Abstract
Introduction
Polymorphic alleles in the human genome have been identified as affecting numerous drug responses. Currently, genotyping of all patients before starting a drug regimen is impractical. Since many polymorphisms occur at varying rates in different racial groups, we investigated whether a patient's race could predict presence of drug-relevant genetic variants well enough to be used as a substitute for individual genotyping.
Methods
We performed hierarchical clustering and principal components analysis on tagSNPs from three pathways (irinotecan, 5-fluorouracil and insulin) across 270 individuals from four racial groups available from the International HapMap Project.
Results
For the drug pathways, irinotecan and 5-fluorouracil, individuals from each race were widely dispersed, although several subclusters consisted entirely of individuals from a single racial group. Principal components analysis confirmed race was not a major contributor to the SNP data variance. Interestingly, individuals tended to cluster more by race across the endogenous insulin signaling pathway SNPs.
Conclusions
Most genetic variation was determined by individual variation, not racial grouping, indicating race is not adequate as a surrogate to individualized therapy.
Keywords: 5-fluorouracil, HapMap, irinotecan, pharmacogenomics, race
One of the most difficult challenges in biomedical research has been defining the mechanisms underlying the interindividual variations in drug response. Among the many demographic (e.g., gender and age) and biochemical (e.g., renal function and liver function) factors often evaluated for influence on drug effects, one of the most commonly included variants is self-declared patient race. Racial differences have been observed for response to β-blockers, mydriatics and opioid pain relievers, among other medications [1-8]. In fact, the US FDA has approved drugs with prescribing guidelines targeted towards a specific racial group [9]. BiDil, a combination of hydralazine and isosorbide dinitrate, was approved for treating heart failure in African-American patients, after results from the African-American Heart Failure Trial (A-HeFT) supported clinical benefit from the drug [10,101].
In addition to demographic and biochemical factors influencing drug response, variants in genes involved in drug metabolism, membrane transport and cellular targets have also been identified. The US FDA has begun including genetic variation in prescribing information when it has been shown to identify elevated risk for toxicity or differential efficacy. Initial examples have focused on single genes, in which variants lead to altered protein function or transcription. Recently, the FDA has made changes to the package insert that support genetic testing of specific genes for all patients prior to prescribing warfarin (CYP2C9 and VKORC1) and for patients of Asian descent before taking carbamazepine (HLA-B*1502) [102,103]. These represent initial examples for what will likely become a common approach to pharmacogenomics: the use of heterogeneity in genes associated with molecular pathways to predict drug effects.
With the completion of the human genome sequencing project, vast amounts of genetic data have become readily accessible from online databases, such as the International HapMap Project [104]. HapMap offers SNP analysis from 270 individuals from four ethnic groups: the Yoruba from Ibadan, Nigeria (YRI); Japanese from Tokyo, Japan (JPT); Han Chinese from Beijing, China (CHB); and CEPH Caucasian from Utah, of Northern and Western European descent (CEU). The HapMap project was created to assist researchers in discovering genetic markers that predispose individuals for diseases, such as cancer, stroke and depression, or affect an individual's response to pharmaceutical treatment [105].
Most specific genetic variants occur in all human populations, but at varying rates. Thus, the availability of large numbers of variants begins to allow the discernment of an individual's racial group. We asked if the reverse is also true: if we know a patient's self-identified race, can we predict genotype at specific, drug-relevant markers? We were not seeking ancestry informative markers. Instead, we were attempting to determine if self-identified patient race has any potential utility as a predictive factor prior to prescribing medications and, therefore, allow for better patient care by improving efficacy and lowering toxicity rates.
We recognize that the term `race' is largely a social construction, and its usage can raise a variety of socio-ethical concerns [11-15]. Furthermore, the definition is apt to evolve and change over time, making long-term research and recommendations difficult [16]. However, in the absence of a better term, we chose to use `race' to signify the diverse population from within a specific geographic locale (Ibadan, Nigeria; Tokyo; Japan; Beijing; China; or Utah, USA), as chosen by the International HapMap Project [17].
