Abstract
The honey bee genome predicts ≈100 peptides from 36 prohormones, but the functions of many of these peptides are unknown. We used differential isotope labeling combined with mass spectrometric analysis to quantify ≈50% of known bee brain peptides in the context of foraging, with 8 showing robust and dynamic regulation. Some showed differences in brain abundance as a function of experience; specifically, nectar and pollen collection led to quick changes in abundance. These changes were related to the act of food collection, not ingestion, because foragers bring food back to the hive for storage rather than eating it themselves. Other peptide differences in brain abundance were seen in bees that either flew to a nectar feeder or a pollen feeder, but did not yet collect any food. These differences likely reflect well-known predispositions of some bees to collect either nectar or pollen, but not both. Tachykinin, PBAN, and sNPF were among the peptides with the strongest changes in association with nectar and pollen foraging. These peptides are known to be involved in regulating food intake in solitary insects, suggesting an evolutionary connection between that behavior and social foraging. These results demonstrate that it is now possible to use quantitative peptidomics to help determine which brain peptides are bioactive and to elucidate their function in the regulation of behavior.
Keywords: Apis mellifera, behavioral maturation, foraging, neuropeptides
Brain peptides play an important role in orchestrating physiological and behavioral processes in animals by functioning as neurohormones, neuromodulators, and neurotransmitters (1). These cell–cell signaling peptides are produced from their corresponding precursor genes by cleavage at specific sites followed by additional posttranslational modifications, a complex process that can make bioactive peptides difficult to predict (2). The availability of genome sequences has led to a new, high-throughput approach for neuropeptide discovery: algorithms that predict cleavage sites in peptide precursors (3, 4) followed by sequencing from brain samples with mass spectrometry (5–7). Applying this methodology to the honey bee genome, Hummon et al. (6) predicted 36 peptide-encoding genes and confirmed 100 endogenous peptides in the bee brain, numbers similar to what is known for other animals such as the fruit fly (Drosophila melanogaster) and the house mouse (Mus musculus) (7, 8).
As with other species, however, the physiological and behavioral functions of most (neuro)peptides in the honey bee are unknown (2, 9–11). Until very recently, most brain peptides were discovered one at a time via biochemical techniques, and functional experiments focused on physiological effects (12). Recent advances in peptidomics have greatly accelerated peptide characterization (13–15). However, corresponding high-throughput approaches to discover peptide function have been much less common. A goal of this study was to determine whether combining a recently established quantitative peptidomic method with compelling behavioral paradigms can help to overcome gaps in our knowledge of peptide function.
One of the newer approaches for quantitative peptidomics involves chemically modifying 2 samples of interest by using a pair of isotopologues (molecules having the same chemical structure but different masses) and quantifying them by comparing peak intensities (16). A technical goal of our study was to modify these protocols to detect quantitative changes in peptide profiles in small (insect brain) samples and employ them to monitor changes in brain peptide profiles in the context of natural behavior. We focused on foraging behavior in honey bees for 2 reasons: it is subject to at least 3 different types of regulation, and the roles of brain peptides in these regulatory processes have yet to be thoroughly examined.
Behavioral maturation in honey bees represents one type of behavioral regulation associated with foraging (17). Brood care and foraging, like all major activities performed by adult worker honey bees, are highly social activities and occur within the framework of the colony's system of age-related division of labor. A bee spends the first 2–3 weeks of adult life working in the hive caring for the brood (“nursing”) and performing other activities, and then spends its final 1–2 weeks collecting food outside as a forager.
The second type of behavioral regulation associated with foraging is the tendency of some bees to collect nectar or pollen, rather than both (18). This tendency is influenced by environmental factors that relate to the needs of the colony; a shortage of either nectar or pollen in the hive will cause some bees to switch to collecting the resource in short supply (19, 20). There also are hereditary influences between a colony's patrilines that arise because of multiple mating by the queen (21, 22). The strong implication emerging from these studies is that many bees leave the hive with a predisposition to preferentially collect nectar or pollen.
The third form of behavioral regulation involves information acquired during foraging. Information bees acquire on food quality and quantity is used to guide subsequent foraging-related behavioral decisions. Each foraging trip involves visiting up to hundreds of flowers to collect a load of nectar and pollen, and usually lasts 10–80 min (up to a few hours) before the bee returns to her hive to unload this material and leave the hive again for another foraging trip (23, 24). Foragers feed on honey in the hive to fuel their foraging activity and do not generally eat the nectar or pollen they collect. Although foragers do technically ingest the nectar, they store it in a portion of their digestive tract modified for this purpose (the foregut or crop) and then expel it on return to the hive (25). They store pollen in specialized “pollen baskets” (corbiculae) on the hind legs (25). A bee decides whether she has collected enough food to return to her hive or whether to continue foraging (26). If she opts to return to the hive, she then decides whether the food is of sufficient quality to recruit others to the same source (27).
