Abstract
We report a serious problem associated with a number of current implementations of Andersen and Langevin dynamics algorithms. When long simulations are run in many segments, it is sometimes possible to have a repeating sequence of pseudorandom numbers enter the calcuation. We show that, if the sequence repeats rapidly, the resulting artifacts can quickly denature biomolecules and are then easily detectable. However, if the sequence repeats less frequently, the artifacts become subtle and easily overlooked. We derive a formula for the underlying cause of artifacts in the case of the Langevin thermostat, and find it vanishes slowly as the inverse square root of the number of time steps per simulation segment. Numerous examples of simulation artifacts are presented, including dissociation of a tetrameric protein after 110 ns of dynamics, reductions in atomic fluctuations for a small protein in implicit solvent, altered thermodynamic properties of a box of water molecules, and changes in the transition free energies between dihedral angle conformations. Finally, in the case of strong thermocoupling, we link the observed artifacts to previous work in nonlinear dynamics and show that it is possible to drive a 20-residue, implicitly solvated protein into periodic trajectories if the thermostat is not used properly. Our findings should help other investigators re-evaluate simulations that may have been corrupted and obtain more accurate results.
Introduction
Molecular simulations of proteins and other complex biomolecules are performed routinely in atomic detail for tens of nanoseconds. A variety of thermodynamic ensembles are available for these simulations, but in virtually all cases, investigators wish to see the dynamics of a system at a particular temperature, corresponding to a Maxwell distribution of momenta for the particles of the molecular model. In simulations of complex biomolecules, the systems typically contain enough inhomogeneity that complete equilibration across all degrees of freedom is not possible over currently achievable simulation timescales, meaning that potential energy will tend to be released as structures relax. This, in addition to the slow but inevitable increase of energy in the system because of the finite time steps taken to propagate the dynamics, leads to an upward drift in the system temperature as the simulation continues. Algorithms, such as SHAKE,1 which apply constraints to a finite degree of precision, can also add to or even dissipate the system’s energy, leading to more temperature drift.
To run simulations on the timescales needed to model chemical processes, a number of algorithms have been developed to maintain a specified system temperature. These include velocity rescaling approaches such as the Berendsen2 and Nose-Hoover3 thermostats and velocity modification approaches such as the Andersen4 and Langevin thermostats.5 In Andersen thermocoupling, particle velocities are periodically reassigned to pseudorandom values so that the resulting momenta follow a Maxwell distribution at the desired temperature. In the Langevin scheme, velocities of the particles in the simulations are modified with pseudorandom forces as if they were undergoing stochastic collisions with imaginary particles whose momenta follow a Maxwell distribution at the desired temperature.
The importance of generating long, decorrelated sequences of random numbers for accurate simulations has been discussed before,6,7 and modern molecular dynamics codes use algorithms8,9 that can generate sequences so long that they would be unlikely to repeat over the course of a simulation even if millions of particles were simulated for trillions of time steps (for example, if a virus capsid were simulated at atomic detail for several milliseconds).
However, modern molecular dynamics codes also offer a large number of options for managing simulations, and it is difficult to anticipate all the permutations of how those options might be used. Long simulations can generate tens of gigabytes of trajectory data and take weeks or months to complete. For this reason, checkpoint files are nearly always used to store the positions and velocities of atoms so that the simulation may be broken up into small segments that make it feasible to run on managed computing resources and easy to recover from a machine crash. However, in several popular molecular dynamics packages, the checkpoint files do not contain information on the state of the random number generator. In such cases, reuse of the same random number generator seed causes a finite sequence of random numbers to appear in every simulation segment. As will be shown, these repeating sequences of random numbers can drastically affect simulations using either Langevin or Andersen thermostats if the simulation segments are short; the effects can be subtle but significant if the segments are longer.
To help determine when this issue may produce significant problems in typical simulations, we have quantified the effects of repeating sequences of pseudorandom numbers in several test systems, including two explicitly solvated proteins, and surveyed existing codes to see which packages are vulnerable. We also show that simply incrementing the random number seed with each simulation segment effectively removes the artifacts.
Theory
The effects of repeating sequences of pseudorandom numbers are straightforward to describe in the case of the Langevin thermostat, as we show in the following formalism. We expect that similar principles hold for the Andersen thermostat.
The Langevin thermostat maintains a desired temperature by application of a friction force with coefficient ζ and a random force Ri to all particles i to simulate random collisions between particles in the simulation and imaginary particles in an external bath held at temperature T. In this framework, collisions with particle i occur at a frequency γi
| (1) |
such that the central equation of motion is
| (2) |
For real water, γ has a value of roughly 50 ps−1. In simulations, smaller values of 2–5 ps−1 are typically used,10 although some investigators have found that the full 50 ps−1 gives better results.11 The random force is related to γ by eq 3
| (3) |
where the angular brackets denote an ensemble average, kb is Boltzmann’s constant, and δ(t) is the Dirac delta function. By eq 3, the components of the instantaneous random force vector at time t follow a Gaussian distribution with zero mean and variance 2miγikbT, henceforth denoted σ2R.
To understand the effect of repeating random number sequences on molecular dynamics simulations, we consider Ψ(N), the “residual” force on a particle causesed by Langevin collisions after N steps of simulation. Each of the three components of Ψ can be expressed as
| (4) |
where α represents x, y, or z. Ψ is an average rather than a sum because each of the random forces is only applied during one of the N steps. By the Central Limit Theorem,12 the distribution of Ψ is also Gaussian with zero mean and variance σ2R/N. Therefore, the magnitude of each component of the residual force on an atom after N Langevin dynamics steps of length Δt can be expressed as
| (5) |
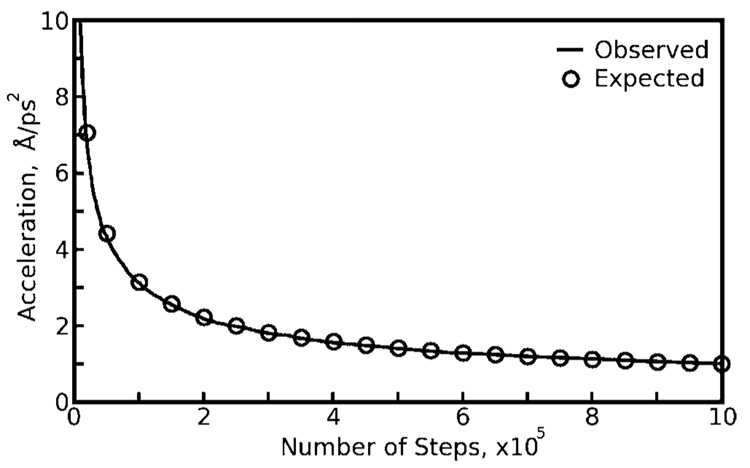
If the same sequences of N pseudorandom forces are used repeatedly in a Langevin dynamics simulation, each atom is exposed to a finite number of forces and therefore a nonvanishing residual force. Over many iterations, this is similar to applying a constant force to each particle, in a particular direction relative to the axes of the simulation cell, with a magnitude given by eq 5. The expected and observed magnitudes of residual forces for a Langevin thermostat with collision frequency 3 ps−1 and a bath temperature of 298 K are plotted as a function of N in Figure 1. (Observations of the residual forces were made with a modified version of the AMBER9 PMEMD software, available upon request.)
Figure 1.
Residual accelerations on atoms observed in Langevin dynamics. Langevin forces on individual atoms were summed over steps of a molecular dynamics run of the Trp-Cage miniprotein using collision frequency 3 ps−1 and a bath temperature of 298 K. Averaging the forces over N previous steps gives a value for the residual force on that atom, a quantity which tends to zero as 1/√N. Residual forces on each atom were normalized by the atom’s mass to give accelerations. The black line shows average residual acceleration for all atoms; circles show the values expected from eq 5.
