Abstract
Data for genome-wide association studies are being collected for a myriad of phenotypes. Many of these studies do not include control samples selected to reflect ancestry similar to the case samples. At the same time “control databases” are becoming available to be utilized as a common resource. These data are often being genotyped using a large-scale SNP array. Human populations exhibit complex structure which can lead to spurious associations if not properly handled. How to couple case and control databases effectively is a pressing question. We review available methods for modeling genetic ancestry based on the information gleaned from the SNP array. Methods for selecting control samples with genetic ancestry similar to the case samples are described.
Keywords: Eigenanalysis, Genome-wide association, Genetic ancestry, Genetic matching, Principal component analysis
Introduction
As the results of more genome-wide association studies are published it is becoming apparent that genetic effects are likely to be smaller than anticipated necessitating even bigger samples to attain good power. At the same time, numerous sources of pre-genotyped control subjects have become available. With limited resources for genotyping, more cases can be genotyped if control samples are already in hand.
How to use these control databases effectively is an open question. A number of problems could potentially arise. Standard epidemiological principles dictate that controls should be well matched to cases on both genetic ancestry and environmental exposure variables. Typically many more controls are available than cases. Can this matching be done effectively post hoc by selectively choosing a subset of the controls? Here we focus on the genetic aspects of this problem, discussing problems of population stratification and study design.
Case-control studies rely on the unrealistic assumption of population homogeneity. In the face of population heterogeneity and differential prevalence, spurious associations can arise. If cases and controls are matched based on detailed records of their ancestry, then this problem can be circumvented, for example [1]. Of course, ancestry data are typically not available or inaccurate. As a response to this problem two statistical approaches to controlling structure arose in the past decade. Genomic control corrects for stratification using an estimate of the inflation factor due to stratification [2]. Structured association uses the sampled genotypes to cluster subjects into more homogeneous subsets prior to analysis [3]. Structured association performs best when the population consists of a mixture of highly diverse subpopulations, or distinct admixed populations, for example, [4]. If the populations vary in subtle ways, such as a continental cline, it is generally not possible for structured association to detect any structure in the sample. Both of these approaches have shortcomings when applied to large samples with huge panels of SNPs. Genomic control exhibits diminishing power because the effect of stratification increases with sample size [5]. Structured association is computationally intensive and quickly becomes intractable for large numbers of SNPs.
Principal Component based approaches
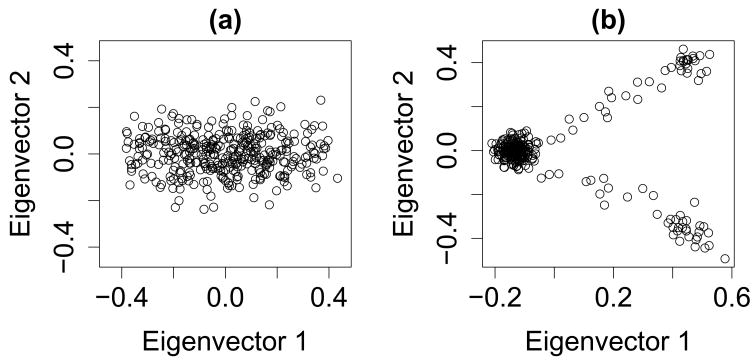
A third approach for population classification, based on principal component analysis [6], known as eigenanalysis, has recently been updated for association testing [7, 8, 9]. This method utilizes an eigenvector decomposition (EVD) to adjust for population structure. Cavalli-Sforza and colleagues [10] show that eigenvectors displayed in two dimensions often reflect the geographical distribution of populations. For populations that are geographically close, they found that genetic and geographic distances are often highly correlated. Consequently, the ancestry can be indirectly estimated based on the EVD. To illustrate the concept, Figure 1a displays simulated data based on a model with 9 subpopulations following a continental cline. The first eigenvector captures this cline and the remaining dimensions show little or no additional information.
Figure 1.
(a) Simulated data with nine subpopulations following a continental cline. One axis is required to capture the gradient structure. (b) Three subpopulations with admixture. The first two eigenvectors are needed to show the structure.
The EVD is obtained from allele counts derived from a sample of individuals measured at a large number of SNPs; see [7] for details. Results are more reliable if only tag SNPs [11] are included. Otherwise some axes of the decomposition will be defined by regions of high linkage disequilibrium. The EVD provides two quantities: the eigenvectors and the corresponding eigenvalues. The eigenvalues are ordered from largest to smallest. The larger the eigenvalue the more successful the eigenvector is in identifying the structure in the data. To determine the number of important dimensions empirically a test can be based on the magnitude of the standardized eigenvalues [8].