In the context of these 3 types of regulation, we designed 2 experiments to examine the role of brain peptides in the regulation of foraging behavior. In Experiment 1, we compared brain peptide levels between nurse bees and foragers. For Experiment 2, we used 2 types of collection strategies. To explore changes in brain peptide levels that might be associated with foraging predisposition, we sampled nectar and pollen foragers that arrived at feeders before they had a chance to collect any food. To explore changes in brain peptide levels that might be associated with food collection behavior itself, we sampled nectar and pollen foragers that were given the opportunity to collect food and were preparing to leave the feeder.
Results
Peptide Identification.
We were able to quantify a total of 51 of 100 peptides from 18 precursors in either Experiment 1 or 2 (Table 1) by using accurate mass, charge state, and retention time obtained in previous analyses (6). There were 19 peptides from 11 precursors that were detected in both experiments. Some of the previously characterized peptides not detected in the current experiments were likely below the detection limits owing to small sample size and others were not isotopically labeled because of N-terminal modifications. More peptides were detected in Experiment 1 than Experiment 2, despite comparable sample sizes. Forty-two peptides from 15 peptide precursors were detected in Experiment 1 and 28 peptides from 15 peptide precursors in Experiment 2 (Table 1). In general, more peptides from the same precursor were identified in Experiment 1 than Experiment 2. An extreme example was allatostatin (5 peptides from this precursor were identified in Experiment 1, and 2 in Experiment 2).
Table 1.
Differences in brain peptide abundance as a function of behavior
| Precursor | Peptide | Mass | Nurse/forager |
Nectar/pollen |
||
|---|---|---|---|---|---|---|
| Raw | FDR | Raw | FDR | |||
| Allatostatin | AYTYVSEY | 994.43 | n.s. | n.s. | – | – |
| GRDYSFGLa | 912.45 | n.s. | n.s. | – | – | |
| GRQPYSFGLa | 1022.53 | n.s. | n.s. | n.s. | n.s. | |
| LPVYNFGIa | 920.51 | n.s. | n.s. | n.s. | n.s. | |
| RQYSFGLa | 868.46 | n.s. | n.s. | – | – | |
| Apidaecin | GNNRPVYIPQPRPPHP | 1837.9 | * | n.s. | n.s. | n.s. |
| GNNRPVYIPQPRPPHPRL | 2107.14 | *** | n.s. | – | – | |
| PVYIPQPRPP | 1162.65 | n.s. | n.s. | – | – | |
| Corazonin | SFSENMINDHRQPAPTNNNY | 2348.02 | n.s. | n.s. | – | – |
| IDLSRFYGHFNT-cont. | DLSRFYGHFN | 1254.58 | ** | n.s. | – | – |
| DLSRFYGHFNT | 1355.63 | n.s. | n.s. | – | – | |
| IDLSRFYGHF | 1253.62 | n.s. | n.s. | * | n.s. | |
| IDLSRFYGHFN | 1367.66 | n.s. | n.s. | *** | *** | |
| IDLSRFYGHFNT | 1468.71 | n.s. | n.s. | *** | *** | |
| ITGQGNRIF-cont. | ITGQGNRIF | 1004.54 | n.s. | n.s. | * | * |