Although Figure 1 clearly shows significant residual forces acting on each atom even for lengthy simulation segments, the forces do not act in any concerted fashion (see Figure S1 of the Supporting Information), and so, their overall effects must be determined by simulations. As we will show in the Results, these residual forces quickly give rise to severe artifacts when short simulation segments are used, but subtle artifacts can occur with greater segment lengths, such as those investigators might use in practice.
Methods
Proteins for molecular dynamics simulations were obtained from the Protein Data Bank (PDB).13 All proteins were protonated using the TLEAP module of AMBER914 and modeled using the AMBER ff99 force field,15,16 with improvements suggested by Simmerling et al.17 SPC/E water18 was used for simulations in explicit solvent. The Generalized Born (GB) model of Onufriev et al.19 combined with the LCPO pairwise surface area approximation20 was used for simulations in implicit solvent. The PMEMD and SANDER modules of AMBER9 were used for simulations in explicit and implicit solvent, respectively.
Molecular dynamics simulations in explicit solvent were initiated by adjusting positions of added water molecules with 2000 steps of steepest-descent energy minimization, while restraining the positions of protein atoms, then performing similar energy minimization of the protein atoms with the solvent held fixed, and finally running energy minimization of the entire system with no restraints. Energy minimization of the protein was also done prior to implicit solvent simulations. Equilibration dynamics in all simulations were performed at a constant temperature of 298 K using a Langevin thermostat with a collision frequency of 3.0 ps−1 (unless otherwise stated, this temperature and collision frequency were used in all simulations in this study). Position restraints were initially used to limit the motion of all heavy atoms; the restraints were gradually relaxed over a period of 500 ps. For simulations in explicit solvent, periodic boundary conditions were applied, and the simulation volume was held constant until the final stages of equilibration, when dynamics were continued in the constant-pressure ensemble. For implicit solvent calculations, no boundary conditions were used. The equilibration phase typically involved about ten restarts; different random seeds were used to initialize the pseudorandom number generator with each restart.
Force calculations for all stages of dynamics in explicit solvent were performed with a 9.0 Å cutoff on real-space interactions, particle-mesh Ewald electrostatics,21 and Lennard-Jones tail corrections. Force calculations in implicit solvent were performed with no cutoff on real-space interactions and a 25 Å cutoff on calculations of the Born radii. The SHAKE algorithm1 was used to constrain all bonds including hydrogen on protein atoms, and the SETTLE algorithm22 was used to constrain the internal geometry of explicit water molecules. A time step of 1.5 fs was used for all production dynamics.
Results
Langevin Artifacts in Explicit Solvent
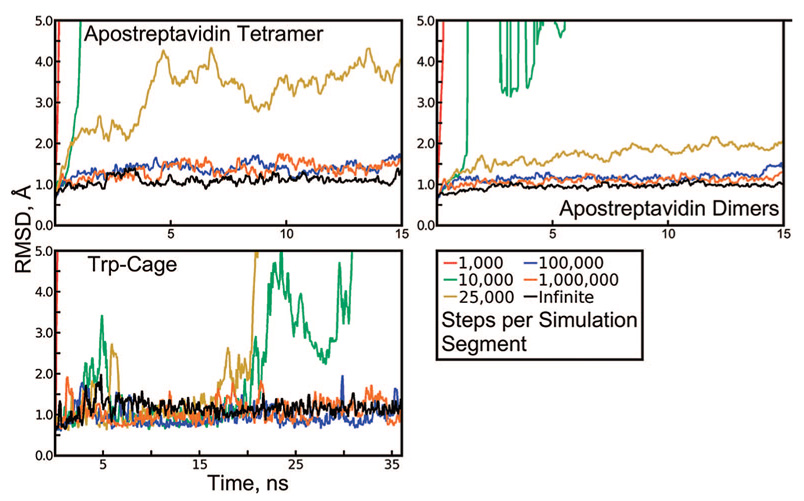
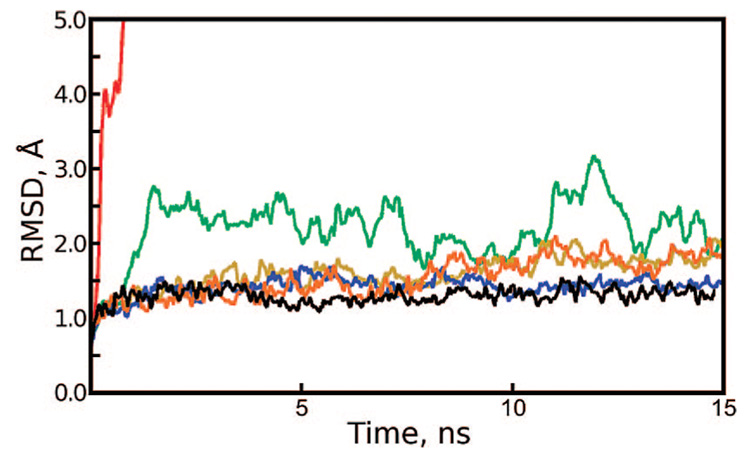
We first became aware of the danger of repeating random number sequences when we noticed that the apostreptavidin tetramer (PDB accession code 1SWA) was relatively stable in explicit solvent when dynamics were propagated at 100 000 steps (150 ps) per segment but rapidly unfolded when dynamics were propagated at 1000 steps per segment. A Langevin thermostat with a collision frequency of 3 ps−1 had been used to maintain the temperature at 298 K, and the same random seed had been provided to initialize the pseudorandom number generator (PRNG) in all cases. To quantify the protein destabilization effect, Figure 2 shows backbone root-mean-squared deviation (rmsd) results, taking the equilibrated conformation of the protein as a reference, for a series of 15 ns simulations of the tetramer using different segment lengths. When the PRNG is repeatedly initialized with the same seed, the segment length corresponds to the parameter N as discussed in Theory. For comparison, a nonrepeating sequence of Langevin forces was generated by running the same simulation with 100 000 steps per segment and changing the PRNG seed with each restart. To demonstrate that the protein is destabilized by repeating sequences of Langevin forces and not some problem with restarting a simulation from a checkpoint file, we performed a 6 ns simulation with 1000 steps per segment, incrementing the PRNG seed with each restart. The results in Figure S3 of the Supporting Information show that this method also results in stable dynamics.
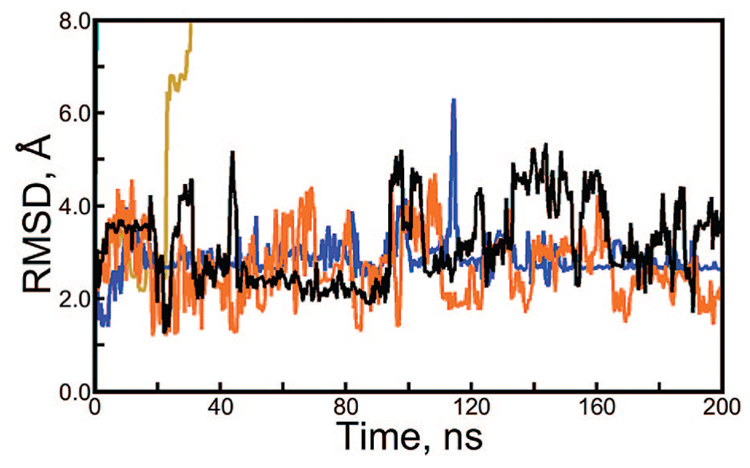
Figure 2.
Backbone rmsd of apostreptavidin and Trp-Cage, revealing artifacts in Langevin dynamics. Each protein was simulated in explicit solvent at 298 K using a Langevin thermostat with a collision frequency of 3 ps−1 and simulation segments with lengths given in the figure legend. The same random seed was used to reinitialize the pseudorandom number generator (PRNG) at the beginning of every segment, except for the “infinite” case, in which segments of 100 000 steps were initiated with different PRNG seeds every time.
