Abstract
JC virus (JCV) is a human polyomavirus that can emerge from a latent state to cause the cytolytic destruction of oligodendrocytes in the brain resulting in the fatal demyelinating disease, Progressive Multifocal Leukoencephalopathy (PML). Previous studies described a cis-acting transcriptional regulatory element in the JCV non-coding control region (NCCR) that is involved in the response of JCV to cytokines. This consists of a 23 base pair GGA/C rich sequence (GRS) near the replication origin (5112 to +4) that contains potential binding sites for Sp1 and Egr-1. Gel shift analysis showed that Egr-1, but not Sp1, bound to GRS. Evidence is presented that the GRS gel shift seen on cellular stimulation is due to Egr-1. Thus, TPA-induced GRS gel shift could be blocked by antibody to Egr-1. Further, the TPA-induced GRS DNA/protein complex was isolated and found to contain Egr-1 by Western blot. No other Egr-1 sites were found in the JCV NCCR. Functionally, Egr-1 was found to stimulate transcription of JCV late promoter but not early promoter reporter constructs. Mutation of the Egr-1 site abrogated Egr-1 binding and virus with the mutated Egr-1 site showed markedly reduced VP1 expression and DNA replication. Infection of primary astrocytes by wild-type JCV induced Egr-1 nuclear expression that was maximal at 5–10 days post-infection. Finally, upregulation of Egr-1 was detected in PML by immunohistochemistry. These data suggest that Egr-1 induction may be important in the life cycle of JCV and PML pathogenesis.
Keywords: JCV, Egr-1, PML, polyomavirus
INTRODUCTION
The fatal demyelinating disease known as progressive multifocal leukoencephalopathy (PML) is caused by the lysis of oligodendrocytes in the brain consequent to lytic replication of the human polyomavirus, JC virus (JCV). While infection is very common in most human populations, this is usually subclinical since the virus is readily controlled by the immune system. After the initial infection is resolved, JCV nonetheless persists in the body and enters a state of latency which is poorly understood. However, under circumstances in which the immune system becomes impaired, e.g., AIDS, the virus reactivates and replicates in the central nervous system (CNS) to cause PML (Berger and Houff, 2006; Del Valle L and Piña-Oviedo S, 2006; Khalili et al., 2006). The mechanisms involved in this reactivation are not known but it is possible that changes in the levels of cytokines and immunomodulators, such as TNF-α, MIP-1α and TGF-β, that are associated with immunosuppression (Enam et al., 2004), elicit changes in intracellular signal transduction pathways that, in turn, modulate the activities of transcription factors that are involved in regulating the expression of JCV genes (Raj and Khalili, 1995).
The circular genome of JCV consists of an early and a late region that are transcribed bidirectionally starting at a common promoter region known as the non-coding control region (NCCR) that also contains the origin of viral DNA replication. The origin of DNA replication contains a region that possesses a series of trinucleotide repeats with the sequence GG(A/C) which has been called the GG(A/C)-rich sequence (GRS). The sequence of the GRS predicts that it contains potential binding sites for GC-box-binding zinc-finger transcription factors such as Sp1 and Egr-1. Previously, we had described an unknown protein activity that bound to GRS and was named GRS-binding protein (GBPi) which is rapidly induced by certain cytokines, including, TNF-α, TGF-β and IFN-γ, and by the phorbol ester, TPA, reaching a maximum after 1.5–3 hrs (Raj and Khalili, 1994). The induction of the GBPi gel shift activity was rapid and could be blocked by cycloheximide or actinomycin D. This would indicate that de novo protein synthesis is required for GBPi induction in response to TPA rather than signaling through modification (e.g., phosphorylation) of pre-existing signaling proteins. Cloning of the GRS into CAT reporter constructs conferred responsiveness to TPA indicating that GBPi was able to regulate transcription by binding to GRS (Raj and Khalili, 1994). Since the sequence within the GRS contains potential binding sites for the GC-box-binding proteins Sp1 and Egr-1, we have now extended our analysis by determining the binding of these proteins to the GRS region of JCV.
Sp1, or Specificity Protein-1, is a transcription factor that is involved in the regulation of the expression of a large number of cellular genes and its activity can be regulated by a variety of protein kinases (Chu and Ferro, 2005). Sp1 is the founding member of a family of zinc-finger-containing transcription factors. The transcription of JCV has been reported to be regulated by Sp1 in glial cells (Henson et al., 1992; Henson, 1994). However these studies utilized the MH1 strain of JCV and Sp1 exerted its effects through a different Sp1 site that is contained within a strain-specific region of DNA that is not present in the original isolate of JC virus (Mad-1 strain) that we use in our experiments. The sequence of the PML-derived Mad-1 strain GRS is predictive of an Sp1 binding site but the binding of Sp1 to this potential Sp1 site in the GRS has not been previously investigated (Raj and Khalili, 1995).
The early growth response-1 protein (Egr-1) is a zinc finger transcription factor that was discovered independently by several laboratories. The Egr-1 gene responds rapidly to stimulation by a variety of environmental stimuli including cytokines, hormones and neurotransmitters by exhibiting a rapid and dramatic induction of expression (reviewed in Thiel and Cibelli, 2002). Egr-1 functions as a convergence point for many signaling cascades and couples short term extracellular signals to longer term changes in the expression of Egr-1 target genes. These genes play important roles in the regulation of cell proliferation and apoptosis (Thiel and Cibelli, 2002). In this study, we show that Egr-1 binds to the GRS region at the JCV origin of replication and is a component of the TPA-inducible GBPi protein complex that was previously demonstrated to be an important regulator of viral transcription in response to cytokine signaling. Further, we now report that Egr-1 is induced by JCV infection of primary astrocytes in culture and in JCV-infected cells in PML clinical samples and that Egr-1 stimulates transcription of the JCV late promoter.
RESULTS
Sp1 does not bind to the GRS element
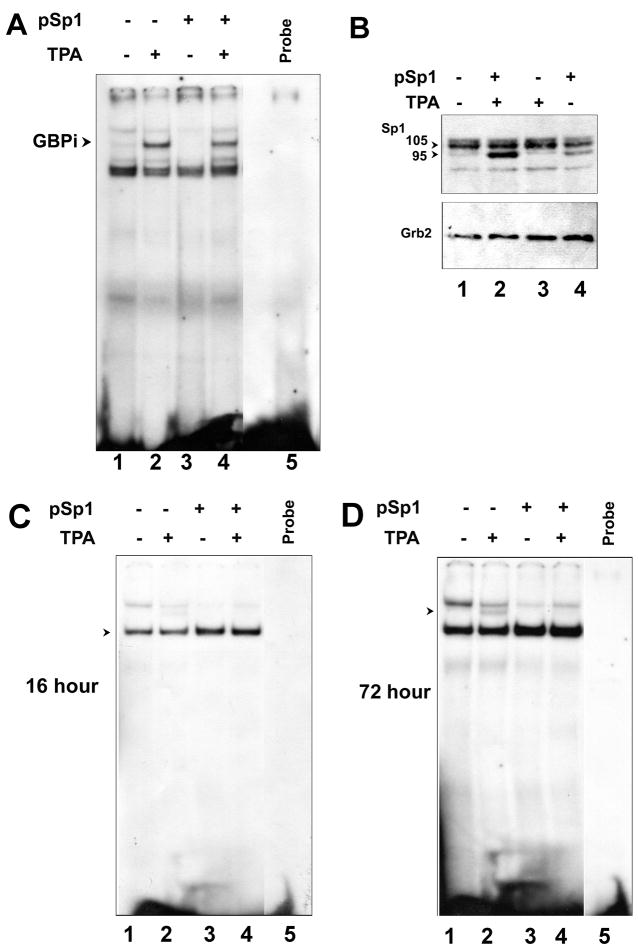
Since the GRS contains a potential binding site for Sp1, we analyzed Sp1 association with GRS by overexpressing Sp1 in cells and performing gel shift analyses. U-87 MG human glioblastoma cells were transfected with pCMV-Sp1 plasmid or with vector control for 24 hours and then treated for 90 min with TPA or vehicle control (DMSO). Nuclear extracts were isolated and incubated with a radiolabeled GRS double-stranded oligodeoxyribonucleotide. TPA strongly induced a band running above the constitutive band seen in all lanes (Fig. 1A compare lane 1 to 2 and 3 to 4). This inducible band is indicated by an arrowhead and is referred to as GBPi as we have previously reported (Raj and Khalili, 1994). In contrast, no change in the gel shift profile was observed in cells that overexpressed Sp1 (Fig. 1A, compare lanes 1 and 2 to lanes 3 and 4). The overexpression of Sp1 p95 protein was confirmed by Western blot (Fig. 1B). Note that the anti-Sp1 antibody recognizes both the p106 and p95 isoforms of Sp1. p106 is the predominant endogenous isoform while the plasmid pCMV-Sp1 expresses only the p95 isoform. Expression of Sp1 p95 in Sp1-transfected cells was higher in TPA-treated cells (lane 2) than in untreated cells (lane 4), presumably because TPA stimulates the CMV promoter of the pCMV-SP1 plasmid. As a further control, we performed gel shift with an oligodeoxyribonucleotide containing an authentic Sp1 site from the mouse cyclin-dependent kinase 5 regulatory subunit p35 promoter which has been shown to be a functional Sp1 control element in vivo (Ross et al., 2002). This oligodeoxyribonucleotide gave a gel shift indicated by the arrowhead in Fig. 1C that was increased in the Sp1-transfected cells (compare lanes 3 and 4 to lanes 1 and 2). Interestingly, at long autoradiography exposure times (72 hours), another very faint band could be seen in the TPA-treated cells (Fig. 1D, lane 2) suggesting that GBPi binds to the SP1 oligodeoxyribonucleotide with a very low affinity.