| LRNQLDIGDLQ-cont. | LRNQLDIGDL | 1155.62 | n.s. | n.s. | – | – |
| LRNQLDIGDLQ | 1283.68 | n.s. | n.s. | ** | *** | |
| MVPV | MVPVPVHHMADELLRNGPDTVI | 2439.24 | – | – | ** | *** |
| VPVPVHHMADELL | 1455.75 | – | – | n.s. | n.s. | |
| Myosuppressin | QDVDHVFLRFa | 1273.66 | n.s. | n.s. | n.s. | n.s. |
| NPLP-1 | GIFLPGSVILRALSRQa | 1725.04 | – | – | n.s. | n.s. |
| NIASLMRDYDQSRENRVPFPa | 2406.19 | – | – | ** | ** | |
| NVASLARTYTLPQNAa | 1616.86 | n.s. | n.s. | – | – | |
| NVGSVAREHGLPYa | 1396.72 | n.s. | n.s. | n.s. | n.s. | |
| NVGTLARDFALPPa | 1368.75 | n.s. | n.s. | – | – | |
| SVSSLAKNSAWPVSL | 1544.82 | n.s. | n.s. | – | – | |
| SVSSLARTGDLPVREQ | 1713.9 | n.s. | n.s. | – | – | |
| YVASLARTGDLPIRGQ | 1715.78 | n.s. | n.s. | n.s. | n.s. | |
| NVPIYQEPRF-cont. | NVPIYQEPRF | 1261.65 | n.s. | n.s. | – | – |
| VPIYQEPRF | 1147.6 | – | – | n.s. | n.s. | |
| Orcokinin | LTNYLATTGHGTNTGGPVLT | 1987 | n.s. | n.s. | – | – |
| NIDEIDRTAFDNFF | 1715.78 | n.s. | n.s. | n.s. | n.s. | |
| NLDEIDRVGWSGFV | 1605.79 | n.s. | n.s. | – | – | |
| PBAN | IYLPLFASRLa | 1190.72 | – | – | ** | *** |
| Perivisc | AYRKPPFNGSIFa | 1394.75 | n.s. | n.s. | – | – |
| PDH | NSELINSLLGLPKNMNNAa | 1940.01 | ** | n.s. | – | – |
| SIFamide | RKPPFNGSIFa | 1160.65 | – | – | n.s. | n.s. |
| sNPF | SPSLRLRFa | 973.59 | n.s. | n.s. | *** | *** |
| Tachykinin | ALMGFQGVRa | 976.53 | n.s. | n.s. | * | n.s. |
| APMGFQGMRG | 1050.47 | n.s. | n.s. | – | – | |
| APMGFQGMRa | 992.47 | n.s. | n.s. | *** | *** | |
| APMGFYGT | 842.36 | n.s. | n.s. | – | – | |
| APMGFYGTRa | 997.48 | – | – | * | * | |
| ARMGFHGMRa | 1060.52 | n.s. | n.s. | – | – | |
| ASFDDEYY | 1008.38 | * | n.s. | – | – | |
| GVMDFQIGLQ | 1106.55 | n.s. | n.s. | – | – | |
| IILDALEELD | 1142.61 | n.s. | n.s. | n.s. | n.s. | |
| NSIINDVKNELFPEDIN | 1972.97 | – | – | n.s. | n.s. | |
| SLEEILDEI | 1059.54 | n.s. | n.s. | n.s. | n.s. | |
| SPFRYLGARa | 1064.59 | * | n.s. | n.s. | n.s. | |
| TWKSPDIVIRFamide | GRNDLNFIRYa | 1265.67 | n.s. | n.s. | *** | *** |
*, P < 0.1;
**, P < 0.05;
***, P < 0.01.
FDR values: *, FDR < 0.2; **, FDR < 0.15; ***, FDR < 0.1.
Experiment 1: Comparison of Peptide Abundances in Nurses and Foragers.
Despite the greater number of peptides identified in this experiment, no peptide showed a significant difference in abundance between nurses and foragers at the false discovery rate (FDR) of <0.10. Three peptides showed significant differences at an unadjusted P < 0.05 (indicated as ** and *** in Table 1). GNNRPVYIPQPRPPHPRL (apidaecin) and NSELINSLLGLPKNMNNAa (PDH, pigment-dispersing hormone) were more abundant in forager brains, and DLSRFYGHFN (IDLSRFYGHFNT-containing peptide) was more abundant in nurse brains.
Experiment 2: Comparison of Peptide Abundances in Nectar and Pollen Foragers.