The streptavidin tetramer is a dimer of dimers.23 The two dimers are each more stable than the tetramer as a whole, as demonstrated by the existence of dimeric streptavidin mutants24 and the mechanism of tetramer stabilization by biotin binding.25 For this reason, we tracked backbone rmsd not just for the tetramer but also for its dimer components. As shown in Figure 2, individual dimers maintained their original backbone conformations better than the tetramer as a whole under cycles of repeating sequences of Langevin forces. rmsd for the individual monomers is not shown, but it closely parallels the dimer rmsd.
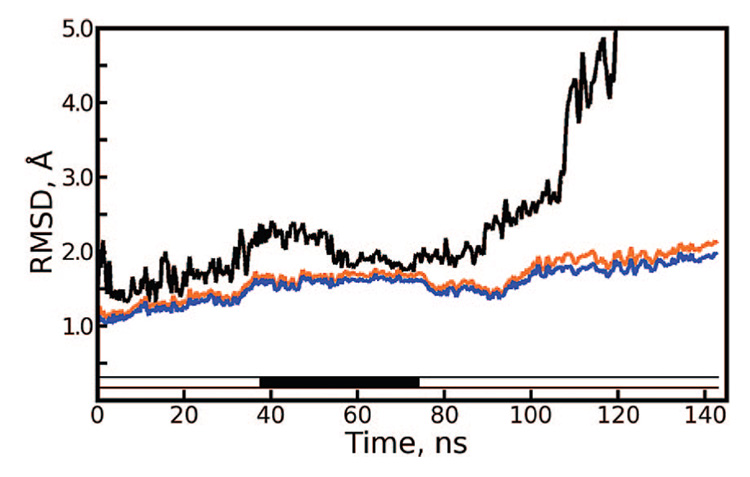
If very long sequences of repeating Langevin forces (100 000 and 1 000 000 steps per segment) are used, artifacts are difficult to detect in simulations of only 15 ns. Before we became aware of the problem with Langevin dynamics, a simulation of the apostreptavidin tetramer was carried out for 145 ns, using 100 000 and 1 000 000 step segments at different times but with the same PRNG seed in all cases. Backbone rmsds for monomers, dimers, and the tetramer in this system are shown in Figure 3. At a glance, the system appears to behave reasonably, except for the dissociation of the tetramer at 110 ns.
Figure 3.
Long-time scale Langevin dynamics of apostreptavidin tetramer under repeating sequences of Langevin forces. Root-mean-sqaured deviation (rmsd) of the tetramer (black line), average dimer rmsd (orange line), and average monomer rmsd (blue line) are plotted over 145 ns. The bar just above the x-axis is solid black when 1 000 000 step segments were used and white when 100 000 step segments were used. The period of simulation using the longer segments shows a slight reduction in the rmsd of the tetramer and stable RMSDs for dimers and monomers. In contrast, all of these rmsd values steadily increase, particularly that of the tetramer, when segments of 100 000 steps are used. Figure S2 of the Supporting Information illustrates the tetramer dissociation at 110 ns.
The apostreptavidin tetramer is known to be highly stable, even in concentrated urea,26 so the dissociation seen in Figure 3 and in Figure S2 of the Supporting Information is not realistic. But because the tetramer is known to be stabilized by biotin binding27 and because we had a 250 ns simulation showing the biotin-liganded tetramer to be stable in solution (data not shown), we initially believed that the dissociation of the unliganded tetramer was qualitatively correct. However, inspection of the rmsd for portions of the trajectory run with 1 000 000 versus 100 000 steps per segment suggests that over a very long simulation the tetramer can be destabilized by 100 000 step segments with identical PRNG seeds in much the same way that shorter segments destabilize it more quickly. Indeed, with sequences of 100 000 pseudorandom Langevin forces acting on each atom, the residual forces described in equ 5 would have been half as strong as those obtained with sequences of 25 000 Langevin forces, which created artifacts immediately.
To further investigate the extent of these artifacts, we conducted 36 ns simulations of the 20-residue Trp-Cage miniprotein in explicit solvent and subjected the system to repeating sequences of Langevin forces in the same manner as was done with the 500-residue apostreptavidin tetramer. The results in Figure 2 show that Trp-Cage also unfolds under rapidly repeating Langevin forces, but remains stable if the Langevin thermostat is used correctly. Notably, whereas residual forces from a repeating sequence of 25 000 Langevin forces caused some instability in the apostreptavidin system, residual forces of the same magnitude denatured the Trp-Cage miniprotein. Moreover, under repeating sequences of 10 000–25 000 Langevin forces, Trp-Cage appeared to be stable for 17–20 ns before suddenly unfolding.
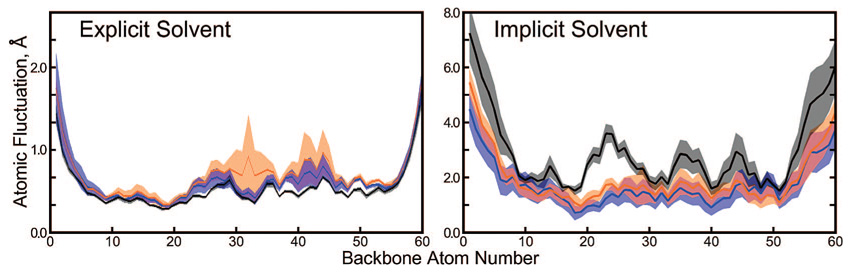
Still, the extent of artifacts in simulations using 100 000 or 1 000 000 steps per segment remains uncertain. Because the residual forces do not act in a concerted fashion, their effects may be more pronounced on local features of the protein structure. We therefore computed atomic root-mean-squared (rms) fluctuations for backbone atoms over the final 30 ns of each simulation as shown in Figure 5. Error bars were determined by computing rms fluctuations over four 7.5 ns subintervals and taking the standard deviation. In explicit solvent, the computed fluctuations do not differ greatly if the thermostat applies sequences of 100 000, 1 000 000, or an infinite number of Langevin forces. Furthermore, there is no apparent trend in the data; atomic fluctuations for nearly all backbone atoms increase slightly if repeating sequences of 1 000 000 Langevin forces instead of 100 000 are used, but they decrease again if an infinite sequence of Langevin forces is used. The amount of sampling in 36 ns of dynamics is rather small, however. In the next section, we sample protein conformations over longer time scales with a Langevin thermostat and an accelerated dynamics method.
Figure 5.
Atomic fluctuations of Trp-Cage backbone atoms in two solvent environments. The numbering of atoms on the x-axis proceeds as (residue 1) N, CA, C, (residue 2) N, CA, C,…, (residue 20) N, CA, C. Fluctuations for simulations with sequences of 105, 106, and an infinite number of Langevin forces are shown as the blue, orange, and black lines, respectively. Error bars are given in the same colors as partially transparent regions surrounding each line. Simulations in explicit solvent were run for 36 ns, and simulations in implicit solvent were run for 200 ns. Note that the y-axis has a different scale in each panel.
Langevin Artifacts in Implicit Solvent
Although a Langevin thermostat can be used with explicitly solvated systems, it is more commonly used to simulate stochastic collisions with imaginary solvent particles in an implicitly solvated system. We therefore conducted simulations of the Trp-Cage miniprotein28 in Generalized Born (GB) solvent. Because the system is so small (300 atoms versus 8000 for the explicitly solvated Trp-Cage versus 40 000 for the explicitly solvated apostreptavidin tetramer), we were able to obtain very long (200 ns) simulations and more convergent estimates of atomic fluctuations.
As shown in Figure 4, the Trp-Cage miniprotein explores conformations with larger backbone rmsd relative to the native state in GB implicit solvent as opposed to SPC/E explicit solvent. Again, with repeating sequences of 100 000 or more Langevin forces acting on each atom, Trp-Cage is stable, but with fewer sequences, it becomes denatured quickly.
Figure 4.
