Abstract
Objective: Genetic linkage studies in schizophrenia (SZ) have primarily focused on the phenotype of disease susceptibility. A limited number of studies, however, have reported suggestive linkage to specific SZ symptom domains including regions on chromosomes 6, 8, and 20. We examined these chromosomal regions for association to positive, negative, and disorganized symptom clusters, using a dense set of single-nucleotide polymorphisms (SNPs). Methods: We ascertained 178 Caucasian patients with SZ for lifetime severity of clinical symptomatology using a structured diagnostic interview. The cohort was genotyped with the Affymetrix 500K microarray, from which we selected, a priori, 4833 intragenic SNPs located within chromosomal regions previously linked to specific SZ symptom clusters. Parametric tests, corrected for multiple testing, were used to compare the effects of allelic variation within these SNPs to the lifetime severity of the specific symptom domain that had been implicated by prior linkage studies. Results: We were able to extend previous reports of linkage between chromosome 6q and both positive and disorganized symptoms. Lifetime severity of positive symptoms was significantly (P = 2.50 × 10−5) associated with a SNP within the origin recognition complex subunit 3–like (ORC3L) gene, a gene implicated in synaptic plasticity. Level of disorganized symptoms was significantly (P < 6.00 × 10−5) associated 2 SNPs within the brain-specific angiogenesis inhibitor 3 (BAI3) gene, which is highly expressed in brain during development. Conclusions: These data point toward specific candidate genes located within previously implicated linkage peaks for clinical symptomatology. Identification of functional variants within these regions and a characterization of the effect of these risk genotypes on the treatment of specific clinical symptoms are needed.
Keywords: schizophrenia, symptoms, quantitative traits, ORC3L, BAI3
The clinical presentation of schizophrenia (SZ) is complex and characterized by a myriad of symptoms that may include positive, negative, and disorganized features. Because no single symptom or symptom cluster is considered pathognomonic, the current categorical/syndromal nosology may not be optimal for elucidating the molecular mechanisms underlying the illness. Consequently, it has been suggested that the current system be replaced1 or supplemented2 by a dimensional, symptom-based approach focused on individual, subsyndromal, and quantitative phenotypes.3–7 Such a nosology is attractive because it provides a mechanism for studying the heterogeneity of SZ and may enhance our ability to identify the underlying pathophysiology of the illness.8
The notion that the diagnostic entity of SZ functions only as a boundary around a collection of heterogeneous pathophysiological processes is consistent with the view that SZ is a polygenic disorder in which many genes of small effect may be contributing to its risk. Moreover, it may provide an explanation for the failures to replicate so often seen in SZ genetics. Specifically, if multiple and variable genetic loci are associated with pathophysiological and phenotypic variability in SZ, substantial genetic overlap between healthy and broadly defined patient groups will exist. Thus, any comparison of these groups would likely mask or minimize the true risk alleles associated with more homogenous phenotypes.
Empirical support for a quantitative, symptom-based approach to elucidating the molecular underpinnings of SZ can be derived from family studies that demonstrate the heritability of symptom factors including positive, negative, and affective symptoms;9 thought withdrawal, thought insertion; thought broadcasting; and delusions of control10 and disorganization11,12 in sibling pairs (including twins) concordant for SZ. To date, however, relatively limited work has been conducted to identify, at the molecular level, the genetic variants associated with specific clinical phenomena. Gene-symptom relationships have emerged primarily from follow-up studies of putative SZ risk genes, with only a handful of replicated findings. For example, significant relationships have been identified between single-nucleotide polymorphisms (SNPs) in the Disrupted in Schizophrenia 1 (DISC1) gene and the severity of delusions in patients with SZ,13,14 as well as between SNPs in the dystrobrevin binding protein gene (DTNBP1) and negative symptoms in patients with SZ.15–17 Based on these data, it seems likely that susceptibility genes for SZ may not only increase the risk for the illness but also influence the clinical presentation of the illness. Moreover, a number of studies have reported data indicating that specific genetic variants may act to modify24 the clinical presentation of illness without increasing the overall risk for the illness. These include reported relations between negative symptoms and variation in BDNF, DAT1,18 and hkCa319; positive symptoms and variation in DRD4,20 DRD2,21 CCK-A,22 and 5-HTT23; and disorganization and variation in DRD2.21
In contrast to candidate gene studies, genetic linkage approaches have the capacity to detect novel loci located within relatively large chromosomal regions. To date, only a few studies have utilized linkage approaches to identify quantitative trait loci (QTLs) for SZ symptoms. Brzustowicz et al25 assessed 28 genetic markers spanning chromosome 6 for linkage to the positive, negative, and general psychopathology subscales of the Positive and Negative Syndrome Scale (PANSS) in 10 moderate sized affected families (n = 183). Despite no significant linkage to diagnosis, significant evidence for linkage to specific PANSS subscales was observed at several loci on chromosome 6p. Kendler et al26 conducted a similar analysis on 4 genomic regions on chromosome 5q, 6p, 8p, and 10p that had been previously linked to the broad diagnostic category of SZ in a large sample of 265 affected families (n = 1408) from the Irish Study of High-Density Schizophrenia Families. Linkage to 8p22-8p21 was reported for a Kraepelinian constellation of symptoms, including negative thought disorder, affective deterioration, and poor outcome. Wilcox et al27 conducted a genome-wide linkage scan of symptoms in SZ, in a study of 51 families (n = 136) derived from the National Institute of Mental Health–funded Genetics Initiative on Schizophrenia. These analyses revealed suggestive linkage to chromosomes 6, 9, and 20 for the disorganized symptoms trait and to chromosome 12 for the negative symptoms trait. Finally, Fanous et al28 recently conducted a genome-wide scan in 270 Irish high-density families (n = 755), using clinically homogeneous phenotypes derived from a latent class analysis of the Operational Criteria Checklist for Psychotic Illness. This analysis implicated several regions of suggestive linkage to clinical phenotypes including a region on chromosome 20 linked to deficit syndrome,2 a clinical phenotype characterized by severe negative symptoms.
