Abstract
Gene expression represents a fundamental interface between genes and environment in the development and ongoing plasticity of the human brain. Individual differences in gene expression are likely to underpin much of human diversity, including psychiatric illness. In the past decade, the development of microarray and proteomic technology has enabled global description of gene expression in schizophrenia. However, it is difficult on the basis of gene expression assays alone to distinguish between those changes that constitute primary etiology and those that reflect secondary pathology, compensatory mechanisms, or confounding influences. In this respect, tests of genetic association with schizophrenia will be instructive because changes in gene expression that result from gene variants that are associated with the disorder are likely to be of primary etiological significance. However, regulatory polymorphism is extremely difficult to recognize on the basis of sequence interrogation alone. Functional assays at the messenger RNA and/or protein level will be essential in elucidating the molecular mechanisms underlying genetic association with schizophrenia and are likely to become increasingly important in the identification of regulatory variants with which to test for association with the disorder and related traits. Once established, etiologically relevant changes in gene expression can be recapitulated in model systems in order to elucidate the molecular and physiological pathways that may ultimately give rise to the condition.
Keywords: psychosis, genetic, functional polymorphism, mechanism, susceptibility
Introduction
Gene expression is the process whereby genomic DNA sequence is converted into RNA and, typically, from this to a specified protein. During early brain development, the complement of genes expressed in each cell, coordinated through complex genetic-developmental cascades, is integral in determining cellular specialization. Later in development, and throughout life, environmental influences on neural gene expression also become important, underlying the physiological adaptations that accompany learning and long-term changes in behavior. Individual differences in gene expression, arising through genetic, epigenetic, and environmental variation, are thus likely to underpin much of human phenotypic diversity, including psychiatric illness.
Measuring Gene Expression in Schizophrenia
Differences in gene expression between individuals suffering schizophrenia and those who do not are likely to be wide ranging and relatively subtle; certainly, there exists no single established biomarker for the disorder. The majority of studies of gene expression in schizophrenia are based on postmortem brain tissue, although peripheral cells (eg, blood lymphocytes) from living subjects have also been used (eg, Vawter et al1 and Bowden et al2). Traditionally, researchers have examined RNA or protein from specific genes, selected on the basis of either a hypothesized role in the disorder (eg, dopamine receptor expression3) or as a proxy measure for postulated pathophysiological defects (eg, messenger RNA [mRNA] for synaptic proteins as an index of synaptic pathology4). With the development of microarray technology, it has become possible to assay the expression level of mRNA transcripts at the genome-wide scale in schizophrenia.5,6 Proteomic studies of schizophrenia, using 2-dimensional gel electrophoresis and mass spectrometric sequencing, are also now possible (eg, Johnston-Wilson et al7, Beasley et al8, and Clark et al9). These large-scale assays allow hypothesis-free expression profiling of schizophrenia tissue and thus have the potential to highlight previously unexplored molecular processes in the disorder. Despite much disparity in the findings from microarray studies carried out thus far (potentially reflecting differences between cohorts, microarray platforms, and analyses), some replicated themes have emerged. These include disturbances in the expression of genes related to synaptic function,10–12 energy metabolism,13–15 and oligodendrocyte function.16–18
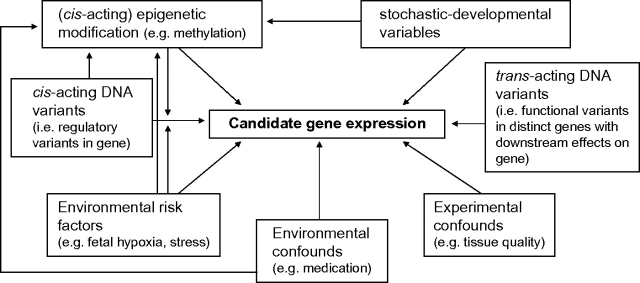
While undoubtedly of great value, it is difficult on the basis of gene expression assays alone to distinguish between those changes that constitute primary etiology and those that reflect secondary pathology, compensatory mechanisms, or confounding influences. Changes in gene expression that play a primary role in schizophrenia etiopathology may arise from sequence variants in regulatory regions of genomic DNA, epigenetic/stochastic variation, or environmental influences (eg, psychosocial stress). Secondary pathological changes in gene expression may be no less important; indeed, they may reflect points of convergence in the action of individual susceptibility genes. In an organ remarkable for its plasticity, other changes in neural gene expression are likely to reflect compensatory mechanisms, which may act at many levels. Lastly, while great care is usually applied in sample matching, measures of gene expression between samples are inevitably susceptible to confounding influences, arising from the characteristics of the tissues themselves (eg, RNA quality, cellular heterogeneity), as well as demographic and environmental confounds (eg, effects of medication). Potential influences on gene expression in postmortem brain are shown in figure 1.