Figure 1. Sp1 fails to bind to GRS.
U-87 MG cells were transfected with plasmid expressing Sp1 and/or treated with TPA as described in Materials and Methods. A. Nuclear proteins were extracted and used in a gel shift assay. B. Overexpression of Sp1 was confirmed by Western blot. Grb2 was used as a loading control. C and D. Binding of Sp1 and GBPi to an authentic Sp1 site from the mouse cyclin-dependent kinase 5 regulatory subunit p35 promoter was evaluated with autoradiography exposure times of 16 and 72 hours respectively.
Egr-1 is induced by TPA and binds to the GRS element
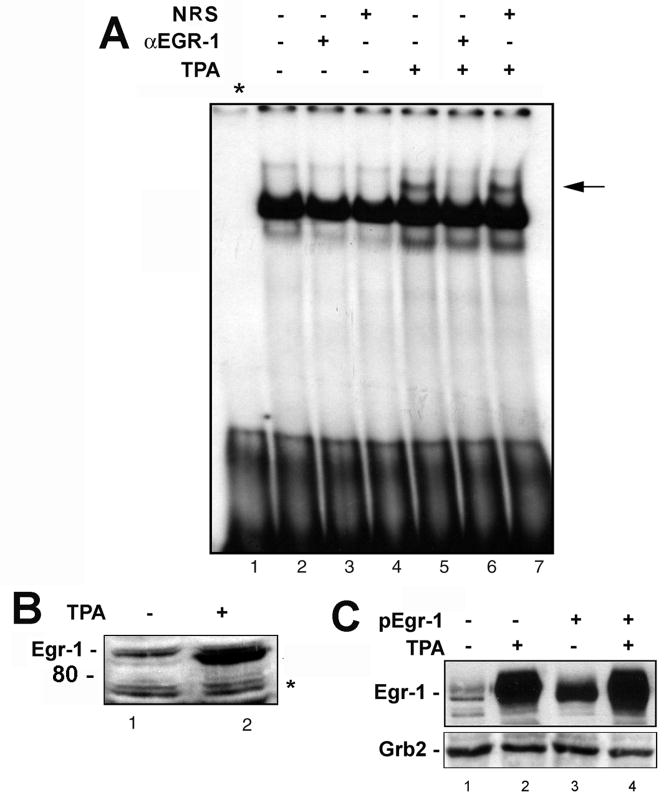
Analysis of the GRS with the new enhanced version of the program MatInspector, which identifies transcription factor binding site in nucleotide sequences using a large library of weight matrices (Cartharius et al., 2005), revealed that it contained a potential binding site for Egr-1. For this reason, we next analyzed the association of Egr-1 with GRS by performing gel shift analyses in the presence or absence of antibody to Egr-1 or normal mouse serum control. U-87 MG cells were treated for 90 min with or without TPA, nuclear extracts were isolated and incubated with GRS probe in gel shift incubations. The GRS-binding activity (GBPi) was induced in the cells by TPA (Fig. 2A, lane 5). The GBPi band was abrogated by antibody to Egr-1 (lane 6) but not non-immune serum (lane 7). This provides strong evidence that the GBPi band shift involves Egr-1. Fig. 2B shows that TPA increased expression of Egr-1 measured by Western blot in the same extracts as were used in Fig 2A. The asterisk indicates a nonspecific band showing equivalent protein loading. In another experiment (Fig. 2C), U-87 MG cells were transfected with pCMV-Egr-1 plasmid or vector control for 24 h followed by treatment for 90 min with or without TPA. Transfection of cells with pCMV-Egr-1 increased the level of Egr-1 (lane 3) relative to control (lane 1) while TPA caused a very large increase (lane 2). The largest increase was seen in cells that were both transfected with Egr-1 and stimulated with TPA (lane 4). Taken together, these data provide evidence that TPA induces expression of Egr-1 which binds to the GRS element and is responsible for the activity that was originally designated GBPi. Next we sought to directly prove the presence of Egr-1 in this protein complex by isolating and analyzing the GBPi:GRS protein DNA complex.
Figure 2. Egr-1 binds to the GRS element.
A. U-87 MG cells were treated with TPA as described in Materials and Methods and gel shifts were performed with nuclear extracts in the presence and absence of antibody to Egr-1 (α-Egr-1) or non-immune rabbit serum as indicated. Lane 1 - free probe. The arrow indicates the GBPi gel shift band. B. The nuclear extracts from Panel A were analyzed by Western blot for Egr-1 expression. The asterisk indicates a nonspecific band that indicates equivalent protein loading. C. In a separate experiment, U-87 MG cells were transfected with plasmid expressing Egr-1 and/or treated with TPA as described in Materials and Methods. Expression of Egr-1 was evaluated by Western blot with Grb2 as loading control.
The GBPi:GRS protein-DNA complex contains Egr-1
The observation that TPA induces a gel shift that is inhibited by antibody to Egr-1 strongly suggests that Egr-1 is responsible for this gel shift. Next, we directly investigated the association of Egr-1 with this gel shift band. The GRS-GBPi complex was isolated by treating U-87 MG cells with TPA, extracting nuclear proteins, incubating with double-stranded GRS DNA and running the resulting DNA:protein complexes on a non-denaturing 5% polyacrylamide gel as described in Materials and Methods. In essence, this is an EMSA gel except that it contains increased amounts of protein and unlabeled DNA. The DNA contained some end-labeled probe allowing the position of the DNA:protein complexes to be determined by autoradiography (Fig. 3A). The position of the GBPi band is labeled “i” and was excised from the gel. The corresponding “i” area from lane from control cells untreated with TPA was also excised. As a further control, we also isolated the band from each lane labeled “b” which is found constitutively in all lanes and does not respond to TPA. The excised bands were stored at −80°C in order to allow the 32P to decay and then electrophoresed on denaturing 7% polyacrylamide gel and analyzed for the presence of Egr-1 by immunoblotting (Fig. 3B). Only band “i” from the TPA-treated cells (lane 5) was positive for Egr-1, i.e., Egr-1 in this band was induced by TPA (compare lanes 3 and 5). We conclude that the GBPi:GRS complex contains Egr-1 but that the protein:GRS complex designated “b”, which is not inducible by TPA, does not contain Egr-1 (lanes 2 and 4).
Figure 3. Detection of Egr-1 in the GBPi:GRS protein-DNA complex.
A. Nuclear extracts from U-87 MG cells treated with or without TPA were incubated with cold double-stranded GRS oligodeoxyribonucleotide probe that had been spiked with [32P]-labeled GRS probe, electrophoresed on a non-denaturing PAGE and subject to autoradiography. * - Free probe. After autoradiography, the developed X-ray film was used for alignment to allow gel slices to be excised from the gel for each lane at the positions labeled “b” and “i”. B. Gel slices corresponding to bands at positions b and i in the −TPA and +TPA lanes (Panel A) were stored at −80°C for several months to allow the 32P to decay. To determine if the DNA:protein complexes contained in these bands contained Egr-1, the gel slices were boiled in Laemmli sample buffer containing SDS and subject to SDS PAGE followed by Western blotting with antibody to Egr-1.