Unfortunately, follow-up linkage disequilibrium studies have not been reported, and no genes in these previously identified linkage regions have been identified that significantly influence the clinical presentation of SZ. Thus, the present study was designed to densely map the previously implicated linkage regions to detect the specific genes in these regions that may be acting to modify the clinical presentation of SZ.
Methods
Participant Sample
The study sample was comprised of 178 patients who met criteria for a SZ-spectrum disorder (SZ, n = 158; schizoaffective disorder, n = 13; or schizophreniform disorder, n = 7), based on the Structured Clinical Interview for the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, (SCID).29 All participants were recruited from the Zucker Hillside Hospital (ZHH), a division of the North Shore-Long Island Jewish Health System (NSLIJHS), in Glen Oaks, NY, and provided written informed consent to a protocol approved by the Institutional Review Board of the NSLIJHS.
Clinical Assessment
Each subject was assessed with a SCID administered by trained and reliable raters. Information obtained from the SCID was supplemented by a review of medical records and interviews with family informants when possible. The interview, clinical information from available medical records, and information from family informants were then compiled into a narrative case summary and presented at a diagnostic consensus conference. Diagnoses were then determined by a consensus among a minimum of 3 expert diagnosticians from the ZHH faculty.
Because symptom severity often varies during the course of illness, a lifetime symptom severity rating rather than a cross-sectional rating was used.1,30 Lifetime symptom ratings were derived from SCID data and included overall ratings on negative symptoms, positive symptoms, and disorganized symptoms. The overall negative symptom rating included ratings on avolition, alogia, and affective flattening.16 The overall positive symptom rating included ratings on delusions (referential, paranoid, grandiose, somatic, control, thought broadcasting, bizarre, and other delusions) and hallucinations (auditory, visual, tactile, and other hallucinations).14 Finally, the overall disorganization symptom rating included ratings on disorganized speech and disorganized behavior. Each SCID item used in these ratings were rated on a continuous scale where 1 = absent, 2 = subthreshold, and 3 = present. The overall rating for each symptom cluster was calculated by summing the scores of all items within a cluster.
Genotyping
Genomic DNA was extracted from whole blood, and genotype data were derived using the Affymetrix 500K array (Affymetrix, Santa Clara, CA) and are described in detail by Lencz et al.31 We restricted our analyses to SNPs located within the chromosomal regions previously linked to quantitative symptom traits in SZ. Moreover, allelic variation at each SNP within a region was only tested for association to symptom severity in the specific domain to which it had been previously linked.
Selection of SNPs within linkage regions was based on the statistical results reported for each microsatellite marker that had been assessed in the individual linkage studies. We selected SNPs within regions encompassed by neighboring microsatellite markers that were nominally associated with a specific symptom domain (P < .0513; P < .0126; logarithm of the odds (LOD) > 225,27,28). Each nominally associated marker was entered into the University of California at Santa Cruz genome browser (UCSC) database to identify the corresponding location of that marker on the physical chromosome map (Build 35, May 2004). The physical positions of the most telomeric and centromeric boundaries were then used to limit the region of the chromosome we investigated.
