry. 2005;10:213–219. doi: 10.1038/sj.mp.4001562. [DOI] [PubMed] [Google Scholar]
- 44.Buckholtz JW, Meyer-Lindenberg A, Honea RA, et al. Allelic variation in RGS4 impacts functional and structural connectivity in the human brain. J Neurosci. 2007;27:1584–1593. doi: 10.1523/JNEUROSCI.5112-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]