Abstract
Purpose
Helical alanine-rich polypeptides with functional groups displayed along the backbone can display desired molecules such as saccharides or therapeutic molecules at a prescribed spacing. Because these polypeptides have promise for application as biomaterials, the conformation and association of these molecules have been investigated under biologically relevant conditions.
Methods
Three polypeptide sequences, 17-H-3, 17-H-6, and 35-H-6, have been produced through recombinant techniques. Circular dichroic (CD) spectroscopy was used to monitor the secondary structure of the polypeptides in PBS (phosphate buffered saline, pH 7.4). The aggregation behavior in PBS was monitored via analytical ultracentrifugation and non-denaturing polyacrylamide gel electrophoresis.
Results
The three polypeptides adopt a highly helical structure at low and ambient temperatures, and when heated, undergo a helix-to-coil transition, typical of other alanine-rich peptide sequences. The melting temperatures and van’t Hoff enthalpies, extracted from the CD data, suggest similar stability of the sequences. Although alanine-rich sequences can be prone to aggregation, there is no indication of aggregation for the three polypeptides at a range of concentrations relevant for possible biological applications.
Conclusions
The helical polypeptides are monomeric under biologically relevant conditions enabling application of these polypeptides as useful scaffolds for ligand or drug display.
Keywords: aggregation, conformational behavior, multivalent scaffolds, polypeptides
INTRODUCTION
Biologically directed methods of polymer synthesis have enabled the production of macromolecules with specific amino acid sequences and molecular weights. These strategies have been employed in the production of natural protein mimics including silk (1–5), elastin (6–12), and collagen (13,14), which, although having great potential in materials-based applications, can be difficult to obtain in large quantities from their natural sources. Artificial proteins have also been designed to take advantage of the absolute control over amino acid sequence and molecular weight and the secondary structural preferences of specific amino acids and amino acid consensus sequences (15–21). Accordingly, the versatility of these synthetic methods has permitted the production of a variety of protein polymers with well-defined secondary structures designed for a specific function or application. Numerous studies have focused on the synthesis of repetitive amino acid sequences that adopt β-sheet, coiled-coil or random coil structures (17,19,20,22–29), although there have been fewer reports about sequences designed to adopt monomeric α-helical secondary structures (15,16,18,30).
Recombinant approaches of protein and polypeptide synthesis have also been employed in pharmaceutical applications in which the polypeptide acts as the delivery vehicle for a therapeutic molecule or is itself a therapeutic molecule. For example, recombinant proteins such as insulin serve a therapeutic purpose, and polypeptide sequences derived from other natural proteins are also used in various therapeutic approaches (31). Elastin-based polypeptides have been widely employed for this purpose and have been conjugated to sequences derived from natural proteins including cell penetrating peptides, which enable the internalization of the molecule into eukaryotic cells (32), as well as to peptides that inhibit the production of genes involved in tumor proliferation, such as the oncogene c-Myc (32,33). Drug molecules, such as the anticancer drug doxorubicin, can also be attached to elastin-based polypeptides for delivery of the drug to targeted sites without decreasing the efficacy of the drug (34,35). In addition to the design of polypeptidic drug carriers, the sequence specificity of recombinant methods also offers opportunities in the design of polymers that may be useful in controlled multivalent display of ligands or drugs on the basis of their architecture. In the work described here, recombinant helical polypeptides have been designed as scaffolds in the production of multivalent inhibitors of protein—saccharide binding events. The α-helical structure is desired for its potential in controlling functional group placement, thus affording opportunities to manipulate binding affinity via changes polypeptide sequence, to either enhance or diminish the effectiveness of the polypeptide-based inhibitor. The multivalent display of functional groups along the helical polypeptide backbone also lends itself to possible utility in therapeutic applications in which multiple, controlled attachment sites for therapeutic molecules is desired.
Because α-helical structures are ubiquitous in naturally occurring proteins, there has been a great deal of research into the stability and behavior of α-helical peptides (36–42). Alanine-rich sequences have been the primary focus in such studies owing to their chemical inertness, which permits investigation of helical propensities of individual amino acids without complication. The inclusion of charged amino acids has also been shown to reduce the aggregation behavior of the peptides through charge repulsion. In the alanine-rich polypeptides described here, glutamine has been employed for improved solubility, while glutamic acid has been included for subsequent chemical modification. The thermal and conformational behaviors of these longer repetitive alanine-rich polypeptides in solution at low pH generally follow trends predicted by the study of shorter alanine-rich sequences (15,16). In addition, the modification of the glutamic acid residues with amine-functionalized monosaccharides does not significantly affect the conformational behavior of the polypeptides, and specific helical glycopolypeptides have shown promise as toxin inhibitors (30). Because of these encouraging results and the potential general use of these polypeptides in other biological applications, we present here the characterization of the conformational and aggregation behaviors of an expanded set of two of the alanine-rich polypeptide sequences, with essentially identical compositions, under physiologically relevant conditions. Our results demonstrate the lack of aggregation and well-behaved conformational properties of these macromolecules, and also suggest slight differences in properties as a result of sequence differences.
MATERIALS AND METHODS
Materials
General reagents for protein expression and purification were obtained from Sigma (St. Louis, MO), ISC Bioexpress (Kaysville, UT), and Fisher Scientific (Fairlawn, NJ). Nickel-chelated sepharose resin for protein purification was obtained from Qiagen (Valencia, CA). The sequences of the polypep-tides discussed here are listed in Table I with their respective abbreviations and pertinent molecular information.
Table I.
Sequence, Molecular Weight, and Nominal Distance Between Glutamic Acid Residues for each of the Sequences Studied
| Designationa | Repeat Sequence | MW (Da)b | Approx. Spacingc(Å) | A:Q | # of E |
|---|---|---|---|---|---|
| 17-H-3 | [AAAQEAAAAQAAAQAEAAQAAQ]3 | 8,875 | 17 | 3:1 | 6 |
| 17-H-6d | [AAAQEAAAAQAAAQAEAAQAAQ]6 | 14,770 | 17 | 3:1 | 12 |
| 35-H-6 | [AAAQAAQAQAAAEAAAQAAQAQ]6 | 14,159 | 35 | 5:2 | 6 |
The designation, Y-H-X, indicates spacing between E residues (Y), α-helical backbone structure (H), and number of sequence repeats (X).
Theoretical molecular weight based on molecular weights of individual amino acids.
Spacing between E residues in an α-helical structure estimated via energy minimization calculations as previously reported (15,16).
