Abstract
O6-Alkylguanine-DNA alkyltransferase (AGT) plays an important role protecting cells from alkylating agents. This reduces carcinogenesis and mutagenesis initiated by such agents but AGT also provides a major resistance mechanism to some chemotherapeutic drugs. In order to improve understanding of the AGT-mediated repair reaction and to increase understanding of the spectrum of repairable damage, we have studied the ability of AGT to repair interstrand cross-link DNA damage where the two DNA strands are joined via the guanine-O6 in each strand. An oligodeoxyribonucleotide containing a heptane cross-link was repaired with initial formation of an AGT-oligo complex and further reaction of a second AGT molecule yielding a hAGT dimer and free oligo. However, an oligodeoxyribonucleotide with a butane cross-link was a very poor substrate for AGT-mediated repair and only the first reaction to form an AGT-oligo complex could be detected. Models of the reaction of these substrates in the AGT active site show that the DNA duplex is forced apart locally to repair the first guanine. This reaction is greatly hindered with the butane cross-link, which is mostly buried in the active site pocket and limited in conformational flexibility. This limitation also prevents the adoption of a conformation for the second reaction to repair the AGT-oligo complex. These results are consistent with the postulated mechanism of AGT repair that involves DNA binding and flipping of the substrate nucleotide and indicate that hAGT can repair some types of interstrand cross-link damages.
O6-Alkylguanine-DNA alkyltransferase (AGT1) is a widely distributed DNA repair protein to maintain genomic integrity (1–4). It acts on adducts at the O6-position of guanine and transfers the alkyl group to an internal Cys residue restoring the DNA in a single step. The S-alkyl-Cys formed is not regenerated and the protein can act only once. Human O6-alkylguanine-DNA alkyltransferase (hAGT1) can repair not only methyl groups but longer alkyl groups including ethyl-, 2-chloroethyl-, and butyl and more bulky cyclic adducts such as benzyl- and pyridyloxobutyl- (5–7). In addition to beneficial effects of protecting from carcinogens and mutagens such as N-methyl-N'-nitro-N-nitrosoguanidine, dimethylnitrosamine, N-ethyl-N-nitrosourea, methylbenzylnitrosamine and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, hAGT has the deleterious effect of providing resistance to killing of cancer cells by therapeutic alkylating drugs such as the methylating agent temozolomide and the chloroethylating agent 1,3-bis(2-chloroethyl)-1-nitrosourea (8).
The resistance to such chloroethylating agents is caused by the efficient repair of O6-chloroethylguanine adducts. This prevents the formation of the primary toxic lesion, which is a 1-(3-cytosinyl)-2-(1-guanyl)-ethane interstrand cross-link (9, 10). This is formed spontaneously in a two step process from the O6-(2-chloroethyl)guanine that undergoes an internal cyclization to form 1, O6-ethanoguanine which, in turn, reacts with the opposite strand cytosine to yield the interstrand cross-link (11, 12). The final cross-link does not involve the O6-position of guanine and is unaffected by AGT. However, it was shown that the intermediate 1, O6-ethanoguanine was a substrate for AGT and its repair leads to a DNA-protein cross-link in which the AGT is joined by an ethano linkage to N1 of guanine (13, 14). Structural and biochemical studies of the repair reaction brought about by hAGT have provided a plausible preliminary model for understanding the mechanism of DNA repair (4, 15–19). Available crystal structures for human hAGT include those with the inactive C145s mutant bound to DNA containing O6-methylguanine (16), wild type AGT crosslinked to the mechanistic inhibitor N1, O6-ethanoxanthosine (16) and wild type protein bound to a DNA containing a modified cytosine residue (17). These structures indicate that hAGT binds substrate DNA via the minor groove using a helix-turn-helix motif. The alkylated guanine deoxynucleotide is flipped out from the base stack into the AGT active site pocket via a 3′ phosphate rotation, which is promoted by Tyr114, either by steric (16) or by electrostatic effects (19), and stabilized by an Arg finger (Arg128). An asparagine hinge formed by Asn137 couples the helix-turn-helix DNA binding and active site motifs. The extruded O6-alkylguanine is positioned for repair in a hydrophobic cleft made up of the Met134 side chain and Val155-Gly160 of the active site loop. Cys145 and Val148 carbonyls accept hydrogen bonds from guanine's exocyclic amine and Tyr114 hydroxyl and Ser159 N atoms donate hydrogen bonds to guanine's N3 and O6 atoms, respectively. These interactions position the alkyl group for attack by Cys145, which has a very high reactivity (20) due to its activation to a thiolate anion by a Glu172-His146-water-Cy145 hydrogen bond network (4, 15). The reaction may also be facilitated by reduction of the negative charge on the repaired guanine via the hydrogen bond from Tyr114. The description given above (and amino acid numbering) relates to the hAGT but it is likely to be universally applicable since all of the key residues mentioned above are highly conserved and the structures of three other AGTs from the bacterium Escherichia coli (Ada-C) (21), the archaebacterium Pyrococcus kodakaraensis (22), and the thermophile Methanococcus jannaschii (23) are generally similar particularly in the active site and DNA binding domains. Further details of the mechanism by which lesions are identified and the DNA structure is altered to allow repair by AGTs remain to be identified.
In the experiments presented here, we have examined the repair of DNA interstrand cross-links formed between the O6-position of two guanine residues. These experiments had two aims. Firstly, to establish whether the repair of such cross-links that may be formed by bifunctional agents such as 1,4-butanediol dimethanesulfonate (busulfan1) and 1,7-heptanediol dimethanesulfonate (hepsulfam1), which are developmental anticancer agents, (24, 25), could influence response to these drugs and secondly to obtain more information on the AGT-mediated repair of structurally-restricted substrates. Studies were carried out with oligodeoxyribonucleotide (oligo1) substrates containing alkyl cross-links with 7 carbons and 4 carbons and using wild type hAGT and mutants at key active site residues and with E. coli AGTs. It was found that the human but not E. coli AGTs were competent to repair the longer cross-link but that the shorter cross-link was highly resistant to repair. Modelling studies based on the crystal structure of hAGT and Ada-C provide a plausible explanation for these results and further support was generated by the result of studies with mutants of hAGT at positions Arg128 and Tyr114.
EXPERIMENTAL PROCEDURES
Materials
All the control unmodified oligos were synthesized and purified by the Macromolecular Core Facility, Hershey Medical Center. Ampicillin, isopropyl β-D-thiogalactopyranoside (IPTG1), hemocyanin, calf thymus DNA, and most other biochemical reagents were purchased from Sigma Chemical Co. (St. Louis, MO). Talon Metal Affinity IMAC Resin was obtained from BD Bioscience Clontech (Palo Alto, CA), liquid acrylamide/bisacrylamide (37.5:1) and urea were purchased from Roche Diagnostic Corporation (Indianapolis, IN). Nitrocellulose filters (0.45 µm) were obtained from Millipore (USA). PVDF membrane was purchased from Pall Life Sciences (Pensacola, FL). T4 polynucleotide kinase (PNK1) was purchased from New England Biolabs (Beverly, MA), [γ-35S]ATPγS was purchased from Amersham (Piscataway, NJ). Sequence grade modified trypsin was obtained from Promega (Madison, WI). Tobacco etch virus (Tev) protease (26) was generously provided by Dr. J. M. Flanagan (Penn State Univ, Dept Biochem & Mol Biol). Penta His antibody was obtained from Qiagen (Chatsworth, CA). HRP linked anti-mouse IgG, anti-rabbit IgG and lumiGLO reagent were purchased from Cell signal technology (Danvers, MA).
Cross-linked Oligonucleotide Preparation and Purification
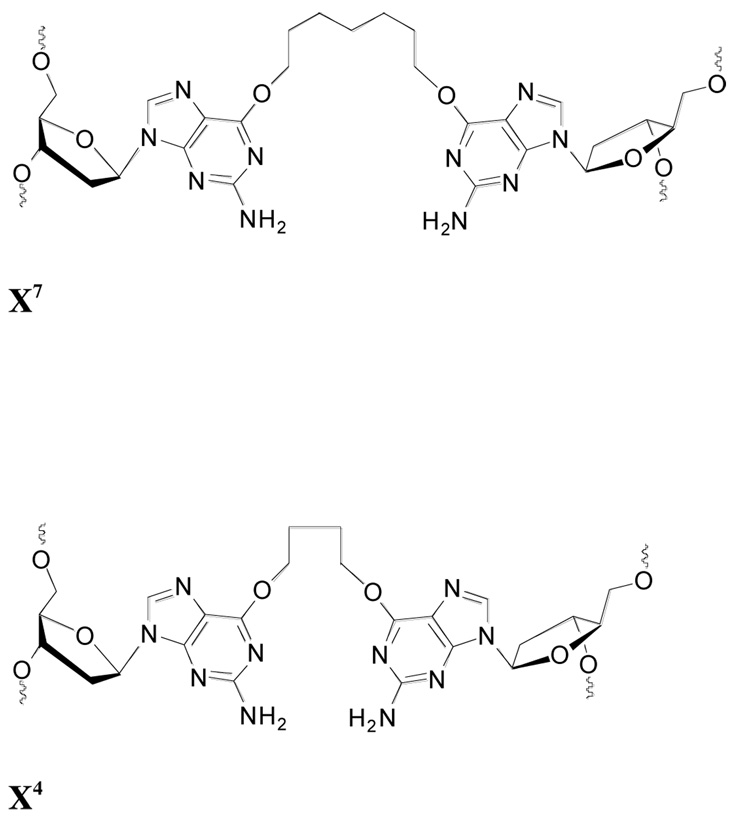
The oligos used are shown in Table 1 and the structure of the cross-link is shown in Figure 1. Studies were carried out with double stranded 10-base-pair (BP1) substrate (CJW 81), 11-BP substrates (CJW80 and CJW 119) and the 17-BP CJW98. The CJW119 substrate contained a butane-cross-link and the CJW80, CJW 81 and CJW98 substrates contained a heptane-cross-link. The linked guanine residues were opposite one another in all substrates except CJW81 where the linked guanines were paired with a cytosine in the opposite strand.
Table 1.
The sequence and structure of oligodeoxyribonucleotides used
| Oligodeoxyribonucleotide | Nucleotide Sequence |
|---|---|
| CJW80 | 5′-CGAAAX7TTTCG-3′ |
| 3′-GCTTTX7AAAGC-5′ | |
| 80-C | 5-CGAAAGTTTCG-3′ |
| CJW81 | 5′-CGAAX7CTTCG-3′ |
| 3′-GCTTCX7AAGC-5′ | |
| 81-C | 5′-CGAAGCTTCG-3′ |
| CJW98 | 5′-CGACTAAAX7TTTAGTCG-3′ |
| 3′-GCTGATTTX7AAATCAGC-5′ | |
| 98-C | 5′-CGACTAAAGTTTAGTCG-3′ |
| CJW119 | 5′-CGAAAX4TTTCG-3′ |
| 3′-GCTTTX4AAAGC-5′ |
The structures of X7 and X4 are shown in Figure 1
Figure 1. Structure of cross-links.