No Egr-1 sites are found within the 98 bp repeat control region or in the NF-κB binding region
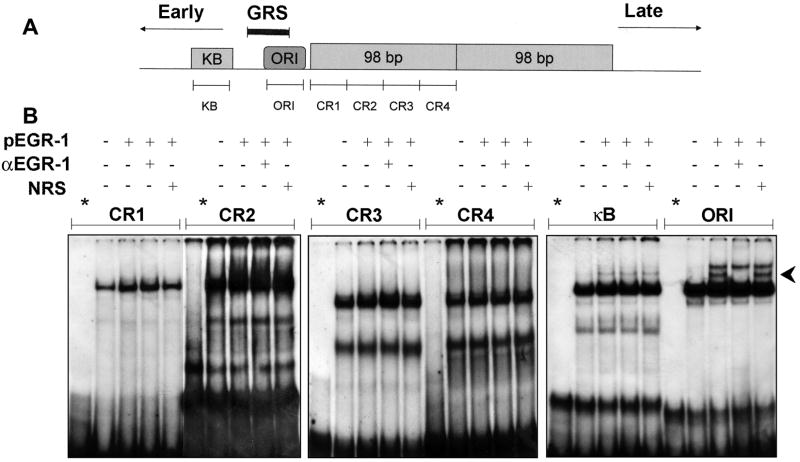
The NCCR of the Mad-1 strain of JCV contains a 98 bp repeat region on the late side of the origin of replication and a region containing an NF-κB site that is located on the early side of the origin. These regions contain the binding sites for the transcription factors that regulate JCV early and late gene expression (Raj and Khalili, 1995). To investigate Egr-1 binding to the various areas of the JCV NCCR, we synthesized a number of spanning double-stranded oligodeoxyribonucleotide probes (Fig. 4A). U-87 MG cells were transfected with plasmid expressing Egr-1, nuclear extracts prepared and incubated in gel shift reactions with each of these six probes in the presence and absence of antibody to Egr-1 (Fig. 4B). With the probes CR1, CR2, CR3, CR4 and κB, there was no change in the pattern DNA:protein bands with Egr-1 or α-Egr-1 antibody. However, a band appears for the cells expressing Egr-1 (Fig. 4B, indicated by the arrowhead). This band is abrogated by α-Egr-1 antibody but not by normal rabbit serum (NRS). We can conclude that ORI contains a binding site for Egr-1. This is consistent with the earlier data since the ORI probe overlaps at its 5′ end with the GRS probe and this region of overlap contains the GG(A/C) repeat motif.
Figure 4. Gel shift analysis for Egr-1 sites found within the JCV NCCR 98 bp repeat and the NF-κB binding region.
A. Schematic depiction of NCCR of Mad-1 strain of JCV showing positions of gel shift probes. B. U-87 MG cells were transfected with or without plasmid expressing Egr-1 (pEGR-1), nuclear extracts harvested and gel shift assays performed with JCV probes and antibody to Egr-1 as indicated.
Egr-1 binds to the JCV NCCR in vivo
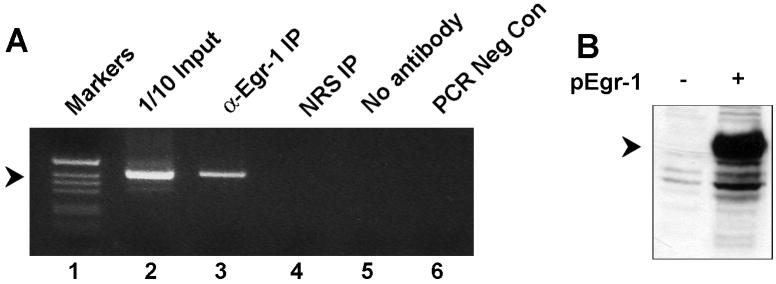
To assess binding of Egr-1 to the NCCR in vivo, we employed a Chromatin ImmunoPrecipitation (ChIP) assay. U-87 MG cells were transfected with plasmid expressing Egr-1, cross-linked and immunoprecipitation was performed with α-Egr-1. DNA associated with IPs was amplified using primers spanning the JCV NCCR. A band is seen for α-Egr-1 (lane 3) but not for NRS (lanes 4) indicating that Egr-1 can bind to the JCV NCCR in vivo (Fig. 5A). Expression of Egr-1 was confirmed by Western blot (Fig. 5B).
Figure 5. Egr-1 binds to the JCV NCCR in vivo.
U-87 MG cells were transfected with plasmid containing the JCV NCCR and plasmid expressing Egr-1. After crosslinking, ChIP assays were performed as described in Materials and Methods. 1/10 of the input extract was used as a positive control.
Egr-1 stimulates transcription of the JCV late promoter
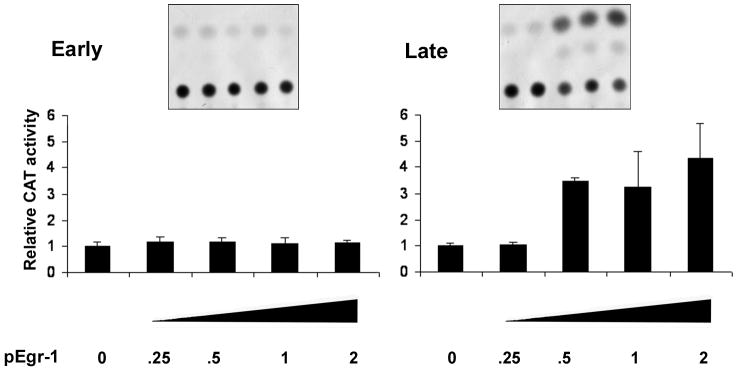
Previous studies indicated that the GRS element functions in the regulation of JCV transcription (Raj and Khalili, 1994). Since we have now found that GRS binds Egr-1, we next analyzed if Egr-1 could function to modulate JCV transcription. To investigate the effect of Egr-1 on the transcription of the JCV early and late promoters, U-87 MG cells were transfected either with JCVE-CAT or JCVL-CAT alone or in combination with different amounts of pCMV-Egr-1. Egr-1 had no effect on transcription from the early promoter (Fig. 6A). However, transcription from the late promoter was stimulated by Egr-1 (Fig. 6B).
Figure 6. Egr-1 stimulates the JCV late promoter.
U-87 MG cells were transfected with either JCVE-CAT (left panel) or JCVL-CAT (right panel) together with various amounts (μg) of plasmid expressing Egr-1. CAT activities were measured and normalized relative to cells expressing reporter plasmid alone. Triplicate analyses were performed and error bars represent the standard deviation.
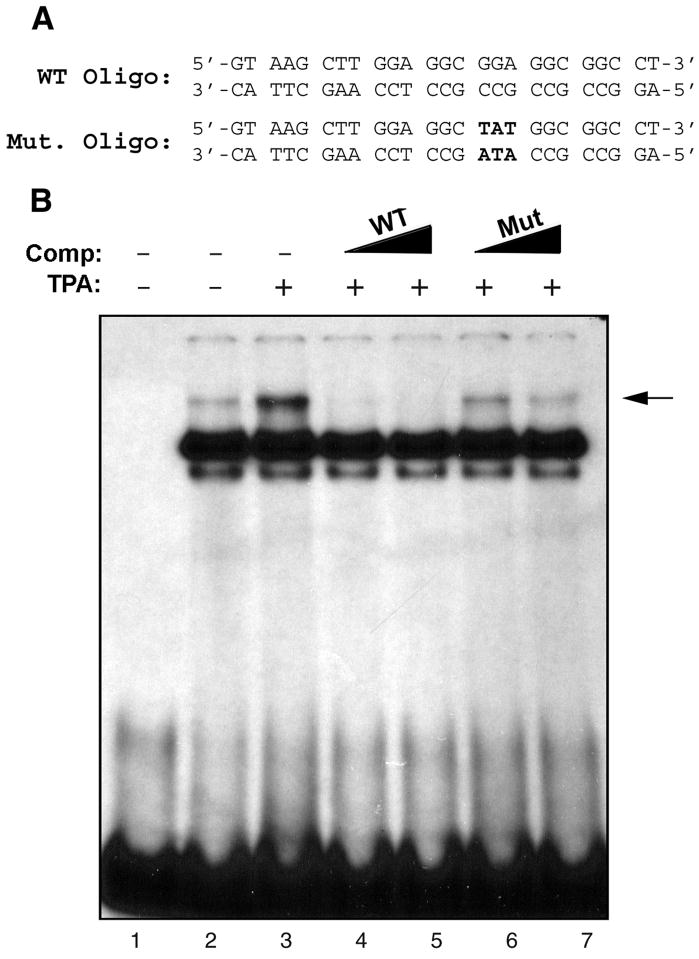
Mutation of the Egr-1 site abrogates Egr-1 binding
To establish the importance of the Egr-1 site, we engineered a trinucleotide mutation that removed the consensus Egr-1 binding sequence (Fig. 7A). Double-stranded oligonucleotide containing this mutation was included in gel shift experiments with radiolabeled wild-type GRS probe (Fig. 7B). For these gel shifts, nuclear extracts were prepared from U-87 MG cells that were untreated or treated with TPA. The Egr-1 gel shift band was induced by TPA treatment (compare lanes 2 and 3). Addition of increasing amounts of unlabeled wild-type GRS oligonucleotide competed away the Egr-1 band (lanes 4 and 5). However the intensity of the band was reduced to a much lesser extent by unlabeled mutant GRS oligonucleotide (lanes 6 and 7) indicating that the binding of Egr-1 to GRS is impaired in the mutant.
Figure 7. Mutation of the Egr-1 binding site abrogates Egr-1 binding.