Amino acid analysis, MALDI and HPLC results for 17-H-6 shown in the Electronic Supplementary Material.
Gene Construction and Polypeptide Expression
Protocols for construction of DNA plasmids and expression of 17-H-3 and 35-H-6 have been reported previously (15); these protocols were employed in the synthesis of 17-H-6. Briefly, the DNA sequence encoding the amino acid sequence, [AAAQEAAAAQAAAQAEAAQAAQ], was ligated into a modified expression plasmid, pET28b-JS1 (15). The length of the target artificial repetitive gene inserted into the expression plasmid was confirmed via restriction digest analysis of recombinant plasmids with the enzymes BamH I and Nco I and subsequent analysis via agarose gel electrophoresis. The expression plasmid, pET28-JS1-A6, which contains six repeats of the target sequence, was used to transform chemically competent E. coli bacterial cells. As previously described, cultures of the expression host were chemically induced to begin expression of the polypeptide (15). The protein polymer was purified from the cell lysate via Ni—NTA affinity chromatography with stepwise pH gradient elution under denaturing conditions (Qiagen). The expression yield for each polypeptide was 15–30 mg/l of culture, more than sufficient for chemical modification and assays of biological activity. After purification, 17-H-6 was characterized via SDS-PAGE analysis, MALDI-TOF mass spectrometry, HPLC, and amino acid analysis, as previously described (15,16). Results from the various characterization methods, shown in the Electronic Supplementary Material, indicate that this polypeptide, as well as the other previously reported polypeptides, has the expected composition and a purity of greater than 95%.
General Polypeptide Characterization
Amino acid analysis of purified polypeptides was performed at the Molecular Analysis Facility at the University of Iowa (Iowa City, IA). Protein polymer concentrations used in circular dichroic spectroscopy were confirmed via quantitative amino acid analysis employing an internal standard, norvaline. MALDI-TOF analysis of purified protein polymers was performed at the Mass Spectrometry Facility in the Department of Chemistry and Biochemistry at the University of Delaware on a Biflex III (Bruker, Billerica, MA). Polypeptide samples and calibration standards [bovine insulin (MW=5,734.59), thioredoxin from E. coli (MW=11,647.48), and horse apomyoglobin (MW=16,952.56)] were prepared in a 3,5-dimethoxy-4-hydroxycinnamic acid matrix. Data were recorded using the OmniFLEX program and subsequently analyzed in the Xmass Omni program.
Circular Dichroic Spectroscopy
Circular dichroic spectra were recorded on an AVIV 215 spectrophotometer (Proterion Corporation, Piscataway, NJ) or a Jasco J-810 spectrophotometer (Easton, MD) in a 1-mm pathlength quartz cuvette in the single-cell mount setup. Background scans of the buffer [pH 7.4, phosphate buffered saline (PBS)] were subtracted from the sample spectra. Samples were produced at a concentration of approximately 10–20 μM in pH 7.4 PBS. Data points for wavelength dependent CD spectra were recorded at a continuous scan rate of 50 nm/min (Jasco J-810) or recorded as a step scan with data being taken every nanometer (AVIV 215). Samples used for temperature dependent CD spectra were heated at a rate of 60°C/h, with data points taken every 1°C. The mean residue ellipticity, [θ]MRW (deg cm2 dmol-1), was calculated using the molecular weight of the protein polymer and cell pathlength. Reported fractional helicities and thermal denaturation parameters are based on at least duplicate measurements of a given polypeptide.
Analytical Ultracentrifugation
Equilibrium analytical ultracentrifugation (AUC) experiments were performed in a ProteomeLab XL-1 Protein Characterization Ultracentrifuge (Beckman Coulter, Fullerton, CA) using an An-60Ti rotor. Cells were assembled using quartz windows and six-channel centerpieces. All three polypeptides were dissolved in PBS buffer (pH 7.4) at concentrations of approximately 7, 15 and 30 μM. Samples were subjected to ultracentrifugation at sequentially increasing speeds (17-H-3 was centrifuged at 25,000, 27,500, 30,000, 32,500, and 35,000 rpm; 17-H-6 and 35-H-6 were centrifuged at 22,500, 25,000, and 28,500 rpm) with an equilibration period of 22 h at each speed prior to recording the absorbance at 230 nm. The data were analyzed using SEDEQ, a freeware program designed by Allen Minton. In the data analysis, a specific volume of 0.71 ml/g, calculated from the amino acid composition (43), and a solvent density of 1.02 g/ml, were employed. Samples were analyzed in duplicate.
Non-denaturing Gel Electrophoresis
Non-denaturing polyacrylamide gel electrophoresis (PAGE) was performed in a 15% (w/v) acrylamide separating gel and a 3.9% (w/v) acrylamide stacking gel, using a running buffer comprising 12.5 mM Tris and 96 mM glycine at a pH of 8.3. Samples of all three polypeptides were prepared at concentrations ranging from approximately 5 to 100 μM in pH 7.4 PBS buffer. A volume of 5 μl of sample buffer [100 mM Tris·Cl pH 6.8, 20% (w/v) glycerol, 0.001% (w/v) bromphenol blue] was added to 20 μl of polypeptide solution, and 5 μl of that solution was loaded onto the gel. Samples were subjected to electrophoresis at 50 mA for 20 min at room temperature. Gels were stained for 30 min in a 50% methanol, 10% acetic acid solution with 0.5 g brilliant blue and 0.5 g cupric sulfate. Excess stain was removed via successive washes with a 5% methanol, 7% acetic acid solution.
RESULTS AND DISCUSSION
We have previously reported the synthesis and conformational characterization of three families of alanine-rich artificial polypeptide sequences, with the sequences [AAA QEAAAAQAAAQAEAAQAAQ]x, [(AAAQ)(AAQA Q)(AAAE)(AAAQ)(AAQAQ)]x, and [(AAAQ)5(AAAE)(AAAQ)5]x (15,16). The use of alanine and gluta-mine residues imparts an α-helical secondary structure while the glutamic acids provide chemical reactivity in select and specific positions along the backbone. Sequence variations were specifically chosen to control the nominal spacing of the glutamic acid residues, while variations in the x values control the molecular weight of the polypeptide and the number of glutamic acids that are displayed. The sequences were designed to display glutamine residues around the circumference of the α-helix, in order to reduce hydrophobic patches along the polypeptide backbone (Electronic Supplementary Material). The main design feature is the presentation of the glutamic acid residues on approximately the same face of the helix, with different nominal distances between Glu residues that are dependent on the sequence and were determined via energy minimization calculations (15,16). The sequences were determined for polypeptides of essentially 100% helicity, so the exact placement of functional groups on polypeptides in solution may differ from theoretically predicted. Sequences are designated with the general form Y-H-X, where Y is the spacing between glutamic acid residues, H indicates the helical backbone, and X is the number of monomer sequence repeats. This designation is different than that used in previous reports in which the 17-H-X family was designated as AX, 35-H-X as BX, and 65-H-X as CX.