Cross-linked phosphoramidite dimers containing butyl and heptyl linkers were synthesized and the oligo substrates prepared by solid phase synthesis using an ABI 3400 oligonucleotide synthesizer and purified as described below. 5′-O-Dimethoxytrityldeoxyribonucleoside-3′-O-(β-cyanoethyl-N,N'-diisopropyl) phosphoramidites and controlled pore glass (CPG) supports (500 Å) derivatized with protected 2′-deoxynucleosides were purchased from ChemGenes Inc. (Wilmington, MA). Bis-3′-O-phosphoramidites containing 4 and 7 carbon linkers between the O6 atoms of two 2′-deoxyguanosines were synthesized from N2-phenoxyacetyl-2′-deoxyguanosine according to published procedures (27). The cross-linked duplexes CJW 80, 81, 98 and 119 were assembled on an Applied Biosystems Model 3400 synthesizer on a 1 µmole synthesis scale using standard β-cyanoethylphosphoramidite chemistry. The nucleoside phosphoramidites were dissolved in anhydrous acetonitrile at a concentration of 0.1 M for the 3’-O-deoxyphosphoramidites and 0.05 M for the cross-linked bis-3’-O-deoxyphosphoramidites. Assembly of the sequences was carried out as follows: (a) detritylation: 3% trichloroacetic acid in dichloromethane; (b) nucleoside phosphoramidite coupling of 2 minutes for commercial 3′-O-deoxyphosphoramidites and 30 minutes for the cross-linked bis-phosphoramidites; (c) capping: phenoxyacetic anhydride/pyridine/tetrahydrofuran 1:1:8 (v/v/v; solution A) and 1-methyl-1 H-imidazole/tetrahydrofuran 16:84 (w/v; solution B); (d) oxidation: 0.02 M iodine in tetrahydrofuran/water/pyridine 2.5:2:1. The 5′-O-terminal trityl groups were removed on the synthesizer.
The oligomer-derivatized supports were transferred to screw cap microfuge tubes lined with teflon caps and the protecting groups removed with concentrated ammonium hydroxide: ethanol (0.3mL: 0.1mL) treatment for 4 h at 55°C. The oligomers were then purified by SAX HPLC using a Dionex DNAPAC PA-100 column (0.4 cm × 25 cm, Dionex Corp, Sunnyvale, CA.) with a linear gradient of 0–50% of buffer B over 30 minutes (where buffer A: 0.1 M Tris HCl, pH 7.5, 10% acetonitrile and buffer B: 0.1 M Tris HCl, pH 7.5, 1 M NaCl, 10% acetonitrile) at 30°C. The detector was set at 260 nm for analytical runs and 290 nm for preparative runs. The purified oligomers were desalted using C-18 SEP PAK cartridges (Waters Inc.).
The cross-linked oligomers (0.1 A260 units) were characterized by digestion with a combination of snake venom phosphodiesterase (0.28 units) and calf intestinal phosphatase (5 units) in a buffer containing 10 mM Tris (pH 8.1) and 2 mM magnesium chloride for 16 hours at 37°C. The resulting mixture of nucleosides was analyzed by reversed phase HPLC carried out using a Symmetry® C-18 5µm column (0.46 × 15 cm) purchased from Waters Inc, Milford, MA. The C-18 column was eluted with a linear gradient of 0–60% buffer B over 30 minutes (buffer A: 50 mM sodium phosphate, pH 5.8 and buffer B: 50 mM sodium phosphate, pH 5.8, 50% acetonitrile). The resulting peaks were identified by co-injection with the corresponding standards and the ratio of nucleosides was determined. The molecular weights of the cross-linked oligomers were determined by ESI MS and these were in agreement with the calculated values.
Protein Purification
Plasmids for the production of: N-terminal His6-tagged hAGT and the N-terminal His6-tagged hAGT mutants R128K, R128A, R128G, C145A, P140K, Y158H, G160R and E. coli Ogt; the C-terminal His6-tagged hAGT and the C-terminal His6-tagged hAGT mutants Y114F, Y114A, C145S, E. coli Ada-C; and the Tev-hAGT with a Tev protease cleavable His6 tag were prepared as described previously (28–32). Studies with the wild type hAGT showed that the presence or positioning of the His6-tag did not affect the results. Proteins were expressed in XL-1 blue cells and purified using Talon IMAC resin. In order to remove the His6 tag from Tev-hAGT, the protein was dialyzed in 50mM Tris-HCl, pH 7.6, 1 mM dithiothreitol (DTT1) and 0.1 mM EDTA, and then concentrated to <1.0 ml with Centrion Ultracel YM-10 of Millipore corporation (Bedford, MA) before digestion by Tev protease (1:20 ratio) as described (26, 32). The purified protein was analyzed by SDS-PAGE on 12% gels.
Alkyltransferase Activity Assays
The purified AGT proteins were assayed by measuring the ability of the protein to transfer methyl groups from a [3H]-methylated DNA substrate as previously described (29, 32). The protein was incubated with 10 µg of [3H]-methylated DNA substrate in 1.0 ml of 50 mM Tris-HCl buffer (pH 7.6) containing 5 mM DTT, 0.1 mM EDTA (AGT buffer) and 50 µg of hemocyanin for 2 h at 37 °C. 0–2.5 pmol hAGT, Ogt, Ada-C, G160R or R128K, 0–9 pmol Y114F, P140K or Y158H, 0–700 pmol R128A or R128G or Y114A was used in the assay. Nitrocellulose filters were used to collect the formed [3H]-methylated protein and counted in scintillation liquid. At the counting efficiency used, 1 fmol of transferred methyl groups was equal to 20 cpm.
Polyacrylamide Gel Electrophoresis Assay of O6-G-alkyl-O6-G Interstrand Cross-link Repair
The cross-link-containing oligos, CJW80, CJW81, CJW98 and CJW119 and corresponding controls were labeled at the 5′-end with [γ-35S]ATPγS. Labeling was carried out in 50 µL reaction buffer composed of 1 × T4 PNK buffer (New England Biolabs), 4 µL [γ-35S]ATPγS (10µCi/µL), 10 units T4 PNK and 500 pmol oligo. The mixture was incubated at 37°C for 1 h, then the reaction stopped by heating at 65°C for 20 min. The labeled oligos were passed through a Sephadex MicroSpin G-25 column to remove unincorporated [γ-35S]ATPγS. The purified and labeled oligos were heated to 100°C and allowed to cool to room temperature.
The repair of these substrates was studied by incubation with the AGT protein as indicated (0–60 pmol) in 15 µL AGT buffer with the labeled substrate oligo (2–4 pmol) for up to 2 h at 37°C. The product of the reaction were then analyzed either by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE1) or by denaturing-PAGE in 7M urea (33). For analysis by SDS-PAGE the reaction was terminated by the addition of 10 µL loading buffer (containing 125 mM Tris-HCl (pH 7.6), 5% SDS, and 25% glycerol (v/v)) and the samples loaded on 20% mini-SDS-PAGE gels with 0.125 M Tris-HCl buffer containing 0.1% SDS and run for 60–80 min with voltage 100–120v. The gels were fixed, dried and quantified on a Molecular Dynamics PhosphorImager SI system (Molecular Dynamics, Mountain View, CA) using the program ImageQuant (Amersham Biosciences). The band of each lane on the gel representing different products or substrate was quantitated and calculated using the control band as reference. A control of AGT covalently linked to an oligo by reaction with 1,2-dibromoethane (DBE1) was prepared by incubation of 60 pmol hAGT and 4 pmol oligo with 300 pmol of DBE in 15 µL for 2 h (34) and loaded on to the gels.
For analysis by denaturing-PAGE (33), the samples were prepared as described above but the reaction was stopped by the addition of 1.6 µL 10% SDS and 16.6 µL loading buffer containing 80% formamide in 1×TBE (89 mM Tris-HCl, 89 mM boric acid and 2 mM EDTA, pH 8.0) buffer. Samples were heated 5 min in boiling water and loaded on to 17% 18 cm × 16 cm polyacrylamide gels with 7M urea in 1×TBE buffer and run 3–4 h with 200–250v. The gels were treated and the products were quantitated as described above.
Detection of the DNA-AGT(peptide) Cross-link Product by Mass Spectrometry (MS)
hAGT (400–600 pmol, N-his tagged or C-his tagged) was incubated with CJW98 (200 pmol) in 15 µL AGT reaction buffer for 3 h at 37°C. Reactions were terminated by the addition of 10 µL loading buffer (containing 125 mM Tris-HCl pH 7.6, 2% SDS and 25% glycerol). Samples were subsequently heated at 100°C for 5 min and separated on 20% mini-SDS-PAGE gels with 0.125M Tris-HCl, 0.1% SDS and run for 60–80 min with a voltage of 100–120v. Gel slices containing the Coomassie blue-stained bands corresponding to the hAGT bound to the oligo were excised, washed with 50% acetonitrile containing 0.1% trifluoroacetic acid (TFA1), destained twice with 200 µl of 100 mM ammonium bicarbonate pH 8.0, 50% acetonitrile for 45min at 37°C and dried in a SpeedVac. The gel slices were then incubated with 0.02 µg/µL trypsin in digestion buffer (1.5 times the volume of the original gel slice) containing 10% acetonitrile, 40 mM NH4HCO3 pH 8.0, 0.1% w/v n-octylglucoside and 0.1% TFA for 1 h at room temperature. The buffer was removed and another 100–200 µL of digestion buffer was added and incubated for 4–5 h at 48°C. The solution was removed and kept in a new pre-washed tube. After addition of 100 µL 0.1% TFA to the gel slice, it was incubated for an additional 1 h at 37°C. The solution was removed and added to the first extraction. The combined sample was then dried and resuspended in 200 µL water three times to remove volatile materials. A final volume of c. 10 µL was used for analysis.