Fig. 1.
Influences on Measured Expression of Candidate Genes in Human Postmortem Brain. A gene can be subject to cis-acting genetic influences on its expression, arising from DNA sequence variation in the gene itself, which may show genetic association with a disorder. Gene expression can also be influenced by epigenetic modification, such as DNA methylation, which can be influenced by stochastic and environmental factors, and also sequence variation in the gene itself. Environmental effects on gene expression can be risk factors as well as confounding influences and may modulate genetic effects on gene expression (ie, gene-environment interaction). Gene expression may also be influenced by genetic variation with functional consequences on a distinct gene (through effects on protein structure or expression), which then has a downstream effect on the measured gene (trans-acting genetic influences). Measures of gene expression in brain are additionally prone to a host of experimental confounds, such as differences in tissue and RNA quality or cellular composition between samples.
Insights From Psychiatric Genetics
Establishing etiological mechanisms in schizophrenia will be crucial in advancing neurobiological models of the disorder and, ultimately, in the development of novel therapies. Molecular genetic studies are likely to be vital in this pursuit for 2 reasons; first, schizophrenia has a substantial genetic component19 and second, unlike RNA and protein, the primary genomic DNA sequence that encodes it is generally impervious to extrinsic influences. Establishing genetic association between a particular gene and schizophrenia indicates that the gene is in some way involved in the primary causation of the disorder. This is of course far from trivial task; schizophrenia is a complex, multifactorial condition, likely involving multiple, interacting genes of small effect, in addition to environmental factors.20 Very large, well-characterized samples will be essential in establishing true association with any given candidate gene. However, evidence is currently accumulating in support of several such candidates.21
There are 2 broad (and overlapping) mechanisms by which genetic variation in a candidate gene may influence susceptibility to a disorder. One, exemplified by several simple Mendelian disorders, is where the DNA variant alters the structure of the encoded protein (through, eg, an amino acid substitution or frameshift mutation). The other is where the variant in some way impacts on the expression of the gene product (such as by altering transcription, stability or splicing of RNA, or through effects on the translation or stability of the protein). Notably, for several of the best supported schizophrenia susceptibility genes identified to date (eg, DTNBP1, encoding dysbindin-1, and NRG1, encoding neuregulin-1), genetic association does not appear to reflect DNA variants with an obvious effect on protein structure. Therefore, by default, it is assumed that, if genetic association is genuine, it is mediated by sequence variants that affect expression of these genes.
Several putative schizophrenia susceptibility genes have been reported to show altered expression in schizophrenic postmortem brain compared with unaffected controls. These include DTNBP1,22–24 G72 (DAOA),25 NRG1,26,27 and RGS4,28,29 the latter initially implicated through a microarray study of the disorder.10 Given that these case-control cohorts were not selected for genotypes associated with schizophrenia, it is unlikely that observations of altered expression result from DNA variation in the candidate genes alone (cis-acting genetic influences). Rather, changes in the expression of even genuine susceptibility genes are likely to additionally reflect the downstream effects of other susceptibility genes (trans-acting genetic influences), environmental risk factors, or epigenetic variation. Indeed, it is possible that, for some pathogenic changes in gene expression, the causal mechanism will be entirely environmental and/or stochastic-epigenetic. However, where altered gene expression can be shown to result, at least in part, from genetic variation that is associated with the disorder, it is with some confidence that etiological significance can be ascribed.
Difficulties in Identifying Regulatory Variation
At present, the DNA variants that affect expression of any given candidate gene, and their relationship to schizophrenia susceptibility, are largely unknown. DNA variation can take several forms, including single nucleotide polymorphisms (SNPs), deletions/insertions, variable repeat sequences, rearrangements, or duplications (copy number variations). Such variants are common in the genome, and the majority are likely to be of no functional importance. However, unlike DNA variants that affect protein structure (which typically occur within exons, with predictable effects on the amino acid sequence), those that affect gene expression are extremely difficult to recognize as such. Cis-acting regulatory variants may be located in the transcribed sequence itself, promoters, intronic sequence, as well as distant control elements.30 Although bioinformatic prediction tools are continually developing,31 these sequences remain poorly characterized for any gene.
Currently, tests of association between candidate genes and schizophrenia rely, in general, on sets of polymorphic DNA markers (eg, SNPs) that are designed to detect association through linkage disequilibrium (LD) with the “true” functional susceptibility variants. Such studies have frequently differed in the pattern of markers showing association with schizophrenia.21 However, interpretation of these data is complicated by the possibility that, for any of these candidate genes, there may exist multiple functional variants, occurring at different frequencies and showing different patterns of LD across populations.32 Given that LD can extend long distances, it may not be possible to resolve disease association beyond multimarker haplotypes spanning large genomic regions, even for genes that contain only a single functional variant. Moreover, in order to establish whether observed genetic associations are indeed mediated by effects on gene expression, functional assays at the RNA and/or protein level are essential.