These three alanine-rich polypeptide sequences were designed as scaffolds for the regular display of functional groups along a helical protein polymer backbone (15,16). Structural and thermal characterization of three polypeptides, 17-H-3, 35-H-6, and 65-H-2, was performed under acidic pH conditions to mimic the behavior these polypep-tides might display after the chemical attachment of neutral ligands to the chemically reactive glutamic acid residues. All three polypeptides displayed α-helical character at low and ambient temperatures, but upon heating, the three sequences exhibited a sequence-dependent structural transition. The protein polymers with higher densities of glutamic acid residues, 17-H-3 and 35-H-6, displayed a helix-coil transition with increasing temperature (15), while the protein polymer with the lowest density, 65-H-2, displayed a helix-coil transition when heated to intermediate temperatures, but irreversibly adopted a β-sheet structure at elevated temperatures (16). The polypeptides all demonstrated high helicities at ambient temperatures, consistent with their design, in which adoption of a helical structure is intended to control presentation of functional groups at distances commensurate with, or different from, receptor sites on protein receptors. Owing to our interests in employing specific architectures in areas of biological significance, both the conformational and aggregation properties of 17-H-3, 17-H-6, and 35-H-6 have been characterized in phosphate buffered saline (PBS) at a pH of 7.4. The characterization of 17-H-6 has permitted more direct comparison of the length and sequence-dependent properties of these polypeptides (15,16).
Characterization of Conformational Behavior
The conformational behavior of the three polypeptides was monitored via circular dichroic spectroscopy. Fig. 1 shows the wavelength scans recorded at 5°C for 17-H-3, 17-H-6, and 35-H-6 in PBS. The spectra for all three polypep-tides display double minima at 222 and 208 nm, characteristic of an α-helical structure. The average mean residue ellipticity values at 222 nm, [θ]222, at 5°C from multiple samples of 17-H-3, 17-H-6, and 35-H-6 were -30,300, -39,025, and -42,250 deg cm2 dmol-1, respectively (generally 5–15% error based on multiple measurements; differences in sample preparation can cause significant changes in the MRE values for these polypeptides that in isolated cases exceed 25%). These [θ]222 values follow the same trends as those observed in pH 2.3, 10 mM phosphate with 150 mM salt (-29,000, -38,600, and -50,000 deg cm2 dmol-1, respectively), despite the negative charge of the glutamic acids (pKa=4.1) at pH 7.4. Slight differences in helicity may result from some electrostatic repulsion between the charged glutamates at pH 7.4, but this effect is not pronounced, as might be expected given that the Debye length under these isotonic solution conditions (7.8Å (44)) is less than the nominal distance between the glutamic acids in these sequences. Short alanine rich peptides containing both glutamine (Q) and glutamic acid (E), which provide stabilizing interactions when located at i and i+4 positions, display a slight loss in helicity with increasing pH (45). The differences in the [θ]222values of the 35-H-6 at low and neutral pH (given the lack of such a difference for the other sequences) may, therefore, result from a similar loss of such interactions between the glutamic acid and glutamine residues at i and i+4 positions in the polypeptides (the 17-H-X sequences have no such arrangement of Q and E residues). The stabilization of the secondary structure for 35-H-6 at low pH may also arise from differences in its association state, triggered by protonation of the glutamic acid residues; in some experiments there is indication of stabilization of 17-H-6 at lower pH via association, as well. A more detailed characterization and treatment of the association behavior of the sequences in their unionized state is underway and will be the subject of future reports.
Fig. 1.
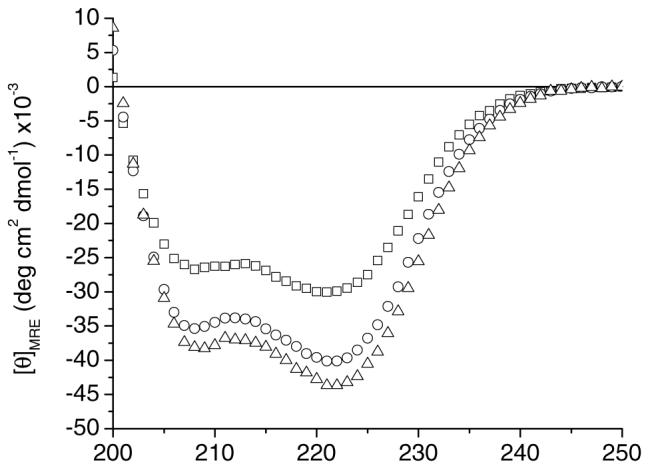
Circular dichroic spectra of 17-H-3 (square) (17 μM), 17-H-6 (circle) (10 μM), and 35-H-6 (triangle) (16 μM) in pH 7.4 PBS at 5°C.
The fractional helicity can be determined for these three polypeptides sequences by comparing the experimental [θ]222 values with a theoretical [θ]222 value for a 100% helical peptide of a given length (Electronic Supplementary Material) (40,46). The fractional helicity values of 17-H-3, 17-H-6, and 35-H-6 were calculated to be 51, 65 and 70%, respectively. The fractional helicity of these polypeptides is greater for the polypeptides of higher theoretical molecular mass (8,875, 14,770 and 14,159 Da for 17-H-3, 17-H-6, and 35-H-6, respectively), as expected (15,41). 17-H-6 and 35-H-6 have similar molecular weights and were expected to display similar helical content; however, 35-H-6, of slightly lower molecular mass, displays slightly higher helicity (70%) in comparison to 17-H-6 (65%). Computational estimation of helix content of the full sequences (short N- and C- terminal fusions included) indicate a similar difference in helicity of 17-H-6 relative to that of 35-H-6 (68 and 76% at 25°C). Estimates for the alanine-rich portion (no terminal tags) of 17-H-6 and 35-H-6 predict the two sequences have more comparable and higher helical content, indicating the addition of the slightly different fusion sequences to the polypeptides differentially reduces the overall helicity given the lower helix propensities of the amino acids found in those tags in comparison to Ala and Gln (see Electronic Supplementary Material) (47). Slight differences in helicity owing to the sequence differences of the polypeptides are also possible but are not suggested to be as significant as the differences caused by the fusion tags. These data indicate that alanine- and glutamine-rich sequences of slightly varying compositions and sequences can be equally useful in the production of chemically reactive helical polypeptide-based templates.