If the EVD approach is applied carefully, the eigenvectors can be interpreted as axes of ancestry. The number of dimensions required to capture the key features in the data vary, depending on the nature of the structure; however, some simple rules apply. If the sample consists of K distinct subpopulations, then K-1 axes will be required to differentiate these subpopulations. If a population has a gradient or cline, such as observed in Europe from the northwest to the southeast, then an axis is required for this feature. With admixed populations, such as African Americans, a single axis tends to separate the individuals based on their admixture proportions. Figure 1b displays simulated data from 3 subpopulations with some admixture. For this data, the number of significant dimensions is two. Generally, when data are collected for an association study, the sample is stratified by continental ancestry group before analysis. Consequently the remaining structure is subtle and the number of eigenvectors required to capture the key dimensions of ancestry are estimated empirically.
The Eigenstrat approach put forth by Price and colleagues [7] adjusts for estimated ancestry by regressing out the leading eigenvectors before testing for association between SNPs and phenotypes. Alternatively the effects of ancestry could be removed by genetic matching. This idea was explored in [12] and developed fully in [13]. Cases and controls need not be matched on a SNP by SNP basis. The EVD offers a convenient metric for genetic matching, followed by logistic regression conditional on the matched strata. This approach is implemented in GEM [13]. The distance between individuals can be calculated as the Euclidean distance defined by the leading D eigenvectors, scaled by the eigenvalues. This distance is equivalent to metric multidimensional scaling and can be shown to be optimal [14].
Although “pair matching” is naturally appealing, an alternative form of matching called “full matching” has notable advantages [15]. With the latter approach each strata consists of one case paired with one or more controls or one control paired with one or more cases. When case and control subjects have different distributions of ancestry, this form of matching permits the inclusion of more subjects without including poor matches.
As the sample size grows, the effect of structure grows so that even subtle population structure can lead to spurious associations [5]. At the same time, subtle structure can be difficult to detect using EVD unless a sufficient amount of information is available in the matrix of allele counts. The usual parameter used to express the variability in allele frequencies between subpopulations is Fst. For a world population this quantity is large, say (0.1 to 0.15), but for a continental population it is much smaller [10]. Within a continent, the amount of variability varies depending on the history of the populations. In Europe Fst is estimated to be only 0.006, while AmerIndians have a much higher level of diversity, typically assessed at 0.04 or greater [10]. Patterson and colleagues [8] studied the sensitivity of EVD to detecting the presence of two subpopulations as a function of Fst and found that structure is detectable in the first eigenvalue with high probability provided the product of the sample size and the number of tag SNPs is greater than 1/Fst 2. Fortunately, the amount of information for detecting structure is increased by either sampling more people or more SNPs.
Another way to increase the chance of detecting subtle structure via EVD is to choose SNPs that have been under differential selective pressures. An example is the lactase variant, which facilitates the digestion of milk in adults. This variant is present in 96% of individuals from northwestern Europe, but only 36% of those from southeastern Europe [16]. SNPs under selective pressure tend to be much more informative about clinal variation than randomly selected SNPs. A large number of SNPs have been identified that successfully differentiate between European and African ancestry [17]; these are labeled “ancestry informative markers”.
A similar way to control for population substructure is based on propensity scores [15]. For genetic association studies this quantity is obtained by modeling the odds of disease given a panel of genetic markers [18]. Cases and controls can be clustered into a handful of strata based on having similar scores. The data can then be tested for association, conditional on these strata. This approach uses both case/control status and the panel of genetic markers to stratify the subjects. In contrast, EVD uses only the structure apparent in the genetic SNP panel to stratify subjects.
Selecting a suitable sample
With a sufficient number of markers, the EVD is remarkably effective in capturing subtle structure in the data. For example, Luca and colleagues [13], combined the PopGen sample of 500 northern Germans [19] and the KORA sample of 1644 southern Germans [20]. Ignoring labels these populations show remarkable differentiation between samples using only two eigenvectors, but this subtle structure is obscured if the samples include outliers (individuals with notably different ancestry). Price and colleagues [7] define outliers as those individuals differing from the mean by at least 6 standard deviations in at least one key dimension of the EVD. In the German samples, Luca and colleagues removed 53 individuals with ancestry that were determined to be clearly outlying. Many of these were cryptic relative pairs. Before removing these outliers, 22 dimensions were required to illustrate key axes of variation; in each of these axes, a handful of individuals are highlighted who differ markedly from the rest of the sample.