Using data from individuals in the four populations available in HapMap, we examined SNPs in genes from two drug pathways, irinotecan and 5-fluorouracil (5-FU). The genes included in the irinotecan pathway have little endogenous biological interaction with each other, outside of irinotecan processing, while the 5-FU pathway genes include both 5-FU specific genes (e.g., MTHFR and DHFR), as well as genes that interact in several endogenous pathways (e.g., pyrimidine biochemistry). In addition, a parallel analysis was performed to compare the results obtained from the drug pathways with a collection of random SNPs and an entirely endogenous pathway (insulin signaling). Using hierarchical clustering and principal components analysis (PCA), we assessed whether race, and the attendant race-dependent genetic variation, is sufficient to act as a surrogate for genotyping individuals. Although there is some clustering within racial groups for the genes of the drug pathways, the majority of the genetic variation between individuals genotyped by the International HapMap Project is due to individual variation, not race.
Materials & methods
Genes involved in the pharmacokinetics and pharmacodynamics of irinotecan (n = 13) and 5-FU (n = 21) were identified using the drug pathway diagrams on the PharmGKB website [106]. Irinotecan and 5-FU were chosen because the metabolic pathways involved in processing the drugs have relatively clear delineation. Ten genes involved in insulin signaling were chosen from the diagram on the National Cancer Institute/Nature Pathway Interaction Database [107]. Insulin signaling was chosen to represent a biological pathway containing a similar number of genes to the irinotecan and 5-FU pathways. A total of 362 SNPs were randomly chosen from the Affymetrix 500K SNP chip to use as a control. Supplementary Table 1, available online [108] lists all of the genes and SNPs used in this analysis.
SNP genotype data was downloaded for all of the individuals from the four populations from HapMap release NCBI build 35, dbSNP b125 [104]. The four HapMap populations were included in the analysis: YRI, JPT, CHB and CEU. TagSNPs were chosen for each population using Haploview (version 3.32), using a minimum minor allele frequency (MAF) of 10%, a haplotype frequency of 10%, and an r2 threshold of 0.8. The tagSNPs from each population were merged to generate the list of tagSNPs for each gene. This resulted in 173 SNPs covering 13 genes in the irinotecan pathway, 362 SNPs covering 21 genes in the 5-FU pathway and 172 SNPs covering ten genes in the insulin signaling pathway. The genotype frequency for all of the SNPs used in this study is available on the HapMap website [104].
For each dataset (irinotecan pathway, 5-FU pathway, insulin signaling pathway, random SNPs), the genotype data were transformed to numerical values for the purpose of clustering. Genotypes for individuals were converted to 1 (reference allele homozygous), 0 (heterozygous) or -1 (nonreference allele homozygous). The reference allele was determined from HapMap. Any missing data was left blank. For each dataset, we performed hierarchical clustering on the individuals and SNPs. Clustering was performed using Eisen's cluster program [18], using uncentered correlation and complete linkage clustering. The clusters were viewed using Java TreeView (version 1.1.0). PCA was performed for each data set using the `prcomp' algorithm in R (version 2.4.1). Before running the PCA, missing data were imputed using a k-nearest neighbor approach (k = 10). The resulting heat maps from the cluster analysis and the results of the PCA across the first three principal components are available in Supplemental File 2, can be found online [108].