Based on the report of Brzustowicz et al,25 SNPs on chromosome 6 encompassed by markers D6S477–D6S1050 were assessed for association to negative symptoms, and SNPs encompassed by markers D6S1280–D6S10566 were assessed for association to positive symptoms. Based on the report of Kendler et al,26 SNPs on chromosome 20 encompassed by markers D8S552–D8S283 were assessed for association to negative symptoms. Based on Wilcox et al,27 SNPs on chromosome 6 encompassed by markers D6S426–D6S1270 were assessed for association to disorganized symptoms. Finally, SNPs on chromosome 20 encompassed by markers D20S115–D20S481 were assessed for association to negative symptoms based on the findings reported by Fanous et al.28
For each linkage region identified by the aforementioned studies, we included an additional 100 kb on each side of the marker boundaries to ensure optimal coverage of the linkage region. Further, to reduce the number of statistical tests being carried out, only intragenic SNPs were selected for analysis. Of note, although Wilcox et al27 identified single markers on chromosome 9, 12, and 20, with LOD scores greater than 2, these regions were not examined in the present study due to the lack of a second significant boundary to delineate a chromosomal region.
Based on the aforementioned methodology of SNP selection and the availability of SNPs on the Affymetrix array, we assessed a total of 5747 SNPs in the region on chromosome 6p linked to positive symptoms and 1399 SNPs in the region on chromosome 6p linked to negative symptoms by Brzustowicz et al.25 Further, the region on chromosome 8p derived from the Kendler et al26 study yielded a total of 3979 SNPs for analysis. The Wilcox et al27 linkage region on chromosome 6pq yielded 5574 SNPs for analysis. Finally, 2211 SNPs on chromosome 20pq were assessed to test the Fanous et al28 linkage region. Overall, a total of 12 438 SNPs were available on the Affymetrix array for the present analyses of which 4833 were intragenic and tested for association to symptom severity. The number of intragenic SNPs, as well as information about the chromosomal regions, source citations (including associated symptom cluster), and number of SNPs assessed in each region, are shown in table 1.
Table 1.
Regions Analyzed for Association to Symptom Domains
| Source | Chr | Symptom Association | Physical Position | Size (Mb) | Genes Region | SNPs in Region | Significance Threshold |
| Brzustowicz et al25 | 6q | Positive | 48857800–94254488 | 45.4 | 257 | 791 | 6.32 × 10−5 |
| Brzustowicz et al25 | 6p | Negative | 5985381–22551196 | 16.57 | 124 | 493 | 1.01 × 10−4 |
| Wilcox et al27 | 6pq | Disorganized | 40650161–85740355 | 45.09 | 322 | 771 | 6.48 × 10−5 |
| Kendler et al26 | 8p | Negative | 12686453–33847805 | 21.16 | 153 | 2,073 | 2.41 × 10−5 |
| Fanous et al28 | 20pq | Negative | 7607966–43201778 | 35.59 | 135 | 705 | 1.78 × 10−5 |
Note: The first 3 columns represent the findings of previous linkage studies and include reference to the study, the chromosome identified as suggestive of linkage to symptom domains, and the symptom cluster to which suggestive linkage was found, respectively. The fourth column indicates the start and stop positions that correspond to the linkage regions reported in these prior studies. The next 3 columns provide information regarding the size of the region analyzed, the number of known genes in the region, and the number of intragenic SNPs in the Affymetrix 500K microarray in the region of interest. The last column indicates the Bonferroni-corrected P value threshold for significance used in each region (see text for details). SNP, single-nucleotide polymorphism.
Statistical Analyses
As detailed in table 1, a total of 5 hypotheses were tested in the present study. Specifically, we tested for association between positive symptoms and allelic variation in SNPs from a region of chromosome 6q, disorganized symptoms and allelic variation in SNPs from chromosome 6q, and negative symptoms and allelic variation in SNPs on chromosomes 6p, 8p, and 20pq. The significance of each association was assessed using allelic t tests conducted within HelixTree 6.2 software (GoldenHelix, Bozeman, MT) and were corrected using a strict Bonferroni procedure based on the total number of SNPs analyzed within each region (for Bonferroni-corrected P value thresholds see table 1). For any gene containing SNPs that met criteria for significance, all SNPs within that gene were further assessed for association at a nominal level of significance (P < .05). To assess the advantage of the QTL approach over the case-control approach, we also compared genetic variation in these regions between our SZ sample and a sample of 144 healthy controls that have been previously described.31
Results
We tested regions on chromosome 6, 8p, and 20pq for association with specific SZ symptoms domains and identified 3 intragenic SNPs that met Bonferroni-corrected criteria for association to symptom severity. Specifically, we identified a single SNP associated with lifetime severity of positive symptoms in our cohort that was located within the region of 6q that had previously been linked to positive symptoms.25 In addition, we also identified 2 SNPs in our cohort that were associated with lifetime severity of disorganized symptoms that were located within the region of 6q that had previously been linked to disorganized symptoms.27 These data are shown in table 2. No significant associations were found between negative symptom severity and SNPs within chromosome 6p, 8p, or 20pq. Moreover, no Bonferroni-corrected differences in allele frequencies were found between cases and controls in any of the regions assessed.