Characterization of the Thermal Behavior
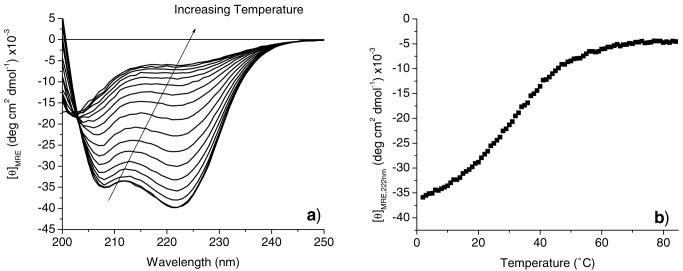
The 17-H-3 and 35-H-6 polypeptides were previously reported to display a transition from an α-helical to a non-α-helical conformation with increasing temperature in 10 mM phosphate buffer at pH 2.3, followed by an irreversible transition to β-sheet conformation with incubation at elevated temperatures (15). The conformational behavior of 17-H-3, 17-H-6, and 35-H-6 was monitored with increasing temperature in PBS at pH 7.4 to reveal any differences in conformational behavior changes relative to those seen at low pH values. Fig. 2a shows full wavelength spectra collected for 17-H-6 in pH 7.4 PBS in 5°C increments from 5°C to 80°C. As the temperature increases, there is a loss in ellipticity at 222 nm and a blue shift of the minimum at 208 nm, indicating the transition to a non-α-helical structure. The same transition to a non-α-helical structure is observed for the 17-H-3 and 35-H-6 polypeptides (data not shown). The isodichroic point observed at 203 nm suggests the two-state nature of this conformational transition, as previously observed. The thermal transition observed for these three polypeptides is completely reversible upon cooling from 80°C, with [θ]222 values recovering back to values observed prior to heating (data not shown). Irreversible conformation changes in 17-H-6 and 35-H-6 can, however, be induced through incubation at elevated temperatures for long periods of time (∼hours, conversion to β-sheet with aggregation, data not shown), and the tendency for this conversion to β-sheet is greater at the lower pH value (15,16). Taken together with results from the studies at low pH, the data indicate that the polypeptides, at both low and neutral pH, undergo similar conversions from helix to non-α-helical and then β-sheet with increasing temperature, although the specific temperatures and apparent rates of the transitions are different at the two pH values. β-sheet formation at elevated temperature is readily observed for the uncharged polypeptides at low pH but is significantly reduced for the polypeptides at neutral pH.
Fig. 2.
Circular dichroic spectroscopic characterization of the thermal denaturation of 17-H-6 (10 μM) in pH 7.4 PBS buffer. a Full wavelength spectra collected in increments of 5°C at temperatures ranging from 5 to 80°C. b Temperature dependence of [θ]222 recorded at a heating rate of 60°C/h.
The thermal denaturation of the secondary structure was also investigated via CD experiments in which the intensity of the minimum at 222 nm is monitored as a function of temperature. Fig. 2b shows a plot of [θ]222 for a representative sample of 17-H-6 in PBS (pH 7.4) (thus the difference in exact MRE values relative to the average values given above) as the sample is heated from 5 to 80°C at a rate of 60°C/h. The conformational transition occurs over a broad temperature range, as reported for other alanine-rich peptides (40,41); broad transitions were also observed for 17-H-3 and 35-H-6 (see Electronic Supplementary Material). Fits of these data to standard thermodynamic models of two-state transitions yield estimates of the van’t Hoff enthalpy and melting temperature of the transition (see Electronic Supplementary Material); the values determined for 17-H-3, 17-H-6, and 35-H-6 from multiple experiments are displayed in Table II. The greater error in the values for the 17-H-3 are likely caused by increased scatter in the data and the lack of a well-defined lower-temperature baseline in comparison to the longer macromolecules. The transition temperatures determined for the three polypeptides are comparable, all near 35°C, and are all slightly higher than the melting temperatures near 30°C that have been observed for shorter alanine-rich peptides (41), suggesting the improved thermal stability of these helical polypeptides (see below). Longer sequences should therefore also show improved stability for maintenance of higher helicity at physiological temperatures. The van’t Hoff enthalpy of the transition for each of the polypeptides is approximately 20 kcal/mol, suggesting that the cooperative unit in each of the polypeptides displays similar energetic behavior. These values are slightly higher than the van’t Hoff enthalpy (∼11 kcal/mol) reported by Scholtz et al. for a similar, but shorter peptide sequence (4,703 Da, containing lysine and glutamic acid residues) (48), which is consistent with the higher molecular weight of the polypeptides. The van’t Hoff enthalpy is approximately four times smaller, however, than values measured for collagen-like polypeptides (49), which is consistent with expected differences in cooperativity between the monomeric polypeptides and the collagen triple-helix. The melting temperatures and van’t Hoff enthal-pies for the three polypeptides are indicated to be statistically similar, as assessed via ANOVA analysis (p>0.05).
Table II.
Transition Temperature and van’t Hoff Enthalpy for each of the Polypeptide Sequences
| Sequence | Tm (°C)a | ΔHvH (kcal/mol) | |
|---|---|---|---|
| 17-H-3 | [AAAQEAAAAQAAAQAEAAQAAQ]3 | 33.5±3.4 | 20.3±2.7 |
| 17-H-6 | [AAAQEAAAAQAAAQAEAAQAAQ]6 | 34.8±0.2 | 21.4±0.8 |
| 35-H-6 | [AAAQAAQAQAAAEAAAQAAQAQ]6 | 36.5±1.9 | 18.8±0.4 |
Errors are estimated on the basis of at least duplicate measurements.
Characterization of the Aggregation Behavior
The aggregation behavior of polypeptides plays an important role in the function and application of macromolecules in biological applications. In the case of these alanine-rich polypeptides, the formation of aggregates could compromise the control over functional group placement, and may therefore also compromise the interaction of the functional groups with receptor sites on toxins, lectins or cell surfaces. Polyalanine sequences are hydrophobic and aggregate in aqueous solutions; however, the incorporation of helix caps or polar residues into the polyalanine sequences has been shown to increase the solubility of the polypeptides (15,16,40,41,48,50). Glutamine and glutamic acid residues were incorporated into these three polypeptide sequences to reduce the tendency of these molecules to aggregate (15,16); the other domains were not included owing to our interests in having a limited and prescribed number of chemically reactive groups along the chain.