Results
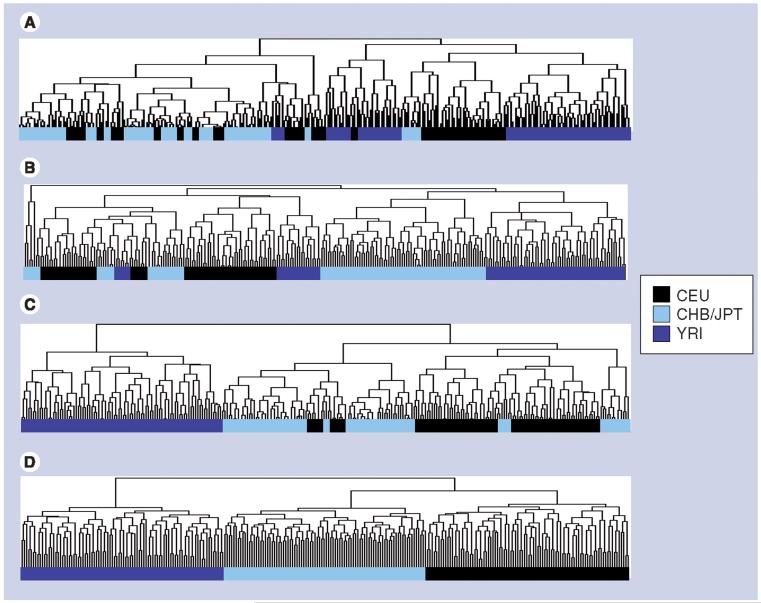
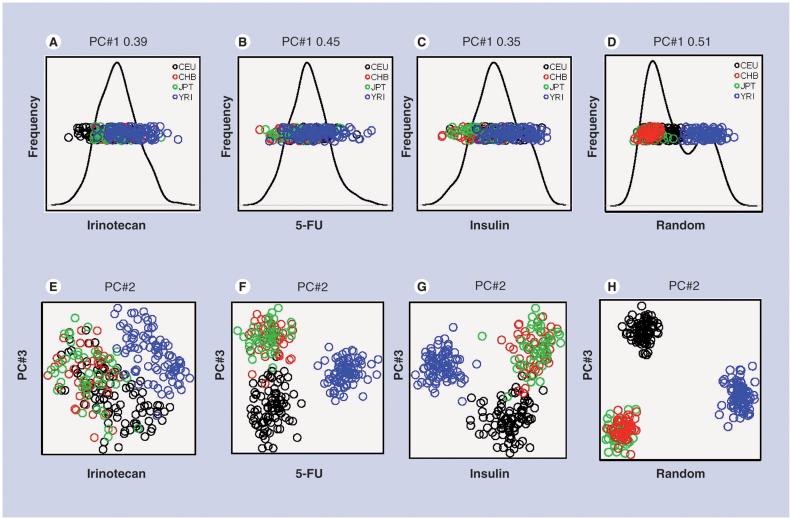
Using genotype data from the HapMap project, we examined the genetic groupings among individuals in all four populations (YRI, JPT, CHB and CEU; n = 270) across the irinotecan and 5-FU pathway genes. The genes in the irinotecan pathway come from many different functional classes and have little endogenous interaction. In total, 173 tagSNPs from 13 genes involved in irinotecan cell membrane transport and metabolism were analyzed. Hierarchical clustering performed to examine the similarities in genotypes across the irinotecan pathway demonstrated that race contributed little to the separation of individuals (Figure 1A). Subclusters were relatively narrow, and individuals of different race were intermingled, although a subcluster consisting of over 60% of the individuals in the YRI population was identifiable. Individuals from the CHB and JPT populations were intermingled, while small clusters of CEU individuals were scattered throughout the CHB/JPT, and among the YRI. We also performed PCA for the irinotecan pathway genes to determine how much of the variance of the SNP data could be attributed to race. Principal component (PC)#1 accounted for 39% of the variance in the data, but failed to separate individuals on the basis of race (Figure 2A). In fact, individuals of all races were found to be completely overlapping in PC#1, in agreement with the hierarchical clustering analysis. However, by plotting PC#2 (14% of variance) against PC#3 (7% of variance), the YRI individuals tended to separate themselves from the other three populations, but the CEU individuals still significantly overlapped with the CHB/JPT individuals (Figure 2E).
Figure 1. Hierarchical clustering of 270 HapMap individuals across SNPs.
(A) irinotecan, (B) 5-FU, and (C) insulin pathway genes, and (D) 362 randomly chosen SNPs. Racial groupings are indicated in the dendrogram. Full clustering figures are available in Supplemental Figure 2 available online [108].
YRI: Yoruba from Ibadan; JPT: Japanese from Tokyo; CHB: Han Chinese from Beijing;
CEU: CEPH Caucasian.
Figure 2. Principal components analysis of the 270 HapMap individuals.
PC#1 (A-D) and PC#2 versus PC#3 (E-H) across SNPs from (A, E) irinotecan, (B, F) 5-FU, and (C, G) insulin pathway genes and (D, H) 362 randomly chosen SNPs. Full PCA figures are available in Figure 2 of the Supplemental material available at [108].