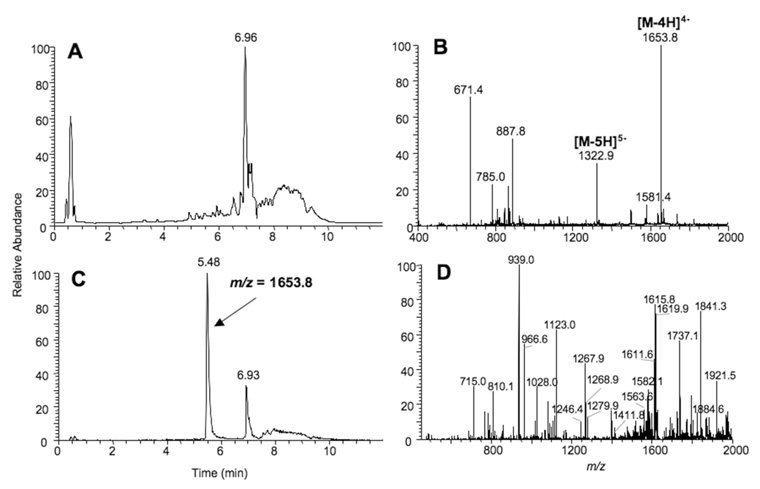
Samples were analyzed in an ESI linear trap ion mass spectrometer (LTQ, Thermo-Fisher, San Jose, CA) connected to a Waters Acquity UPLC system (Waters, Milford, MA) using an Aquity UPLC BEH C18 column (1.7 µm, 1.0 mm × 100 mm). DNA-peptide cross-links were separated using buffer A containing 10 mM NH4CH3CO2/2% CH3CN (v/v) and buffer B containing 10 mM NH4CH3CO2/95% CH3CN (v/v) following a gradient program with a flow rate of 150 µL min−1: 0–3.0 min, linear gradient from 100% A to 97% A; 3.0–4.5 min, linear gradient to 80% A; 4.5–5.0 min, linear gradient 100% B; 5.0–5.5 min, hold at 100% B; 5.5–6.5 min, linear gradient to 100% A; 6.5–9.5 min, hold at 100% A. The temperature of the column was maintained at 50 °C and samples (10 µL) were infused with an auto-sampler. ESI conditions were as follows: source voltage 4 kV, source current 100 µA, auxiliary gas flow rate setting 20, sweep gas flow rate setting 5, sheath gas flow setting 34, capillary voltage −49 V, capillary temperature 350 °C, tube lens voltage −90 V. MS/MS conditions were as follows: normalized collision energy 35%, activation Q 0.250, and activation time 30 ms. The quadruply charged species (m/z 1653.8) were used for CID analysis. The m/z values of the CID fragments corresponding to the peptide-DNA cross-link were calculated using a program linked to the Mass Spectrometry Group of Medicinal Chemistry at the University of Utah (medlib.med.utah.edu/masspec/).
Western blot analysis
In order to detect the cross-linked AGT dimer, hAGT was incubated with potential substrate oligos (unlabeled) in AGT buffer for up to 2 h at 37°C and the reaction was terminated by the addition of loading buffer containing 2% SDS. The samples were heated at 100°C for 5 min and separated by 15% SDS-PAGE gels and electrotransferred to PVDF membranes. Detection was carried out using ATO-1 antibody (35), which is a polyclonal antiserum raised in rabbits to a peptide corresponding to residues Lys8-Glu20, or anti-Penta His antibody, which recognizes the His6-tag as the primary antibody and HRP linked anti-rabbit IgG or anti-mouse IgG as the secondary antibody and the chemiluminescent reagent lumiGLO used to visualize the immunoreactive bands.
Generation of Models for Alkyl Interstrand Cross-link Repair by hAGT
Initial models of oligonucleotides containing alkyl interstrand cross-links with 4 or 7 carbons were built and manually placed in the hAGT active site (pdb 1t38), using AGT crystal structures (pdb 1eh6, 1eh8, and 1t39) as guides for DNA and Cys145 side chain placement. Initial models were subjected to conjugate gradient minimization, simulated annealing, and torsion angle dynamics by Crystallography & NMR System (CNS) (36), which employs molecular dynamics with CHARMM (Chemistry at HARvard Macromolecular Mechanics) (37, 38) force field parameters. One hundred steps of conjugate gradient minimization were followed by forty steps of simulated annealing. Simulated annealing was carried out at a starting temperature of 1000K and slow cooled with a drop in temperature of 25K per cycle of dynamics. Human AGT (pdb 1t38) active site analysis was done with the CASTp (39) (http://sts-fw.bioengr.uic.edu/castp/index.php) and MOLEOnline (40) (http://mole.chemi.muni.cz/index.php) servers. The models were validated using the rigorous structure validation programs ProCheck, WhatCheck, Errat and Prove of the JCSG Protein Structure Validation Suite (http://www.jcsg.org/prod/scripts/validation/sv3.cgi), the Verify3D Structure Evaluation Server (http://nihserver.mbi.ucla.edu/Verify_3D), and MolProbity (http://molprobity.biochem.duke.edu). These programs, which are routinely used for checking quality of models in theoretical studies, indicated that models for 4- and 7-carbon alkyl interstrand cross-link repair by hAGT were acceptable and comparable to pdb 1t38 from which they were derived.
RESULTS
Repair of Alkyl Interstrand Cross-Link with 7 Carbons by hAGT
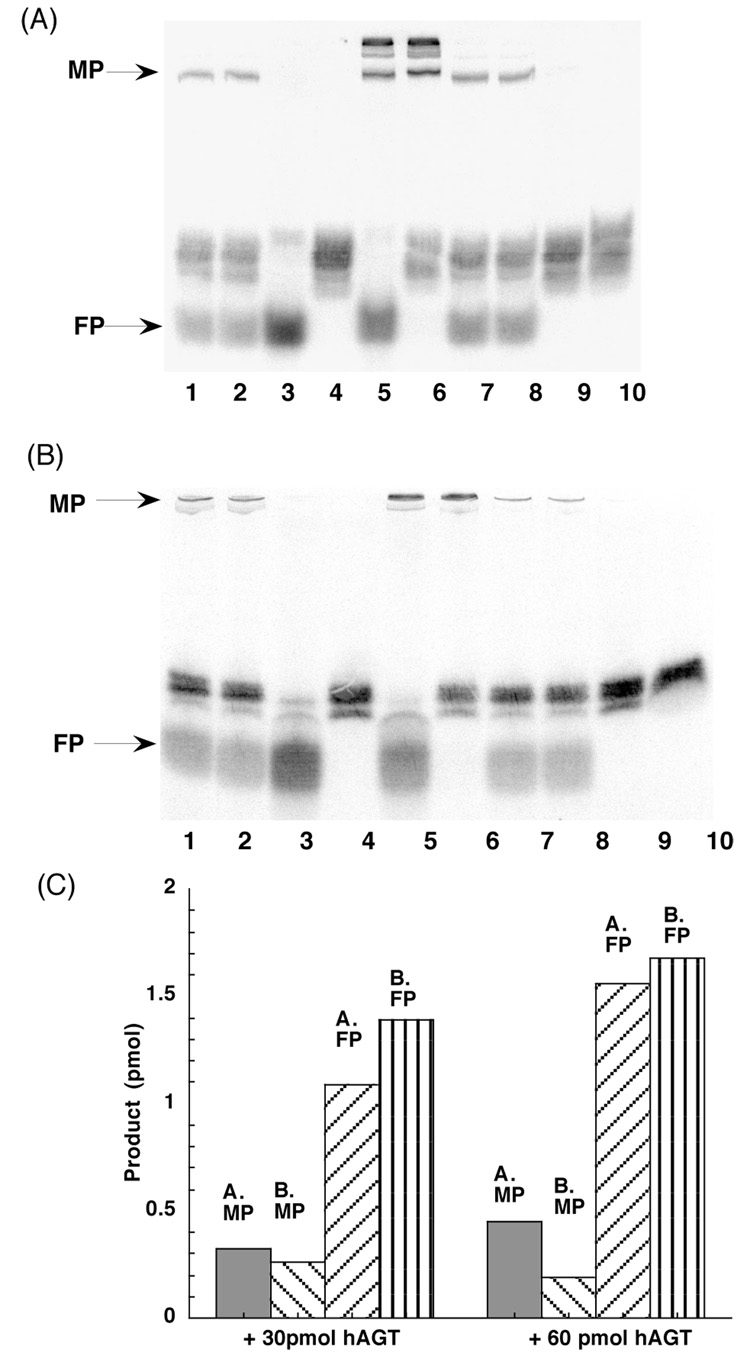
The labeled double stranded 11-mer substrate CJW80 was incubated with hAGT and the products were determined with SDS-PAGE (Figure 2A). As can be seen by comparing lanes 1 and 2 with lane 3, the incubation with wild type hAGT led to a loss of the substrate oligo and the appearance of two new product bands. A larger amount of the substrate was converted when the amount of hAGT added was increased (lanes 7 and 8). These product bands were not formed when hAGT that had been inactivated with SDS (Lane 10) or the C145S mutant (Lane 9) were used showing that active AGT was needed. The most rapidly migrating product band corresponds to the free oligo marker 80-C (Lane 3). The more slowly migrating band corresponds approximately to the lower band of a marker in which the labeled oligo was covalently attached to AGT by reaction with DBE (Lanes 5 and 6). Previous studies in which this reaction was fully characterized (34, 41) show that the adduct is formed by a ethane bridge between the Cys145 side-chain of hAGT and the N7 or N2 positions of a guanine residue in the added oligo. Multiple hAGT molecules can be added to an oligo containing multiple guanine residues in this reaction and the upper band in this marker is likely to represent the attachment of two AGT molecules. The very slight difference in mobility of the product of the repair of CJW80 by AGT and the lower marker band formed by DBE is likely to be due to either the longer alkane bridge or the fact that the hAGT-oligo cross-link formed in the repair reaction would be expected to be attached at the O6-position of guanine.
Figure 2. Repair of 11-mer 7-carbon-cross-linked oligo CJW80 by hAGT.
Panel A. Oligos were incubated with hAGT at 37°C for 2h, and the products were separated by SDS-PAGE as described under materials and methods. Lanes 1–2: 4 pmol CJW80 + 30 pmol hAGT; Lane 3: 4 pmol 80-C (FP marker); Lane 4: 4 pmol CJW80; Lane 5: 4 pmol 80-C + 60 pmol hAGT + 300 pmol DBE (MP marker); Lane 6: 4 pmol CJW80 + 60 pmol hAGT + 300 pmol DBE (MP marker); Lanes 7–8: 4 pmol CJW80 + 60 pmol hAGT. Lane 9: 4 pmol CJW80 + 60 pmol C145S; Lane 10: 4 pmol CJW80 + 60 pmol inactivated hAGT (hAGT inactivated by SDS). Panel B. The products were separated by denaturing PAGE as described under materials and methods. Lanes as in panel A. Panel C. The amount of products was determined by ImageQuant software and the amount of product formed is shown. MP: hAGT-DNA complex. FP: free 11-mer oligo.