Analytical ultracentrifugation (AUC) experiments were conducted to assess the tendency of these sequences to associate under various solution conditions. The aggregation behavior of 17-H-3, 17-H-6, and 35-H-6 in PBS was monitored at concentrations similar to those of the CD experiments (19, 17 and 18 μM, respectively). At equilibrium, the polypeptide concentration (cr) is related to the radial distance, r, through the following equation:
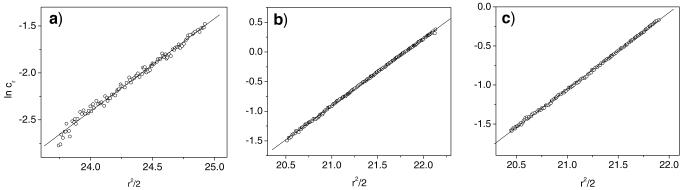
where, v̄ is the polypeptide specific volume, ρ is the solvent density, ω is the rotational velocity, and R is the gas constant (51). If the sample is composed of a single molecular weight species, the ln cr has a linear relationship with r2/2, and the slope of the line is a function of the polypeptide molecular weight. Data taken for 17-H-3 (19 μM), 17-H-6 (17 μM), and 35-H-6 (18 μM) after 22 h of equilibration at 30,000, 25,000 and 25,000 rpm, respectively, are plotted as ln cr vs r2/2 in Fig. 3. The rotor speeds were chosen based on optimum speeds reported for other molecules of similar molecular weights (52). The linear trend between ln cr and r2/2 indicates that all of the polypeptides exist as a single species under these conditions. The molecular mass values calculated from the slopes are 9,006, 14,860, and 12,880 g/mol for 17-H-3, 17-H-6, and 35-H-6, respectively, which are within 10% of both the theoretical molecular masses (8,875, 14,770, 14,159 g/mol) and the molecular masses determined by mass spectrometry (8,770, 14,659, and 14,023 g/mol), indicating the monomeric nature of the polypeptides under these conditions.
Fig. 3.
Plots of ln cr (expressed as natural log of absorbance units at 230 nm) vs r2/2 derived from analytical ultracentrifugation results measured at 230 nm for alanine-rich polypeptides. a 17-H-3 (19 μM, 30,000 rpm), b 17-H-6 (17 μM, 25,000 rpm), and c 35-H-6 (18 μM, 25,000 rpm) in pH 7.4 PBS.
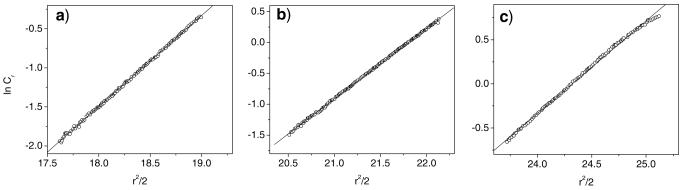
The reproducibility of the measurement and monomeric nature of the polypeptides was confirmed over multiple concentrations, as well. In Fig. 4, AUC results are shown for 17-H-6 in pH 7.4 PBS at concentrations of 4, 9, and 17 μM. As was seen for the individual polypeptides at a single concentration, the plot of ln cr vs r2 yields a linear trend for multiple polypeptide concentrations indicating only one species is present in the solution over this concentration range. The molecular weight values calculated for the three concentrations averaged 14,784 g/mol, which is within 1% error of the expected molecular weight and within 9% error of each other. Similar results were obtained for 17-H-3 and 35-H-6 (Electronic Supplementary Material). The average molecular weight values for 17-H-3 and 35-H-6 were 9,029 g/ mol and 13,172 g/mol, and both values are within 7% of their respective expected molecular weights.
Fig. 4.
Plots of ln cr (expressed as natural log of absorbance units at 230 nm) vs r2/2 from analytical ultracentrifugation results for 17-H-6 at 25,000 rpm with peptide concentrations of a 4 μM, b 9 μM, and c 17 μM in pH 7.4 PBS.
It has been shown previously that alanine-rich peptides can be driven toward aggregation by increasing polypeptide concentration in a 5 mM MOPS buffer at pH 7.0 at 65°C (53). At low concentrations, the alanine-rich polypeptides discussed here were shown to be monomeric, however, the application of these polypeptides may require polypeptide concentrations that are higher than those that can be reliably monitored via AUC. For example, in addition to applications in toxin inhibition, these polypeptides have also found use as templates for biomineralization of silicon or iron; the polypeptide concentration necessary for these applications ranges from 30 to 140 μM. Knowledge of the aggregation state of the polypeptides at these higher relevant concentrations would therefore aid in understanding the mechanisms of their activities.
Non-denaturing PAGE was employed to investigate the aggregation behavior of the polypeptides at higher concentrations. Fig. 5 shows the non-denaturing gels of 17-H-3, 17-H-6, and 35-H-6 with polypeptide concentrations ranging from approximately 5 to 140 μM. Although the relative positions of the bands in these images (which come from different gels) do not capture the molecular mass differences between polypep-tides, a single gel of the three polypeptides shows relative band positions that are generally consistent with the molecular mass differences (see Electronic Supplementary Material). Molecular mass standards were not included in these measurements owing to their lack of relevance to the polypeptides, which are not of the composition and globular structure of the standards. Although the polypeptide concentrations are not identical in the three gels, they cover a similar concentration range, and permit rapid and facile assessment of the changes in the aggregation state of a given polypeptide as a function of concentration. Of particular relevance for the aggregation assessment is that all three polypeptides migrate as a single band at all concentrations, with no variation in the location of the band, indicating a lack of aggregation at the elevated concentrations and corroborating the lack of aggregation observed in the AUC data. The complete absence of higher-order oligomers at higher molecular mass values in these gels suggests that there is no significant association, although very low concentrations of oligomers/aggregates may not be detected under these assay conditions. In addition, the range of polypeptide concentrations monitored suggests that these polypeptides will not aggregate under a range of concentrations useful for targeted applications.
Fig. 5.