Backbone rmsd of the Trp-Cage miniprotein revealing artifacts in Langevin dynamics. The Trp-Cage miniprotein was simulated in Generalized Born solvent using a Langevin thermostat. Simulations with different segment lengths are plotted in different colors following the legend in Figure 2. Simulations with 1000 and 10 000 steps per segment unfolded within 2 ns and, so, are not visible on the plot.
Atomic fluctuations for backbone C atoms obtained from the final 180 ns of trajectories with 100 000 and 1 000 000 step segments using repeating random seeds are compared to those obtained from a trajectory generated with constantly changing random seeds in Figure 5. As before, error bars were created by splitting the data into four 45 ns segments and computing standard deviations. In implicit solvent, the atomic fluctuations generally increase as the length of the repeating sequence of random forces goes from 100 000 to infinity. Because more than six times as many conformations were used to calculate these fluctuations, the results are somewhat more certain than those from the explicit solvent simulations. Although the error bars look larger in the implicit solvent case, as a fraction of the corresponding fluctuations the error bars in implicit solvent are in fact roughly two times smaller. While short repeating sequences of Langevin forces acting on each atom denatured the protein, sequences of 100 000 forces appeared to reduce its mobility relative to much longer ones. This apparent contradiction may be explained by looking at the backbone rmsd obtained for shorter repeating sequences of Langevin forces, as shown in Figure S4 of the Supporting Information. In such cases, the rmsd may climb to very high values, but then hovers around particular values for extended periods of time, suggesting that the denatured conformations do not fluctuate very much.
The atomic fluctuations only appear to diminish in the absence of explicit solvent particles, however (see Figure S5, Supporting Information). When solvent is represented explicitly, denatured protein structures tend to fluctuate more even after the native conformation is lost. This dichotomy likely arises as the individual water molecules can migrate to different regions of the protein even if subjected to repeating sequences of Langevin forces. (Indeed, as will be discussed later in the Results, if the sequences are very short, the water molecules are all being propelled in particular directions and the polypeptides are literally showered with rapidly moving water molecules.) These solvent interactions impart instability on the polypeptide motion, increasing the atomic fluctuations, whereas in implicit solvent the polypeptide moves only according to the Langevin forces acting on its own atoms and thereby becomes trapped in a particular conformation.
The dissociation of the apostreptavidin tetramer over very long simulations in explicit solvent and reduced atomic fluctuations of the Trp-Cage miniprotein in implicit solvent give indications that simulations run with repeating sequences of 100 000 Langevin forces are not safe from artifacts. However, different PRNG seeds will create unique repeating sequences of Langevin forces that will affect the system in different ways, whereas our results thus far have shown the effects of only one sequence of a given length on each system tested. To precisely quantify the microscopic effects of residual forces as a function of the simulation segment length, we needed to be able to thoroughly sample the entire conformational space of a system and run many simulations with different sequences of Langevin forces.
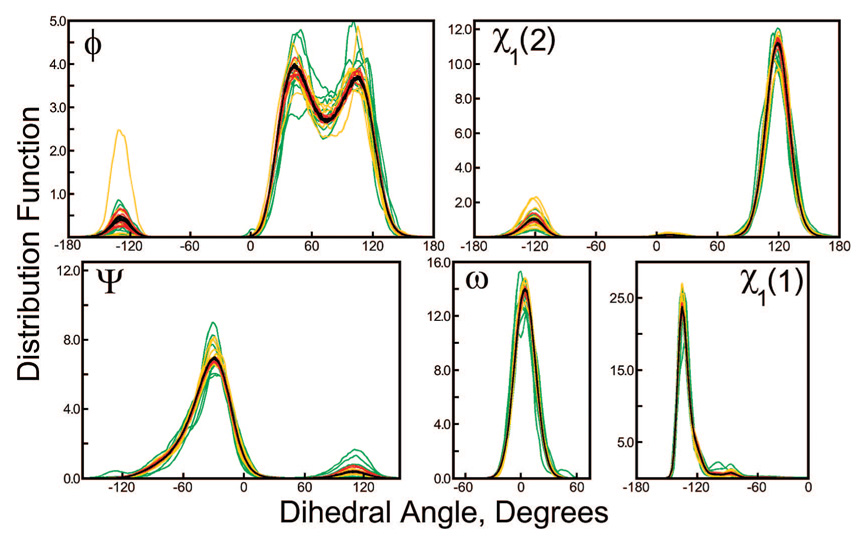
For this purpose, we chose to study the seryl-serine peptide in implicit solvent. Because the serine side-chain is so small, residual forces acting on it will not be averaged over many atoms, and therefore, its χ1 angle should be very prone to reorientation due to these forces. Eight independent simulations of 1 µs were done using segments of 25 000, 100 000, and 1 000 000 steps with repeating random seeds, as well as segments of 100 000 steps with changing random seeds. Distributions of χ1 angles for each serine side-chain, as well as three backbone dihedral angles, are shown in Figure 6. While nonrepeating sequences of Langevin forces consistently generate the same distribution for each of the dihedral angles, unique repeating sequences of Langevin forces each impart their own bias on the system, causing the distribution of dihedral angles to converge differently in each case. As expected, the distortions grow larger as the simulation segment length decreases.
Figure 6.
Distributions of five dihedral angles in the seryl-serine system under finite sequences of Langevin forces. Eight independent trajectories of the seryl-serine system were computed with 25 000 (green lines), 100 000 (yellow lines), 1 000 000 (red lines), and an infinite sequence of Langevin forces (black lines) acting on each atom. The distributions above are normalized by the expected population of each dihedral angle value if the potential energy surface were completely flat. The distributions obtained for infinite sequences of Langevin forces are mutually convergent, demonstrating the thoroughness of the sampling from these 1000 ns simulations. However, finite sequences of Langevin forces tend to perturb the distributions. These perturbations are quantified in terms of transition free energies in Table 1.
The thoroughness of equilibrium sampling in the seryl-serine system permitted direct calculation of transition free energies, ΔG, by comparing the probability of finding each of the five dihedral angles at two values ν and η in the unbiased ensemble (the trajectory computed with a nonrepeating sequence of Langevin forces). We also computed changes in the transition free energies, ΔΔG, between the unbiased ensemble and each of the biased ensembles generated with finite sequences of Langevin forces. These quantities are defined mathematically as
| (6) |
| (7) |
In the above equations, T represents the temperature (298 K), and R represents the gas constant. Results from this analysis are given in Table 1. The table only reports average values of ΔΔG, but individual cases showed changes in the transition free energies in excess of 1 kcal/mol for some of the biased ensembles obtained with 100 000 steps per segment. Contrary to our expectations, the largest ΔΔG values were obtained in the backbone ϕ angle of the second residue; residual forces on many atoms exert torques about this dihedral, yet the distortion resulting from an average of all these torques remains large. Although the distributions of each dihedral angle in the unbiased ensemble may not be totally accurate, the computed values of ΔG and ΔΔG provide precise measurements of the degree to which simulations using finite sequences of Langevin forces are biased, as well as the degree of bias present in short simulations (40 ps to 1.5 ns) using Langevin dynamics.
Table 1.