Table 2.
SNPs Within Previously Identified Linkage Regions That Were Significantly Associated With Symptom Severity
| Gene | Location | Position | dbSNP | Alleles | Symptom | Associated Allele | t | P |
| ORC3L | 6q15 | 88403068 | rs16879646 | G/A | Positive | A | 4.28 | 2.50 × 10−5 |
| BAI3 | 6q12 | 69757118 | rs1415031 | G/C | Disorganized | C | 4.08 | 5.69E × 10−5 |
| BAI3 | 6q12 | 69768622 | rs9446083 | G/A | Disorganized | A | 4.08 | 5.90 × 10−5 |
Note: The first 5 columns indicate the gene symbol, its chromosomal location, physical position, the SNP within the gene that was significantly associated with a symptom cluster, and the major and minor alleles at that SNP (minor allele is in bold). Column 6 indicates the symptom cluster to which the SNP was associated, and column 7 indicates the allele associated with increased severity on the symptom cluster. Statistical comparisons are presented in columns 8 and 9. SNP, single-nucleotide polymorphism; BAI3, brain-specific angiogenesis inhibitor 3.
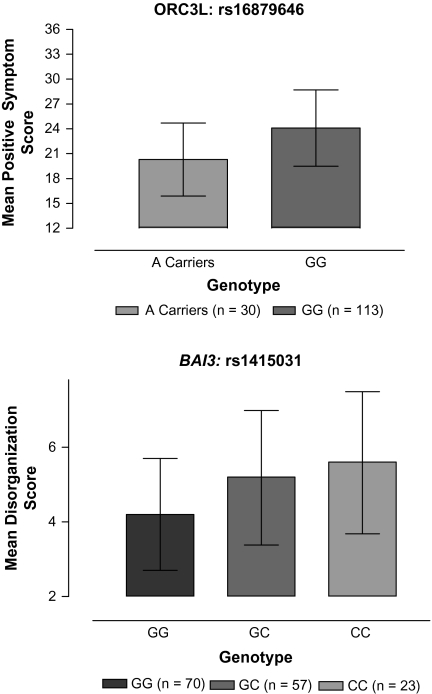
At chromosome 6q15, the major allele (A) at rs16879646 was significantly associated with a higher lifetime severity of positive symptoms (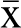 A=23.72 ± 0.29 vs
A=23.72 ± 0.29 vs 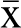 G = 20.06 ± 0.79; t = 4.28; P = 2.50 × 10−5). A comparison of positive symptom severity by genotype is presented in the upper panel of figure 1. Because only 3 AA homozygotes were identified in our sample, the data presented are for A carriers vs G homozygotes. This SNP is located in the intronic region 3’ to exon 12 of the origin recognition complex subunit 3–like (ORC3L) gene. Three additional SNPs in ORC3L that were available for analysis were tested but were not associated with the severity of positive symptoms.
G = 20.06 ± 0.79; t = 4.28; P = 2.50 × 10−5). A comparison of positive symptom severity by genotype is presented in the upper panel of figure 1. Because only 3 AA homozygotes were identified in our sample, the data presented are for A carriers vs G homozygotes. This SNP is located in the intronic region 3’ to exon 12 of the origin recognition complex subunit 3–like (ORC3L) gene. Three additional SNPs in ORC3L that were available for analysis were tested but were not associated with the severity of positive symptoms.
Fig. 1.
Comparison of Lifetime Severity of Psychotic Symptoms in Patients With Schizophrenia by Genotype. The upper panel represents the data for the single-nucleotide polymorphism in ORC3L that is significantly associated with lifetime severity of positive symptoms. The lower panel represents the data for the rs1415031 in brain-specific angiogenesis inhibitor 3 (BAI3) that was significantly associated with lifetime severity of disorganized symptoms. Error bars indicate SDs.
Further, at 6q12, the minor alleles at 2 SNPs, rs1415031 and rs9446083, were significantly associated with lifetime severity of disorganized symptoms (rs1415031: 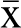 C = 5.37 ± 0.1780 vs
C = 5.37 ± 0.1780 vs 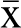 G = 4.520 ± 0.12; t = 4.08; P = 5.69 × 10−5; and rs9446083:
G = 4.520 ± 0.12; t = 4.08; P = 5.69 × 10−5; and rs9446083: 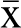 A = 5.22 ± 0.15 vs
A = 5.22 ± 0.15 vs 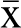 G = 4.41 ± 0.13; t = 4.08; P = 5.90 × 10−5). A comparison of disorganized symptom severity by genotype is presented in the bottom panel of figure 1. Because of the high linkage disequilibrium (LD) between these 2 SNPs (D‘ = 0.83) and to avoid redundancy, only the data for rs1415031 are shown. Both of these SNPs are located in the intronic region 5′ to exon 9 in the gene encoding brain-specific angiogenesis inhibitor 3 (BAI3). Eighty-nine additional SNPs in BAI3 that were available for analysis were tested for association to disorganized symptoms. Of these, 18 (20%), spanning almost the entire coding region of the gene, were nominally associated with disorganized symptoms (P < .05). These SNPs, along with their physical position and associated P values are shown in table 3.