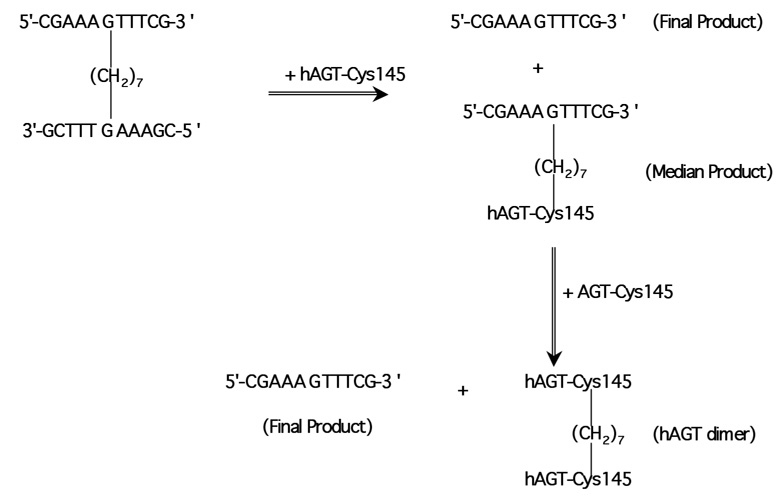
These experiments indicate that the cross-link of CJW80 and CJW98 could be repaired by hAGT and forms a hAGT-DNA complex (median product, MP1) and free oligo (final product, FP1) (Figure 2 and Figure 3). As both strands of CJW80 were 5′-end labelled, if only one side of alkyl cross-link was repaired, the ratio of MP/FP-1 should be close to 1. However, the yield of free oligo was much more than the MP (Figure 2C) suggesting that both sides of cross-link are efficiently repaired by hAGT. Essentially the same results were obtained when the products of the reaction with CJW80 were analyzed using gels that contained 7M urea (denaturing PAGE) (Figure 2B) but in this case the amount of FP was increased slightly and the amount of MP decreased (Figure 2C). This difference is likely to be due to the formation of a double stranded product after the cross-link is repaired. If the released strand is not dissociated from the hAGT-oligo MP, radioactivity that should be associated with the FP will remain with the MP. The denaturing PAGE conditions provide a more complete dissociation of the complex. However, the resolution in the area of the gel occupied by the hAGT-DNA complex is better with the SDS-PAGE conditions (compare MP in Figures 2A and 2B).
Figure 3. Repair of cross-links by hAGT.
The scheme is shown with an 11-mer substrate such as CJW80 but similar reactions would occur with the 17-mer substrate CJW98 and 10-mer substrate CJW81.
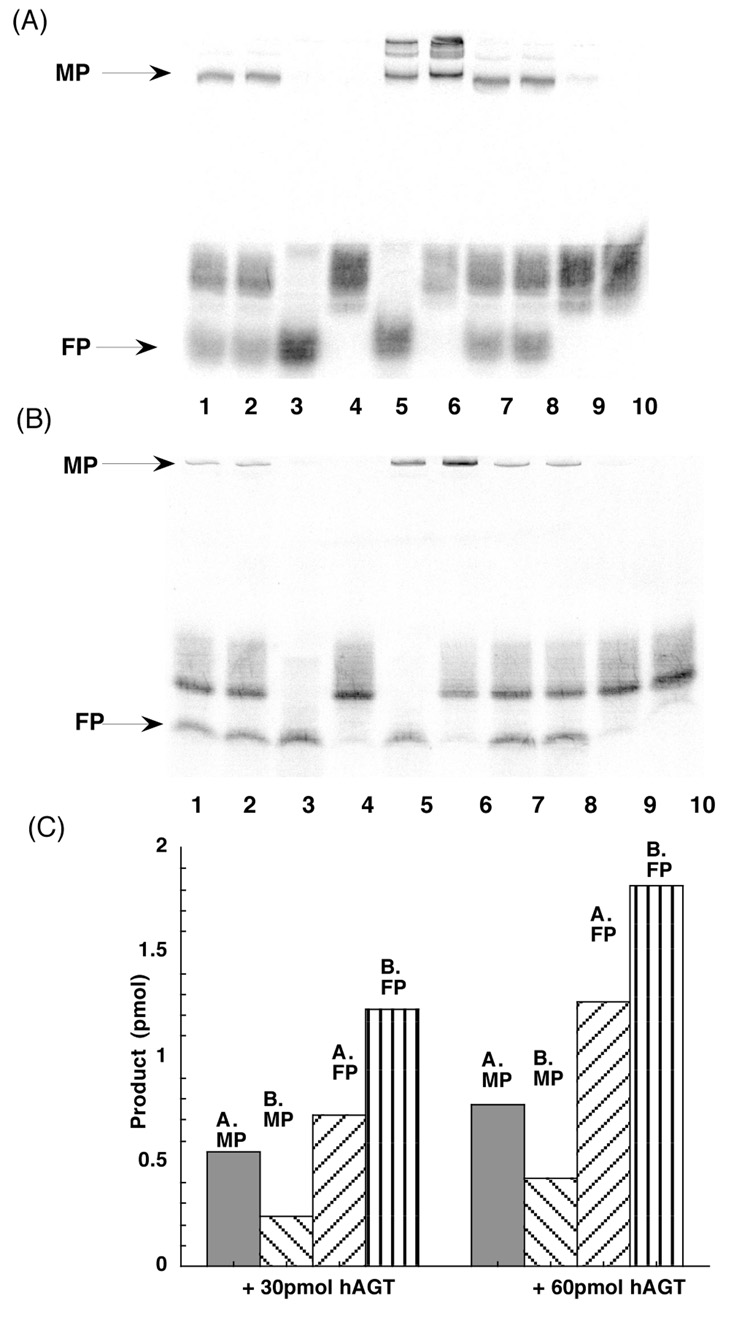
These results were confirmed by studies in which the labeled double stranded 17-mer substrate CJW98 was incubated with hAGT (Figure 4). Irrespective of whether the products were separated by SDS-PAGE (Figure 4A) or denaturing PAGE (Figure 4B), there was clear formation of a FP corresponding to the single stranded 17-mer and an intermediate product corresponding to the hAGT-oligo complex. Quantification of these results is shown in Figure 4C. There was a greater discrepancy between the relative amounts of MP and FP assayed under SDS-PAGE and denaturing PAGE conditions with the 17-mer cross-linked substrate compared to the results with 11-mer cross-linked substrate shown in Figure 2. The amount of the AGT-oligo complex (MP) found was substantially reduced when separated by denaturing PAGE and there was a corresponding increase in the free 17-mer FP (Figure 4C). This result is consistent with the explanation given above since the repaired 17-mer strand is less likely to dissociate from the AGT-oligo complex than the 11-mer product under the SDS-PAGE conditions.
Figure 4. Repair of 17-mer 7-carbon-cross-linked oligo CJW98 by hAGT.
Panel A. Oligos were incubated with hAGT at 37°C for 2h, and the products were separated by SDS-PAGE as described under materials and methods. Lanes 1–2: 4 pmol CJW98 + 30 pmol hAGT; Lane 3: 4 pmol 98-C (FP marker); Lane 4: 4 pmol CJW98; Lane 5: 4 pmol 98-C + 60 pmol hAGT + 300 pmol DBE (MP marker); Lane 6: 4 pmol CJW98 + 60 pmol hAGT + 300 pmol DBE (MP marker); Lanes 7–8: 4 pmol CJW98 + 60 pmol hAGT. Lane 9: 4 pmol CJW98 + 60 pmol C145S; Lane 10: 4 pmol CJW98 + 60 pmol inactivated hAGT (hAGT inactivated by SDS). Panel B. The products were separated by denaturing PAGE as described under materials and methods. Lanes as in panel A. Panel C. The amount of products was determined by ImageQuant software and the amount of product formed is shown. MP: hAGT-DNA complex. FP: free 17-mer oligo.
The identity of the MP formed in the reaction with CJW98 was confirmed by LC-MS analysis. After separation by SDS-PAGE, the band corresponding to the AGT-oligo complex (MP) was eluted from the gel and digested with trypsin. The digest was analyzed by LC-MS (Figure 5 and supporting information Table S1) which confirmed the presence of a peptide corresponding to the oligo linked via a (CH2)7 moiety to a peptide derived from residues Gly136-Arg147 of hAGT which contains the Cys145 acceptor site.
Figure 5. LC-MS analysis of the MP: detection of the DNA-hAGT cross-link.
hAGT was incubated with CJW98 (200 pmol) in AGT reaction buffer for 3 h at 37 °C. The reaction mixture was then separated by SDS-PAGE using a 20% gel. Bands corresponding to the DNA-hAGT cross-links were excised, digested with trypsin, and analyzed by: (A) Total ion chromatogram (TIC) of the trypsin digested band (Median Product). (B) ESI mass spectrum of the products eluting at ~5.48 min. Peaks with m/z 1653.8 and 1322.9 corresponds to the −4 and −5 charged species of the CJW98-hAGT(peptide) product. (C) Chromatogram of the m/z 1653.8 species. (D) CID mass spectrum of the m/z 1653.8 species.
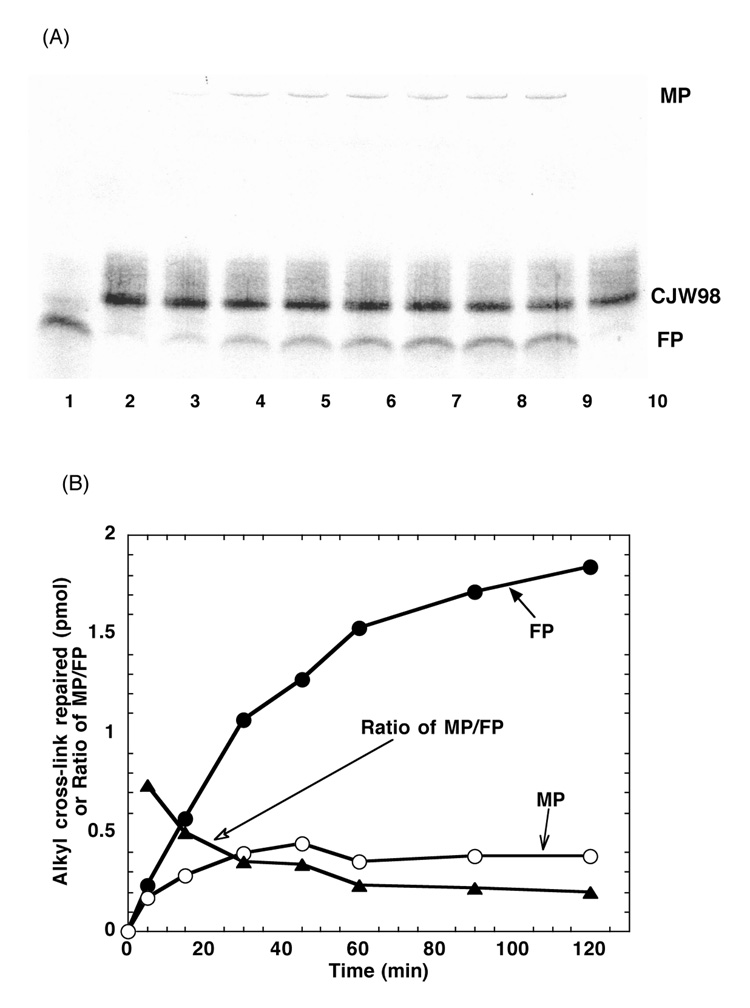
A time course of the repair of the double stranded 17-mer substrate CJW98 by hAGT with analysis carried out using the denaturing PAGE conditions is shown in Figure 6. The production of the free 17-mer FP continued to increase with time over a 2 h incubation whereas the hAGT-oligo MP peaked at about 45 min. The ratio of MP to FP continuously declined from a maximum at 5 min. This experiment supports the interpretation that both O6-adducts in the substrate could efficiently be repaired with initial formation of an hAGT-oligo complex and further reaction yielding a hAGT dimer and free oligo.