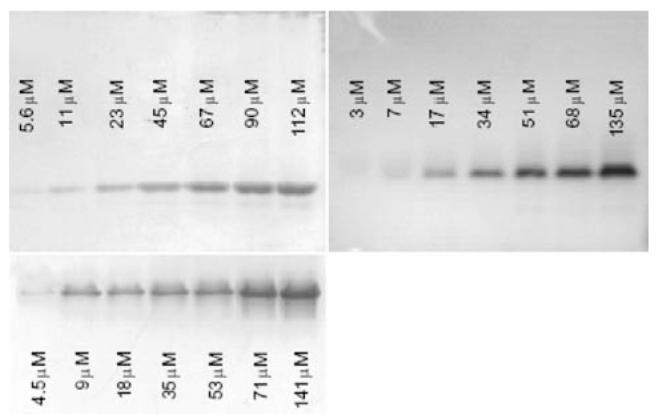
Non-denaturing polyacrylamide gel electrophoresis of 17-H-3 (top left), 17-H-6 (top right), and 35-H-6 (bottom left) samples at various concentrations.
Given the well-behaved conformational and association properties of the polypeptides under neutral aqueous conditions as described above, these macromolecules may offer unique opportunities for therapeutic applications. By utilizing recombinant techniques to produce well-defined polymeric sequences, delivery of an exact number of drug molecules per polypeptide could be achieved. Multiple molecules of the anticancer drug, doxorubicin, have been attached to the amine-terminated sidechains of linear elastin-based polypep-tides (54). Branched polypeptides are widely used to display multiple drug molecules; branched polypeptides with sequences containing glutamic acid, lysine, and alanine have been conjugated with the antitumor drug daunomycin in an attempt to probe the role structure plays in delivery (55,56). The helical polypeptides discussed in this manuscript could be similarly employed for delivery of a given number of drug molecules at a specific density. In addition to the advantages of longer circulation times and reduced drug toxicity that occur with conjugation of drugs to polymers and may be afforded by the conjugation of drugs to these polypeptides, the control of ligand or drug density on the scaffold may offer additional advantages. Multivalent ligands of designed architectures are known to have specific influences on cellular responses (57), and the delivery of a multivalent polypeptide-drug conjugate with a designed spacing between drug or ligand molecules may improve interactions at the cell surface and improve the uptake or effects of the drug.
CONCLUSIONS
The conformational and aggregation behavior of ala-nine-rich polypeptides have been investigated for three molecules, 17-H-3, 17-H-6, and 35-H-6, under isotonic buffer conditions. The synthesis of a new molecule, 17-H-6, has allowed the differentiation of the effect of molecular weight and composition on the conformational behavior of these families of polypeptides. An increase in molecular weight between 17-H-3 and 17-H-6 (8,875 to 14,770, respectively) results in increased helicity, and slight changes in composition of the alanine-rich domains between 17-H-6 and 35-H-6 are not suggested to have large effects on polypeptide helicity. Although the helicity of the molecules differs slightly with molecular weight and composition, the thermal stability of the helix is not significantly affected by these variables, as suggested by the comparable values of the Tm and van’t Hoff enthalpies for each of the polypeptides. These alanine-rich polypeptides do not show any signs of aggregation under physiologically relevant conditions. The combination of these results suggests the utility of these polypeptide templates for various applications in molecular recognition, biomineralization, or in multivalent display of therapeutic agents.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded in part by grant 1-P20-RR017716-01 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), by grant NA68-01923 from the National Aeronautics and Space Administration, and by grant DMR-0210223 from the National Science Foundation. This work was also supported by grant P20-RR015588 (instrument facilities) from NCRR. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH. Jared Sharp is acknowledged for the construction of genes encoding the 17-H-X polypeptides.
REFERENCES
- 1.Fahnestock SR, Irwin SL. Synthetic spider dragline silk proteins and their production in Escherichia coli. Appl. Microbiol. Biotechnol. 1997;47:23–32. doi: 10.1007/s002530050883. [DOI] [PubMed] [Google Scholar]
- 2.Qu Y, Payne SC, Apkarian RP, Conticello VP. Self-assembly of a polypeptide multi-block copolymer modeled on dragline silk proteins. J. Am. Chem. Soc. 2000;122:5014–5015. [Google Scholar]
- 3.Szela S, Avtges P, Valluzzi R, Winkler S, Wilson D, Kirschner D, Kaplan DL. Reduction—oxidation control of beta-sheet assembly in genetically engineered silk. Biomacro-molecules. 2000;1:534–542. doi: 10.1021/bm0055697. [DOI] [PubMed] [Google Scholar]
- 4.Wilson D, Valluzzi R, Kaplan D. Conformational transitions in model silk peptides. Biophys. J. 2000;78:2690–2701. doi: 10.1016/S0006-3495(00)76813-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winkler S, Szela S, Avtges P, Valluzzi R, Kirschner DA, Kaplan D. Designing recombinant spider silk proteins to control assembly. Int. J. Biol. Macromol. 1999;24:265–270. doi: 10.1016/s0141-8130(98)00088-9. [DOI] [PubMed] [Google Scholar]
- 6.Betre H, Setton LA, Meyer DE, Chilkoti A. Characterization of a genetically engineered elastin-like polypeptide for cartilaginous tissue repair. Biomacromolecules. 2002;3:910–916. doi: 10.1021/bm0255037. [DOI] [PubMed] [Google Scholar]
- 7.Chilkoti A, Dreher MR, Meyer DE. Design of thermally responsive, recombinant polypeptide carriers for targeted drug delivery. Adv. Drug Deliver. Rev. 2002;54:1093–1111. doi: 10.1016/s0169-409x(02)00060-1. [DOI] [PubMed] [Google Scholar]
- 8.Girotti A, Reguera J, Rodriguez-Cabello JC, Arias FJ, Alonso M, Testera AM. Design and bioproduction of a recombinant multi(bio)functional elastin-like protein polymer containing cell adhesion sequences for tissue engineering purposes. J. Mater. Sci Mater. M. 2004;15:479–484. doi: 10.1023/b:jmsm.0000021124.58688.7a. [DOI] [PubMed] [Google Scholar]
- 9.McMillian RA, Lee TAT, Conticello VP. Rapid assembly of synthetic genes encoding protein polymers. Macromolecules. 1999;32:3643–3648. [Google Scholar]