G = 4.41 ± 0.13; t = 4.08; P = 5.90 × 10−5). A comparison of disorganized symptom severity by genotype is presented in the bottom panel of figure 1. Because of the high linkage disequilibrium (LD) between these 2 SNPs (D‘ = 0.83) and to avoid redundancy, only the data for rs1415031 are shown. Both of these SNPs are located in the intronic region 5′ to exon 9 in the gene encoding brain-specific angiogenesis inhibitor 3 (BAI3). Eighty-nine additional SNPs in BAI3 that were available for analysis were tested for association to disorganized symptoms. Of these, 18 (20%), spanning almost the entire coding region of the gene, were nominally associated with disorganized symptoms (P < .05). These SNPs, along with their physical position and associated P values are shown in table 3.
Table 3.
SNPs Within BAI3 That Are Significantly (Indicated by Asterisk) or Nominally Associated With Severity of Disorganized Symptoms in Schizophrenia
| dbSNP | Position | P Value |
| rs544398 | 69632524 | 0.04 |
| rs1880177 | 69646677 | 0.03 |
| rs7760666 | 69678581 | 0.01 |
| rs2184723 | 69703807 | 0.02 |
| rs1932615 | 69713729 | 0.04 |
| rs3823064 | 69738338 | 0.02 |
| rs3798979 | 69738381 | 0.02 |
| rs1415031* | 69757118 | 5.70 × 10−5 |
| rs3798995 | 69757481 | 0.02 |
| rs9446083* | 69768622 | 5.90 × 10−5 |
| rs9454674 | 69777487 | 0.01 |
| rs9446085 | 69783083 | 0.01 |
| rs1889878 | 69793014 | 0.01 |
| rs7743332 | 69803749 | 0.01 |
| rs688606 | 69840895 | 0.04 |
| rs7759645 | 69874050 | 0.03 |
| rs1147510 | 69912418 | 0.01 |
| rs7739401 | 69915651 | 0.02 |
| rs779467 | 69988140 | 0.01 |
| rs779462 | 70001377 | 0.02 |
Note: SNP, single-nucleotide polymorphism; BAI3, brain-specific angiogenesis inhibitor 3.
Discussion
We report an association between a SNP on chromosome 6q and positive symptoms and 2 SNPs on chromosome 6q and disorganized symptoms in SZ. On chromosome 6q, rs16879646 was significantly associated with positive symptoms in SZ. These results are consistent with the findings of Brzustowicz et al25 who reported significant linkage of this region to positive symptom severity in SZ. Moreover, on chromosome 6q, rs1415031 and rs9446083 were significantly associated with the severity of disorganization in SZ. These results are consistent with the findings of Wilcox et al27 who reported suggestive linkage of this region to disorganization in SZ.
The SNP identified in the region of chromosome 6q that had been linked to positive symptoms by Brzustowicz et al25 is located within the ORC3L gene. ORC3L is a component of the 6-subunit protein complex known as the ORC. ORC3L is a 77-kb gene comprised of 20 exons and is located at 6q15. Although the most well-documented aspect of ORC function is related to the formation of prereplication complexes upon exit from mitosis, recent data implicate ORC in sister chromatin cohesion, cytokinesis, and neural dendritic branching.32
Although little is known about the function of human ORC3L, the drosophila homolog, Latheo, codes for a protein that is localized to the presynaptic terminals of differentiated motor neurons and appears to modulate mechanisms necessary for behavioral plasticity.33 Mutations in Latheo have been shown to disrupt associative learning through its effect on Ca2+ and activity-dependent synaptic plasticity.34 Alterations in synaptic plasticity have long been suggested to play a role in both the development of schizophrenic symptoms, particularly positive symptoms, and the deterioration process in SZ.35 Thus, these data suggest that variation in ORC3L may influence the lifetime severity of positive symptoms by mediating Ca2+ and activity-dependent synaptic plasticity.