Functional Assays of Putative Regulatory Variation
There are 2 ways in which functional assays of DNA sequence variation will be vital in establishing the relevance of gene expression to schizophrenia etiology. Most immediately, they may be used to assess the effects on gene expression of DNA variants/haplotypes that have been implicated through genetic association with the disorder. However, functional assays are likely to become increasingly important in the identification of genetic regulatory variation, which can then be used for direct tests of association with schizophrenia and related traits/endophenotypes.
A variety of methods are available with which to assay the effect of DNA variation on gene expression. One approach is to compare the total amount or distribution of the candidate gene, at the RNA or protein level, in tissue from individuals of different genotypes. In relation to schizophrenia, this approach has yielded evidence for a regulatory effect of putative risk variants in several candidate genes. Of particular note, Law et al27 found a putative schizophrenia risk variant within the NRG1 gene to be associated with increased expression of a novel NRG1 transcript in human hippocampal tissue.27 For the purposes of identifying variants associated with gene expression, large-scale analyses of regulatory variation are also now possible, through the combined analysis of genome-wide expression and SNP genotyping microarrays.33,34 An alternative approach for assessing the effects of cis-acting regulatory variation involves comparing the RNA levels of the 2 alleles of a given gene in individual heterozygotes.35 Under this “within-subjects” approach, each allele acts as an internal control for the other, allowing assessment of the net cis-acting influences on a gene's expression, while controlling for the trans-acting factors that can confound measures of total expression between samples. Because these assays expose only the variance in gene expression resulting from cis-acting variation, they are particularly powerful for examining the influence of specific haplotypes on expression of the target gene. For example, this method has been employed to show that putative schizophrenia risk haplotypes in the DTNBP1 gene reduce DTNBP1 expression in human brain,36 thus implicating reduced DTNBP1 expression as a primary etiological mechanism in the disorder. It has also proven possible to define regulatory haplotypes using this method.37 Haplotypes that are associated with effects on RNA transcription can also be identified using the “haploChIP” method.38 For directly testing the effect of particular DNA variants on gene transcription, the most widely used method is the in vitro reporter gene assay, in which a putative regulatory sequence is transfected into cells along with a reporter gene in order to assess its transcriptional activity. For example, using this method, Tan et al (2007) found increased transcriptional activity of the putative NRG1 risk allele that showed association with type IV NRG1 expression in the study of Law et al.27,39
The methods described above do have certain limitations. Most seriously, genetic effects on gene expression can be highly context dependent. It may be particularly difficult to extrapolate from in vitro assays, in which the effects of DNA variation are assessed out of their normal chromosomal and physiological context.40 Assays using human brain tissue are advantageous in this regard but may be limited in that genetic effects on gene expression can show cellular specificity,41 as well as potential developmental/environmental sensitivity. In order to address possible regional/cellular differences in the effects of regulatory variation, methods for selecting discrete neural populations, such as laser-capture microdissection, will be extremely valuable. Indeed, this may indicate brain regions and cell populations that are particularly affected by schizophrenia risk variation. However, possible changes in gene regulation arising from interaction with developmental and/or environmental trans-influences may go undetected. Despite these caveats, the potential of these assays for ascribing function to putative risk variants, and in identifying functional genetic variation with which to test for disease association, should not be understated. The combination of psychiatric genetics and functional genomics will be an essential first step in advancing our understanding of the molecular mechanisms commonly involved in schizophrenia.