A. GRS wild-type (WT) and mutant (Mut) oligonucleotide sequences spanning nucleotides 5112 to 4 of JCV Mad-1 regulatory region. Nucleotides in bold font indicate the base substitution within the Mut oligonucleotide relative to the WT oligonucleotide. B. WT and Mut oligonucleotides were end-labeled with [γ]-32ATP with T4 polynucleotide kinase and purified. Nuclear extracts (10 μg/lane) prepared from U-87MG cells either untreated (lane 2) or treated (lane 3) or with TPA (75 ng/ml) for 1.5 h were incubated labeled wild-type GRS probe (50,000 cpm/lane) in a binding buffer. In addition, probe plus nuclear extract mixture was incubated either with unlabeled WT (lanes 4 and 5, in 50- and 150-fold molar excess respectively) or Mut (lanes 6 and 7, in 50- and 150-fold molar excess respectively) oligonucleotide, which serve as competitors. DNA-protein complexes were then resolved on a 6% polyacrylamide gel under native conditions and visualized by autoradiography. An arrow indicates the specific DNA-protein complexes. Oligo: Oligonucleotide, Comp: Competitor.
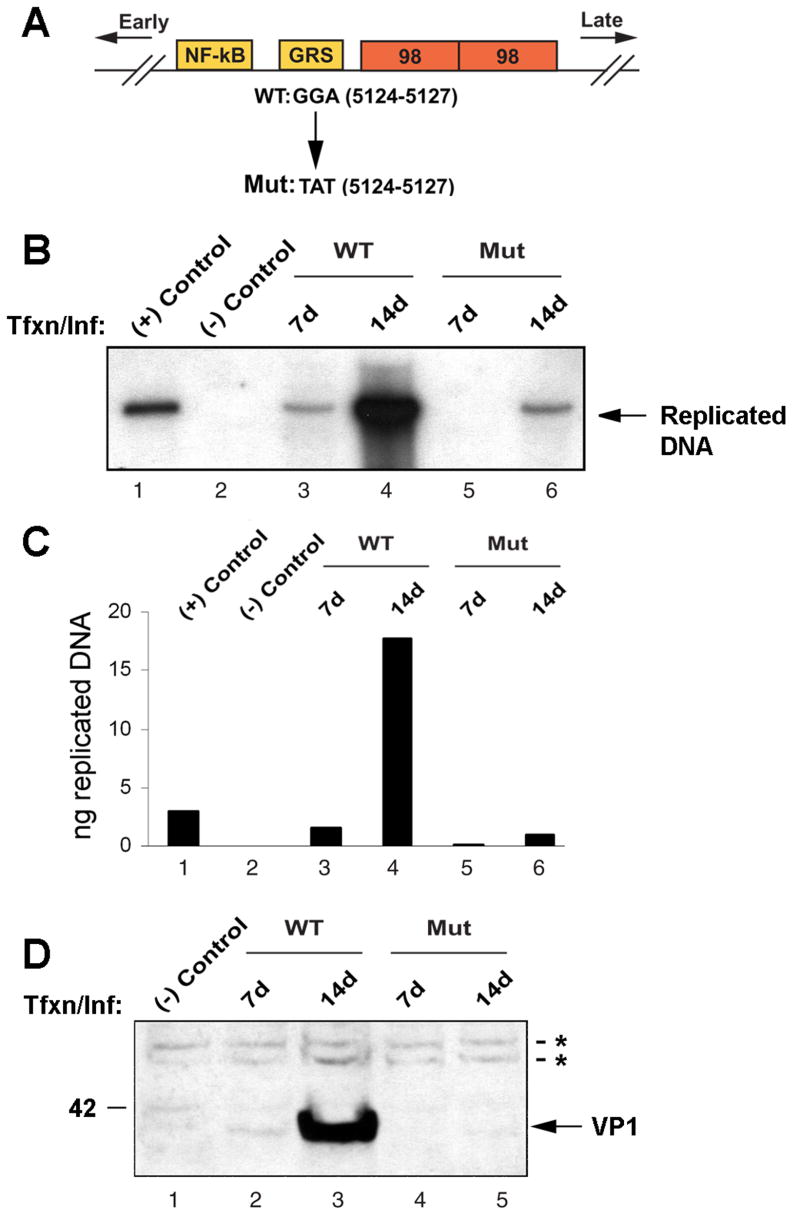
JCV with the mutated Egr-1 site showed markedly reduced late gene expression and DNA replication during the course of viral infection
To investigate the functional importance of the Egr-1 binding site for the life cycle of JCV, we introduced the Egr-1 binding site mutation described above into the viral backbone. The structure of the viral NCCR and the position of the Egr-1 binding site mutation is shown diagrammatically in Fig. 8A. Mutant virus and wild-type control were introduced into SVGA cells by transfection/infection and monitored for DNA replication by DpnI assay and for late gene expression by Western blot for the late capsid protein VP1. As shown in Fig. 8B, wild-type virus replicated efficiently with replicated DNA detectable as a 5.2 Kb DpnI-resistant band on day 7 (lane 3). By day 14, a very intense band of replicated DNA was present for wild-type virus (lane 4). In contrast, no replicated DNA could be detected for the mutant virus after 7 days (lane 5) and a band, which was much reduced compared to the wild-type, was observed after 14 days (lane 6). As a positive control, 3 ng of BamH1-digested Mad-1JCV genome was loaded (lane 1) while uninfected cells provided the negative control. To quantify these data, the intensities of the bands in Panel B were measured with a phosphorimager and compared to the positive control (lane 1, 3 ng) to calculate the amount of replicated DNA in each sample (Fig. 8C). The amount of replicated viral DNA at day 14 was reduced 17-fold for the mutant relative to the wild-type (compare lanes 4 and 6).
Figure 8. Virus with mutation of the Egr-1 binding site is defective in DNA replication and late gene expression.
A. A schematic representation of the JCV Mad-1 regulatory region indicating position of the trinucleotide mutation in the mutant virus. Orientation of early and late gene expression is indicated. The relative location of NF-κB, GRS and 98 bp tandem repeats are also indicated. GGA sequences in WT virus (JCV Mad-1, 5124–5127) were substituted with TAT sequences in the mutant virus. B. Analysis of DNA replication for the mutant virus. SVG-A cells were transfected/infected with either Mad-1 WT genome or its Egr-1 binding mutant genome (Mut, 8 μg/2 × 106 cells/75cm2 flask) using lipofectin as described in Materials and Methods. At 7d and 14d after transfection, low molecular weight DNA was isolated by a Qiagen spin columns, digested with Bam HI and Dpn I restriction enzymes, resolved on a 0.8% agarose gel and analyzed by Southern blotting. In lane 1, Mad-1 wild-type genome (3 ng) digested with Bam HI was loaded as a positive control. In lane 2, DNA isolated from uninfected cells was loaded as a negative control. C. A quantitative analysis of the data from panel B. The nitrocellulose filter from Panel B was analyzed using a Bio-Rad Molecular Imager FX phosphorimager and the intensity of each band measured using the Bio-Rad Quantity One Quantitation Analysis Software. The intensity of each band relative to the positive control (3 ng) was used to calculate the equivalent amount of replicated DNA in each sample. D. In parallel to the studies described for Panel B, whole cell lysates were also prepared at the time points indicated and analyzed by Western blot using an anti-VP1 antibody (AB597, kindly provided by W. Atwood, Brown University, Rhode Island). In lane 1, whole cell extract from untransfected/uninfected cells was loaded as a negative control. Tfxn: transfection, Inf: infection. The migration pattern of a molecular weight marker is shown on the left side of the panel in kilodalton. The asterisks indicate nonspecific bands present in uninfected and infected cells that serve to show equivalent protein loading.
In parallel to the DpnI experiment, the U-87 MG cell cultures were also harvested for total cell protein extracts. These were analyzed by Western blot for expression of the late capsid protein VP1. As shown in Fig. 8D, expression of VP1 was detectable by day 7 in the wild-type virus-infected cells (lane 2) and was robust by day 14 (lane 3). In contrast, in the mutant virus-infected cells was undetectable at day 7 (lane 4) and barely detectable by day 14 (lane 5) indicating that late gene expression is markedly reduced by the mutation of the Egr-1 binding site.