Even after removing these gross outliers, some of the individuals may still be outliers in a more subtle sense. Some regions of the space may be occupied exclusively by controls (or cases). In this instance, the sample can not be successfully matched. If the control sample is chosen by convenience this mismatch of case and control samples is quite likely to occur and can lead to spurious associations.
Individuals who cannot be successfully matched should not be included in association analyses because it leads to an excess of spurious findings. In addition a more heterogeneous sample often leads to reduced power [13]. The question remains: if a large number of well-matched controls are available, is there an advantage in including them? The power of the test will be increased if the variance is reduced; however, variance is largely determined by the size of the smaller sample. For instance, in a standard case-control analysis with 100 cases, 1000 controls is better than 100 controls, but 10,000 controls is not measurably better than 1000. Hence there is a limit to how much a large control sample can help. If the samples are not well matched, it can hurt.
Examples
Height and a Lactase variant
With population stratification the most challenging problem occurs when some candidate SNPs are highly differentiated, but the majority of SNPs in the panel have similar allele frequencies across subpopulations. This situation arises when a SNP is under strong population specific selection and is aggravated if the phenotype is also differentially distributed. If the allele frequency cline matches the phenotype cline it maximizes the opportunity for confounding. The data in [1] provides an example of this scenario: the phenotype is height (short vs. tall) and the genotype is a lactase variant. These data include only a modest number of SNPs for calculating the EVD: 111 missense and noncoding SNPs and 67 ancestry informative markers. Ignoring the inherent structure in these data, one obtains a significant association between the lactase variant and height (p-value = 0.0037). Neither genomic control nor structured association were able to correct for this spurious association [7].
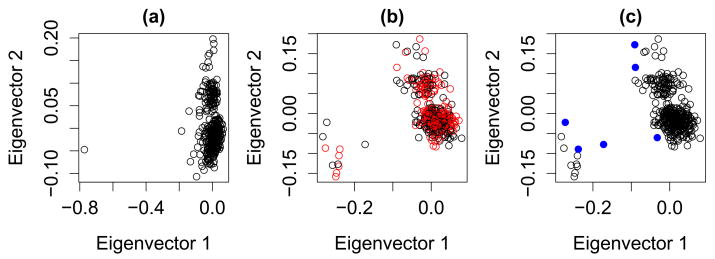
EVD applied to the 67 ancestry informative markers (excluding the two lactase SNPs) reveals one outlier (Fig. 2a) and three significant axes of variation (Fig. 2a). After removal of this outlier there are two significant axes (Fig. 2b). It is worth noting that these axes do not separate the observations in a way that reveals a gradient in height. GEM detects 6 observations that appear to be unmatchable (4 short and 2 tall; Fig. 2c). After removing these 6 observations only one axis of variation continues to be significant. Applying the regression and matching procedures to the remaining sample we find that neither method was fully successful in removing the signal; however, matching reduces the signal by a full order of magnitude (Table 1). The theory supporting this analysis assumes that the number of SNPs is much larger than the number of samples [21]. This assumption doesn’t hold in this application. Moreover, based on the calculations in Patterson and colleagues [8] our panel of SNPs is not likely to yield sufficient information to fully remove subtle structure from a European sample. Presumably results would improve with a larger number of SNPs.
Figure 2.
(a) EVD applied to AIM markers reveals one outlier. (b) First two eigenvectors for AIM markers after removing the outlier; short (red) and tall (black) appear uniformly spread across the clusters. (c) GEM detects 6 unmatchable observations (blue).
Table 1.
P-values for the height/lactase association example, using logistic regression, Eigenstrat, and GEM. EVD are calculated using ancestry informative markers, before and after removing the unmatchables. Results are for the LCT -13910C → T (rs4988235) variant. Outliers and unmatchable observations were identified using GEM. Because the sample size is small we used a smaller critical value to determine significance of ancestry dimensions than the default (α=0.05).
| With unmatchables | Without unmatchables | |
|---|---|---|
| Logistic | .0032 | .0034 |
| Eigenstrat | .0033 | .0033 |
| GEM | .0028 | .0200 |
A propensity score approach also achieved partial success with these data. The scores were used to define five strata. Unlike the EVD, the propensity strata show a strong gradient in the height distribution. The first strata includes 4 tall and 78 short individuals, while the last strata includes 71 tall and 3 short individuals. Next the data were analyzed using the Mantel-Haenzel test and the stratified logistic regression test with resulting p-values of 0.039 and 0.44, respectively. Although these tests usually perform similarly, the former failed to fully correct for the spurious association even with the benefit of the propensity strata. In practice we have found that this method fails to scale to large panels of SNPs.