PC: Principal component; PCA: Principal components analysis.
The 5-FU pathway combines genes from several endogenous pathways into an artificial drug-processing pathway. We analyzed 362 tagSNPs, encompassing 21 genes involved in 5-FU catabolism, anabolism and activity. Hierarchical clustering generated five predominant clusters of individuals (Figure 1B). In general, individual genotypes across the 5-FU pathway influenced the clustering more than race, although a few subclusters consisting of individuals from a single population were present in the data. Over 70% of the YRI were located in a single subcluster, whose individuals were most similar to a subcluster consisting of JPT and CHB individuals. Another prominent subcluster consisted of approximately 50% of the CEU, along with 20% of the YRI individuals. PCA indicated no clear race differences across PC#1, which accounted for 45% of the variance within the data (Figure 2B). However, the YRI completely separated from the other populations across PC#2, which accounted for 7% of the variance. Additionally, PC#3 separated the CEU individuals from the JPT/CHB mixture, accounting for 4% of the variance. Plotting the individuals across PC#2 and PC#3 almost totally separated them into three distinct groups: YRI, CEU and the JPT/CHB mixture (Figure 2F).
We were surprised that race was not the primary contributor to the variance observed in the hierarchical clustering and PCA results. To ensure we had a sufficient number of SNPs to perform the analysis properly, we selected 362 SNPs at random from the Affymetrix 500K SNP chip and performed the same hierarchical clustering and PCA. We found that individuals clustered exclusively with members of the same racial group (although the CHB and JPT populations overlapped entirely) across the 362 random SNPs (Figure 1D). Similarly, PC#1, which accounted for 51% of the variance, mostly separated the YRI from the other three populations (Figure 2D). As was seen with the 5-FU pathway analysis, the YRI were completely isolated across PC#2 (7% of variance), while the CEU were separated from the other groups across PC#3 (3% of variance). Again, plotting PC#2 against PC#3 separated the YRI, CEU and CHB/JPT mixture into three distinct populations (Figure 2H). The complete separation of the populations is consistent with recently published findings from Rosenberg et al., who found that by using a sufficient number of genetic markers, sampled individuals can be separated into distinct genetic clusters along geographic boundaries [19].
We also performed analysis of SNPs from genes involved in an endogenous pathway, which has been under evolutionary pressure. Using ten genes involved in insulin signaling, we obtained individual genotypes for 172 tagSNPs and performed hierarchical clustering. Using SNP genotypes for insulin-signaling pathway genes containing polymorphisms, the YRI individuals clearly separated from the individuals in the three other populations (Figure 1C). The rest of the three populations were interspersed, although a majority of the CEU individuals tended to cluster together. As with the drug pathways, PCA analysis showed no clear racial differences across PC#1, accounting for 35% of the variance (Figure 2C). PC#2, accounting for 14% of the variance in the data, separated the YRI individuals from the other three populations. As with the 5-FU pathway PCA, plotting individuals across PC#2 and PC#3 (7% of variance) separated the YRI individuals from the other three populations, while also tending to separate the CEU individuals from the mixture of JPT/CHB individuals (Figure 2G).
Discussion
The completion of the Human Genome Project was supposed to lead to an era of personalized medicine, where patients would be treated with medications based on their individual genotype. Although progress has been made in identifying gene polymorphisms that influence the response to several drugs, the promise of personalized medicine has not been realized as of yet because personal genotyping is cost prohibitive and most drug-genotype interactions remain unknown. Since individual causative alleles usually have distinct frequencies across the `Old World' populations, there is potential utility in using race labels as a surrogate for genetic information, as a means to the ultimate goal of individualized therapy.