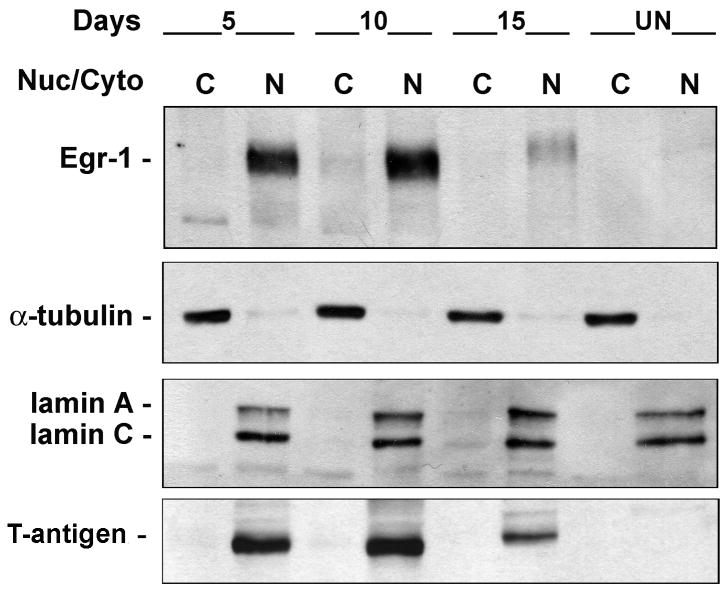
The level of Egr-1 is upregulated in the nucleus of JCV-infected astrocytes
To examine Egr-1 expression during the course of wild-type JCV infection, primary human fetal astrocytes were infected with Mad-1 JCV (MOI = 1) and harvested for cytoplasmic and nuclear fractions at 5, 10 and 15 days post-infection together with uninfected controls (Fig. 9). Egr-1 was confined to the nucleus at all times. Egr-1 was barely detectable in uninfected cells but was greatly induced at 5 and 10 days post-infection and to a lesser extent at 15 days. Lamin A/C and α-tubulin were used as markers for the purity of the nuclear and cytoplasmic fractions respectively. T-antigen was used as a positive control for infection.
Figure 9. Expression of Egr-1 during JCV infection of primary astrocytes.
Primary human astrocytes were infected with JCV and cytoplasmic and nuclear fractions isolated from cells 5, 10 and 15 days post-infection and from uninfected controls. Western blot was performed for Egr-1, α-tubulin (cytoplasmic marker), lamin A/C (nuclear marker) and T-antigen (control for infection).
Egr-1 is upregulated in PML clinical samples examined by immunohistochemistry
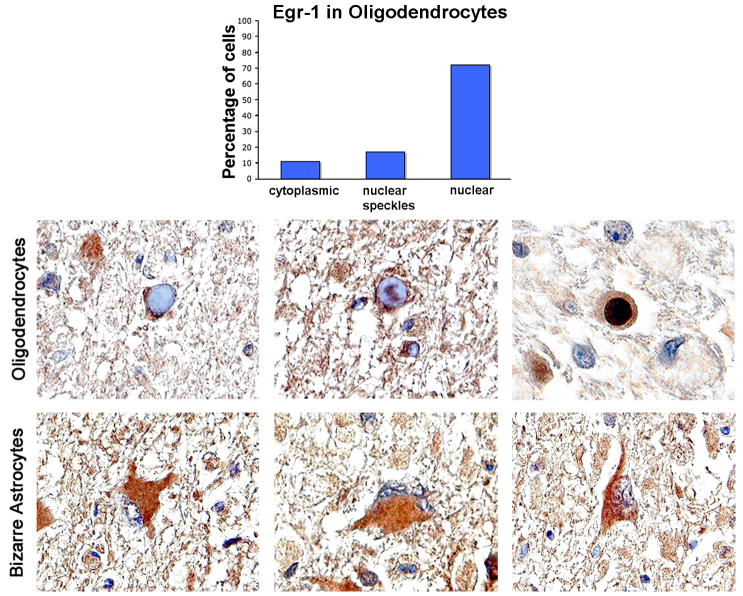
To evaluate the importance of Egr-1 upregulation in the pathophysiology of PML, we examined clinical samples of PML lesions by immunohistochemistry using antibody to Egr-1. As shown in Fig. 10, robust expression of Egr-1 was observed mainly in the nuclei of oligodendrocytes bearing inclusion bodies where JCV is actively replicating. When 100 inclusion body-bearing oligodendrocytes were scored, all were positive for Egr-1. Of these 72% showed robust nuclear labeling, 17% speckled nuclear labeling and 11% showed cytoplasmic labeling. No immunolabeling for Egr-1 was observed in normal brain tissue surrounding the PML lesion (data not shown). The bizarre astrocytes that are characteristic of PML also showed robust expression of Egr-1 (Fig. 10, bottom panel). Interestingly, Egr-1 was expressed predominantly in the cytoplasm of these cells. This is in contrast to the subcellular distribution in JCV-infected astrocytes in culture where Egr-1 is localized to the nucleus (Fig. 9). In this regard, it is important to note that primary cultures of human fetal astrocytes (Fig. 9) actively support the replication of JCV, while adult astrocytes in vivo (Fig. 10) can become infected with JCV within PML lesions, assume a bizarre morphology and express large T-antigen but do not replicate the virus (Del Valle and Piña-Oviedo, 2006; Khalili et al, 2007). Rather, the sites of viral replication in PML are the oligodendrocytes, which show expression of Egr-1 mainly in the nucleus (Fig. 10).
Figure 10. Detection of Egr-1 in JCV-infected cells of PML cases.
Immunohistochemistry was performed for Egr-1 as described in Materials and Methods. Three representative fields each are shown for oligodendrocytes and astrocytes. Egr-1 shows robust labeling of the inclusion body-harboring oligodendrocytes, within plaques of demyelination in PML cases. Three patterns of labeling were observed; cytoplasmic (left-hand oligodendrocyte panel), nuclear speckled (middle oligodendrocyte panel) and nuclear (right-hand oligodendrocyte panel). One hundred cells were counted and scored for their labeling pattern. 11% showed cytoplasmic labeling, 17% nuclear speckled labeling and 72% nuclear labeling. These data are depicted as a histogram. Bizarre astrocytes, also within demyelinated lesions demonstrated 100% cytoplasmic immunoreactivity (lower panels). All panels are original magnification (x1000).
DISCUSSION
Reciprocal interactions between the cells of the immune system and cells of the CNS are of paramount importance in viral infection and disease in the brain. After JCV infection during childhood, the virus remains in an ill-defined latent state with no apparent clinical symptoms. However, under immunocompromised conditions, the virus can reactivate and enter the lytic cycle resulting in the cytolytic destruction of glial cells and the clinical manifestation of PML (Berger and Houff, 2006; Del Valle and Piña-Oviedo, 2006). It is possible that this reactivation involves transcription factors that bind and regulate the JCV NCCR and that these transcription factors respond to intracellular signaling pathways downstream of cytokines and immunomodulators found in the CNS. For example, replication of JCV DNA in the presence of PMA-stimulated T-cell supernatant is substantially decreased in transfected glial cells (Chang et al., 1996). Further, mitogen-responsive transcription factors such as NF-κB, c-fos and c-jun have been found to bind and regulate the JCV NCCR (Ranganathan and Khalili, 1993; Sadowska et al., 2003). In this regard, we previously reported an unknown JCV NCCR binding-protein that we designated GBPi which was rapidly induced by certain cytokines and by TPA (Raj and Khalili, 1994). This induction was rapid, reaching a maximum after 1.5–3 hrs, and required de novo protein synthesis since it could be blocked by cycloheximide or actinomycin D. GBPi binds to a region at the origin of DNA replication which contains a GG(A/C)-rich sequence (GRS) that is 100% conserved between JCV and another human polyomavirus, BK virus (BKV). The sequence of this region indicated that it contains potential binding sites for the zinc finger transcription factors Sp1 and Egr-1.
Our data indicate that Sp1 does not bind to the GRS. Other studies have reported that the transcription of JCV is regulated by Sp1 in glial cells (Henson et al., 1992; Henson, 1994). However, the strain of JCV used in these studies differed from the Mad-1 strain that we used in our experiments. The Mad-1 strain differs from the prototypical JCV sequence in the region that lies to the late side of GRS that has been designated the A-F box region (Frisque et al., 1984). In the Mad-1 strain of JCV, there is a 23 and a 66 base pair deletion followed by a duplication of the entire region to generate a 98 base pair repeat region. These deletions in the Mad-1 strain remove the only two Sp1 sites found in the A-F box region. The GRS lies outside of the 98 base pair repeat and had not been previously analyzed for Sp1 binding. Our data indicate that GRS does not bind Sp1 and thus does not constitute a functional Sp1 regulatory site in the Mad-1 JCV NCCR. With respect to Sp1 regulation of JCV, it is likely that strains of JCV in which the 23 and a 66 base pair deletions have not occurred (unlike Mad-1), are regulated by Sp1 (Henson et al., 1992; Henson, 1994). Recently, it has been postulated, from sequence data of the A-F box region from rearranged JCV NCCRs from isolated by PCR from PML patients, that the Sp1 sites from the A-F box region may be important in the development of PML in non-AIDS patients since non-AIDS PML isolates usually contain both Sp1 sites intact whereas in AIDS PML, they are often deleted (Mischitelli et al., 2005).