Cytogenetic Effects on Gene Expression
So far, this review has focused on genetic influences on gene expression that may commonly influence schizophrenia susceptibility but which are likely to be associated with only small elevations in risk. Cytogenetic abnormalities may constitute a less common, but potentially stronger, influence on risk for schizophrenia, and these are likely to have direct effects on gene expression. For example, very high rates of schizophrenia are observed in velo-cardio-facial syndrome (VCFS),42 a disorder caused by heterozygous microdeletions of chromosome 22q11. The greatly increased risk for schizophrenia in VCFS could be conferred by heterozygous loss of a single gene or, perhaps more likely, the combined haploinsufficient expression of multiple genes in the 22q11 region.43 Genes within the deleted region (which include COMT, ZDHHC8, PRODH, and GNB1L) may also predispose to schizophrenia in the wider population.44–47 While association with schizophrenia is far from established for any of the candidates in the region, several of these genes are reported to contain common regulatory variation that could potentially impact on general risk for the disorder.45,47,48 A second cytogenetic abnormality of particular interest is a balanced chromosomal translocation t(1;11)(q42.1; q14.3), observed in a large Scottish family, which strongly cosegregates with schizophrenia and other major psychiatric illnesses.49 The chromosome 1 break point has been found to directly interrupt 2 novel genes, which were named Disrupted-In-Schizophrenia-1 (DISC1) and -2 (DISC2).50 For DISC1—the focus of interest because it has protein-coding potential—5 exons at the 3′ end of the gene are translocated to chromosome 11, potentially giving rise to a truncated protein. However, no such product was observed in lymphoblastoid cells derived from translocation carriers, where total DISC1 levels were reduced by approximately half, suggesting haploinsufficiency as the pathogenic mechanism in these individuals.51 There is accumulating evidence that variation at the DISC1 locus may also increase susceptibility to schizophrenia (and other psychiatric disorders) in general populations,52 although current evidence suggests that, if genuine, this is unlikely to be mediated by direct effects on DISC1 mRNA abundance.53,54
Gene Expression as a Schizophrenia Endophenotype
Psychiatric endophenotypes are measurable, subclinical aspects of brain function (be they neuropsychological, neuroanatomical, neurophysiological, or biochemical/molecular) that might more directly reflect the actions of individual susceptibility genes. As such, endophenotypes may be of considerable utility in assisting the genetic dissection of complex psychiatric disorders.55 As potentially the most immediate “downstream” effect of genetic risk variation, expression of individual susceptibility genes can be considered the ultimate endophenotype.56 Thus, where sequence variation in a given candidate gene influences susceptibility through (cis-acting) effects on its expression, other genes containing polymorphism with a downstream (trans-acting) effect on expression of that gene are also likely to confer risk. For complex disorders such as schizophrenia, these loci are likely to confer a greater effect on the intermediate expression phenotype than on the clinical phenotype and should therefore be more easily detected. This provides the rationale for a recent study in which expression of the DTNBP1 gene was assayed in large sibships in order to map polymorphic regulatory loci through genome-wide quantitative linkage analysis.57 DTNBP1 expression was found to be influenced not only by genetic variation at the DTNBP1 locus itself, which appears to mediate association with schizophrenia,36 but also by a polymorphic trans-acting locus on chromosome 8p. Intriguingly, the locus on 8p is contiguous with linkage findings based upon the clinical schizophrenia phenotype,58 suggesting that a gene (or genes) in this region influences risk, at least in part, through downstream effects on DTNBP1 expression. It has proven possible to define loci influencing gene expression phenotypes more narrowly using genetic association.59 Approaches based on gene expression endophenotypes may therefore prove valuable both in the identification of further susceptibility genes and in establishing the functional relationship between them.
Modeling Etiological Changes in Gene Expression
Where etiological changes in gene expression can be established, these can be modeled in whole animal or cell systems in order to elucidate their functional consequences. Downregulation of individual susceptibility genes can be mimicked through gene knockout or, perhaps to a more realistic level, by methods such as RNA interference (RNAi). Conversely, upregulation of candidate gene transcripts can be emulated through gene knock-in techniques or, potentially, though methods based on the recently described phenomenon of RNA activation.60 Given that, at present, few (if any) schizophrenia susceptibility genes have been convincingly identified, much less the mechanism underlying association, discussion of potentially relevant data will be limited.
For DTNBP1, where reduced expression has been observed in schizophrenia cohorts,22–24 as well as a consequence of putative risk haplotypes,36 knock-down assays have already yielded some highly intriguing findings. For example, RNAi-induced reductions in DTNBP1 expression have been found to result in lower presynaptic protein expression and a decrease in glutamate release in neuronal culture.61 Again, a recent study found that, in human neuroblastoma and rat primary cortical neurons, knockdown of DTNBP1 expression resulted in increased cell surface dopamine D2 (DRD2) receptor expression.62 Parenthetically, for DRD2, for which disturbances in schizophrenia have long been postulated,63,64 transient overexpression in the striatum of transgenic mice has been found to result in abnormal prefrontal cortex function.65 The well-supported schizophrenia susceptibility gene NRG1 gene is known to participate in a diverse array of functions in brain, partly reflecting the multitude of isoforms it encodes.66 Mice that are heterozygous for knockout of the major NRG1 isoforms display behavioral abnormalities67,68 and altered hippocampal plasticity,69 although it is currently unclear how this relates to the effects of genetic susceptibility variation in schizophrenia. Several studies have modeled potential changes in DISC1 expression arising from the t(1;11) translocation, reporting neuroanatomical and cytoarchitectural anomalies reminiscent of those described in schizophrenia. In mice, downregulation of DISC1 by RNAi has been found to alter neuronal migration, leading to cell mispositioning.70 Transgenic mice that express the putative human truncated DISC1 product (on a background of endogenous DISC1 expression) show enlargement of lateral ventricles.71,72 In cell culture, both downregulation of DISC1 by RNAi and transfection of truncated DISC1 have been reported to inhibit neurite outgrowth.72–74 Future studies allowing hypothesis-free assessment of the effects of modeled changes in gene expression (eg, through microarray) will be particularly valuable in elucidating molecular pathways in schizophrenia pathogenesis and may suggest novel targets for therapeutic intervention.