Figure 6. The time course of repair of 17-mer seven-carbon-cross-linked oligo CJW98 by hAGT.
Panel A. The products were separated by denaturing PAGE. Lane 1: 4 pmol 98-C + 45 pmol hAGT. Lanes 2–9: 4 pmol CJW98 was incubated with 45 pmol hAGT for 0, 5, 15, 30, 45, 60, 90 and 120 min respectively. Lane 10: 4 pmol CJW98 incubated with 45pmol C145S for 120 min. Panel B. The amount of products was determined by ImageQuant software and the amount of product formed is shown. MP, hAGT-DNA complex (open circles); FP, free 17-mer oligo (filled circles); ratio MP/FP (filled triangles).
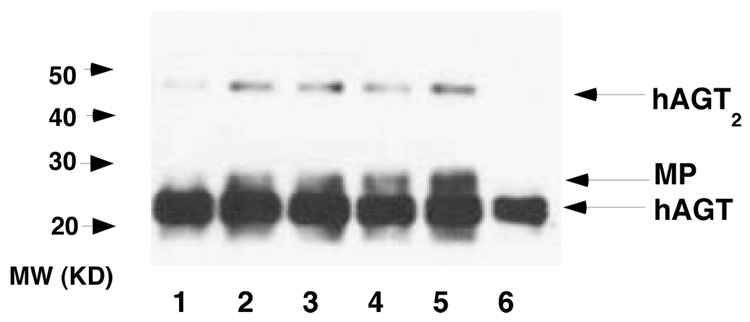
The formation of an hAGT dimer was confirmed by Western blotting analysis shown in Figure 7. After incubation of active hAGT with CJW98 and separation of the products by SDS-PAGE, a band of c. 44 kDa was produced that reacts with an antibody to hAGT was detected (Lanes 5–8 in Figure 7). The MP with hAGT-linked to the 17-mer (c. 23.7 kDa) can also be seen just above the band corresponding to the hAGT monomer (c.22 kDa).
Figure 7. Formation of hAGT dimer after incubation with CJW98.
hAGT was incubated with CJW98 for 2h at 37°C in 15 µL reaction buffer. The samples were then separated by SDS-PAGE using 15% gels and AGT protein detected using antiserum ATO-1. Lanes 1–5: 1, 2, 3, 4 and 5 pmol CJW98 +10 pmol hAGT respectively; Lane 6, 5 pmol hAGT. MP: hAGT-DNA complex. Similar results (not shown) were observed using anti-Penta His antibody for detection.
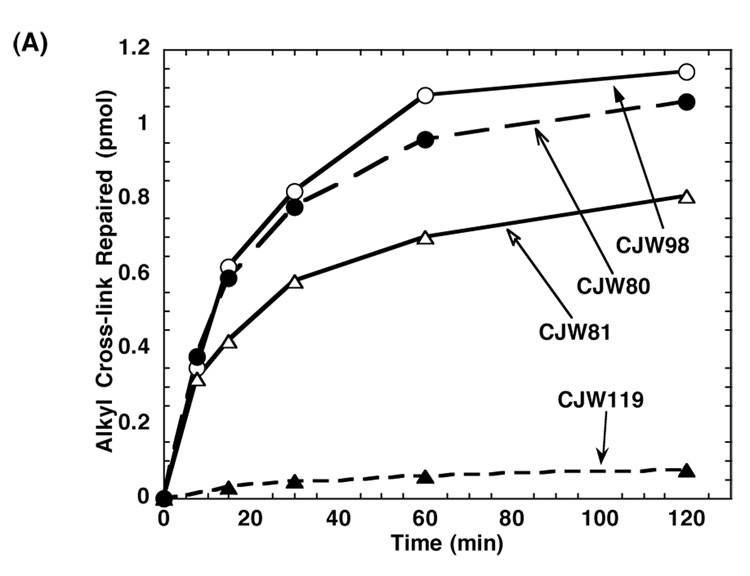
The oligos CJW98 and CJW80 contain a 7-carbon alkyl interstrand cross-link between two opposing guanine residues, which would not normally be paired in a double stranded DNA. However, the possible distortion associated with this linkage is not a critical factor in allowing repair since oligo CJW81 (Figure 1), which is a 10-mer oligo containing a central 7-carbon alkyl interstrand cross-link in which the cross-linked guanines are paired with cytosine was also repaired well by hAGT. The repair rate of this oligo was only slightly less than that of CJW80 or CJW98 (Figure 8).
Figure 8. Relative rates of repair of CJW80, CJW81, CJW98 and 11-mer four-carbon-cross-linked oligo CJW119 by hAGT.
2 pmol of each substrate was incubated with 30 pmol hAGT for the time shown. The samples were separated by denaturing PAGE and the products were determined as described in Materials and Methods. The repair of CJW98 (open circle), CJW80 (filled circle), CJW81(open triangle) and CJW119 (filled triangle) are shown.
Repair of Alkyl Interstrand Cross-link with 4 Carbons by hAGT
In contrast to the efficient repair of the oligos with a 7-carbon cross-link, a 4-carbon alkyl interstrand cross-link (CJW119, Figure 1) was repaired by hAGT very poorly (Figure 8). A very small amount of covalent hAGT-DNA complex and of the free oligo product could be detected but the ratio of MP/FP was close to 1 and no hAGT dimer could be detected on Western blots (results not shown). Therefore, it appears that the hAGT is only very weakly active in attacking the 4-carbon alkyl interstrand cross-link and is totally unable to repair the hAGT-oligo complex when this is linked to the guanine-O6- by a short 4-carbon linker.
Model of alkyl interstrand cross-link repair by hAGT
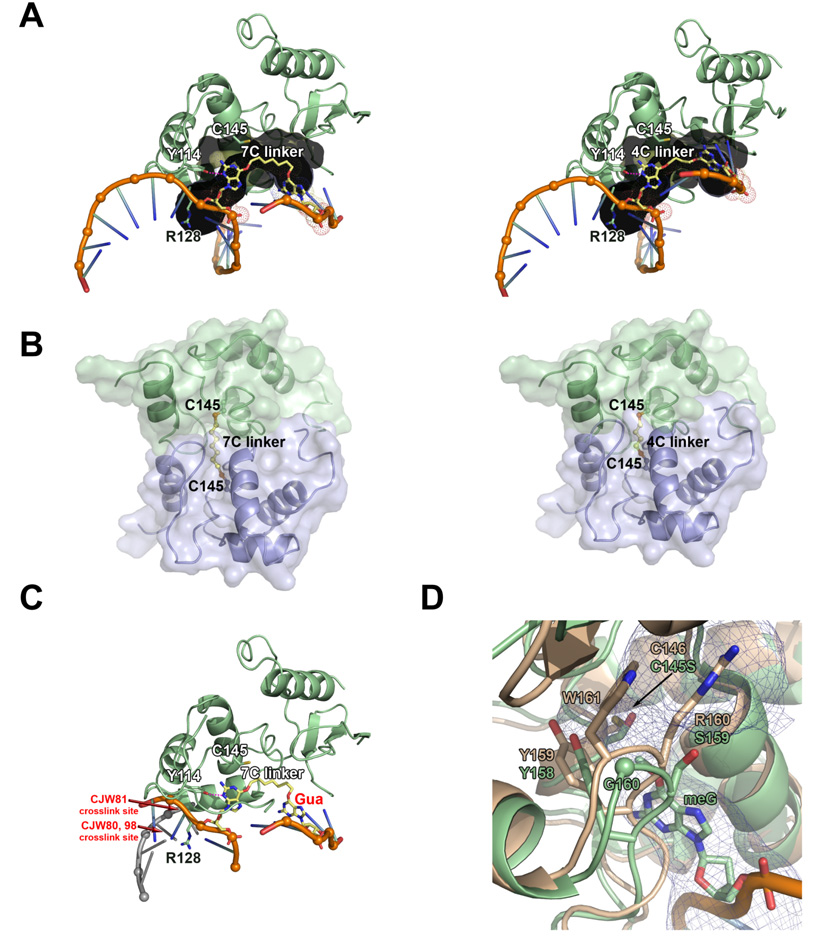
To understand differences in the ability of hAGT to repair alkyl interstrand cross-link substrates with 4 and 7 carbon linkers, we modeled the resulting hAGT-DNA complex and hAGT dimer products (Figure 9). Coordinates for these models are given in Supporting information Tables S2 and S3. These models reveal several insights into alkyl interstrand cross-link repair by hAGT. In the hAGT-DNA complex models, the guanine targeted for repair is rotated into the active site and the associated carbon linkers are nestled in the active site channel (Figure 9A), which is lined with Glu30, Ile31, Lys32, Leu33, Tyr114, Met134, Arg135, Gly136, Asn137, Pro138, Pro140, Ser145, Val148, Asn157, Tyr158, Ser159, Gly160, and Lys165. The 7-carbon linker spans the entire length of the channel, placing the non-target guanine on the exterior of the protein. In contrast, this non-target guanine is mostly buried in the active site channel with the shorter 4-carbon linker, thereby hindering it from adopting a suitable conformation for its subsequent repair by a second AGT molecule. In agreement with these observations, an AGT C-terminal domain dimer linked at Cys145 by a 7-carbon linker, but not a 4-carbon linker, can reasonably be modeled (Figure 9B). The short 4- carbon linker results in dimer protein-protein steric clash, which explains why this product is not observed experimentally. Finally, in models for both substrates, the DNA duplex would have to be forced apart locally to repair the target guanine (Figure 9C). Thus, repair of these cross-links by AGT likely disrupts the DNA double helix.
Figure 9. Model of O6-G-alkyl-O6-G interstrand cross-link repair by AGT.
(A 7-carbon (left) and 4-carbon (right) cross-linked substrate (yellow) modeled within the AGT active site channel (gray). (B) Model of AGT-AGT dimer resulting from repair of 7-carbon (left) and 4-carbon (right) cross-linked substrates. The protein-protein steric clash in the 4-carbon model could explain why this product is not observed experimentally. The N-terminal domain of AGT has been omitted from these models. (C) Model of AGT bound to 7-carbon cross-linked substrate and dsDNA, showing that repair of these cross-links by AGT likely disrupts the DNA double helix. Guanine labeled in red would have to be positioned at one of the cross-link sites (labeled in red) to maintain dsDNA. For clarity, helix 8 is not shown. (D) Ada-C structure (tan; pdb 1sfe) overlayed with AGT bound to DNA containing O6-methylguanine structure (green; pdb 1t38). The AGT active site channel is shown as a blue mesh.
Repair of Alkyl Interstrand Cross-link with 7 Carbons by hAGT Mutants and E. coli AGTs
A number of mutants of hAGT with alterations in the active site were examined for the ability to repair the cross-linked oligo substrates. In many cases the repair was too slow for accurate measurement of the rate of repair. Therefore the results, which are shown in Table 2 are expressed as the amount of protein needed to obtain 25% repair in the standard assay for either methylated DNA or the cross-linked oligos. As indicated in the footnote to this Table, the values for 25% repair of methylated DNA by each of the proteins tested do reflect the rate constant for repair of O6-methylguanine by these proteins.