Our data also indicate that, unlike Sp1, Egr-1 is able to bind to GRS and that Egr-1 is responsible for the protein activity that we had previously reported to be induced in cells treated with certain cytokines or TPA and which was designated GBPi (Raj and Khalili, 1994). The properties of GBPi are consistent with it being Egr-1, i.e., it is rapidly induced by certain cytokines and TPA with rapid kinetics, reaching a maximum after 1.5–3 hrs, and induction is directed at the transcriptional level requiring de novo RNA and protein synthesis. We now have provided direct evidence to identify this GBPi activity as being due to Egr-1, i.e., antibody to Egr-1 abrogates the GRS/GBPi gel shift produced by TPA treatment of cells and GRS/GBPi complex isolated from TPA-treated cells was demonstrated to contain Egr-1 by Western blot.
The binding of Egr-1 to the JCV promoter functions to enhance viral gene expression. Thus, co-expression of Egr-1 stimulates the transcription of a reporter construct containing the JCV late promoter. These data are consistent with earlier experiments where it was shown that GRS confers TPA-inducibility on heterologous reporter constructs (Raj and Khalili, 1994). Importantly, we found that mutation of this site, which abrogated the binding of Egr-1 resulted in a large reduction of late gene expression and in viral DNA replication.
Interestingly, the expression of Egr-1 is upregulated early in the course of JCV infection of primary human astrocytes (days 5 and 10 postinfection). This induction may be a result of expression of the transforming activities of the viral early proteins, large-T and small-t antigens. For example, SV40 small-t antigen binds and inhibits protein phosphatase 2A which is a potent negative regulator of growth promoting signal transduction pathways including the centrally important mitogen-activated protein kinase (MAPK) pathway (Sontag et al., 1993). Since activation of MAPK stimulates Egr-1 gene expression (Hipskind et al., 1994), it is possible that the Egr-1 induction that occurs upon JCV infection is mediated by activation of the MAPK pathway by JCV small-t. Since Egr-1 accumulates during the early phase of JCV infection (Fig. 9), Egr-1 stimulates JCV late transcription (Fig. 6), and ablation of the Egr-1 site strongly inhibits late transcription, it is very possible that Egr-1 may be important in the transition from the early to the late phase of JCV infection.
Induction of Egr-1 expression has also been reported to occur upon infection of cells by other viruses including HTLV-I (Fujii et al., 1991;Sakamoto et al., 1992), HTLV-II (Sakamoto et al., 1992), Japanese encephalitis virus (Saha and Rangarajan, 2003), rabies virus (Fu et al., 1993; Saha and Rangarajan, 2003), borna disease virus (Fu et al., 1993) and human foamy virus (Wagner et al., 2000). Infection of CD4(+) T-cell lines with HIV-1 increases cell activation, signaling and expression of c-Jun and Egr-1 (van ‘t Wout et al., 2003). Mouse hepatitis virus infection results in Egr-1 induction, while it was found that knockdown of Egr-1 by an siRNA inhibited viral propagation suggesting the biological relevance of Egr-1 induction to virus replication (Cai et al., 2006). Studies with Egr-1 siRNA also suggested a functional role for Egr-1 in Epstein-Barr virus reactivation and indicated a positive feedback loop involving Egr-1 and the viral Zta transactivator protein (Chang et al., 2006). The orthopoxviruses vaccinia virus (VV) and cowpox virus (CPV) activate the MAPK pathway and stimulate Egr-1 expression. Knockdown of Egr-1 with siRNA decreased VV production by one log and caused a small virus plaque phenotype showing the importance of Egr-1 for VV biology while Egr-1 siRNA had no effect on CPV multiplication or plaque size (Silva et al., 2006).
It is possible that Egr-1 induction in response to changes in the extracellular cytokine milieu is involved in JCV reactivation through binding of Egr-1 to GRS in co-ordination with other cytokine-modulated transcription factors that bind to the JCV NCCR such as AP-1 and NF-κB. Once JCV enters the early phase of activation, high levels of Egr-1 are produced that may co-operate with T-antigen and act at the GRS to modulate late transcription. Additionally production of Egr-1 may function indirectly by stimulating the transcription of cellular genes that are involved in the process of cellular transformation or interact in other ways with the viral life cycle.
It should be noted that GRS likely has other important functions besides Egr-1 binding. This region is involved in the activation of JCV late transcription by HIV-1 Tat which is thought to be important in the development of AIDS PML (Chowdhury et al., 1993) and is part of a functional element designated “upTAR” (upstream transactivation region) to which Tat binds in conjunction with the cellular protein Purα (Krachmarov et al., 1997). The upTAR element (GGAGGCGGAGGC) is contained within the JCV GRS. Gel shift experiments with oligodeoxynucleotides containing this region demonstrated that at least five different DNA:protein complexes can form in this region (Chowdhury et al., 1993; Krachmarov et al., 1997). In addition, the GRS is near the origin of viral DNA replication and may be involved in the binding of JCV T-antigen and cellular proteins involved in DNA replication such as DNA polymerase-α and replication protein-A. The multifunctional importance of GRS is indicated by the fact that it is 100% conserved between JCV and BKV, and that the region is almost never involved in the multiple DNA rearrangement events that can occur in other parts of the JCV NCCR during PML pathogenesis (Pietropaolo et al., 2003). Thus Egr-1 represents a new participant that is involved in the interplay of many proteins that regulates the highly conserved GRS core element that controls JCV transcription and replication.
Finally, the pathological importance of Egr-1 induction by JCV is indicated by the strong expression of Egr-1 in PML clinical samples where it is found in the nuclei of most of the oligodendrocytes that are harboring inclusion bodies, which represent the site of JCV infection and replication. Interestingly, cultured primary fetal astrocytes, which are infected by JCV and replicate the virus, also show nuclear Egr-1 expression (Fig. 9) while adult astrocytes, which are infected by JCV in PML samples (bizarre astrocytes) do not productively replicate the virus and show cytoplasmic localization of Egr-1 (Fig. 10). Thus sequestration of Egr-1 in the cytoplasm may be associated with the non-permissive nature of adult astrocytes for JCV replication. This is an unusual finding since most reports on Egr-1 show a nuclear subcellular localization (reviewed in Thiel and Cibelli, 2002). However, it has been reported that Egr-1 can localize to the cytoplasm of prostate cells and that this is mediated through binding of Egr-1 to the microtubules (Mora et al., 2004). In conclusion, the induction of Egr-1 that we have reported may be important for the life cycle of JCV and the pathogenesis of PML.
MATERIALS AND METHODS
Cell culture, transfection and plasmids
U-87 MG human glioblastoma cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and were transfected using the calcium phosphate precipitation method (Graham and van der Eb, 1973) except for the ChIP experiments where Fugene6 was used according to the manufacturer’s instruction (Roche). Primary cultures of human fetal astrocytes were prepared and grown as we have previously reported (Radhakrishnan et al., 2003). Reporter constructs, JCVE-CAT and JCVL-CAT contained the JCV promoter from the Mad-1 strain linked to the chloramphenicol acetyltransferase (CAT) gene in the early and late orientations respectively (Chen and Khalili, 1995). pCMV-Sp1 expresses Sp1 from the CMV promoter (Sawaya et al., 1998) and pCMV-Egr-1 expresses Egr-1 from the CMV promoter (Papanikolaou and Sabban, 2000).
Antibodies
Rabbit polyclonal anti-Sp1 (PEP2, sc-59, Santa Cruz Biotechnology, Inc., Santa Cruz) recognizes both the p95 and p106 Sp1 proteins and was used at a dilution of 1:1,000. Rabbit polyclonal Egr-1 antibody was used for Western blot (1:1000) and gel shift experiments (sc-110, and sc-110x respectively, Santa Cruz Biotechnology).
Gel Shift Assays
Cells were transfected with and without pCMV-Sp1 or pCMV-Egr-1 for 48 hours and then treated with 100 ng/ml tetradecanoyl phorbol acetate (TPA) for 90 min except for the experiment in Fig. 7 where 75 ng/ml TPA was used. Nuclear proteins were then extracted and 10 μg were incubated with 50,000 cpm of a [32P]-labeled double-stranded oligodeoxyribonucleotide probe as previously described (Raj and Khalili, 1994). The following JCV-specific double-stranded oligodeoxyribonucleotide probes were used in these gel shift experiments. Nucleotide numbers refer to the Mad-1 reference strain of JCV (GenBank # NC_001699, formerly J02226) which is a 5130 base pair circular DNA genome. Note that the GRS and ORI nucleotides span the numerical origin.
GRS: 5′-gtaagcttggaggcggaggcggcct-3′ (5112–4).
κB: 5′-aaaacaagggaatttccctggcctc-3′ (5052–5078).
ORI: 5′-ggaggcggaggcggcctcggcctcc-3′ (5118 to 2).