Conclusions
The combination of psychiatric genetics and functional genomics will be essential in establishing primary etiological changes in gene expression in schizophrenia. In the short term, functional assays of gene expression can be used to elucidate the mechanism by which putative susceptibility variation in known candidates influences risk to the disorder. Functional assays are likely to become increasingly important in the identification of genetic regulatory variation, which can then be used for direct tests of association with schizophrenia and related traits/endophenotypes, as has been demonstrated in other psychiatric disorders.75 Once established, etiologically relevant changes in gene expression can be modeled in animal and cell systems in order to elucidate the molecular and physiological pathways that may ultimately give rise to this devastating condition.
References
- 1.Vawter MP, Ferran E, Galke B, Cooper K, Bunney WE, Byerley W. Microarray screening of lymphocyte gene expression differences in a multiplex schizophrenia pedigree. Schizophr Res. 2004;67:41–52. doi: 10.1016/s0920-9964(03)00151-8. [DOI] [PubMed] [Google Scholar]
- 2.Bowden NA, Weidenhofer J, Scott RJ, et al. Preliminary investigation of gene expression profiles in peripheral blood lymphocytes in schizophrenia. Schizophr Res. 2006;82:175–183. doi: 10.1016/j.schres.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Tallerico T, Novak G, Liu IS, Ulpian C, Seeman P. Schizophrenia: elevated mRNA for dopamine D2(Longer) receptors in frontal cortex. Brain Res Mol Brain Res. 2001;87:160–165. doi: 10.1016/s0169-328x(00)00293-x. [DOI] [PubMed] [Google Scholar]
- 4.Eastwood SL, Harrison PJ. Hippocampal synaptic pathology in schizophrenia, bipolar disorder and major depression: a study of complexin mRNAs. Mol Psychiatry. 2000;5:425–432. doi: 10.1038/sj.mp.4000741. [DOI] [PubMed] [Google Scholar]
- 5.Mirnics K, Levitt P, Lewis DA. Critical appraisal of DNA microarrays in psychiatric genomics. Biol Psychiatry. 2006;60:163–176. doi: 10.1016/j.biopsych.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Iwamoto K, Kato T. Gene expression profiling in schizophrenia and related mental disorders. Neuroscientist. 2006;12(4):349–361. doi: 10.1177/1073858406287536. [DOI] [PubMed] [Google Scholar]
- 7.Johnston-Wilson NL, Sims CD, Hofmann JP, et al. Disease-specific alterations in frontal cortex brain proteins in schizophrenia, bipolar disorder, and major depressive disorder. The Stanley Neuropathology Consortium. Mol Psychiatry. 2000;5:142–149. doi: 10.1038/sj.mp.4000696. [DOI] [PubMed] [Google Scholar]
- 8.Beasley CL, Pennington K, Behan A, Wait R, Dunn MJ, Cotter D. Proteomic analysis of the anterior cingulate cortex in the major psychiatric disorders: evidence for disease-associated changes. Proteomics. 2006;6:3414–3425. doi: 10.1002/pmic.200500069. [DOI] [PubMed] [Google Scholar]
- 9.Clark D, Dedova I, Cordwell S, Matsumoto I. A proteome analysis of the anterior cingulate cortex gray matter in schizophrenia. Mol Psychiatry. 2006;11:423, 459–470. doi: 10.1038/sj.mp.4001806. [DOI] [PubMed] [Google Scholar]
- 10.Mirnics K, Middleton FA, Marquez A, Lewis DA, Levitt P. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron. 2000;28:53–67. doi: 10.1016/s0896-6273(00)00085-4. [DOI] [PubMed] [Google Scholar]
- 11.Vawter MP, Crook JM, Hyde TM, et al. Microarray analysis of gene expression in the prefrontal cortex in schizophrenia: a preliminary study. Schizophr Res. 2002;58:11–20. doi: 10.1016/s0920-9964(01)00377-2. [DOI] [PubMed] [Google Scholar]
- 12.Hemby SE, Ginsberg SD, Brunk B, Arnold SE, Trojanowski JQ, Eberwine JH. Gene expression profile for schizophrenia: discrete neuron transcription patterns in the entorhinal cortex. Arch Gen Psychiatry. 2002;59:631–640. doi: 10.1001/archpsyc.59.7.631. [DOI] [PubMed] [Google Scholar]
- 13.Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci. 2002;22:2718–2729. doi: 10.1523/JNEUROSCI.22-07-02718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prabakaran S, Swatton JE, Ryan MM, et al. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol Psychiatry. 2004;9:643, 684–697. doi: 10.1038/sj.mp.4001511. [DOI] [PubMed] [Google Scholar]