The SNPs identified in the region of chromosome 6q that had been linked to disorganization by Wilcox et al27 are located within the gene encoding BAI3. BAI3 is a p53-target gene encoding a 7-span transmembrane protein and is thought to be a member of the secretin, g-protein–coupled receptor family.36 BAI3 is located at chromosome 6q12 and is a 750-kb gene comprised of 31 exons. Very little is known about the function of human BAI3, and to date, a relatively limited amount of data regarding the function of BAI3 homologs have been reported in other species. Recent work in mice, however, indicates that the protein coded for by mouse BAI3 is expressed almost exclusively in the brain with the highest levels of expression of BAI3 during the early neonatal period. Approximately 10 days after birth, BAI3 decreases steadily until adulthood and relatively little expression of BAI3 is seen in the adult brain.37 Thus, it seems likely that BAI3 plays a role in the neurodevelopmental process and may influence the clinical symptomatology of SZ through its involvement in neurodevelopment. This analysis is consistent with the prevailing explanatory model of SZ, which posits that the disease is the behavioral outcome of an aberration in neurodevelopmental processes that begins long before the onset of clinical symptoms.38
Several limitations of the current study should be noted. First, the method of genotyping employed in the present study limited the markers within ORC3L and BAI3 that could be assessed, and thus, it is possible that the SNPs we have identified are not directly associated with symptom severity. Rather, the SNPs we have identified may be in high LD with SNPs within the coding regions of these genes that are directly associated with symptom severity. Further, as with any genetic association study, the present findings may represent false positives. Although we believe that the use of a strict Bonferroni correction has substantially limited the potential for type I errors, additional independent replications will be required to confirm the present findings.
The present data support previous reports of linkage between 6q and both positive and disorganized symptoms and suggest that there are specific candidate genes located within these previously implicated linkage peaks that are associated with clinical symptomatology. These data add to the accumulating evidence demonstrating the potential utility of a symptom-based approach to the genetics in SZ. Given the clinical heterogeneity and molecular complexity of the illness, however, it is unlikely that any single genetic paradigm will account for a large degree of the variance to be explained. With the advent of large-scale genotyping platforms, hypothesis-free genome-wide searches using traditional association methods31 as well as more contemporary methods such as Whole Genome Homozygosity Association39 and copy number variation40 should permit discovery of additional symptom QTLs, although larger samples may be required to overcome the necessary corrections for multiple testing.
Funding
Donald and Barbara Zucker Foundation; internal funding from the NSLIJHS; Stanley Foundation (to A.K.M.); National Alliance for Research on Schizophrenia and Depression (to A.K.M.), National Institutes of Health (MH065580 to T.L., MH074543 to J.M.K., MH001760 to A.K.M.).
Acknowledgments
Data presented in part at the Society of Biological Psychiatry 62nd Annual Scientific Convention and Meeting, San Diego, CA, in May 2007.
References
- 1.Craddock N, Owen MJ. Rethinking psychosis: the disadvantages of a dichotomous classification now outweigh the advantages. World Psychiatry. 2007;6:20–27. [PMC free article] [PubMed] [Google Scholar]
- 2.Carpenter WT. Deconstructing and reconstructing illness syndromes associated with psychosis. World Psychiatry. 2007;6:28–29. [PMC free article] [PubMed] [Google Scholar]
- 3.Anttila V, Kallela M, Oswell G, et al. Trait components provide tools to dissect the genetic susceptibility of migraine. Am J Hum Genet. 2006;79:85–99. doi: 10.1086/504814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamet P, Merlo E, Seda O, et al. Quantitative founder-effect analysis of French Canadian families identifies specific loci contributing to metabolic phenotypes of hypertension. Am J Hum Genet. 2005;76:815–832. doi: 10.1086/430133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Potash JB, Willour VL, Chiu YF, et al. The familial aggregation of psychotic symptoms in bipolar disorder pedigrees. Am J Psychiatry. 2001;158:1258–1264. doi: 10.1176/appi.ajp.158.8.1258. [DOI] [PubMed] [Google Scholar]