CR1: 5′-cctgtatatataaaaaaaagggaag-3′ (11 to 35)
CR2: 5′-ggatagctgccagccaagcatgagc-3′ (36 to 60)
CR3: 5′-tcatacctagggagccaaccagcta-3′ (61 to 85)
CR4: 5′-acagccagtaaacaaagcacaaggc-3′ (86 to 110)
The authentic Sp1 binding site probe sequence corresponds to that of the Mus musculus cyclin-dependent kinase 5 regulatory subunit p35 gene promoter: 5′–gggcgggggcggagccg–3′. (S82819: nts 761 to 777)(Ross et al., 2002). For gel shifts in the presence of antibody, 2μl of 1μg/μl of anti-Egr-1 or non-immune rabbit serum was added to the gel shift reaction. For competition assays with cold oligonucleotides, two unlabeled oligonucleotides were used: wild-type GRS: (as above), mutant GRS: 5′-gtaagcttggaggctatggcggcct-3′
Isolation and analysis of the GBPi:GRS protein-DNA complex
Nuclear proteins were prepared from U-87 MG cells treated with TPA or vehicle control for 90 min. 25 μg of nuclear proteins were incubated in 20 μl reactions with 2 μg of unlabelled double-stranded GRS double-stranded oligodeoxyribonucleotide in binding buffer as used for the gel shift reactions. 100,000 cpm of GRS probe, which had been end-labeled with 32P and gel-purified, was added so that the GRS/protein complexes could be detected by autoradiography. After a 1 hour incubation at 37°C, samples were loaded onto a 5% native polyacrylamide gel and resolved by electrophoresis in 0.5X TBE. The gel was dried and subject to autoradiography. Using the autoradiograph to align their positions, bands were excised from the gel. After boiling in 6X Laemmli sample buffer, the proteins from the bands were separated by SDS PAGE and Western blotted for Egr-1.
Transient transfection assays
U-87 MG cells were transfected with the JCVE-CAT or JCVL-CAT reporter constructs alone (5 μg) or in combination with pCMV-Egr-1 plasmid (0.25, 0.5, 1 or 2 μg). The total amount of DNA transfected into the cells was normalized with relevant empty vector DNA. Chloramphenicol acetyl transferase (CAT) activity of samples was determined by utilizing 100 μg of protein for each sample as previously described (Safak et al., 2001).
ChIP assay
106 U-87 MG cells were transfected with 3 μg of plasmid containing the JCV NCCR (JCVE-CAT) in combination with 3 μg of plasmid expressing Egr-1 (pCMV-Egr-1) using Fugene6 according to the manufacturer’s instructions (Roche). ChIP was performed 48 hours after transfection as we have previously described (Amini et al., 2005) using the ChIP assay kit (Upstate Cell Signaling Solutions). Briefly, cross-linking was performed with formaldehyde and the DNA sheared by sonication. The cells were lysed and immunoprecipitation was performed with antibodies as indicated. The following primers spanning the JCV NCCR were used for PCR: 5′-cctccctattcagcactttgtcc - 3′ (Mad-1, 4989 to 5011), 5′ – ggccagctggtgacaagcc – 3′ (276-258). PCR was 30 cycles (95°C for 30 sec, 60°C for 1 min, 72°C for 1.5 min) followed by 72°C for 7 min.
Site-directed mutagenesis
JCV Mad-1 genome was subcloned into pBluescript(KS) vector at the Bam HI site and a base substitution (GGA to TAT, JCV Mad-1 5124–5127) within the GRS binding site was performed using the “quick change site-directed mutagenesis kit” (Stratagene, La Jolla, CA). The presence of the mutated nucleotides was verified by DNA sequencing.
Transfection/infection
SVG-A cells (2 × 106 cells/75cm2 flask) were transfected with either wild-type Mad-1 (WT) or Mutant Mad-1 (Mut, the Egr-1 binding site mutant), using 8μg in each case and the lipofectin transfection reagent according to manufacturer’s recommendations (Invitrogen, Carlsbad, CA). At five hours after transfection, cells were washed twice with PBS and fed with DMEM supplemented with 5% FBS and antibiotics (penicillin/streptomycin, 100 μg/ml). Cells were then maintained at 37°C in a humidified atmosphere with 7% CO2 until they were processed for extract preparation or DNA isolation.
Replication assay
Replication assays were carried out as previously described (Safak et al, 2001). Briefly, SVG-A cells either with Mad-1 WT or Mad-1 Mut1 (8 μg each) as described above. At the time points indicated after transfection, low molecular weight DNA containing JC viral DNA was isolated by Qiagen spin columns as described by Ziegler et al (2004), digested with BamHI and DpnI restriction enzymes, resolved on 1% agarose gel and analyzed by Southern blotting using probe prepared from whole Mad-1 genome. BamHI linearizes the viral genome (5.2 Kb) while the methylation-sensitive DpnI restriction enzyme digests the input DNA.
JCV infection of astrocytes
Primary cultures of human astrocytes were infected with JCV as described previously (Radhakrishnan et al., 2003) at an moi = 1. At 0, 5, 10 and 15 days after infection, cells were harvested and extracted to give nuclear and cytoplasmic fractions using the NE-PER nuclear and cytoplasmic reagents according to the manufacturers protocol (Pierce Biotechnology, Rockford, IL).
Immunohistochemistry for brain samples
Immunohistochemistry was performed as we have previously described (Enam et al., 2005) using the avidin-biotin-peroxidase system according to the manufacturer’s instructions (Vector Laboratories) with a rabbit polyclonal antibody to Egr-1 (Santa Cruz Biotechnology, sc-110, 1:500 dilution). Our modified protocol includes deparaffination in xylene, re-hydration in alcohol up to water, non-enzymatic antigen retrieval in citrate buffer, pH 6.0 at 95°C for 30 min. After a cooling period, endogenous peroxidase quenching was performed with 0.3% H2O2 in methanol for 20 min. Sections were rinsed with PBS and blocked with normal goat serum and incubated with primary antibody overnight at room temperature in a humidified chamber. After rinsing with PBS, biotinylated secondary antibody was incubated for 1 hour, then avidin-biotin-peroxidase complexes (ABC Kit, Vector Laboratories), then sections were developed with diaminobenzidine (Sigma), and finally sections were counterstained with hematoxylin and mounted.
Acknowledgments
We thank past and present members of the Center for Neurovirology for their insightful discussion and sharing of ideas and reagents. We also wish to thank C. Schriver for editorial assistance and Jennifer Gordon for helpful advice. This work was supported by grants awarded by the NIH to MS, LDV and KK.