- 10.Meyer DE, Chilkoti A. Genetically encoded synthesis of protein-based polymers with precisely specified molecular weight and sequence by recursive directional ligation: Examples from the elastin-like polypeptide system. Biomacromolecules. 2002;3:357–367. doi: 10.1021/bm015630n. [DOI] [PubMed] [Google Scholar]
- 11.Panitch A, Yamaoka T, Fournier MJ, Mason TL, Tirrell DA. Design and biosynthesis of elastin-like artificial extracellular matrix proteins containing periodically spaced fibronectin CS5 domains. Macromolecules. 1999;32:1701–1703. [Google Scholar]
- 12.Wright ER, Conticello VP. Self-assembly of block copolymers derived from elastin-mimetic polypeptide sequences. Adv. Drug Deliver. Rev. 2002;54:1057–1073. doi: 10.1016/s0169-409x(02)00059-5. [DOI] [PubMed] [Google Scholar]
- 13.Arnold WV, Sieron AL, Fertala A, Bachinger HP, Mechling D, Title DJ. A cDNA cassette system for the synthesis of recombinant procollagens. Variants of procollagen II lacking a D-period are secreted as triple-helical monomers Prockop. Matrix Biol. 1997;16:105–116. doi: 10.1016/s0945-053x(97)90039-1. [DOI] [PubMed] [Google Scholar]
- 14.Pakkanen O, Hamalainen ER, Kivirikko KI, Myllyharju J. Assembly of stable human type I and III collagen molecules from hydroxylated recombinant chains in the yeast Pichia pastoris—Effect of an engineered C-terminal oligomerization domain foldon. J. Biol. Chem. 2003;278:32478–32483. doi: 10.1074/jbc.M304405200. [DOI] [PubMed] [Google Scholar]
- 15.Farmer RS, Argust LM, Sharp JD, Kiick KL. Conformational properties of helical protein polymers with varying densities of chemically reactive groups. Macromolecules. 2006;39:162–170. doi: 10.1021/ma051534t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farmer RS, Kiick KL. Conformational behavior of chemically reactive alanine-rich repetitive protein polymers. Biomacromolecules. 2005;6:1531–1539. doi: 10.1021/bm049216+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goeden-Wood NL, Conticello VP, Muller SJ, Keasling JD. Improved assembly of multimeric genes for the biosynthetic production of protein polymers. Biomacromolecules. 2002;3:874–879. doi: 10.1021/bm0255342. [DOI] [PubMed] [Google Scholar]
- 18.Koltover I, Sahu S, Davis N. Genetic engineering of the nanoscale structure in polyelectrolyte-lipid self-assembled systems. Angew. Chem. Int. Ed. 2004;43:4034–4037. doi: 10.1002/anie.200460164. [DOI] [PubMed] [Google Scholar]
- 19.Krejchi MT, Atkins EDT, Waddon AJ, Fournier MJ, Mason TL, Tirrell DA. Chemical sequence control of beta-sheet assembly in macromolecular crystals of periodic polypeptides. Science. 1994;265:1427–1432. doi: 10.1126/science.8073284. [DOI] [PubMed] [Google Scholar]
- 20.McGrath KP, Fournier MJ, Mason TL, Tirrell DA. Genetically directed syntheses of new polymeric materials—Expression of artificial genes encoding proteins with repeating (AlaGly)3ProGluGly elements. J. Am. Chem. Soc. 1992;114:727–733. [Google Scholar]
- 21.Yu SM, Conticello V, Zhang G, Kayser C, Fournier MJ, Mason TL, Tirrell DA. Smectic ordering in solutions and films of a monodisperse derivative of poly(g-benzyl α,L-glutamate) Nature. 1997;389:187–190. doi: 10.1038/38254. [DOI] [PubMed] [Google Scholar]
- 22.Krejchi MT, Cooper SJ, Deguchi Y, Atkins EDT, Fournier MJ, Mason TL, Tirrell DA. Crystal structures of chain-folded antiparallel b-sheet assemblies from sequence-designed periodic polypeptides. Macromolecules. 1997;30:5012–5024. [Google Scholar]
- 23.Petka WA, Hardin JL, McGrath KP, Wirtz D, Tirrell DA. Reversible hydrogels from self assembling artificial proteins. Science. 1998;281:389–392. doi: 10.1126/science.281.5375.389. [DOI] [PubMed] [Google Scholar]
- 24.Shen W, Lammertink RGH, Sakata JK, Kornfield JA, Tirrell DA. Assembly of an artificial protein hydrogel through leucine zipper aggregation and disulfide bond formation. Macromolecules. 2005;38:3909–3916. [Google Scholar]
- 25.Shen W, Zhang K, Kornfield JA, Tirrell DA. Tuning the erosion rate of artificial protein hydrogels through control of network topology. Nature Materials. 2006;5:153–158. doi: 10.1038/nmat1573. [DOI] [PubMed] [Google Scholar]
- 26.Topilina NI, Higashiya S, Rana N, Ermolenkov VV, Kossow C, Carlsen A, Ngo SC, Wells CC, Eisenbraun ET, Dunn KA, Lednev IK, Geer RE, Koaloyeros AE, Welsh JT. Bilayer fibril formation by genetically engineered polypeptides: preparation and characterization. Biomacromolecules. 2006;7:1104–1111. doi: 10.1021/bm0509016. [DOI] [PubMed] [Google Scholar]
- 27.Barron JI, Won AE. A new cloning method for the preparation of long repetitive polypeptides without a sequence requirement. Macromolecules. 2002;35:8281–8287. [Google Scholar]
- 28.Won JI, Meagher RJ, Barron AE. Protein polymer drag-tags for DNA separations by end-labeled free-solution electrophoresis. Electrophoresis. 2005;26:2138–2148. doi: 10.1002/elps.200410042. [DOI] [PubMed] [Google Scholar]
- 29.Farmer RS, Charati MB, Kiick KL. Biosynthesis of Protein-based Polymeric Materials. In: Matyjaszewski K, Gnanou Y, Leibler L, editors. Macromolecular Engineering. Precise Synthesis, Materials Properties, Applications. Vol. 1. Wiley-VCH; New York: 2007. [Google Scholar]
- 30.Wang Y, Kiick KL. Monodisperse protein-based glycopolymers via a combined biosynthetic and chemical approach. J. Am. Chem. Soc. 2005;127:16392–16393. doi: 10.1021/ja055102+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Talmadge JE. The pharmaceutics and delivery of therapeutic polypeptides and proteins. Adv. Drug Deliver. Rev. 1993;10:247–299. [Google Scholar]