Examples of individuals of different races having different responses to cancer chemotherapy treatment have been well documented. A recent study reported that African-American patients with stage III colon cancer had a lower 5-year survival rate than their non-Hispanic white counterparts, when treated with a 5-FU-based therapy [20]. A pharmacokinetic study demonstrated that clearance of tegafur, a 5-FU derivative, as part of the S-1 formulation (tegafur plus potassium oxonate and gimeracil) was faster in Western (European and American) cancer patients than in Japanese patients [21]. By contrast, a separate report found that 31 out of 45 (68.9%) American colorectal cancer patients suffered from diarrhea on uracil/tegafur (UFT) plus leuvocorin (LV), compared with only 17 out of 44 (38.6%) Japanese patients (p = 0.006), although both groups exhibited similar pharmacokinetics in processing tegafur [22]. The discrepancy between the studies with regard to tegafur processing in Western and Japanese populations may be due to the other components of the formulations (S-1 vs UFT/LV) affecting metabolism of tegafur. Distinct responses have also been oberseved towards other cancer chemotherapeutics. Non-small-cell lung cancer patients of Asian ancestry were found to have a significantly improved response rate to docetaxel and carboplatin when compared with Caucasians [23].
The results from our hierarchical clustering and PCA of the irinotecan, 5-FU and insulin signaling pathways demonstrated that race is not a robust predictor of the genetic subgroups within the human population. Clustering individuals across gene SNPs from the irinotecan pathway (which do not interact with one another in the absence of drug) yielded very little grouping by race, indicating that individuals within a racial group display heterogenous genotypes for genes in the irinotecan pathway. Clustering individuals across SNPs from the 5-FU pathway (which consists of a mixture of genes involved solely in drug metabolism and genes involved in endogenous pyrimidine metabolism), showed that the majority of YRI individuals grouped together, and possibly indicated more genotype homogeneity within these individuals. Examination of the clustering based on insulin-signaling SNPs (consisting exclusively of genes involved in endogenous biological processes), found even greater clustering by racial group, with all of the YRI in a separate subcluster. The other individuals tended to be grouped in relatively larger racial blocks than the other two pathways, which were interspersed with one another.
We acknowledge that this study has limitations. Variability in response to drug therapies between populations probably is not solely due to SNP differences. Most drug interactions are polygenic, indicating that multiple genetic variants may act together to influence drug response. Other factors, such as diet or other cultural practices, likely play equally significant roles. Ideally, a large-scale clinical study of the interactions between drug treatments and race, among different global populations, should be conducted to answer the fundamental question of how to provide the best medical care to every individual. However, it is not feasible to do such a study. Thus, we sought to identify an alternative method, using the data that is readily available from the HapMap website. We attempted to determine whether the methodology of using tagSNP genetic markers from well-defined drug pathways to separate samples from self-identified racial groups was viable. Because this method is relatively inexpensive and easy to perform, it has great potential benefit for future studies, as additional drug pathways are elucidated and more SNPs are associated with response to treatment. Our results clearly demonstrate that racial generalizations for treatment recommendations are not valid and are consistent with other recent publications regarding the suitability of using patient race to determine medical treatment [24,25].
Executive summary.
Human genomic variants that affect patient response to drugs have been identified. However, it remains unfeasible to individually genotype all patients prior to drug treatment.
Genomic SNP data for 270 individuals from four racial groups is available in the online HapMap database.
Using HapMap SNP data, the ability of race to be substituted for individual genotyping, with regards to predicting the presence of drug-relevant genetic markers, was determined. SNPs were selected from genes located in irinotecan and 5-fluorouracil drug pathways.
Methods
Hierarchical clustering and principal component analysis were performed to analyze the data.
Results
The majority of genomic heterogeneity in pharmacologic genes was determined to be due to individual variation rather than racial grouping. Thus, race should not be used as a predictive substitute for individual patient genotyping.
Conclusion
Our results indicate that the heterogeneity in genotypes in individuals from the examined racial groups (YRI, CHB, JPT and CEU) increases, going from a totally endogenous biological pathway to a totally exogenous pathway. However, it should be noted, that even for the insulin-signaling pathway, which exhibited the least variability among individuals in a racial group, race contributed only a small percentage to the variance across individuals. Since only a small percentage of the variance of the two-drug pathway genotypes across individuals is accounted for by race, the use of race as a substitute for genotyping would not appear to be clinically beneficial. Therefore, until the knowledge surrounding the interactions of race, genotype and drug response improves, individual genotyping of patients for known drug-influencing polymorphisms is the best course for optimizing drug treatment.