References
- Amini S, Mameli G, Del Valle L, Skowronska A, Reiss K, Gelman BB, White MK, Khalili K, Sawaya BE. p73 Interacts with human immunodeficiency virus type 1 Tat in astrocytic cells and prevents its acetylation on lysine 28. Mol Cell Biol. 2005;25:8126–8138. doi: 10.1128/MCB.25.18.8126-8138.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger JR, Houff S. Progressive multifocal leukoencephalopathy: lessons from AIDS and natalizumab. Neurol Res. 2006;28:299–305. doi: 10.1179/016164106X98198. [DOI] [PubMed] [Google Scholar]
- Cai Y, Liu Y, Zhang X. Induction of transcription factor Egr-1 gene expression in astrocytoma cells by Murine coronavirus infection. Virology. 2006;355:152–163. doi: 10.1016/j.virol.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- Chang CF, Otte J, Kerr DA, Valkkila M, Calkins CE, Khalili K. Evidence that the soluble factors secreted by activated immune cells suppress replication of human neurotropic JC virus DNA in glial cells. Virology. 1996;221:226–231. doi: 10.1006/viro.1996.0369. [DOI] [PubMed] [Google Scholar]
- Chang Y, Lee HH, Chen YT, Lu J, Wu SY, Chen CW, Takada K, Tsai CH. Induction of the early growth response 1 gene by Epstein-Barr virus lytic transactivator Zta. J Virol. 2006;80:7748–7755. doi: 10.1128/JVI.02608-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen NN, Khalili K. Transcriptional regulation of human JC polyomavirus promoters by cellular proteins YB-1 and Pur alpha in glial cells. J Virol. 1995;69:5843–5848. doi: 10.1128/jvi.69.9.5843-5848.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury M, Kundu M, Khalili K. GA/GC-rich sequence confers Tat responsiveness to human neurotropic virus promoter, JCVL, in cells derived from central nervous system. Oncogene. 1993;8:887–892. [PubMed] [Google Scholar]
- Chu S, Ferro TJ. Sp1: regulation of gene expression by phosphorylation. Gene. 2005;348:1–11. doi: 10.1016/j.gene.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Del Valle L, Piña-Oviedo S. HIV disorders of the brain: pathology and pathogenesis. Front Biosci. 2006;11:718–32. doi: 10.2741/1830. [DOI] [PubMed] [Google Scholar]
- Enam S, Sweet TM, Amini S, Khalili K, Del Valle L. Evidence for involvement of transforming growth factor beta1 signaling pathway in activation of JC virus in human immunodeficiency virus 1-associated progressive multifocal leukoencephalopathy. Arch Pathol Lab Med. 2004;128:282–291. doi: 10.5858/2004-128-282-EFIOTG. [DOI] [PubMed] [Google Scholar]
- Frisque RJ, Bream GL, Cannella MT. Human polyomavirus JC virus genome. J Virol. 1984;51:458–469. doi: 10.1128/jvi.51.2.458-469.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZF, Weihe E, Zheng YM, Schafer MK, Sheng H, Corisdeo S, Rauscher FJ, Koprowski H, Dietzschold B. Differential effects of rabies and borna disease viruses on immediate-early- and late-response gene expression in brain tissues. J Virol. 1993;67:6674–6681. doi: 10.1128/jvi.67.11.6674-6681.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M, Niki T, Mori T, Matsuda T, Matsui M, Nomura N, Seiki M. HTLV-1 Tax induces expression of various immediate early serum responsive genes. Oncogene. 1991;6:1023–1029. [PubMed] [Google Scholar]
- Graham FL, van der Eb AJ. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Henson J, Saffer J, Furneaux H. The transcription factor Sp1 binds to the JC virus promoter and is selectively expressed in glial cells in human brain. Ann Neurol. 1992;32:72–77. doi: 10.1002/ana.410320112. [DOI] [PubMed] [Google Scholar]
- Henson JW. Regulation of the glial-specific JC virus early promoter by the transcription factor Sp1. J Biol Chem. 1994;269:1046–1050. [PubMed] [Google Scholar]
- Hipskind RA, Buscher D, Nordheim A, Baccarini M. Ras/MAP kinase-dependent and -independent signaling pathways target distinct ternary complex factors. Genes Dev. 1994;8:1803–1816. doi: 10.1101/gad.8.15.1803. [DOI] [PubMed] [Google Scholar]
- Khalili K, Gordon J, White MK. The polyomavirus, JCV and its involvement in human disease. Adv Exp Med Biol. 2006;577:274–287. doi: 10.1007/0-387-32957-9_20. [DOI] [PubMed] [Google Scholar]
- Khalili K, Safak M, Del Valle L, White MK. JC virus molecular biology and the human demyelinating disease, progressive multifocal leukoencephalopathy. In: Shoshkes Reiss C, editor. Neurotropic virus infections. Cambridge University Press; Cambridge, UK: 2007. [Google Scholar]
- Krachmarov CP, Chepenik LG, Barr-Vagell S, Khalili K, Johnson EM. Activation of the JC virus Tat-responsive transcriptional control element by association of the Tat protein of human immunodeficiency virus 1 with cellular protein Pur alpha. Proc Natl Acad Sci USA. 1996;93:14112–14117. doi: 10.1073/pnas.93.24.14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischitelli M, Fioriti D, Videtta M, Degener AM, Antinori A, Cinque P, Giordano A, Pietropaolo V. Investigation on the role of cell transcriptional factor Sp1 and HIV-1 TAT protein in PML onset or development. J Cell Physiol. 2005;204:913–918. doi: 10.1002/jcp.20375. [DOI] [PubMed] [Google Scholar]
- Mora GR, Olivier KR, Cheville JC, Mitchell RF, Lingle WL, Tindall DJ. The cytoskeleton differentially localizes the early growth response gene-1 protein in cancer and benign cells of the prostate. Mol Cancer Res. 2004;2:115–128. [PubMed] [Google Scholar]
- Pallas DC, Shahrik LK, Martin BL, Jaspers S, Miller TB, Brautigan DL, Roberts TM. Polyoma small and middle T antigens and SV40 small t antigen form stable complexes with protein phosphatase 2A. Cell. 1990;12:167–176. doi: 10.1016/0092-8674(90)90726-u. [DOI] [PubMed] [Google Scholar]
- Papanikolaou NA, Sabban EL. Ability of Egr1 to activate tyrosine hydroxylase transcription in PC12 cells. Cross-talk with AP-1 factors. J Biol Chem. 2000;275:26683–26689. doi: 10.1074/jbc.M000049200. [DOI] [PubMed] [Google Scholar]
- Pietropaolo V, Videtta M, Fioriti D, Mischitelli M, Arancio A, Orsi N, Degener AM. Rearrangement patterns of JC virus noncoding control region from different biological samples. J Neurovirol. 2003;9:603–611. doi: 10.1080/13550280390246507. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan S, Otte J, Enam S, Del Valle L, Khalili K, Gordon J. JC virus-induced changes in cellular gene expression in primary human astrocytes. J Virol. 2003;77:10638–10644. doi: 10.1128/JVI.77.19.10638-10644.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj GV, Khalili K. Identification and characterization of a novel GGA/C-binding protein, GBP-i, that is rapidly inducible by cytokines. Mol Cell Biol. 1994;14:7770–7781. doi: 10.1128/mcb.14.12.7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj GV, Khalili K. Transcriptional regulation: lessons from the human neurotropic polyomavirus, JCV. Virology. 1995;213:283–291. doi: 10.1006/viro.1995.0001. [DOI] [PubMed] [Google Scholar]
- Ranganathan PN, Khalili K. The transcriptional enhancer element, kappa B, regulates promoter activity of the human neurotropic virus, JCV, in cells derived from the CNS. Nucleic Acids Res. 1993;21:1959–1964. doi: 10.1093/nar/21.8.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S, Tienhaara A, Lee MS, Tsai LH, Gill G. GC box-binding transcription factors control the neuronal specific transcription of the cyclin-dependent kinase 5 regulator p35. J Biol Chem. 2002;277:4455–4464. doi: 10.1074/jbc.M110771200. [DOI] [PubMed] [Google Scholar]
- Sadowska B, Barrucco R, Khalili K, Safak M. Regulation of human polyomavirus JC virus gene transcription by AP-1 in glial cells. J Virol. 2003;77:665–672. doi: 10.1128/JVI.77.1.665-672.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safak M, Barrucco R, Darbinyan A, Okada Y, Nagashima K, Khalili K. Interaction of JC virus agno protein with T antigen modulates transcription and replication of the viral genome in glial cells. J Virol. 2001;75:1476–1486. doi: 10.1128/JVI.75.3.1476-1486.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Rangarajan PN. Common host genes are activated in mouse brain by Japanese encephalitis and rabies viruses. J Gen Virol. 2003;84:1729–1735. doi: 10.1099/vir.0.18826-0. [DOI] [PubMed] [Google Scholar]
- Sakamoto KM, Nimer SD, Rosenblatt JD, Gasson JC. HTLV-I and HTLV-II tax trans-activate the human EGR-1 promoter through different cis-acting sequences. Oncogene. 1992;7:2125–2130. [PubMed] [Google Scholar]
- Sawaya BE, Khalili K, Mercer WE, Denisova L, Amini S. Cooperative actions of HIV-1 Vpr and p53 modulate viral gene transcription. J Biol Chem. 1998;273:20052–20057. doi: 10.1074/jbc.273.32.20052. [DOI] [PubMed] [Google Scholar]
- Silva PN, Soares JA, Brasil BS, Nogueira SV, Andrade AA, de Magalhaes JC, Bonjardim MB, Ferreira PC, Kroon EG, Bruna-Romero O, Bonjardim CA. Differential role played by the MEK/ERK/EGR-1 pathway in orthopoxviruses vaccinia and cowpox biology. Biochem J. 2006;398:83–95. doi: 10.1042/BJ20060509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag E, Fedorov S, Kamibayashi C, Robbins D, Cobb M, Mumby M. The interaction of SV40 small tumor antigen with protein phosphatase 2A stimulates the map kinase pathway and induces cell proliferation. Cell. 1993;75:887–897. doi: 10.1016/0092-8674(93)90533-v. [DOI] [PubMed] [Google Scholar]
- Thiel G, Cibelli G. Regulation of life and death by the zinc finger transcription factor Egr-1. J Cell Physiol. 2002;193:287–292. doi: 10.1002/jcp.10178. [DOI] [PubMed] [Google Scholar]
- van’t Wout AB, Lehrman GK, Mikheeva SA, O’Keeffe GC, Katze MG, Bumgarner RE, Geiss GK, Mullins JI. Cellular gene expression upon human immunodeficiency virus type 1 infection of CD4(+)-T-cell lines. J Virol. 2003;77:1392–1402. doi: 10.1128/JVI.77.2.1392-1402.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A, Doerks A, Aboud M, Alonso A, Tokino T, Flugel RM, Lochelt M. Induction of cellular genes is mediated by the Bel1 transactivator in foamy virus-infected human cells. J Virol. 2000;74:4441–4447. doi: 10.1128/jvi.74.10.4441-4447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler K, Bui T, Frisque RJ, Grandinetti A, Nerurkar VR. A rapid in vitro polyomavirus DNA replication assay. J Virol Methods. 2004;122:123–127. doi: 10.1016/j.jviromet.2004.08.012. [DOI] [PubMed] [Google Scholar]