Type I diabetes (T1D)
As mentioned in the previous subsection, a genome wide association study (Affymetrix 500K GeneChip) was conducted on a sample of 416 T1D cases of European descent and 2144 German controls [13]. To reconstruct ancestry, 23,552 nearly independent tag SNPs were used. Outlying cases and controls were removed, based on the six standard deviation rule [7]. Applying Eigenstrat to the remaining sample, 25 SNPs had p-values under 10−15. Although some of these p-values correspond to SNPs in the HLA region, most are false positives.
Using GEM’s criterion 13.2% of the cases and 4.5% of the controls were determine to be unmatchable and removed from the sample to obtain homogeneous strata. On this sample, GEM found 9 SNPs with p-values under 10−15. Closer inspection revealed that most small p-values were due to poor genotyping calls (except those in the HLA region).
To facilitate further comparison of the two methods, Eigenstrat was applied to the data after removal of the unmatchable samples. Results of GEM and Eigenstrat were compared for those SNPs with p-values <10−4 and good genotype calls. GEM’s false positive rate was nearly on target, but Eigenstrat’s was more than twice the desired level.
Only two principal axes are required to model ancestry of these data, after removal of outliers and unmatchable observations. A strong continental gradient differentiates the northern and southern German samples [13]. Surprisingly few cases were unmatchable considering that the controls were all sampled from Germany.
Concluding Remarks
In our experience a large panel of SNPs reveals subtle structure that can be controlled by matching or regression, provided outlying and unmatchable individuals are removed prior to analysis. Recent literature suggests that a careful selection of a moderate number of ancestry informative loci can achieve the same purpose ([22],[23]). It is not surprising that none of the available methods fully succeeded in removing the effects of the cline in Europe on the lactase/height data. Only 67 ancestry informative loci were available, and these were not chosen to optimally reveal the European gradient.
If a large panel of SNPs is available it is advisable to choose the SNP panel from this collection, rather than relying exclusively on a subset of ancestry informative SNPs. In the lactase/height data we used only the ancestry informative SNPs because so few other SNPs were measured. The available SNPs must be pruned to remove SNPs in strong linkage disequilibrium; otherwise the principal component analysis will discover spurious axes that differentiate the subjects based on their genotype in a single region. A rule of thumb that seems to work is to include no pair of SNPs with r2>.04.
Eigenstrat and GEM represent two choices for modeling population substructure using estimated axes of ancestry. Following the classic epidemiological approaches for removing the effects of confounding in observational studies Eigenstrat uses regression and the GEM uses matching. When the ancestry space is not sparse the regression approach tends to be slightly more powerful [13]. In contrast, when small clusters of observations are separated from the bulk of the sample, the matching approach is better able to remove the confounding effects. An open question is how to quantify sparseness of the ancestry space.
In general, EVD is not robust to outliers. Gross outliers in the ancestry space are easily identifiable. Subtle outliers that arise due to a disparate distribution of cases and controls are more difficult to identify (see Fig. 3). An algorithm for identifying these unmatchable observations is described in [13] and implemented in GEM. This algorithm involves clustering and rescaling the data so that distances are interpretable. It works well in practice, but as is typical of clustering problems, the process requires a user specified choice for the minimum cluster size. Further refinement of this algorithm is desirable.
The matching approach has clear advantages over the regression approach for formulating rules for identifying subtle outliers and unmatchable observations. A powerful hybrid approach for modeling structure which involves (i) using a matching criterion to identify unmatchable observations, and (ii) using a regression approach to remove the remaining effects of structure, is worth considering. For data that are not sparse in the ancestry space, this approach would incorporate the best features of Eigenstrat and GEM.
Even after controlling for stratification, typically a slight excess of false positives remains [24]. This could be due to subtle structure that is not successfully removed by statistical modeling, but in our experience this problem arises from genotype calling problems not detected in the initial quality control measures. When using samples of convenience we believe there are additional chances for quality control issues to arise. If control genotypes are obtained from a repository, then laboratory effects and phenotypes are completely confounded. Thus we conclude that laboratory issues may be a concern when controls are obtained from samples of convenience.
Nearly all investigations of the confounding effect of population structure on association studies focus on investigations of common variants. Very little is known about the potential for confounding that may exist for studies of rare variants. This is another question that merits further study.