Free Energies for Transitions between Two Values, ν and η, of Various Dihedral Angles in the Seryl-Serine Systema
| dihedral angle | ν (deg) | η (deg) | 〈ΔG〉 | 〈|ΔΔG|〉 (2.5 × 104) | 〈|ΔΔG|〉 (105) | 〈|ΔΔG|〉 (106) |
|---|---|---|---|---|---|---|
| residue 1, χ1 | −123 | −76 | −2.07 ± 0.01 | 0.37 ± 0.25 | 0.28 ± 0.18 | 0.10 ± 0.07 |
| residue 2, χ2 | 120 | −120 | −1.42 ± 0.02 | 0.31 ± 0.16 | 0.32 ± 0.17 | 0.07 ± 0.07 |
| residue 1, ψ | 107 | −29 | 1.72 ± 0.04 | 0.47 ± 0.31 | 0.20 ± 0.11 | 0.14 ± 0.14 |
| residue 2, ϕ | 43 | 105 | −0.04 ± 0.01 | 0.11 ± 0.09 | 0.08 ± 0.08 | 0.03 ± 0.02 |
| residue 2, ϕ | 105 | 231 | −1.27 ± 0.05 | 0.77 ± 0.50 | 0.70 ± 0.50 | 0.20 ± 0.11 |
| residue 2, ϕ | 43 | 231 | −1.31 ± 0.06 | 0.74 ± 0.48 | 0.69 ± 0.52 | 0.21 ± 0.12 |
The chosen values of ν and η correspond to relative maxima indentified in the unbiased ensemble (see Figure 6) generated with an infinite sequence of Langevin forces. ΔG values, reported with standard deviations in kcal/mol, refer to the transition free energy for the unbiased ensemble (see eq 6); angular brackets 〈 〉 refer to averages from eight independent trajectories. Similarly, |ΔΔG| values refer to absolute changes in the transition free energy if a finite sequence of Langevin forces (of length specified in parentheses) is used (see eq 7).
Severity of Artifacts As a Function of the Langevin Collision Frequency
As was predicted in Theory and shown in the preceding results, the severity of artifacts from the Langevin thermostat diminishes as the length of the repeating seqeunce of pseudorandom forces grows. However, by eq 5, the magnitude of residual forces and thus the severity of artifacts is also proportional to the square root of the collision frequency γi, and different values of this parameter have been used in the past.10,11 We therefore repeated some of the simulations of Trp-Cage in implicit solvent with γi set to 50 ps−1 rather than 3 ps−1. By eq 5, we would expect the higher collision frequency to increase the average residual force on each atom roughly by a factor of 4. With a collision frequency of 3 ps−1, a segment length of 6000 steps would be needed to obtain residual forces of comparable magnitude (this was verified with the modified AMBER9 PMEMD code used to generate Figure 1).
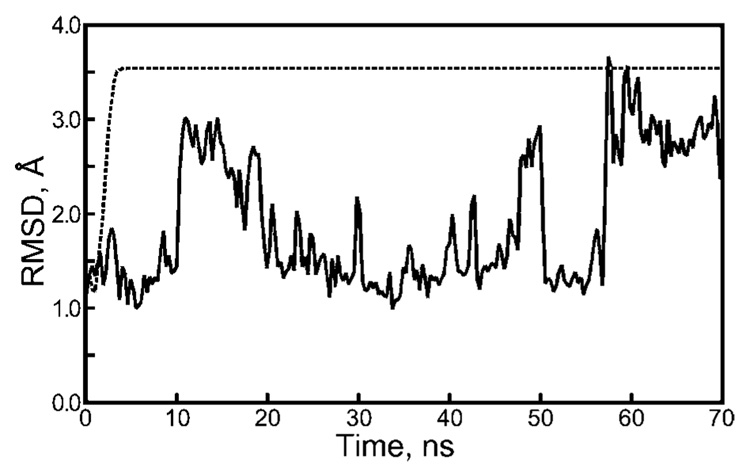
The results in Figure 7 confirm that, even with relatively long 100 000 step segments, the 50 ps−1 Langevin collision frequency can generate a striking artifact when combined with repeating sequences of pseudorandom forces. As indicated by the system’s convergent backbone rmsd, the Trp-Cage miniprotein is driven to a very small set of structures under these conditions. Examination of the checkpoint files from each segment of the simulation shows that, within 12 ns, the coordinates and velocities are converged to 1.0 × 10−7 Å and 1.0 × 10−7 Å ps−1, respectively, and the trajectory segments are identical thereafter. Although it was surprising to obtain periodic behavior over such long (150 ps) intervals in such a complex system, we also observed periodic behavior for 50 000 step segments and 200 000 step segments (data not shown). Periodicity was not observed in the trajectory if the random seed was changed with each restart (see Figure 7) or if a collision frequency of 3 ps−1 was used (see Figure 4).
Figure 7.
Backbone rmsd of the Trp-Cage miniprotein during Langevin dynamics with strong thermocoupling. When each segment is initiated with the same random seed (dashed line), the repeating sequence of 100 000 Langevin forces drives the protein into a periodic trajectory (see Results, Severity of Artifacts As a Function of the Langevin Collision Frequency section). No such behavior is seen if an infinite sequence of Langevin forces is used instead (solid line).
These observations led us to consider the possibility that the periodic behavior observed with strong thermocoupling was related to the protein unfolding seen in previous sections. If so, the fact that the strongly thermocoupled Trp-Cage system run in long segments did not unfold to the same extent as the weakly thermocoupled Trp-Cage system run in short segments (see Figure 4) needed further investigation. We emphasize that, as discussed in the preceding Langevin Artifacts in Implicit Solvent section, different repeating sequences of Langevin forces may drive the system into different conformations, and it is conceivable that occasionally these conformations would fall close to the native state. We therefore ran three additional simulations with 100 000 steps per segment, repeating random seeds, and a collision frequency of 50 ps−1. All trajectories eventually became periodic, but the time to obtain this behavior varied for each different sequence of Langevin forces (see Figure S6 of the Supporting Information), and the length of the period was five simulation segments, rather than just one, in one of the cases. Although each periodic trajectory displayed a different level of backbone rmsd relative to the native state, all of the backbone RMSDs were much lower than the backbone RMSDs eventually seen in similar runs with repeating sequences of 10 000–25 000 Langevin forces (see Figure 4).
In summary, a Langevin thermostat with a collision frequency of 50 ps−1 drove the Trp-cage miniprotein into periodic trajectories 100 000–500 000 steps long. Under such strong thermocoupling, repeating sequences of Langevin forces did not denature the system to the extent seen before, but a periodic trajectory does represent an extreme restriction of the protein’s conformational space.
Artifacts in a Simulation of Pure Water
With an explicitly solvated protein system, the motions of atoms in the protein are tightly coupled, but the motions of solvent particles are not. In the previous section, we tested the effects of repeating sequences of Langevin forces if the system contains only the tightly coupled degrees of freedom; conversely, we can look for artifacts in the thermodynamic properties of a system containing many small, unconnected particles.