Table 2.
Repair of O6-methylguanine and O6-G-alkyl-O6-G interstrand cross-links by hAGT mutants and bacterial AGTs
| Protein | Amount of protein needed for 25% removal of DNA adduct (pmol) | ||
|---|---|---|---|
| Methylated DNA Substrate |
Interstrand cross-linked oligo substrate | ||
| CJW80 | CJW81 | ||
| hAGT* | 0.08 | 2.2 | 4.6 |
| R128K* | 0.12 | 1.6 | 1.8 |
| R128A* | 62 | 65 | Not tested |
| R128G* | 91 | 140 | 86 |
| Y114F* | 0.84 | >200 | >200 |
| Y114A* | 90 | No reaction | No reaction |
| Ogt | 0.08 | No reaction | No reaction |
| Ada-C | 0.08 | No reaction | No reaction |
| P140K | 1.20 | >200 | >200 |
| Y158H | 1.30 | >200 | >200 |
| G160R | 0.18 | 5.4 | 9.6 |
The values are the mean of 2–3 experiments.
The rate constant for repair of O6-methylguanine in methylated DNA were measured for these proteins and were: 7 × 107 M−1 min −1 for wild type hAGT, 4.3 × 107 M−1 min−1 for R128K; 0.0007 × 107 M−1 min−1 for R128A; 0.0004 × 107 M−1 min−1 for R128G; 0.39 × 107 M−1 min−1 for Y114F and 0.0006 × 107 M−1 min−1 for Y114A.
Arg128 is postulated as the residue that replaces the target nucleotide when this is flipped from DNA into the active site pocket by hAGT binding (4, 15, 16). Replacement of this Arg by Lys reduces the rate of repair of methylated DNA slightly as previously reported (15, 28) but actually increased the rate of cross-link repair by about 2-fold (Table 2). Replacement of this residue by Ala or Gly, which profoundly reduced the rate of repair of methylated DNA in agreement with published studies (15, 28), also reduced the repair of cross-links to an approximately similar degree. The results are compatible with the model shown in Figure 9C. Arg128 is presumably needed to open the duplex to start the repair reaction and since Arg128 stabilizes the open duplex for repair, the faster R128K repair rate suggests a faster off-rate for this mutant.
Tyr114 is thought to play a dual role in the repair of methylated DNA by hAGT. Interaction of the target nucleotide with the aromatic side chain causes rotation of the nucleotide phosphate bond which is key for the flipping and its -OH then interacts with N3 of the target guanine to facilitate the alkyl transfer (4, 15, 16, 19). Therefore, alteration of Tyr114 to Phe, which affects only the latter, reduces the rate constant for methyl repair by about 30-fold but alteration to Ala has a much greater effect (c. 600-fold) as previously published (30). However, both of these mutations greatly reduced the repair of cross-links (Table 2). One possible explanation for this decreased activity of the Phe and Ala mutants for cross-link repair is that both lack the Tyr114 OH hydrogen bond with the damaged guanine N3, which likely serves to position alkyl-O6-guanine and lower its negative charge. The absence of this hydrogen bond could result in a slightly different distance from the alkyl group to Cys145, which could lead to a decreased repair rate. This effect would likely be more pronounced for alkyl interstrand cross-link substrates since the damage is large in size. In support of this idea, the distance from Tyr114 -OH to guanine N3 is ~2.5 Å for methylguanine in pdb 1t38 and ~2.9 Å for 4- and 7-carbon cross-link substrate models (Figure 9), and the Cys145 -SH to alkyl damage C1 distance is 2.7 Å, 3.6 Å, and 3.9 Å for O6-methylguanine, 4-carbon cross-link, and 7-carbon cross-link substrates, respectively. Another possible explanation for mutant decreased activity is that steric effects of large cross-link substrates result in their limited conformational flexibility, which could prevent the alkyl group from easily adopting an orientation amenable to repair. Longer alkyl groups are repaired less efficiently than shorter ones by hAGT (7) and this effect may be greatly enhanced when combined with Tyr114 mutants.
Several mutations at the active site for hAGT greatly reduce the ability to react with the low M.W. pseudosubstrate drugs O6-benzylguanine (42–44) and O6-(4-bromothenyl)guanine (45) but have only a modest effect on the ability to repair methylated DNA. Three of these mutations G160R (a naturally occurring polymorphic variant) and P140K and Y156H (which were discovered in a screen for O6-benzylguanine resistant mutants) were tested for the ability to repair cross-links. Repair by G160R was reduced only slightly and to the same extent as repair of methylated DNA but no repair of cross-links was detected by mutants P140K and Y156H (Table 2). It is likely that this is due to restrictions in the space available and flexibility of the active site. This interpretation is consistent with the finding that neither of the E. coli AGTs tested, Ogt and Ada-C were able to repair O6-G-alkyl-O6-G cross-links. These E. coli repair proteins are also resistant to O6-benzylguanine (46–48). Full details of the structure of the Ogt active site are not available but the Ada-C crystal structure is known (49). The Ada-C active site channel is partially blocked by side chains of bulky residues Arg and Trp (corresponding residues in hAGT are Ser and Gly, respectively) and thus, is unable to accommodate the cross-link, as shown in Figure 9D.
DISCUSSION
It is well established that hAGT can prevent interstrand cross-link formation after exposure to chloroethylating agents but this effect is due to the facile repair of the precursors O6-(2-chloroethyl)guanine (50, 51) or 1, O6-ethanoguanine (14). Our results provide the first evidence that hAGT can actually directly break DNA interstrand cross-links if these involve attachment to the guanine O6-position. Although we have only demonstrated such reaction with an O6-O6 cross-link, it is very likely that a similar transfer would occur with any other cross-link in which one side involves the guanine O6- recognized by hAGT. However, efficient reaction with hAGT requires that the cross-link be long enough to ensure that the structural alteration of the DNA substrate brought about by hAGT binding, which is needed to place the thiolate anion at Cys145 in close proximity to the CH2 attached to the guanine O6, can take place. The seven-carbon linker is clearly long enough for this even when the two linked bases were offset by one nucleotide and not directly opposite but the four-carbon linker is too short.
These experimental observations are entirely compatible with the modelling shown in Figure 9C which is based on the crystal structure of hAGT bound to DNA and the mechanism for alkyl transfer proposed on the basis of this structure (4, 16). A recent paper providing theoretical simulations from pair wise potentials suggested the possibility of slightly revised roles for Tyr114 and Arg128 in a two -step kinetic process for lesion recognition by AGT (19). Our results are also consistent with this mechanism.
It is noteworthy that the AGTs from E. coli, Ada-C and Ogt, were unable to carry out the repair of the O6-O6 cross-link even when it did contain the seven-carbon bridge. Modelling based on the Ada-C structure and DNA binding domain (49, 52) shown in Figure 9D shows that the limited space in the active site of the Ada-C cannot accommodate the cross-link. This steric restriction also accounts for the inability of Ada-C to react with the pseudosubstrate inhibitor O6-benzylguanine (15, 47). The structure of Ogt is not known but this protein also reacts very poorly with O6-benzylguanine (47) and its active site is also likely to be more restricted than hAGT. The correlation between the inability to react with O6-benzylguanine is also seen with the P140K and Y156H mutants of hAGT (42, 43) whereas the G160R variant, which is much less resistant to this inhibitor, was able to repair the cross-link (44). Busulfan, a strongly cytotoxic immunosuppressive drug, which has been used for preparation for bone marrow transplantation (24) and for the treatment of tumors particularly chronic myelogenous leukemia (25) forms a variety of DNA adducts causing mutations, sister-chromatid exchanges and cell death (53–55). The presence of an unrepaired interstrand cross-link during S phase is likely to be highly toxic to mammalian cells. Although such an O6-O6 cross-link might be expected to occur to only a limited extent in cells treated with busulfan, this minor adduct could still contribute significantly to its cell-killing ability. It was reported that the toxicity of busulfan was not affected by the activity of hAGT in cultured human erythromegakaryocytic cell line (LAMA-84) and normal human bone marrow cells in culture (56). This is consistent with our finding that the short four-carbon O6-O6 cross-link that could be derived from busulfan is not repairable by AGT. The inability to repair such cross-links may contribute to the potent immunosuppressive effects of busulfan. In contrast, similar agents producing longer cross-links such as hepsulfam (24, 57–59), which could form longer seven-carbon O6-O6 cross-links, are more likely to be affected by hAGT status. Since hAGT is expressed in a wide variety of human tumors, this could explain their disappointing lack of activity despite initial promise when tested in comparison to busulfan (25, 59, 60). If this is the case, their combination with hAGT inactivators such as O6-benzylguanine or O6-(4-bromothenyl)guanine may restore utility.
The initial reaction of hAGT with the seven-carbon O6-O6 cross-linked substrate forms an hAGT-oligo cross-link in which a guanine-O6 is linked to Cys145 of hAGT by a seven-carbon bridge. The structure of this DNA-protein cross-link was confirmed by LC-MS analysis as shown in Figure 5. Based on studies in which hAGT-DNA cross-links are formed by reactions of bis-electrophiles such as dibromomethane (61), DBE (34), butadiene diepoxide (62, 63) and nitrogen mustards (64) it is likely that such a protein-DNA cross-link would be both cytotoxic and mutagenic. However these hAGT-DNA adducts are linked at positions such as the guanine-N7 and N2 whereas the protein adduct formed in the repair of the O6-O6 cross-linked substrate described in the current paper is linked at the guanine-O6. Therefore, it can also be repaired by a second molecule of hAGT as shown in Figure 2, Figure 4 and Figure 6. This repair, which generates an inactive hAGT dimer and restores the DNA structure, would prevent any toxicity. The remarkable ability of the hAGT to accommodate such a large substrate as a hAGT-DNA protein cross-link in its active site pocket is also predictable from the modelling studies shown in Figure 9B. The seven-carbon linker is long enough to allow for the guanine adduct to be placed in the active site pocket in proximity to the Cys145 acceptor site with the covalently attached hAGT protein sufficiently far away that there is no steric clash between the protein molecules. This displacement could not occur with the four-carbon linker so the very slow reaction that occurs with this O6-O6 substrate stops at the formation of the hAGT-oligo step.
In summary, our results add to the spectrum of adducts that are repaired by hAGT. This direct repair reaction requires the attachment of the adduct to the O6-position (5–7) but short alkyl adducts such as methyl- and ethyl- (65), longer linear adducts such as n-propyl-, n-butyl- and 2-chloroethyl- (50, 51, 66), bulky adducts such as benzyl- (7, 18) and pyridyloxobutyl- (7, 67), cyclic intermediates such as N1-O6-ethanoxanthine (68) and 1, O6-ethanoguanine (14), and, as shown here, DNA interstrand cross-links can be repaired.