- 15.Altar CA, Jurata LW, Charles V, et al. Deficient hippocampal neuron expression of proteasome, ubiquitin, and mitochondrial genes in multiple schizophrenia cohorts. Biol Psychiatry. 2005;58:85–96. doi: 10.1016/j.biopsych.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 16.Hakak Y, Walker JR, Li C, et al. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci U S A. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tkachev D, Mimmack ML, Ryan MM, et al. Oligodendrocyte dysfunction in schizophrenia and bipolar disorder. Lancet. 2003;362:798–805. doi: 10.1016/S0140-6736(03)14289-4. [DOI] [PubMed] [Google Scholar]
- 18.Sugai T, Kawamura M, Iritani S, et al. Prefrontal abnormality of schizophrenia revealed by DNA microarray: impact on glial and neurotrophic gene expression. Ann N Y Acad Sci. 2004;1025:84–91. doi: 10.1196/annals.1316.011. [DOI] [PubMed] [Google Scholar]
- 19.Cardno AG, Marshall EJ, Coid B, et al. Heritability estimates for psychotic disorders: the Maudsley twin psychosis series. Arch Gen Psychiatry. 1999;56:162–168. doi: 10.1001/archpsyc.56.2.162. [DOI] [PubMed] [Google Scholar]
- 20.Gottesman II. Schizophrenia genesis: the origins of madness. New York, NY: W.H. Freeman; 1991. [Google Scholar]
- 21.Norton N, Williams HJ, Owen MJ. An update on the genetics of schizophrenia. Curr Opin Psychiatry. 2006;19:158–164. doi: 10.1097/01.yco.0000214341.52249.59. [DOI] [PubMed] [Google Scholar]
- 22.Talbot K, Eidem WL, Tinsley CL, et al. Dysbindin-1 is reduced in intrinsic, glutamatergic terminals of the hippocampal formation in schizophrenia. J Clin Invest. 2004;113:1353–1363. doi: 10.1172/JCI20425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weickert CS, Straub RE, McClintock BW, et al. Human dysbindin (DTNBP1) gene expression in normal brain and in schizophrenic prefrontal cortex and midbrain. Arch Gen Psychiatry. 2004;61:544–555. doi: 10.1001/archpsyc.61.6.544. [DOI] [PubMed] [Google Scholar]
- 24.Weickert CS, Rothmond DA, Hyde TM, Kleinman JE, Straub RE. Reduced DTNBP1 (dysbindin-1) mRNA in the hippocampal formation of schizophrenia patients. Schizophr Res. 2008;98:105–110. doi: 10.1016/j.schres.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korostishevsky M, Kaganovich M, Cholostoy A, et al. Is the G72/G30 locus associated with schizophrenia? Single nucleotide polymorphisms, haplotypes, and gene expression analysis. Biol Psychiatry. 2004;56:169–176. doi: 10.1016/j.biopsych.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 26.Hashimoto R, Straub RE, Weickert CS, Hyde TM, Kleinman JE, Weinberger DR. Expression analysis of neuregulin-1 in the dorsolateral prefrontal cortex in schizophrenia. Mol Psychiatry. 2004;9:299–307. doi: 10.1038/sj.mp.4001434. [DOI] [PubMed] [Google Scholar]
- 27.Law AJ, Lipska BK, Weickert CS, et al. Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5′ SNPs associated with the disease. Proc Natl Acad Sci U S A. 2006;103:6747–6752. doi: 10.1073/pnas.0602002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowden NA, Scott RJ, Tooney PA. Altered expression of regulator of G-protein signalling 4 (RGS4) mRNA in the superior temporal gyrus in schizophrenia. Schizophr Res. 2007;89:165–168. doi: 10.1016/j.schres.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 29.Erdely HA, Tamminga CA, Roberts RC, Vogel MW. Regional alterations in RGS4 protein in schizophrenia. Synapse. 2006;59:472–479. doi: 10.1002/syn.20265. [DOI] [PubMed] [Google Scholar]
- 30.Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 31.Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- 32.Neale BM, Sham PC. The future of association studies: gene-based analysis and replication. Am J Hum Genet. 2004;75:353–362. doi: 10.1086/423901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dixon AL, Liang L, Moffatt MF, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 34.Myers AJ, Gibbs JR, Webster JA, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 35.Bray NJ, O'Donovan MC. Investigating cis-acting regulatory variation using assays of relative allelic expression. Psychiatr Genet. 2006;16:173–177. doi: 10.1097/01.ypg.0000218612.35139.84. [DOI] [PubMed] [Google Scholar]
- 36.Bray NJ, Preece A, Williams NM, et al. Haplotypes at the dystrobrevin binding protein 1 (DTNBP1) gene locus mediate risk for schizophrenia through reduced DTNBP1 expression. Hum Mol Genet. 2005;14:1947–1954. doi: 10.1093/hmg/ddi199. [DOI] [PubMed] [Google Scholar]