- 32.Massodi I, Bidwell GL, Raucher D. Evaluation of cell penetrating peptides fused to elastin-like polypeptide for drug delivery. J. Control Release. 2005;108:396–408. doi: 10.1016/j.jconrel.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 33.Bidwell GL, Raucher D. Application of thermally responsive polypeptides directed against c-Myc transcriptional function for cancer therapy. Mol. Cancer Ther. 2005;4:1076–1085. doi: 10.1158/1535-7163.MCT-04-0253. [DOI] [PubMed] [Google Scholar]
- 34.Dreher MR, Raucher D, Balu N, Colvin OM, Ludeman SM, Chilkoti A. Evaluation of an elastin-like polypeptide—doxorubicin conjugate for cancer therapy. J. Control Release. 2003;91:31–43. doi: 10.1016/s0168-3659(03)00216-5. [DOI] [PubMed] [Google Scholar]
- 35.Furgeson DY, Dreher MR, Chilkoti A. Structural optimization of a “smart” doxorubicin—polypeptide conjugate for thermally targeted delivery to solid tumors. J. Control Release. 2006;110:362–369. doi: 10.1016/j.jconrel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 36.Armstrong KM, Baldwin RL. Charged histidine affects a-helix stability at all positions in the helix by interacting with the backbone charges. Proc Natl Acad Sci U.S.A. 1993;90:11337–11340. doi: 10.1073/pnas.90.23.11337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chakrabartty A, Baldwin RL. Stability of alpha-helicies. Adv. Protein Chem. 1995;46:141–176. [PubMed] [Google Scholar]
- 38.Chakrabartty A, Kortemme T, Baldwin RL. Helix propensities of the amino-acids measured in alanine-based peptides without helix-stabilizing side-chain interactions. Protein Sci. 1994;3:843–852. doi: 10.1002/pro.5560030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huyghues-Despointes BMP, Scholtz JM, Baldwin RL. Effect of a single asparate on helix stability at different positions in a neutral alanine-based peptide. Protein Sci. 1993;2:1604–1611. doi: 10.1002/pro.5560021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kennedy RJ, Tsang K-Y, Kemp DS. Consistent helicities from CD and template t/c data for N-templated polyalanines: Progress toward resolution of the alanine helicity problem. J. Am. Chem. Soc. 2002;124:934–944. doi: 10.1021/ja016285c. [DOI] [PubMed] [Google Scholar]
- 41.Miller JS, Kennedy RJ, Kemp DS. Solubilized, spaced polyalanines: A context-free system for determining amino acid α-helix propensities. J. Am. Chem. Soc. 2002;124:945–962. doi: 10.1021/ja011726d. [DOI] [PubMed] [Google Scholar]
- 42.Blondelle SE, Forood B, Houghten RA, PerezPaya E. Secondary structure induction in aqueous vs membrane-like environments. Biopolymers. 1997;42:489–498. doi: 10.1002/(SICI)1097-0282(19971005)42:4<489::AID-BIP11>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 43.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Royal Society of Chemistry; Cambridge: 1992. [Google Scholar]
- 44.Israelachvili JN. Intermolecular and Surface Forces: With Applications to Colloidal and Biological Systems. Academic; New York: 1992. [Google Scholar]
- 45.Scholtz JM, Qian H, Robbins VH, Baldwin RL. The energetics of ion-pair and hydrogen-bonding interactions in a helical peptide. Biochemistry. 1993;32:9668–9676. doi: 10.1021/bi00088a019. [DOI] [PubMed] [Google Scholar]
- 46.Wallimann P, Kennedy RJ, Miller JS, Shalongo W, Kemp DS. Dual wavelength parametric test of two-state models for circular dichroism spectra of helical polypeptides: Anomalous dichroic properties of alanine-rich peptides. J. Am. Chem. Soc. 2003;125:1203–1220. doi: 10.1021/ja0275360. [DOI] [PubMed] [Google Scholar]
- 47.Pace CN, Scholtz JM. A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 1998;75:422–427. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scholtz JM, Marqusee S, Baldwin RL, York EJ, Stewart JM, Santoro M, Bolen DW. Calorimetric determination of the enthalpy change for the α-helix to coil transition of an alanine peptide in water. Proc. Natl. Acad. Sci. U.S.A. 1991;88:2854–2858. doi: 10.1073/pnas.88.7.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Persikov AV, Xu Y, Brodsky B. Equilibrium thermal transitions of collagen model peptides. Protein Sci. 2004;13:893–902. doi: 10.1110/ps.03501704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scholtz JM, Qian H, York EJ, Stewart JM, Baldwin RL. Parameters of helix-coil transition theory for alanine-based peptides of varying chain lengths in water. Biopolymers. 1991;31:1463–1470. doi: 10.1002/bip.360311304. [DOI] [PubMed] [Google Scholar]
- 51.Ralston G. Introduction to Analytical Ultracentrifugation. Beckman; California: 1993. [Google Scholar]
- 52.McRorie DK, Voelker PJ. Self-associating Systems in the Analytical Ultracentrifuge. Beckman; California: 1993. [Google Scholar]
- 53.Blondelle SE, Forood B, Houghten RA, PerezPaya E. Polyalanine-based peptides as models for self-associated β-pleated-sheet complexes. Biochemistry. 1997;36:8393–8400. doi: 10.1021/bi963015b. [DOI] [PubMed] [Google Scholar]
- 54.Kaufmann D, Weberskirch R. Efficient synthesis of protein—drug conjugates using a functionalizable recombinant elastin-mimetic polypeptide. Macromol. Biosci. 2006;6:952–958. doi: 10.1002/mabi.200600117. [DOI] [PubMed] [Google Scholar]
- 55.Gaal D, Hudecz F. Low toxicity and high antitumour activity of daunomycin by conjugation to an immunopotential amphoteric branched polypeptide. Eur. J. Cancer. 1998;34:155–161. doi: 10.1016/s0959-8049(97)00338-9. [DOI] [PubMed] [Google Scholar]
- 56.Remenyi J, Csik G, Kovacs P, Reig F, Hudecz F. The effect of the structure of branched polypeptide carrier on intracellular delivery of daunomycin. Biochim. Biophys. Acta Biomembr. 2006;1758:280–289. doi: 10.1016/j.bbamem.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 57.Kiessling LL, Gestwicki JE, Strong LE. Synthetic multivalent ligands as probes of signal transduction. Angew. Chem. Int. Edit. 2006;45:2348–2368. doi: 10.1002/anie.200502794. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.