Supplementary Material
Acknowledgements
We would likely to gratefully acknowledge Javier Revollo and Michael Wagner for their assistance during preparation of the manuscript.
This work was supported in part by the Pharmacogenetics Research Network (U01 GM63340), the Lineberger Comprehensive Cancer Center (P30CA016086) and the UNC GI SPORE (5P50 CA106991). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Footnotes
Financial & competing interests disclosure No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Bibliography
- 1.Investigators of the β-Blocker Evaluation of Survival Trial: A trial of the β-blocker bucindolol in patients with advanced chronic heart failure. N. Engl. J. Med. 2001;344:1659–1667. doi: 10.1056/NEJM200105313442202. [DOI] [PubMed] [Google Scholar]
- 2.Exner DV, Dries DL, Domanski MJ, Cohn JN. Lesser response to angiotensin-converting-enzyme inhibitor therapy in black as compared with white patients with left ventricular dysfunction. N. Engl. J. Med. 2001;344:1351–1357. doi: 10.1056/NEJM200105033441802. [DOI] [PubMed] [Google Scholar]
- 3.Carson P, Ziesche S, Johnson G, Cohn JN. Racial differences in response to therapy for heart failure: Analysis of the vasodilator-heart failure trials. J. Card. Fail. 1999;5:178. doi: 10.1016/s1071-9164(99)90001-5. [DOI] [PubMed] [Google Scholar]
- 4.Zhou HH, Koshakji RP, Silberstein DJ, Wilkinson GR, Wood AJ. Altered sensitivity to and clearance of propranolol in men of Chinese descent as compared with American whites. N. Engl. J. Med. 1989;320:565–570. doi: 10.1056/NEJM198903023200905. [DOI] [PubMed] [Google Scholar]
- 5.Greenlee EC, Kwon YH. Ethnic differences in the systemic pharmacology for ophthalmologists. Int. Ophthalmol. Clin. 2003;43:27–38. doi: 10.1097/00004397-200343040-00005. [DOI] [PubMed] [Google Scholar]
- 6.Zhou HH, Sheller JR, Nu H, Wood M, Wood AJ. Ethnic differences in response to morphine. Clin. Pharmacol. Ther. 1993;54:507–513. doi: 10.1038/clpt.1993.182. [DOI] [PubMed] [Google Scholar]
- 7.Caraco Y, Sheller J, Wood AJ. Impact of ethnic origin and quinidine coadministration on codeine's disposition and pharmacodynamic effects. J. Pharmacol. Exp. Ther. 1999;290:413–422. [PubMed] [Google Scholar]
- 8.Cepeda MS, Farrar JT, Roa JH, et al. Ethnicity influences morphine pharmacokinetics and pharmacodynamics. Clin. Pharmacol. Ther. 2001;70:351–361. [PubMed] [Google Scholar]
- 9.Temple R, Stockbridge NL. BiDil for heart failure in black patients: The US Food and Drug Administration perspective. Ann. Intern. Med. 2007;146:57–62. doi: 10.7326/0003-4819-146-1-200701020-00010. [DOI] [PubMed] [Google Scholar]
- 10.Taylor AL, Ziesche S, Yancy C, et al. Combination of isosorbide dinitrate and hydralazine in Blacks with heart failure. N. Engl. J. Med. 2004;351:2049–2057. doi: 10.1056/NEJMoa042934. [DOI] [PubMed] [Google Scholar]
- 11.Bhopal R, Donaldson L. White, European, Western, Caucasian, or what? Inappropriate labeling in research on race, ethnicity, and health. Am. J. Public Health. 1998;88:1303–1307. doi: 10.2105/ajph.88.9.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fullilove MT. Comment: Abandoning “race” as a variable in public health research - an idea whose time has come. Am. J. Public Health. 1998;88:1297–1298. doi: 10.2105/ajph.88.9.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodman AH. Why genes don't count (for racial differences in health) Am. J. Public Health. 2000;90:1699–1702. doi: 10.2105/ajph.90.11.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lieberman L, Stevenson BW, Reynolds LT. Race and anthropology: core concept with concensus. Anthropol. Educ. Q. 1989;20:67–73. [Google Scholar]