- 37.Pastinen T, Ge B, Gurd S, et al. Mapping common regulatory variants to human haplotypes. Hum Mol Genet. 2005;14:3963–3971. doi: 10.1093/hmg/ddi420. [DOI] [PubMed] [Google Scholar]
- 38.Knight JC, Keating BJ, Rockett KA, Kwiatkowski DP. In vivo characterization of regulatory polymorphisms by allele-specific quantification of RNA polymerase loading. Nat Genet. 2003;33:469–475. doi: 10.1038/ng1124. [DOI] [PubMed] [Google Scholar]
- 39.Tan W, Wang Y, Gold B, et al. Molecular cloning of a brain-specific, developmentally regulated neuregulin 1 (NRG1) isoform and identification of a functional promoter variant associated with schizophrenia. J Biol Chem. 2007;282:24343–24351. doi: 10.1074/jbc.M702953200. [DOI] [PubMed] [Google Scholar]
- 40.Cirulli ET, Goldstein DB. In vitro assays fail to predict in vivo effects of regulatory polymorphisms. Hum Mol Genet. 2007;16:1931–1939. doi: 10.1093/hmg/ddm140. [DOI] [PubMed] [Google Scholar]
- 41.Wilkins JM, Southam L, Price AJ, Mustafa Z, Carr A, Loughlin J. Extreme context specificity in differential allelic expression. Hum Mol Genet. 2007;16:537–546. doi: 10.1093/hmg/ddl488. [DOI] [PubMed] [Google Scholar]
- 42.Murphy KC, Jones LA, Owen MJ. High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch Gen Psychiatry. 1999;56:940–945. doi: 10.1001/archpsyc.56.10.940. [DOI] [PubMed] [Google Scholar]
- 43.Meechan DW, Maynard TM, Gopalakrishna D, Wu Y, LaMantia AS. When half is not enough: gene expression and dosage in the 22q11 deletion syndrome. Gene Expr. 2007;13:299–310. doi: 10.3727/000000006781510697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shifman S, Bronstein M, Sternfeld M, et al. A highly significant association between a COMT haplotype and schizophrenia. Am J Hum Genet. 2002;71:1296–1302. doi: 10.1086/344514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mukai J, Liu H, Burt RA, et al. Evidence that the gene encoding ZDHHC8 contributes to the risk of schizophrenia. Nat Genet. 2004;36:725–731. doi: 10.1038/ng1375. [DOI] [PubMed] [Google Scholar]
- 46.Liu H, Heath SC, Sobin C, et al. Genetic variation at the 22q11 PRODH2/DGCR6 locus presents an unusual pattern and increases susceptibility to schizophrenia. Proc Natl Acad Sci U S A. 2002;99:3717–3722. doi: 10.1073/pnas.042700699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams NM, Glaser B, Norton N, et al. Strong evidence that GNB1L is associated with schizophrenia. Hum Mol Genet. 2008;17:555–566. doi: 10.1093/hmg/ddm330. [DOI] [PubMed] [Google Scholar]
- 48.Bray NJ, Buckland PR, Williams NM, et al. A haplotype implicated in schizophrenia susceptibility is associated with reduced COMT expression in human brain. Am J Hum Genet. 2003;73:152–161. doi: 10.1086/376578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.St Clair D, Blackwood D, Muir W, et al. Association within a family of a balanced autosomal translocation with major mental illness. Lancet. 1990;336:13–16. doi: 10.1016/0140-6736(90)91520-k. [DOI] [PubMed] [Google Scholar]
- 50.Millar JK, Wilson-Annan JC, Anderson S, et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- 51.Millar JK, Pickard BS, Mackie S, et al. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science. 2005;310:1187–1191. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- 52.Chubb JE, Bradshaw NJ, Soares DC, Porteous DJ, Millar JK. The DISC locus in psychiatric illness. Mol Psychiatry. 2008;13:36–64. doi: 10.1038/sj.mp.4002106. [DOI] [PubMed] [Google Scholar]
- 53.Lipska BK, Peters T, Hyde TM, et al. Expression of DISC1 binding partners is reduced in schizophrenia and associated with DISC1 SNPs. Hum Mol Genet. 2006;15:1245–1258. doi: 10.1093/hmg/ddl040. [DOI] [PubMed] [Google Scholar]
- 54.Hayesmoore JB, Bray NJ, Owen MJ, O'Donovan MC. DISC1 mRNA expression is not influenced by common cis-acting regulatory polymorphisms or imprinting. Am J Med Genet B Neuropsychiatr Genet. 2008 doi: 10.1002/ajmg.b.30715. doi:10.1002/ajmg.b.30715. [DOI] [PubMed] [Google Scholar]
- 55.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 56.Braff D, Schork NJ, Gottesman II. Endophenotyping schizophrenia. Am J Psychiatry. 2007;164:705–707. doi: 10.1176/ajp.2007.164.5.705. [DOI] [PubMed] [Google Scholar]