Footnotes
This research was supported by grants CA-018137 (AEP), CA-097209 (JAT), R01 ES10546 and P30 ES000267 (FPG) from the US Public Health Service, Bethesda, MD and by funding from the Natural Sciences and Engineering Research Council of Canada (NSERC), the Canada Foundation for Innovation (CFI) and the Canada Research Chair (CRC) program. We also acknowledge a Merck Fellowship for financial support (GC).
Abbreviations: AGT, O6-alkylguanine-DNA alkyltransferase; hAGT, human AGT; busulfan, 1,4-butanediol dimethanesulfonate; hepsulfam, 1,7-heptanediol dimethanesulfonate; oligo, oligodeoxyribonucleotide; IPTG, isopropyl β-D-thiogalactopyranoside; T4 polynucleotide kinase (PNK); BP, base pair; DTT, dithiothreitol; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; DBE, 1,2-dibromoethane; MS, mass spectrometry; trifluoroacetic acid, TFA; MP, median product of the repair reaction (hAGT-oligo); FP, final product of the repair reaction (free oligo).
SUPPORTING INFORMATION AVAILABLE
LC-MS analysis of the 17-mer-AGT complex and coordinates for models of AGT with 4-C and 7 C-O6-G-alkyl-O6-G interstrand cross-links are available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Pegg AE. Repair of O6-alkylguanine by alkyltransferases. Mutation Res. 2000;462:83–100. doi: 10.1016/s1383-5742(00)00017-x. [DOI] [PubMed] [Google Scholar]
- 2.Margison GP, Santibáñez-Koref MF. O6-Alkylguanine-DNA alkyltransferase: role in carcinogenesis and chemotherapy. BioEssays. 2002;24:255–266. doi: 10.1002/bies.10063. [DOI] [PubMed] [Google Scholar]
- 3.Margison G, Povey AC, Kaina B, Santibáñez-Koref MF. Variability and regulation of O6-alkylguanine-DNA alkyltransferase. Carcinogenesis. 2003;24:625–635. doi: 10.1093/carcin/bgg005. [DOI] [PubMed] [Google Scholar]
- 4.Tubbs JL, Pegg AE, Tainer JA. DNA binding, nucleotide flipping, and the helix-turn-helix motif in base repair by O6-alkylguanine-DNA alkyltransferase and its implications for cancer chemotherapy. DNA Repair (Amst) 2007;6:1100–1115. doi: 10.1016/j.dnarep.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pegg AE, Dolan ME, Moschel RC. Structure, function and inhibition of O6-alkylguanine-DNA alkyltransferase. Progr. Nucleic Acids Res. Mol. Biol. 1995;51:167–223. doi: 10.1016/s0079-6603(08)60879-x. [DOI] [PubMed] [Google Scholar]
- 6.Mijal RS, Kanugula S, Vu CC, Fang Q, Pegg AE, Peterson LA. DNA sequence context affects repair of the tobacco-specific adduct O6-[4-oxo-4-(3-pyridyl)butyl]guanine by human O6-alkylguanine-DNA alkyltransferases. Cancer Res. 2006;66:4968–4974. doi: 10.1158/0008-5472.CAN-05-3803. [DOI] [PubMed] [Google Scholar]
- 7.Coulter R, Blandino M, Tomlinson JM, Pauly GT, Krajewska M, Moschel RC, Peterson LA, Pegg AE, Spratt TE. Differences in the rate of repair of O6-alkylguanines in different sequence contexts by O6-alkylguanine-DNA alkyltransferase. Chem. Res. Toxicol. 2007;20:1966–1971. doi: 10.1021/tx700271j. [DOI] [PubMed] [Google Scholar]
- 8.Gerson SL. Clinical relevance of MGMT in the treatment of cancer. J. Clin. Oncol. 2002;20:2388–2399. doi: 10.1200/JCO.2002.06.110. [DOI] [PubMed] [Google Scholar]
- 9.Erickson LC, Laurent G, Sharkey NA, Kohn KW. DNA cross-linking and monoadduct repair in nitrosourea-treated human tumor cells. Nature. 1980;288:727–729. doi: 10.1038/288727a0. [DOI] [PubMed] [Google Scholar]
- 10.Erickson LC, Bradley MO, Ducore JM, Ewig RA, Kohn KW. DNA cross-linking and cytotoxicity in normal and transformed human cells treated with antitumor nitrosoureas. Proc. Natl. Acad. Sci. U.S.A. 1980;77:467–471. doi: 10.1073/pnas.77.1.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ludlum DB. The chloroethylnitrosoureas: Sensitivity and resistance to cancer chemotherapy at the molecular level. Cancer Investigation. 1997;15:588–598. doi: 10.3109/07357909709047601. [DOI] [PubMed] [Google Scholar]
- 12.Bodell WJ, Tokuda K, Ludlum DB. Differences in DNA alkylation products formed in sensitive and resistant human glioma cells treated with N-(2-chloroethyl)-N-nitrosourea. Cancer Res. 1988;48:4489–4492. [PubMed] [Google Scholar]
- 13.Gonzaga PE, Harris L, Margison GP, Brent TP. Evidence that covalent complex formation between BCNU-treated oligonucleotides and E. coli alkyltransferases requires the O6-alkylguanine function. Nucleic Acids Res. 1990;18:3961–3966. doi: 10.1093/nar/18.13.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzaga PE, Potter PM, Niu T, Yu D, Ludlum DB, Rafferty JA, Margison GP, Brent TP. Identification of the cross-link between human O6-methylguanine-DNA methyltransferase and chloroethylnitrosourea-treated DNA. Cancer Res. 1992;52:6052–6058. [PubMed] [Google Scholar]
- 15.Daniels DS, Mol CD, Arvai AS, Kanugula S, Pegg AE, Tainer JA. Active and alkylated human AGT structures: a novel zinc site, inhibitor and extrahelical binding. DNA damage reversal revealed by mutants and structures of active and alkylated human AGT. EMBO J. 2000;19:1719–1730. doi: 10.1093/emboj/19.7.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daniels DS, Woo TT, Luu KX, Noll DM, Clarke ND, Pegg AE, Tainer JA. Novel modes of DNA binding and nucleotide flipping by the human DNA repair protein AGT. Nat. Struct. Mol. Biol. 2004;11:714–720. doi: 10.1038/nsmb791. [DOI] [PubMed] [Google Scholar]
- 17.Duguid EM, Rice PA, He C. The structure of the human AGT protein bound to DNA and its implications for damage detection. J. Mol. Biol. 2005;350:657–666. doi: 10.1016/j.jmb.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 18.Zang H, Fang Q, Pegg AE, Guengerich FP. Kinetic analysis of steps in the repair of damaged DNA by human O6-alkylguanine DNA-alkyltransferase. J. Biol. Chem. 2005;280:30873–30881. doi: 10.1074/jbc.M505283200. [DOI] [PubMed] [Google Scholar]
- 19.Hu J, Ma A, Dinner AR. A two-step nucleotide-flipping mechanism enables kinetic discrimination of DNA lesions by AGT. Proc. Natl. Acad. Sci. U. S. A. 2008;105:4615–4620. doi: 10.1073/pnas.0708058105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guengerich FP, Fang Q, Liu L, Hachey DL, Pegg AE. O6-Alkylguanine-DNA alkyltransferase: Low pKa and reactivity of cysteine 145. Biochemistry. 2003;42:10965–10970. doi: 10.1021/bi034937z. [DOI] [PubMed] [Google Scholar]
- 21.Wibley JEA, Pegg AE, Moody PCE. Crystal structure of the human O6-alkylguanine-DNA alkyltransferase. Nucleic Acids Res. 2000;28:393–401. doi: 10.1093/nar/28.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hashimoto H, Inoue T, Nishioka M, Fujiwara S, Takagi M, Imanaka T, Kai Y. Hyperthermostable protein structure maintained by intra- and inter-helix ion pairs in archael O6-methylguanine-DNA methyltransferase. J. Mol. Biol. 1999;292:707–716. doi: 10.1006/jmbi.1999.3100. [DOI] [PubMed] [Google Scholar]
- 23.Roberts A, Pelton JG, Wemmer DE. Structural studies of MJ1529, an O6-methylguanine-DNA methyltransferase. Magn. Reson. Chem. 2006;44 Spec No:S71–S82. doi: 10.1002/mrc.1823. [DOI] [PubMed] [Google Scholar]
- 24.Westerhof GR, Ploemacher RE, Boudewijn A, Blokland I, Dillingh JH, McGown AT, Hadfield JA, Dawson MJ, Down JD. Comparison of different busulfan analogues for depletion of hematopoietic stem cells and promotion of donor-type chimerism in murine bone marrow transplant recipients. Cancer Res. 2000;60:5470–5478. [PubMed] [Google Scholar]
- 25.Berger DP, Winterhalter BR, Dengler WA, Fiebig HH. Preclinical activity of hepsulfam and busulfan in solid human tumor xenografts and human bone marrow. Anticancer Drugs. 1992;3:531–539. doi: 10.1097/00001813-199210000-00014. [DOI] [PubMed] [Google Scholar]
- 26.Nallamsetty S, Kapust RB, Tozser J, Cherry S, Tropea JE, Copeland TD, Waugh DS. Efficient site-specific processing of fusion proteins by tobacco vein mottling virus protease in vivo and in vitro. Protein Expr. Purif. 2004;38:108–115. doi: 10.1016/j.pep.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 27.Wilds CJ, Booth JD, Noronha AM. Synthesis of oligonucleotides containing an O6-G-alkyl-O6-G interstrand cross-link. Tetrahedron Lett. 2006;47:9125–9128. [Google Scholar]
- 28.Kanugula S, Goodtzova K, Edara S, Pegg AE. Alteration of arginine-128 to alanine abolishes the ability of human O6-alkylguanine-DNA alkyltransferase to repair methylated DNA but has no effect on its reaction with O6-benzylguanine. Biochemistry. 1995;34:7113–7119. doi: 10.1021/bi00021a024. [DOI] [PubMed] [Google Scholar]
- 29.Liu L, Xu-Welliver M, Kanugula S, Pegg AE. Inactivation and degradation of O6-alkylguanine-DNA alkyltransferase after reaction with nitric oxide. Cancer Res. 2002;62:3037–3043. [PubMed] [Google Scholar]
- 30.Goodtzova K, Kanugula S, Edara S, Pegg AE. Investigation of the role of tyrosine-114 in the activity of human O6-alkylguanine-DNA alkyltransferase. Biochemistry. 1998;37:12489–12495. doi: 10.1021/bi9811718. [DOI] [PubMed] [Google Scholar]
- 31.Fang Q, Kanugula S, Pegg AE. Function of domains of human O6-alkylguanine-DNA alkyltransferase. Biochemistry. 2005;44:15396–15405. doi: 10.1021/bi051460d. [DOI] [PubMed] [Google Scholar]