- 6.Potash JB, Zandi PP, Willour VL, et al. Suggestive linkage to chromosomal regions 13q31 and 22q12 in families with psychotic bipolar disorder. Am J Psychiatry. 2003;160:680–686. doi: 10.1176/appi.ajp.160.4.680. [DOI] [PubMed] [Google Scholar]
- 7.Potash JB, Toolan J, Steele J, et al. The bipolar disorder phenome database: a resource for genetic studies. Am J Psychiatry. 2007;164:1229–1237. doi: 10.1176/appi.ajp.2007.06122045. [DOI] [PubMed] [Google Scholar]
- 8.Carpenter WT, Jr, Buchanan RW, Kirkpatrick B, Tamminga C, Wood F. Strong inference, theory testing, and the neuroanatomy of schizophrenia. Arch Gen Psychiatry. 1993;50:825–831. doi: 10.1001/archpsyc.1993.01820220081009. [DOI] [PubMed] [Google Scholar]
- 9.Kendler KS, Karkowski-Shuman L, O'Neill FA, Straub RE, MacLean CJ, Walsh D. Resemblance of psychotic symptoms and syndromes in affected sibling pairs from the Irish Study of High-Density Schizophrenia Families: evidence for possible etiologic heterogeneity. Am J Psychiatry. 1997;154:191–198. doi: 10.1176/ajp.154.2.191. [DOI] [PubMed] [Google Scholar]
- 10.Loftus J, Delisi LE, Crow TJ. Factor structure and familiality of first-rank symptoms in sibling pairs with schizophrenia and schizoaffective disorder. Br J Psychiatry. 2000;177:15–19. doi: 10.1192/bjp.177.1.15. [DOI] [PubMed] [Google Scholar]
- 11.Cardno AG, Jones LA, Murphy KC, et al. Dimensions of psychosis in affected sibling pairs. Schizophr Bull. 1999;25:841–850. doi: 10.1093/oxfordjournals.schbul.a033423. [DOI] [PubMed] [Google Scholar]
- 12.Cardno AG, Sham PC, Murray RM, McGuffin P. Twin study of symptom dimensions in psychoses. Br J Psychiatry. 2001;179:39–45. doi: 10.1192/bjp.179.1.39. [DOI] [PubMed] [Google Scholar]
- 13.Hennah W, Varilo T, Kestila M, et al. Haplotype transmission analysis provides evidence of association for DISC1 to schizophrenia and suggests sex-dependent effects. Hum Mol Genet. 2003;12:3151–3159. doi: 10.1093/hmg/ddg341. [DOI] [PubMed] [Google Scholar]
- 14.DeRosse P, Hodgkinson CA, Lencz T, et al. Disrupted in Schizophrenia 1 (DISC1) genotype and positive symptoms in schizophrenia. Biol Psychiatry. 2007;61:1208–1210. doi: 10.1016/j.biopsych.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 15.Fanous AH, van den Oord EJ, Riley BP, et al. Relationship between a high-risk haplotype in the DTNBP1 (dysbindin) gene and clinical features of schizophrenia. Am J Psychiatry. 2005;162:1824–1832. doi: 10.1176/appi.ajp.162.10.1824. [DOI] [PubMed] [Google Scholar]
- 16.DeRosse P, Funke B, Burdick KE, et al. Dysbindin (DTNBP1) genotype and negative symptoms in schizophrenia. Am J Psychiatry. 2006;163:532–534. doi: 10.1176/appi.ajp.163.3.532. [DOI] [PubMed] [Google Scholar]
- 17.Tosato S, Ruggeri M, Bonetto C, et al. Association study of dysbindin gene with clinical and outcome measures in a representative cohort of Italian schizophrenic patients. Am J Med Genet B Neuropsychiatr Genet. 2007;144:647–659. doi: 10.1002/ajmg.b.30484. [DOI] [PubMed] [Google Scholar]
- 18.Fanous AH, Neale MC, Straub RE, et al. Clinical features of psychotic disorders and polymorphisms in HT2A, DRD2, DRD4, SLC6A3 (DAT1), and BDNF: a family based association study. Am J Med Genet B Neuropsychiatr Genet. 2004;125:69–78. doi: 10.1002/ajmg.b.20103. [DOI] [PubMed] [Google Scholar]
- 19.Cardno AG, Bowen T, Guy CA, et al. CAG repeat length in the hkCa3 gene and symptom dimensions in schizophrenia. Biol Psychiatry. 1999;45:1592–1596. doi: 10.1016/s0006-3223(99)00033-5. [DOI] [PubMed] [Google Scholar]
- 20.Serretti A, Lilli R, Lorenzi C, Lattuada E, Smeraldi E. DRD4 exon 3 variants associated with delusional symptomatology in major psychoses: a study on 2,011 affected subjects. Am J Med Genet. 2001;105:283–290. doi: 10.1002/ajmg.1321. [DOI] [PubMed] [Google Scholar]
- 21.Serretti A, Lattuada E, Lorenzi C, Lilli R, Smeraldi E. Dopamine receptor D2 Ser/Cys 311 variant is associated with delusion and disorganization symptomatology in major psychoses. Mol Psychiatry. 2000;5:270–274. doi: 10.1038/sj.mp.4000726. [DOI] [PubMed] [Google Scholar]