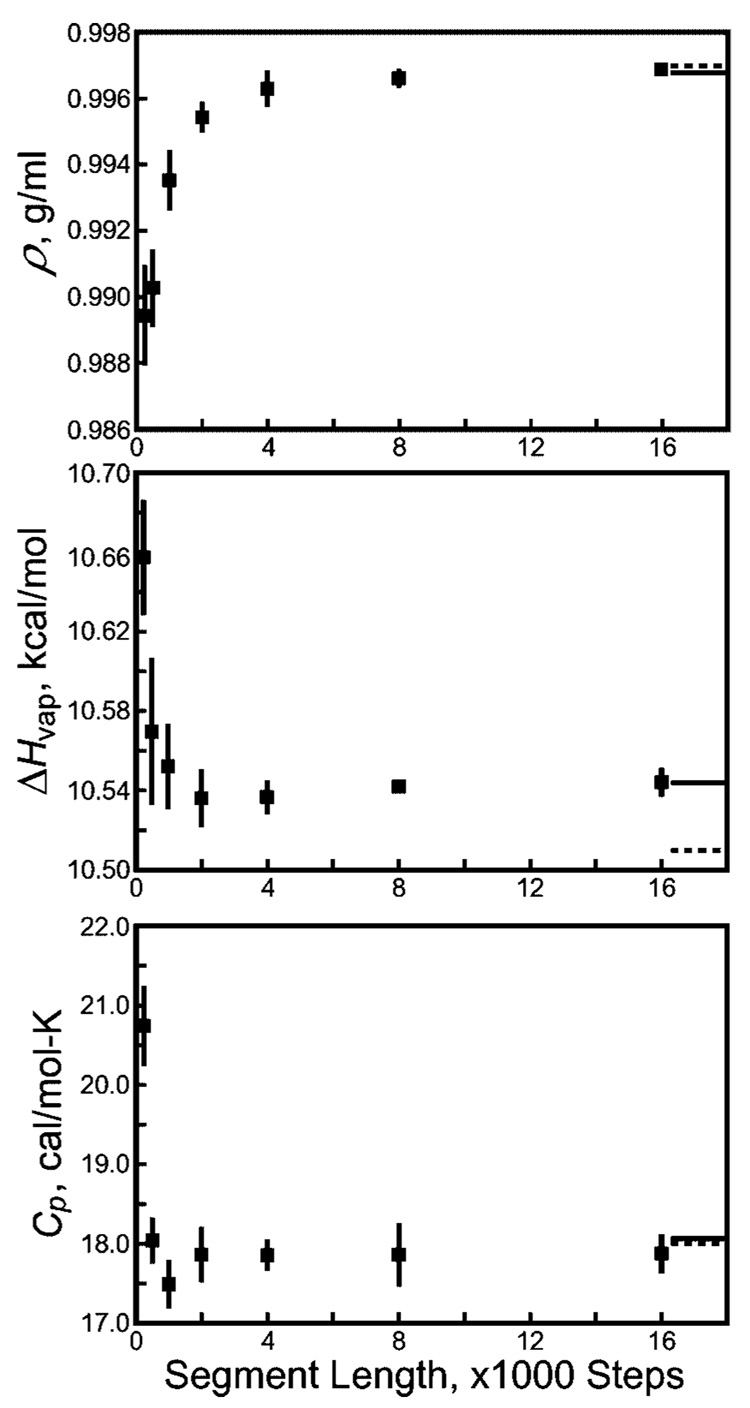
Multiple 6 ns simulations of a box of 512 SPC/E water molecules were conducted at 1 atm pressure and 298 K using a Langevin thermostat (collision frequency 3 ps−1). Constant random seeds were used to restart the simulations in segments ranging from 250–16 000 steps, and four independent simulations were conducted using unique random seeds for each segment length. As shown in Figure 8, the density, heat of vaporization, and heat capacity of SPC/E water all change noticeably for segments with fewer than 4000 steps. In such simulations, one cannot obtain a convergent value of the diffusion coefficient because every water molecule suffers a net displacement along a particular direction during each segment. However, compared to the artifacts observed in solvated proteins, the density, heat of vaporization, and heat capacity of water are not very sensitive to Langevin artifacts.
Figure 8.
Thermodynamic properties of a box of 512 SPC/E water molecules revealing Langevin artifacts. Formulas for the density (ρ), heat of vaporization (ΔHvap), and heat capacity (Cp) can be found in work by Jorgensen and Jenson43 (note that the polarization energy correction18 is invoked in computing ΔHvap). Solid black lines extending from the right border indicate the values of each quantity if a nonrepeating sequence of Langevin forces is used; dashed lines indicate experimental results for water at 298 K. Error bars are obtained from four indpendent simulations.
Artifacts Created by Repeating Random Number Sequences with the Andersen Thermostat
Although we did not provide a formal description of the way repeating sequences of velocity reassignments could create artifacts if the system temperature is maintained by an Andersen thermostat, we expected that this would have similar effects to applying repeating sequences of forces. An array of 15 ns simulations was carried out for the apostreptavidin tetramer with the same repeating random seeds and segment lengths as in the case of the Langevin thermostat. Results are shown in Figure 9. As before, the use of the Andersen thermostat with a repeating PRNG seed destabilized the tetramer, indicating that the Andersen thermostat can create artifacts in much the same manner as the Langevin thermostat.
Figure 9.
Backbone rmsd of the apostreptavidin tetramer revealing artifacts in Andersen dynamics. The apostreptavidin tetramer was simulated in explicit solvent with repeating sequences of Andersen velocity reassignments. The legend in Figure 2 indicates the length of segments in each simulation; velocity reassignment occurred every 1000 steps (e.g., the red line presents backbone rmsd of the tetramer when all atoms are reassigned to the same set of velocities every 1000 steps).
Although the severity of the artifacts appear to be smaller in terms of backbone rmsd than the artifacts created by Langevin dynamics with similar segment lengths, we stress that the strength of thermocoupling in each thermostat is determined differently and that this can also influence the severity of artifacts (see Severity of Artifacts As a Function of the Langevin Collision Frequency section). We did not try to match the degree of thermocoupling in the Andersen dynamics simulations with that used in our other explicit solvent simulations.
Discussion
Common Features of Artifacts Resulting from Repeating Random Number Sequences
In the Results, we identified a number of abnormal behaviors that can be observed in systems run with repeating sequences of Langevin forces. Most of the backbone root-mean-squared deviation (rmsd) artifacts can be explained as consequences of residual forces, which decay slowly as a function of the length of the sequence of Langevin forces as shown in Figure 1. Together, these residual forces do not act in any concerted fashion, but individually they do act in a particular direction relative to the coordinate axes of the simulation box. Each atom of the protein is therefore forced in a unique random direction, and the protein becomes distorted until the forces on each atom are counterbalanced by gradients of the system’s potential energy function. Weak residual forces, such as those encountered with 100 000 steps and a collision frequency of 3 ps−1, appear to be enough to break apart globular domains along their weak interfaces (see Figure S2, Supporting Information), but stronger residual forces can denature the domains themselves (see Figure 2 and Figure 4), regardless of the type of solvent used. Similar artifacts obtained with the Andersen thermostat (see Figure 9) are likely the products of “residual momenta.”
Initially, it would seem that the relative positions of larger groups of atoms would be less prone to artifacts than smaller groups of atoms because the residual forces acting on individual atoms would be averaged such that the net force pulling two groups of atoms apart would be small. However, the larger ΔΔG values observed for the backbone ϕ angle in Figure 6 and the separation of the apostreptavidin tetramer seen in Figure 3 do not support this reasoning. Instead, because each atom of a rigid molecular structure has a different moment arm about some center of rotation, the residual forces on just a few atoms could be amplified, creating the large ΔΔG values between populations of certain dihedral angles and the hinge-bending motion of the apostreptavidin tetramer dissociation (see Figure S2 of the Supporting Information).
In the Theory section, we stated that repeatedly applying a finite sequence of pseudorandom forces to an atom was similar to applying a constant net force on that atom. However, a more precise description is needed to explain the periodicity of trajectories observed in Results, Severity of Artifacts As a Function of the Langevin Collision Frequency section, and the differences in atomic fluctuations observed in Results, Langevin Artifacts in Implicit Solvent section. Separate trajectories initiated from distinct conformations of a system have been observed to synchronize if identical sequences of pseudorandom noise are used to propagate Langevin dynamics.29 This synchronization occurs after the trajectories remain uncorrelated for some amount of time, the length of which depends on the strength of the pseudorandom noise. In the examples given throughout the Results, the checkpoint files written at the end of each segment of a simulation provide distinct conformations of the system, and the collision frequency tunes the strength of the noise. In the Results, Severity of Artifacts As a Function of the Langevin Collision Frequency section, the 100 000-step segments of the trajectory become synchronized as identical sequences of strong pseudorandom noise are repeatedly applied. This offers an explanation of how synchronization of successive trajectory segments could occur in as little as 12 ns with γ set to 50 ps−1 but not in 200 ns if γ is set to 3 ps−1.
An earlier work by Fahy and Hamann30 performed similar calculations on small systems driven with a rudimentary Andersen-like thermostat. In this work, they noted the existence of a critical length of time between velocity reassignments, τc, such that reassigning velocities more frequently resulted in synchronization of the trajectories and reassigning them less frequently resulted in indefinite chaotic behavior. Noting that τc corresponds to the strength of thermocoupling in the Anderson thermostat, we can hypothesize that there exists some critical strength of thermocoupling in the Langevin thermostat above which synchronization of trajectories is guaranteed and below which chaotic behavior will be observed. This is consistent with our results, and knowledge of the value of τc or equivalent γc could help investigators make better choices about how to maintain the temperature of a simulation. However, more studies would be necessary to estimate these critical thresholds for different system sizes and topologies.