- 32.Fang Q, Loktionova NA, Moschel RC, Javanmard S, Pauly GT, Pegg AE. Differential inactivation of polymorphic variants of human O6-alkylguanine-DNA alkyltransferase. Biochem. Pharmacol. 2008;75:618–626. doi: 10.1016/j.bcp.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning. NY: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 34.Liu L, Pegg AE, Williams KM, Guengerich FP. Paradoxical enhancement of the toxicity of 1,2-dibromoethane by O6-alkylguanine-DNA alkyltransferase. J. Biol. Chem. 2002;277:37920–37928. doi: 10.1074/jbc.M205548200. [DOI] [PubMed] [Google Scholar]
- 35.Pegg AE, Wiest L, Mummert C, Dolan ME. Production of antibodies to peptide sequences present in human O6-alkylguanine-DNA alkyltransferase and their use to detect this protein in cell extracts. Carcinogenesis. 1991;12:1671–1677. doi: 10.1093/carcin/12.9.1671. [DOI] [PubMed] [Google Scholar]
- 36.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 37.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comp. Chem. 1983;4 1887-1217. [Google Scholar]
- 38.MacKerell AD, Brooks BP, Brooks I, C L, Nilsson L, Roux B, Won Y, Karplus M. CHARMM: The energy function and its parameterization with an overview of the program. In: Schleyer PVRea., editor. The Encyclopedia of Computational Chemistry. Chichester: John Wiley & Sons; 1998. pp. 271–277. [Google Scholar]
- 39.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petrek M, Kosinova P, Koca J, Otyepka M. MOLE: a Voronoi diagram-based explorer of molecular channels, pores, and tunnels. Structure. 2007;15:1357–1363. doi: 10.1016/j.str.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 41.Liu L, Hachey DL, Valadez G, Williams KM, Guengerich FP, Loktionova NA, Kanugula S, Pegg AE. Characterization of a mutagenic DNA adduct formed from 1,2-dibromoethane by O6-alkylguanine-DNA alkyltransferase. J. Biol. Chem. 2004;279:4250–4259. doi: 10.1074/jbc.M311105200. [DOI] [PubMed] [Google Scholar]
- 42.Xu-Welliver M, Kanugula S, Pegg AE. Isolation of human O6-alkylguanine-DNA alkyltransferase mutants highly resistant to inactivation by O6-benzylguanine. Cancer Res. 1998;58:1936–1945. [PubMed] [Google Scholar]
- 43.Xu-Welliver M, Leitao J, Kanugula S, Pegg AE. Alteration of the conserved residue tyrosine-158 to histidine renders human O6-alkylguanine-DNA alkyltransferase insensitive to the inhibitor O6-benzylguanine. Cancer Res. 1999;59:1514–1519. [PubMed] [Google Scholar]
- 44.Xu-Welliver M, Leitao J, Kanugula S, Meehan WJ, Pegg AE. The role of codon 160 in sensitivity of human O6-alkylguanine-DNA alkyltransferase to O6-benzylguanine. Biochem. Pharmacol. 1999;58:1279–1285. doi: 10.1016/s0006-2952(99)00216-6. [DOI] [PubMed] [Google Scholar]
- 45.Woolford LB, Southgate TD, Margison GP, Milsom MD, Fairbairn LJ. The P140K mutant of human O6-methylguanine-DNA-methyltransferase (MGMT) confers resistance in vitro and in vivo to temozolomide in combination with the novel MGMT inactivator O6-(4-bromothenyl)guanine. J. Gene Med. 2006;8:29–34. doi: 10.1002/jgm.816. [DOI] [PubMed] [Google Scholar]
- 46.Dolan ME, Pegg AE, Dumenco LL, Moschel RC, Gerson SL. Comparison of the inactivation of mammalian and bacterial O6-alkylguanine-DNA alkyltransferases by O6-benzylguanine. Carcinogenesis. 1991;12:2305–2310. doi: 10.1093/carcin/12.12.2305. [DOI] [PubMed] [Google Scholar]
- 47.Pegg AE, Boosalis M, Samson L, Moschel RC, Byers TL, Swenn K, Dolan ME. Mechanism of inactivation of human O6-alkylguanine-DNA alkyltransferase by O6-benzylguanine. Biochemistry. 1993;32:11998–12006. doi: 10.1021/bi00096a009. [DOI] [PubMed] [Google Scholar]
- 48.Elder RH, Margison GP, Rafferty JA. Differential inactivation of mammalian and E. coli O6-alkylguanine-DNA alkyltransferase by O6-benzylguanine. Biochem. J. 1994;298:231–235. doi: 10.1042/bj2980231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore MH, Gulbus JM, Dodson EJ, Demple B, Moody PCE. Crystal structure of a suicidal DNA repair protein: The Ada O6-methylguanine-DNA methyltransferase from E. coli. EMBO J. 1994;13:1495–1501. doi: 10.1002/j.1460-2075.1994.tb06410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gibson N, Zlotogorski C, Erickson LC. Specific DNA repair mechanisms may protect some human tumor cells from DNA interstrand crosslinking by chloroethylnitrosoureas but not from crosslinking by other anti-tumor alkylating agents. Carcinogenesis. 1985;6:445–450. doi: 10.1093/carcin/6.3.445. [DOI] [PubMed] [Google Scholar]
- 51.Brent TP. Inactivation of purified human O6-alkylguanine-DNA alkyltransferase by alkylating agents or alkylated DNA. Cancer Res. 1986;46:2320–2323. [PubMed] [Google Scholar]
- 52.Verdemato PE, Brannigan JA, Dambion C, Zuccotto F, Moody PCE, Lian L-Y. DNA -binding mechanism of the Escherichia coli Ada O6-alkylguanine-DNA alkyltransferase. Nucleic Acids Res. 2000;28:3710–3718. doi: 10.1093/nar/28.19.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanderson BJ, Shield AJ. Mutagenic damage to mammalian cells by therapeutic alkylating agents. Mutat. Res. 1996;355:41–57. doi: 10.1016/0027-5107(96)00021-8. [DOI] [PubMed] [Google Scholar]
- 54.Tong WP, Ludlum DB. Crosslinking of DNA by busulfan. Formation of diguanyl derivatives. Biochim. Biophys. Acta. 1980;608:174–181. doi: 10.1016/0005-2787(80)90145-8. [DOI] [PubMed] [Google Scholar]
- 55.Morales-Ramirez P, Gonzalez-Beltran F. Different behavior of SCE-eliciting lesions induced by low and high doses of busulfan. Environ. Mol. Mutagen. 2007;48:706–714. doi: 10.1002/em.20344. [DOI] [PubMed] [Google Scholar]
- 56.Westerhof GR, Down JD, Blokland I, Wood M, Boudewijn A, Watson AJ, McGown AT, Ploemacher RE, Margison GP. O6-Benzylguanine potentiates BCNU but not busulfan toxicity in hematopoietic stem cells. Exper. Hematol. 2001;29:633–638. doi: 10.1016/s0301-472x(01)00631-2. [DOI] [PubMed] [Google Scholar]
- 57.Sanyal U, Nanda R, Samanta S, Pain A, Dutta S, Verma AS, Rider BJ, Agrawal KC. Evaluation of dimethylaminosulfonates of alkane diols as a novel group of anticancer agents. Cancer Lett. 2000;155:89–97. doi: 10.1016/s0304-3835(00)00409-2. [DOI] [PubMed] [Google Scholar]
- 58.Pacheco DY, Stratton NK, Gibson NW. Comparison of the mechanism of action of busulfan with hepsulfam, a new antileukemic agent, in the L1210 cell line. Cancer Res. 1989;49:5108–5110. [PubMed] [Google Scholar]
- 59.Streeper RT, Cotter RJ, Colvin ME, Hilton J, Colvin OM. Molecular pharmacology of hepsulfam, NSC 3296801: identification of alkylated nucleosides, alkylation site, and site of DNA cross-linking. Cancer Res. 1995;55:1491–1498. [PubMed] [Google Scholar]
- 60.Hendricks CB, Grochow LB, Rowinsky EK, Forastiere AA, McGuire WP, Ettinger DS, Sartorius S, Lubejko B, Donehower RC. Phase I and pharmacokinetic study of hepsulfam (NSC 329680) Cancer Res. 1991;51:5781–5785. [PubMed] [Google Scholar]
- 61.Liu L, Williams KM, Guengerich FP, Pegg AE. O6-alkylguanine-DNA alkyltransferase has opposing effects in modulating the genotoxicity of dibromomethane and bromomethyl acetate. Chem. Res. Toxicol. 2004;17:742–752. doi: 10.1021/tx049958o. [DOI] [PubMed] [Google Scholar]
- 62.Valadez JG, Liu L, Loktionova NA, Guengerich FP, Pegg AE. Activation of bis-electrophiles to mutagenic conjugates by human O6-alkylguanine-DNA alkyltransferase. Chem. Res. Toxicol. 2004;17:972–982. doi: 10.1021/tx049897u. [DOI] [PubMed] [Google Scholar]
- 63.Loeber R, Rajesh M, Fang Q, Pegg AE, Tretyakova N. Cross-Linking of the human DNA repair protein O6-alkylguanine DNA alkyltransferase to DNA in the presence of 1,2,3,4-diepoxybutane. Chem. Res. Toxicol. 2006;19:645–654. doi: 10.1021/tx0600088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Loeber R, Michaelson E, Fang Q, Campbell C, Pegg AE, Tretyakova N. Cross-linking of the DNA repair protein O6-alkylguanine DNA alkyltransferase to DNA in the presence of antitumor nitrogen mustards. Chem. Res. Toxicol. 2008;21:787–795. doi: 10.1021/tx7004508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pegg AE, Scicchitano D, Dolan ME. Comparison of the rates of repair of O6-alkylguanines in DNA by rat liver and bacterial O6-alkylguanine-DNA alkyltransferase. Cancer Res. 1984;44:3806–3811. [PubMed] [Google Scholar]
- 66.Morimoto K, Dolan ME, Scicchitano D, Pegg AE. Repair of O6-propylguanine and O6-butylguanine in DNA by O6-alkylguanine-DNA alkyltransferases from rat liver and E. coli. Carcinogenesis. 1985;6:1027–1031. doi: 10.1093/carcin/6.7.1027. [DOI] [PubMed] [Google Scholar]
- 67.Wang L, Spratt TE, Liu X-L, Hecht SS, Pegg AE, Peterson LA. Pyridyloxobutyl adduct, O6-[4-oxo-4-(3-pyridyl)butyl]guanine, is present in 4-(acetoxymethylnitrosamino -1-(3-pyridyl)-1-butanone-treated DNA and is a substrate for O6-alkylguanine-DNA alkyltransferase. Chem. Res. Toxicol. 1997;10:562–567. doi: 10.1021/tx9602067. [DOI] [PubMed] [Google Scholar]
- 68.Noll DM, Clarke ND. Covalent capture of a human O6-alkylguanine alkyltransferase-DNA complex using N1-O6-ethanoxanthosine, a mechanism-based crosslinker. Nucleic Acids Res. 2001;29 doi: 10.1093/nar/29.19.4025. 4025-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]