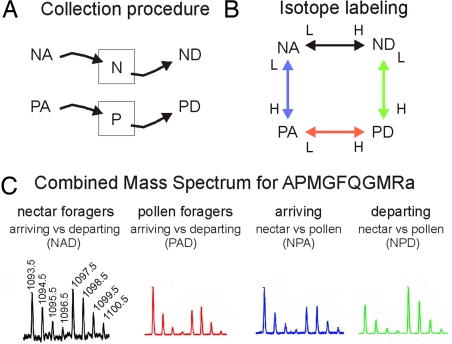
We studied 4 groups of bees: (i) individuals arriving at a feeder containing (artificial) nectar (sugar syrup), before they had a chance to collect any food (nectar arriving, NA); (ii) similarly staged individuals arriving at a feeder containing pollen (pollen arriving, PA); (iii) individuals that collected sugar syrup and were getting ready to depart from the feeder (nectar departing, ND); and (iv) similarly staged individuals at the pollen feeder (pollen departing, PD) (Fig. 1 A and B). The 2 samples per colony for each behavioral group (see Materials and Methods) were either labeled with a light or heavy isotope and compared with samples from the other groups by using a loop design to help control for efficiency of isotope labeling [Fig. 1C and supporting information (SI) Fig. S1]. This comparison scheme allowed 2 measurements for each behavioral group. We compared NA versus PA, NA versus ND, PA versus PD, and ND versus PD for each of the 3 colonies we studied.
Fig. 1.
Experimental design and mass spectrometric results for analyzing changes in peptide profiles during honey bee foraging behavior. Honey bee colonies were kept in outdoor flight cages with separate ad libitum feeders for nectar (N, nectar; i.e., sugar solution) and pollen (P, pollen). (A) Foraging bees were collected at either feeder shortly after they landed and before they started to collect food (NA, PA; A, arriving) or after they had finished food collection and were ready to take off (ND, PD; D, departing). (B) Brain peptide samples from the various behavioral groups were labeled with different isotopes and compared. N and P “squares” refer to the feeders. (C) Mass spectrometric analysis of the isotopically labeled peptides, with the ratio of peak heights used to determine peptide level changes. Bee brain peptides were separated by using HPLC and directly analyzed by using an electrospray ionization quadrupole time-of-flight mass spectrometer. The mass spectrum of APMGFQGMRa for each of the comparisons was obtained by summing the scans in the extracted ion chromatograms: NAD, arriving versus departing nectar foragers; PAD, arriving versus departing pollen foragers; NPA, arriving nectar versus arriving pollen foragers; NPD, departing nectar versus departing pollen foragers.
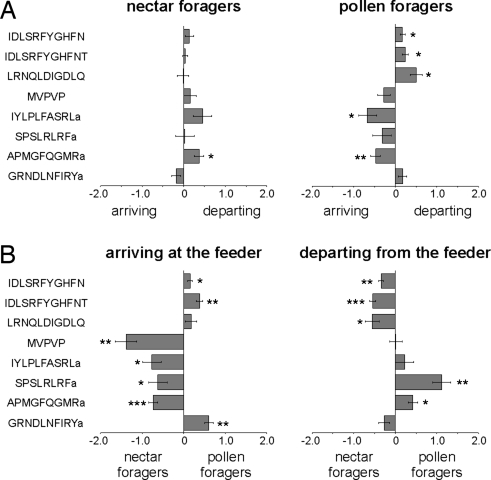
Eight peptides, from 7 peptide precursors, showed significant differences in abundance (indicated in Table 1 as FDR***, and in Fig. 2 as *, **, and ***, respectively) in one or more of the 4 comparisons. Seven of 8 peptides showed differences as a function of foraging predisposition (NA versus PA, Student's t test: P < 0.05; Fig. 2B). These were APMGFQGMRa (tachykinin), GRNDLNFIRYa (TWKSP), IYLPLFASRLa (PBAN), MVPVPVHHMADELLRNGPDTVI (MVPVPVHHMADELLRNGPDTVI-containing), IDLSRFYGHFNT, IDLSRFYGHFN (both IDLSRFYGHFNT-containing), and SPSLRLRFa (sNPF).
Fig. 2.
Differences in brain peptide abundances between 4 behaviorally different honey bee forager groups. (A) Relative differences in peptide abundances between arriving and departing nectar or pollen foragers. (B) Relative differences in peptide abundances between nectar and pollen foragers arriving or departing from the feeder. x axis: normalized, log2 transformed and centered peptide ratios. Negative and positive values indicate the direction of change. MVPVP = MVPVPVHHMADELLRNGPDTVI. Indicated significance levels (Student's t test): *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
Five of 8 peptides showed differences as a function of experience (NA versus ND or PA versus PD; Fig. 2A). For pollen collection these were APMGFQGMRa, LRNQLDIGDLQ (LRNQLDIGDLQ-containing), IYLPLFASRLa, IDLSRFYGHFN, and IDLSRFYGHFNT. For nectar collection, only APMGFQGMRa showed a significant change (P < 0.05), and IYLPLFASRLa (PBAN) showed marginal significance (0.1 > P > 0.05).