- 22.Zhang XY, Zhou DF, Zhang PY, Wei J. The CCK-A receptor gene possibly associated with positive symptoms of schizophrenia. Mol Psychiatry. 2000;5:239–240. doi: 10.1038/sj.mp.4000677. [DOI] [PubMed] [Google Scholar]
- 23.Malhotra AK, Goldman D, Mazzanti C, Clifton A, Breier A, Pickar D. A functional serotonin transporter (5-HTT) polymorphism is associated with psychosis in neuroleptic-free schizophrenics. Mol Psychiatry. 1998;3:328–332. doi: 10.1038/sj.mp.4000412. [DOI] [PubMed] [Google Scholar]
- 24.Fanous AH, Kendler KS. Genetic heterogeneity, modifier genes, and quantitative phenotypes in psychiatric illness: searching for a framework. Mol Psychiatry. 2005;10:6–13. doi: 10.1038/sj.mp.4001571. [DOI] [PubMed] [Google Scholar]
- 25.Brzustowicz LM, Honer WG, Chow EW, Hogan J, Hodgkinson K, Bassett AS. Use of a quantitative trait to map a locus associated with severity of positive symptoms in familial schizophrenia to chromosome 6p. Am J Hum Genet. 1997;61:1388–1396. doi: 10.1086/301623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kendler KS, Myers JM, O'Neill FA, et al. Clinical features of schizophrenia and linkage to chromosomes 5q, 6p, 8p, and 10p in the Irish Study of High-Density Schizophrenia Families. Am J Psychiatry. 2000;157:402–408. doi: 10.1176/appi.ajp.157.3.402. [DOI] [PubMed] [Google Scholar]
- 27.Wilcox MA, Faraone SV, Su J, Van Eerdewegh P, Tsuang MT. Genome scan of three quantitative traits in schizophrenia pedigrees. Biol Psychiatry. 2002;52:847–854. doi: 10.1016/s0006-3223(02)01465-8. [DOI] [PubMed] [Google Scholar]
- 28.Fanous AH, Neale MC, Webb BT, et al. Novel linkage to chromosome 20p using latent classes of psychotic illness in 270 Irish high-density families. Biol Psychiatry. 2008;64:121–127. doi: 10.1016/j.biopsych.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 29.First MB, Spitzer RL, Gibbon M, Williams JBW. Structured Clinical Interview for DSM-IV Axis I Disorders (SCID) New York, NY: American Psychiatric Press; 1997. [Google Scholar]
- 30.Levinson DF, Mowry BJ. Defining the schizophrenia spectrum: issues for genetic linkage studies. Schizophr Bull. 1991;17:491–514. doi: 10.1093/schbul/17.3.491. [DOI] [PubMed] [Google Scholar]
- 31.Lencz T, Morgan TV, Athanasiou M, et al. Converging evidence for a pseudoautosomal cytokine receptor gene locus in schizophrenia. Mol Psychiatry. 2007;12:572–580. doi: 10.1038/sj.mp.4001983. [DOI] [PubMed] [Google Scholar]
- 32.Sasaki T, Gilbert DM. The many faces of the origin recognition complex. Curr Opin Cell Biol. 2007;19:337–343. doi: 10.1016/j.ceb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 33.Pinto S, Quintana DG, Smith P, et al. Latheo encodes a subunit of the origin recognition complex and disrupts neuronal proliferation and adult olfactory memory when mutant. Neuron. 1999;23:45–54. doi: 10.1016/s0896-6273(00)80752-7. [DOI] [PubMed] [Google Scholar]
- 34.Rohrbough J, Pinto S, Mihalek RM, Tully T, Broadie K. Latheo, a Drosophila gene involved in learning, regulates functional synaptic plasticity. Neuron. 1999;23:55–70. doi: 10.1016/s0896-6273(00)80753-9. [DOI] [PubMed] [Google Scholar]
- 35.McGlashan TH. Is active psychosis neurotoxic? Schizophr Bull. 2006;32:609–613. doi: 10.1093/schbul/sbl032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shiratsuchi T, Nishimori H, Ichise H, Nakamura Y, Tokino T. Cloning and characterization of BAI2 and BAI3, novel genes homologous to brain-specific angiogenesis inhibitor 1 (BAI1) Cytogenet Cell Genet. 1997;79:103–108. doi: 10.1159/000134693. [DOI] [PubMed] [Google Scholar]
- 37.Kee HJ, Ahn KY, Choi KC, et al. Expression of brain-specific angiogenesis inhibitor 3 (BAI3) in normal brain and implications for BAI3 in ischemia-induced brain angiogenesis and malignant glioma. FEBS Lett. 2004;569:307–316. doi: 10.1016/j.febslet.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 38.Rapoport JL, Addington AM, Frangou S, Psych MR. The neurodevelopmental model of schizophrenia: update 2005. Mol Psychiatry. 2005;10:434–449. doi: 10.1038/sj.mp.4001642. [DOI] [PubMed] [Google Scholar]
- 39.Lencz T, Lambert C, DeRosse P, et al. Runs of homozygosity reveal highly penetrant recessive loci in schizophrenia. Proc Natl Acad Sci USA. 2007;104:19942–19947. doi: 10.1073/pnas.0710021104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walsh T, McClellan JM, McCarthy SE, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]