- 15.Egalite N, Ozdemir V, Godard B. Pharmacogenomics research involving racial classification: qualitative research findings on researchers' views, perceptions and attitudes towards socio-ethical responsibilities. Pharmacogenomics. 2007;8:1115–1126. doi: 10.2217/14622416.8.9.1115. [DOI] [PubMed] [Google Scholar]
- 16.Olivier C, Williams-Jones B, Godard B, Mikalson B, Ozdemir V. Personalized medicine, bioethics and social responsibilities: re-thinking the pharmaceutical industry to rememdy inequities in patient care and international health. Curr Pharmacogenomics. 2008;6:108–120. [Google Scholar]
- 17.The International HapMap Consortium: Integrating ethics and science in the International HapMap Project. Nat. Rev. Genet. 2004;5:467–475. doi: 10.1038/nrg1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Nat. Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenberg NA, Mahajan S, Ramachandran S, Zhao C, Pritchard JK, Feldman MW. Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet. 2005;1:E70. doi: 10.1371/journal.pgen.0010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jessup JM, Stewart A, Greene FL, Minsky BD. Adjuvant chemotherapy for stage III colon cancer: Implications of race/ethnicity, age, and differentiation. JAMA. 2005;294:2703–2711. doi: 10.1001/jama.294.21.2703. [DOI] [PubMed] [Google Scholar]
- 21.Comets E, Ikeda K, Hoff PM, Fumoleau P, Wanders J, Tanigawara Y. Comparison of the pharmacokinetics of S-1, an oral anticancer agent, in western and Japanese patients. J. Pharmacokinet. Pharmacodyn. 2003;30:257–283. doi: 10.1023/a:1026142601822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shirao K, Hoff PM, Ohtsu A, et al. Comparison of the efficacy, toxicity, and pharmacokinetics of a uracil/tegafur (UFT plus oral leucovorin (LV) regimen between Japanese and American patients with advanced colorectal cancer: Joint United States and Japan study of UFT/LV. J. Clin. Oncol. 2004;22:3466–3474. doi: 10.1200/JCO.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 23.Millward MJ, Boyer MJ, Lehnert M, et al. Docetaxel and carboplatin is an active regimen in advanced non-small-cell lung cancer: a Phase II study in Caucasian and Asian patients. Ann. Oncol. 2003;14:449–454. doi: 10.1093/annonc/mdg118. [DOI] [PubMed] [Google Scholar]
- 24.Barr DA. The practitioner's dilemma: Can we use a patient's race to predict genetics, ancestry, and the expected outcomes of treatment? Ann. Intern. Med. 2005;143:809–815. doi: 10.7326/0003-4819-143-11-200512060-00009. [DOI] [PubMed] [Google Scholar]
- 25.Frank R. What to make of it? The (Re) emergence of a biological conceptualization of race in health disparities research. Soc. Sci. Med. 2007;64:1977–1983. doi: 10.1016/j.socscimed.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 101.FDA approves BiDil heart failure drug for black patients. US FDA press release; 2005. www.fda.gov/bbs/topics/NEWS/2005/NEW01190.html. [Google Scholar]
- 102.FDA approves updated warfarin (coumadin) prescribing information. US FDA press release; 2007. www.fda.gov/bbs/topics/NEWS/2007/NEW01684.html. [Google Scholar]
- 103.Carbamazepine prescribing information to include recommendation of genetic test for patients with Asian ancestry. FDA press release; 2007. www.fda.gov/bbs/topics/NEWS/2007/NEW01755.html. [Google Scholar]
- 104.The International HapMap Project. 2002 http://hapmap.org/
- 105.About the HapMap. 2002 http://hapmap.org/thehapmap.html.en. The HapMap is a worldwide collaborative resource with genotyping data from nearly 4 million SNPs in 270 individuals
- 106.Pharmgkb. www.pharmgkb.org.
- 107.National Cancer Institute: Pathway Interaction Database. http://pid.nci.nih.gov.
- 108.Supplemenatry material available from the Institute for Pharmacogenomics and Individualized Therapy website. www.ipit.unc.edu/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.