On the basis of the above observations, we may extend our description of the artifacts created by repeating sequences of Langevin forces or Andersen velocity reassignments in molecular dynamics simulations and state it loosely as follows: Thermostats operating with repeating finite sequences of random noise will cause incoherent perturbations in a system’s potential energy surface, the strength of the perturbations being inversely proportional to the square root of the length of the noise sequence and directly proportional to the square root of the strength of the noise itself. The incoherent distortions tend to reduce the conformational space available to the system; in the limit of strong noise, the system may be driven into periodic trajectories according to the unique sequence of noise applied.
Unfortunately, the artifacts caused by repeating sequences of Langevin forces or Andersen velocity reassignments seem to be very extensive because of the way the residual forces scale with the sequence length. Many published simulations could potentially have been affected; the results in this study show that, over very long simulations, some observables such as atomic fluctuations in implicit solvent display artifacts if a finite sequence of even 1 000 000 Langevin forces is used to control the temperature. Artifacts in backbone rmsd measurements may be detectable if a repeating sequence of 100 000 Langevin forces is used. With sequences of fewer than 100 000 Langevin forces, the artifacts may take tens of nanoseconds to appear, but they are often dramatic. We would like to offer a general statement such as “simulations performed with sequences of 1 000 000 or more Langevin forces and a weak thermocoupling of 3 ps−1 or less are safe from artifacts,” but certain analyses other than those presented in this study may be more sensitive to thermostat artifacts.
Survey of Current Molecular Dynamics Packages with Respect to Random Number Generation
The potential for artificially distorting a biomolecule by incorrect use of the Langevin or Andersen thermostats represents a serious problem for molecular simulations. This prompted us to make a brief survey of existing molecular dynamics packages to see which implementations could allow users to unwittingly perturb their systems with repeating sequences of pseudorandom numbers. The most robust protection against the artifacts identified in the Results is to pass the state of the random number generator through the molecular dynamics checkpoint files and, by default, to override userspecified random seeds when restarting a molecular dynamics calculation. In this manner, the pseudorandom number generator (PRNG) would produce a single sequence for the entire simulation.
As stated in the results, we discovered this problem while running Langevin dynamics with the AMBER9 software package.14 By default, both of its simulation modules use a random seed of 71277, and users may specify other values. The state of the PRNG is not passed via the checkpoint file, however, so Langevin and Andersen dynamics simulations are prone to artifacts unless the user specifically requests that the random seed be set using the clock time, changes the random seed with a script running outside of the AMBER software, or performs simulations in very long segments. Similarly, the GROMACS (version 3.*)31–34 software runs with a default random seed of 1993 and does not pass the state of the PRNG through its checkpoint files, but users may request that the seed be set using the clock time. Tests with the GROMACS software presented in the Supporting Information confirm that artifacts can be generated in the same manner as was shown for the AMBER code throughout the Results. Robust protection against random number artifacts will be implemented in future versions of both AMBER and GROMACS.
In the DL_POLY package (version 3),35 the random seed is set at compile time, although if segments of a Langevin or stochastic dynamics simulation are run in parallel on a varying number of processors, different series of pseudorandom numbers will be generated.
The NAMD code36 is highly resistant to Langevin artifacts because of the manner in which it generates random numbers. By default, the PRNG seed is set by the clock time, although users may set it to a specific value. The state of the PRNG is not passed through the checkpoint file, but some unique aspects of the NAMD code offer added protection against random number artifacts (see the Supporting Information). These aspects of the code make it very difficult to obtain such artifacts with NAMD.
The CHARMM37 and DESMOND38 packages both implement the robust solution by passing the state of the PRNG through simulation checkpoint files and, so, can be considered safe from the artifacts identified in this study.
Future Directions: Thermostats for Molecular Dynamics Calculations
We have shown that, if used correctly, the Langevin thermostat produces stable dynamics for explicitly solvated proteins over tens of nanoseconds. Other studies39 have reported stable dynamics for tens to hundreds of nanoseconds using the Berendsen “weak-coupling” approach to temperature regulation. However, the results in Figure 1 suggest that a sufficiently large number of random forces (or, by extension, velocity reassignments) must be applied to each atom to ensure that a thermostat based on random numbers has not applied significant net forces or momenta to the individual atoms of the system. Furthermore, data in Results, Severity of Artifacts As a Function of the Langevin Collision Frequency section, corroborate the findings of Ciesla and co-workers,29 suggesting that separate trajectories of large molecular systems can become synchronized if both simulations are run with the same sequence of Langevin forces, even if that sequence is infinite. Investigators should therefore carefully consider the manner in which their thermostat functions, beyond simple qualifications such as stable dynamics.
All molecular dynamics thermostats attempt to simulate coupling of the system to an external bath at the desired temperature, but none of the methods are entirely physically meaningful. In reality, “heating” and “cooling” refer to the equilibration of the momenta of particles in two systems brought into contact with one another. In common biomolecular simulations with explicit solvent, the solvent is typically an accessory, while the analysis focuses on the biomolecule itself. It may therefore be desirable to modify existing thermocoupling schemes to regulate only the temperature of the solvent, or perhaps only the temperature of solvent particles further than some minimum distance from the biomolecule. This method, similar in spirit to stochastic dynamics,40,41 would regulate the temperature of the biomolecule indirectly, hopefully causing very little perturbation to its dynamics. Other modifications of simple thermostats42 should also be considered.
The goal of this study was to expose a serious problem associated with the use of Langevin and Andersen thermostats in molecular simulations and to present in detail possible artifacts that might arise. However, the results from simulations with strong thermocoupling raise questions about the way thermostats affect the dynamical properties of biomolecular models, and whether it would be helpful to modify current thermocoupling schemes as increasing computational resources make it possible to study kinetic properties of events such as protein folding or ligand binding through simulations.
Supplementary Material
Illustration of residual Langevin forces, illustration of the dissociation of the apostreptavidin tetramer, demonstration that rapidly restarting simulations with a changing pseudorandom number generator (PRNG) seed permits stable dynamics, clarification of the fact that Langevin artifacts tend to lead to higher protein backbone rmsd relative to the native state but lower atomic fluctuations thereafter, further demonstration of periodic trajectories obtained with PRNG artifacts and strong thermocoupling, and illustrations of Langevin dynamics artifacts in simulations performed with the GROMACS and NAMD codes. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgment
This research was supported by National Institutes of Health Grant GM080214. D.S.C. thanks the GROMACS development team, Dr. Bernard R. Brooks, Dr. Justin Gullingsrud, and Dr. Ilian Todorov for explanations about the operation of GROMACS, CHARMM, DESMOND, and DL POLY, respectively. Dr. Robert Konecny and Dr. Barrett Abel of the Center for Theoretical Biophysics at the University of California, San Diego, provided part of the computing resources for this study via National Science Foundation Grant PHY-0216576.