- 57.Bray NJ, Holmans PA, van den Bree MB, et al. Cis- and trans- loci influence expression of the schizophrenia susceptibility gene DTNBP1. Hum Mol Genet. 2008 doi: 10.1093/hmg/ddn006. doi:10.1093/hmg/ddn006. [DOI] [PubMed] [Google Scholar]
- 58.Badner JA, Gershon ES. Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry. 2002;7:405–411. doi: 10.1038/sj.mp.4001012. [DOI] [PubMed] [Google Scholar]
- 59.Cheung VG, Spielman RS, Ewens KG, Weber TM, Morley M, Burdick JT. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li LC, Okino ST, Zhao H, et al. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci U S A. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Numakawa T, Yagasaki Y, Ishimoto T, et al. Evidence of novel neuronal functions of dysbindin, a susceptibility gene for schizophrenia. Hum Mol Genet. 2004;13:2699–2708. doi: 10.1093/hmg/ddh280. [DOI] [PubMed] [Google Scholar]
- 62.Iizuka Y, Sei Y, Weinberger DR, Straub RE. Evidence that the BLOC-1 protein dysbindin modulates dopamine D2 receptor internalization and signaling but not D1 internalization. J Neurosci. 2007;27:12390–12395. doi: 10.1523/JNEUROSCI.1689-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seeman P, Chau-Wong M, Tedesco J, Wong K. Brain receptors for antipsychotic drugs and dopamine: direct binding assays. Proc Natl Acad Sci U S A. 1975;72:4376–4380. doi: 10.1073/pnas.72.11.4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cross AJ, Crow TJ, Owen F. 3H-Flupenthixol binding in post-mortem brains of schizophrenics: evidence for a selective increase in dopamine D2 receptors. Psychopharmacology (Berl) 1981;74:122–124. doi: 10.1007/BF00432676. [DOI] [PubMed] [Google Scholar]
- 65.Kellendonk C, Simpson EH, Polan HJ, et al. Transient and selective overexpression of dopamine D2 receptors in the striatum causes persistent abnormalities in prefrontal cortex functioning. Neuron. 2006;49:603–615. doi: 10.1016/j.neuron.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 66.Falls DL. Neuregulins: functions, forms, and signaling strategies. Exp Cell Res. 2003;284:14–30. doi: 10.1016/s0014-4827(02)00102-7. [DOI] [PubMed] [Google Scholar]
- 67.Stefansson H, Sigurdsson E, Steinthorsdottir V, et al. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71:877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O'Tuathaigh CM, Babovic D, O'Sullivan GJ, et al. Phenotypic characterization of spatial cognition and social behavior in mice with ‘knockout’ of the schizophrenia risk gene neuregulin 1. Neuroscience. 2007;147:18–27. doi: 10.1016/j.neuroscience.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 69.Bjarnadottir M, Misner DL, Haverfield-Gross S, et al. Neuregulin1 (NRG1) signaling through Fyn modulates NMDA receptor phosphorylation: differential synaptic function in NRG1+/- knock-outs compared with wild-type mice. J Neurosci. 2007;27:4519–4529. doi: 10.1523/JNEUROSCI.4314-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duan X, Chang JH, Ge S, et al. Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell. 2007;130:1146–1158. doi: 10.1016/j.cell.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hikida T, Jaaro-Peled H, Seshadri S, et al. Dominant-negative DISC1 transgenic mice display schizophrenia-associated phenotypes detected by measures translatable to humans. Proc Natl Acad Sci U S A. 2007;104:14501–14506. doi: 10.1073/pnas.0704774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pletnikov MV, Ayhan Y, Nikolskaia O, et al. Inducible expression of mutant human DISC1 in mice is associated with brain and behavioral abnormalities reminiscent of schizophrenia. Mol Psychiatry. 2008;13:173–186. doi: 10.1038/sj.mp.4002079. [DOI] [PubMed] [Google Scholar]
- 73.Kamiya A, Kubo K, Tomoda T, et al. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol. 2005;7:1167–1178. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
- 74.Shinoda T, Taya S, Tsuboi D, et al. DISC1 regulates neurotrophin-induced axon elongation via interaction with Grb2. J Neurosci. 2007;27:4–14. doi: 10.1523/JNEUROSCI.3825-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lesch KP, Bengel D, Heils A, et al. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science. 1996;274:1527–1531. doi: 10.1126/science.274.5292.1527. [DOI] [PubMed] [Google Scholar]