Acknowledgments
This work was funded by NIH (grant MH057881). We thank Bernie Devlin and Bert Klei for helpful comments and discussion, and Xihong Lin for organizing the “Conference on Design and Analysis Issues in Genomic Studies in Population Sciences” which was the inspiration for this minireview,.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Campbell CD, Ogburn EL, Lunetta KL, Lyon HN, Freedman ML, Groop LC, Altshuler D, Ardlie KG, Hirschhorn JN. Demonstrating stratification in a European American population. Nat Genet. 2005;37:868–872. doi: 10.1038/ng1607. [DOI] [PubMed] [Google Scholar]
- 2.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 3.Pritchard J, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenberg NA, Mahajan S, Ramachandran S, Zhao C, Pritchard JK. Clines, clusters and effect of study design on the inference of human population structure. PLoS Genet. 2005;1:e70. doi: 10.1371/journal.pgen.0010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devlin B, Roeder K, Wasserman L. Genomic Control. a New Approach to Genetic-based Association Studies. Theor Popul Biol. 2001;60:156–166. doi: 10.1006/tpbi.2001.1542. [DOI] [PubMed] [Google Scholar]
- 6.Jolliffe IT. Principal Component Analysis. Springer; NY: 2002. [Google Scholar]
- 7.Price AL, Patterson N, Plenge R, Weinblatt M, Shadick N, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 8.Patterson N, Price AL, Reich D. Population Structure and Eigenanalysis. PLoS Genet. 2006;12:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang S, Zhu X, Zhao H. On a semiparametric test to detect associations between quantitative traits and candidate genes using unrelated individuals. Genet Epidemiol. 2003;24:44–56. doi: 10.1002/gepi.10196. [DOI] [PubMed] [Google Scholar]
- 10.Cavalli-Sforza L, Menozzi P, Piazza A. The history and geography of human genes. Princeton University Press; 1994. [Google Scholar]
- 11.Rinaldo A, Bacanu SA, Devlin B, Sonpar V, Wasserman L, Roeder K. Characterization of Multilocus Linkage Disequilibrium. Genet Epidemiol. 2005;28:193–206. doi: 10.1002/gepi.20056. [DOI] [PubMed] [Google Scholar]
- 12.Hinds DA, Stokowski RP, Patil N, Konvicka K, Kershenobich D, Cox DR, Ballinger DG. Matching strategies for genetic association studies in structured populations. Am J Hum Genet. 2004;74:317–25. doi: 10.1086/381716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luca D, Ringquist S, Klei L, Lee AB, Gieger C, Wichmann HE, Schreiber S, Krawczak M, Lu Y, Styche A, Devlin B, Roeder K, Trucco M. On the Use of General Control Samples for Genome-Wide Association Studies: genetic matching highlights causal variants. Am J Hum Genet. 2008 doi: 10.1016/j.ajhg.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mardia KV, Kent JT, Bibby JM. Multivariate Analysis. London: Academic Press; 1979. [Google Scholar]
- 15.Rosenbaum PR. Observational Studies. New York NY: Springer-Verlag; 1995. [Google Scholar]
- 16.Bersaglieri T, et al. Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Gen. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenberg NA, Lei LM, Ward R, Pritchard JK. Informativeness of Genetic Markers for Inference of Ancestry. Am J Hum Genet. 2003;73:1402 – 1422. doi: 10.1086/380416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Epstein MP, Allen AS, Satten GA. A simple and improved correction for population stratification in case-control studies. Am J Hum Genet. 2007;80:921–930. doi: 10.1086/516842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krawczak M, Nikolaus S, von Eberstein H, Croucher PJ, El Mokhtari NE, Schreiber S. PopGen: population-based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genetics. 2006;9:55–61. doi: 10.1159/000090694. [DOI] [PubMed] [Google Scholar]
- 20.Wichmann H-E, Gieger C, Illig T MONICA/KORA Study Group. KORA-gen–resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen. 2006;67:S26–S30. doi: 10.1055/s-2005-858226. [DOI] [PubMed] [Google Scholar]
- 21.Johnstone I. On the distribution of the largest eigenvalue in principal components analysis. Annals of statistics. 2001;29:295–327. [Google Scholar]
- 22.Price AL, Butler J, Patterson N, Capelli C, Pascali VL, Scarnicci F, Ruiz-Linares A, Groop L, Saetta AA, Korkolopoulou P, Seligsohn U, Waliszewska A, Schirmer C, Ardlie K, Ramos A, Nemesh J, Arbeitman L, Goldstein DB, Reich D, Hirschhorn JN. Discerning the ancestry of European Americans in genetic association studies. PLoS Genet. 2008 Jan;4(1):e236. doi: 10.1371/journal.pgen.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian C, Plenge RM, Ransom M, Lee A, Villoslada P, Selmi C, Klareskog L, Pulver AE, Qi L, Gregersen PK, Seldin MF. Analysis and application of European genetic substructure using 300 K SNP information. PLoS Genet. 2008 Jan;4(1):e4. doi: 10.1371/journal.pgen.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]