References
- 1.Ryckaert JP, Ciccotti G, Berendsen HJC, Hirasawa K. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1997;23:327–341. [Google Scholar]
- 2.Berendsen HJC, Postma PostmaJPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 3.Hoover WG. Canonical dynamics: Equilibrium phase–space distributions. Phys. Rev. A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 4.Andersen HC. Molecular dynamics at constant pressure and/or temperature. J. Chem. Phys. 1980;72:2384–2393. [Google Scholar]
- 5.Izaguirre JA, Catarello DP, Wozniak JM, Skeel RD. Langevin stabilization of molecular dynamics. J. Chem. Phys. 2001;114:2090–2098. [Google Scholar]
- 6.Ferrenberg AM, Landau DP, Wong JY. Monte Carlo simulations: Hidden errors from “good” random number generators. Phys. Rev. Lett. 1992;69:3382–3384. doi: 10.1103/PhysRevLett.69.3382. [DOI] [PubMed] [Google Scholar]
- 7.Vattulainen I. Framework for testing random numbers in parallel calculations. Phys. Rev. E. 1999;59:7200–7204. doi: 10.1103/physreve.59.7200. [DOI] [PubMed] [Google Scholar]
- 8.Holian BL, Percus OE, Warnock TT, Whitlock PA. Pseudorandom number generator for massively parallel molecular-dynamics simulations. Phys. Rev. E. 1994;50:1607–1615. doi: 10.1103/physreve.50.1607. [DOI] [PubMed] [Google Scholar]
- 9.Press WH, Flannery BP, Teukolsky SA, Vetterling WT. Numerical Recipes: The Art of Scientific Computing. 3rd ed. Cambridge, U.K.: Cambridge University Press; 1989. Random Numbers; pp. 380–385. [Google Scholar]
- 10.Wu X, Brooks BR. Self-guided Langevin dynamics simulation method. Chem. Phys. Lett. 2003;381:512–518. [Google Scholar]
- 11.Fan H, Mark AE, Zhu J, Honig B. Comparative study of generalized Born models: Protein dynamics. Proc. Natl. Acad. Sci. U.S.A. 2005;102:6760–6764. doi: 10.1073/pnas.0408857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feller W. The Fundamental Limit Theorems in Probability. Bull. Amer. Math. Soc. 1945;51:800–832. [Google Scholar]
- 13.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Case DA, Cheatham TE, Darden TA, Gohlke H, Luor R, Merz M, Onufriev A, Simmerling C, Wang B, Woods R. The AMBER biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules. J. Comput. Chem. 2000;21:1049–1074. [Google Scholar]
- 16.Cornell WD, Cieplak P, Bayly CI, Gould IR, Jr., Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second-generation force field for the simulations of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 17.Simmerling C, Strockbine B, Roitberg AE. All-atom structure prediction and folding simulations of a stable protein. J. Am. Chem. Soc. 2002;124:11258–11259. doi: 10.1021/ja0273851. [DOI] [PubMed] [Google Scholar]
- 18.Berendsen HJC, Grigera JR, Straatsma TP. The missing term in effective pair potentials. J. Phys. Chem. 1987;91:6269–6271. [Google Scholar]
- 19.Onufriev A, Bashford D, Case DA. Modification of the generalized Born model suitable for macromolecules. J. Phys. Chem. B. 2000;104:3712–3720. [Google Scholar]
- 20.Weiser J, Shenkin PS, Still WC. Approximate atomic surfaces from linear combinations of pairwise overlaps (LCPO) J. Comput. Chem. 1999;20:217–230. [Google Scholar]
- 21.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LH. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 22.Miyamoto S, Kollman PA. SETTLE: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 23.Reznik GO, Vajda S, Smith C, Cantor CR, Sano T. Streptavidins with intersubunit crosslinks have enhanced stability. Nat. Biotechnol. 1996;14:1007–1011. doi: 10.1038/nbt0896-1007. [DOI] [PubMed] [Google Scholar]
- 24.Pazy Y, Eisenberg-Domovich Y, Laitinen OH, Kulomaa MS, Bayer EA, Wilchek M, Livnah O. Dimer–tetramer transition between solution and crystalline states of streptavidin and avidin mutants. J. Bacteriol. 2003;185:4050–4056. doi: 10.1128/JB.185.14.4050-4056.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katz BA. Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH. J. Mol. Biol. 1997;274:776–800. doi: 10.1006/jmbi.1997.1444. [DOI] [PubMed] [Google Scholar]
- 26.Kurzban GP, Bayer EA, Wilcheck M, Horowitz PM. The quaternary structure of streptavidin in urea. J. Biol. Chem. 1991;266:14470–14477. [PubMed] [Google Scholar]
- 27.Sano T, Cantor CR. Cooperative biotin binding by streptavidin. J. Biol. Chem. 1990;265:3369–3373. [PubMed] [Google Scholar]
- 28.Neidigh JW, Fesinmeyer RM, Andersen NH. Designing a 20-residue protein. Nat. Struct. Biol. 2002;9:425–430. doi: 10.1038/nsb798. [DOI] [PubMed] [Google Scholar]
- 29.Ciesla M, Dias SP, Longa L, Oliveira FA. Synchronization induced by Langevin dynamics. Phys. Rev. E. 2001;63:065202. doi: 10.1103/PhysRevE.63.065202. [DOI] [PubMed] [Google Scholar]
- 30.Fahy S, Hamann DR. Transition from chaotic to nonchaotic behavior in randomly driven systems. Phys. Rev. Lett. 1992;69:761–764. doi: 10.1103/PhysRevLett.69.761. [DOI] [PubMed] [Google Scholar]
- 31.Berendsen HJC, van der Spoel D, van Drunen R. GROMACS: A message-passing molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–45. [Google Scholar]
- 32.Daura X, Oliva B, Querol E, Aviles FX, Tapia O. On the sensitivity of MD trajectories to changes in water-protein interaction parameters: The potato carboxypeptidase inhibitor in water as a test case for the GROMOS force field. Proteins. 1996;25:89–103. doi: 10.1002/(SICI)1097-0134(199605)25:1<89::AID-PROT7>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 33.Lindahl E, Hess B, van der Spoel D. GROMACS 3.0: A package for molecular simulations and trajectory analysis. J. Mol. Model. 2001;7:306–317. [Google Scholar]
- 34.van der Spoel D, van Buuren AR, Tieleman P, Berendsen HJC. Molecular dynamics simulations of peptides from BPTI: A closer look at amide-aromatic interactions. J. Biomol. NMR. 1996;8:229–238. doi: 10.1007/BF00410322. [DOI] [PubMed] [Google Scholar]
- 35.Smith W, Yong CW, Rodger PM. DL POLY: application to molecular simulation. Mol. Simulat. 2002;28:385–471. [Google Scholar]
- 36.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kal L, Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 38.Bowers KJ, Chow E, Xu H, Dror RO, Eastwood MP, Gregersen BA, Klepeis JL, Kolossvary I, Moraes MA, Sacerdoti FD, Salmon JK, Shan Y, Shaw DE. Scalable algorithms for molecular dynamics simulations on commodity clusters. In: Buhyan L, editor. Proceedings of the 2006 ACM/IEEE conference on Supercomputing; December 3–6 2006; San Jose, CA. New York: Association for Computing Machinery, Inc.; 2006. [Google Scholar]
- 39.Lei H, Duan Y. Two-stage folding of HP-35 from ab-initio simulations. J. Mol. Biol. 2007;370:196–206. doi: 10.1016/j.jmb.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berkowitz M, McCammon JA. Molecular dynamics with stochastic boundary conditions. Chem. Phys. Lett. 1982;90:215–217. [Google Scholar]
- 41.Brünger A, Brooks CL, III, Karplus M. Stochastic boundary conditions for molecular dynamics simulations of ST2 water. Chem. Phys. Lett. 1984;105:495–500. [Google Scholar]
- 42.Koopman EA, Lowe CP. Advantages of a Lowe–Andersen thermostat in molecular dynamics simulations. J. Chem. Phys. 2006;124:204103. doi: 10.1063/1.2198824. [DOI] [PubMed] [Google Scholar]
- 43.Jorgensen WL, Jenson C. Temperature dependence of TIP3P, SPC, and TIP4P water from NPT Monte Carlo simulations: Seeking temperatures of maximum density. J. Comput. Chem. 1998;19:1179–1186. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Illustration of residual Langevin forces, illustration of the dissociation of the apostreptavidin tetramer, demonstration that rapidly restarting simulations with a changing pseudorandom number generator (PRNG) seed permits stable dynamics, clarification of the fact that Langevin artifacts tend to lead to higher protein backbone rmsd relative to the native state but lower atomic fluctuations thereafter, further demonstration of periodic trajectories obtained with PRNG artifacts and strong thermocoupling, and illustrations of Langevin dynamics artifacts in simulations performed with the GROMACS and NAMD codes. This material is available free of charge via the Internet at http://pubs.acs.org.