Two of 8 peptides, IDLSRFYGHFN and LRNQLDIGDLQ, showed significantly greater abundances in departing versus arriving bees, regardless of food type (Fig. S2B). One peptide, MVPVPVHHMADELLRNGPDTVI, showed significantly greater abundance in nectar foragers than pollen foragers, both arriving and departing (P < 0.05), and 2 additional peptides showed the same trend: IDLSRFYGHFN (P = 0.052) and APMGFQGMR (P = 0.058; see Fig. S2A).
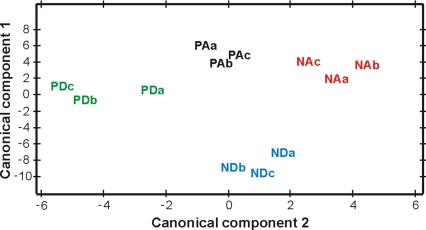
Canonical discriminant analysis revealed that the 4 forager groups were easily distinguishable on the basis of the relative amounts of the 8 peptides that showed significant differences in abundance in one or more of the 4 comparisons. A plot of the first 2 canonical components (which accounted for 98% of the variation in brain abundance across all 4 groups) revealed strong group-specific patterns (Fig. 3). Differences in the first canonical component were mainly due to differences in IDLSRFYGHFNT and MVPVPVHHMADELLRNGPDTVI, and differences in the second canonical component were mainly due to differences in SPSLRLRFa. Separation of ND from the other 3 behavioral groups was due primarily to high levels of IDLSRFYGHFNT and MPVPVPVHHMADELLRNGPDTVI in ND relative to the other 3 groups. Separation of PD from the other 3 behavioral groups was due primarily to high levels of SPSLRLRFa in PD relative to the other groups. These results are consistent with the results of the analyses of individual peptides (Fig. 2; Table 1).
Fig. 3.
Peptide abundances reveal peptide signatures for foraging behavior. (A) A plot of the first 2 canonical components (which accounted for 98% of the variation in brain expression across all 4 groups) revealed strong group-specific expression patterns. (B) Peptide abundance signatures for the different behavioral groups (y axis indicates normalized log2 transformed intensity values). PA, pollen arriving; NA, nectar arriving; PD, pollen departing; ND, nectar departing; a, b, and c refer to three different bee colonies.
Discussion
The principal significance of these results is that they demonstrate that it is now possible to use quantitative peptidomics to help determine which brain peptides are bioactive and to elucidate their function in the regulation of behavior. We used differential isotope labeling combined with mass spectrometric analysis to quantify ≈50% of known bee brain peptides in the context of foraging, with 8 showing dynamic regulation.
There were larger differences in brain peptide profiles between nectar and pollen foragers than between nurses and foragers. This suggests that peptide signaling is important for differences in behavior at short time scales. One peptide system that involves insulin signaling has been shown to be important in regulating honey bee behavioral maturation, based on measurements of gene expression and pharmacological manipulation (28). It may be that some peptide changes during maturation are not localized to the brain but occur in endocrine structures elsewhere, or they are part of the peptidome we did not detect. Another possibility is that changes in peptide signaling during behavioral maturation are mediated through changes in the expression of receptors or downstream components that we did not measure (28). One or more of these speculative ideas may explain why we did not detect any consistent differences in the abundances of any tachykinin peptide between nurses and foragers, in contrast to findings for the prepro-AmTRP (tachykinin) gene (29, 30).
There were differences in brain peptide abundances between bees that arrived at either the nectar or pollen feeders. Because bees were sampled just as they landed, before actually collecting either food resource, these differences are not confounded by the effects of food collection itself. It is therefore likely that these results reflect differences in predispositions to preferentially collect nectar or pollen (see the introduction). sNPF, one of the peptides that differed between NA and PA, is involved in insulin signaling, and several genes related to this molecular pathway are located within quantitative trait loci (QTLs) that influence pollen foraging behavior (31). None of the other peptides (or their respective peptide genes) are located within these QTLs, suggesting that they might be downstream of any hereditary differences between bees, or influenced by environmental factors. Functional analyses must be performed to determine whether the differences we detected exert causal influences on foraging predispositions.
There also were significant differences in brain peptide abundances as a function of experience. Nectar and pollen collection led to changes in brain peptide profiles. These effects were rapid; under our experimental conditions it took bees ≈2 or 9 min to collect the sugar syrup or pollen, respectively (nectar collection: 80 ± 15 sec, N = 10; pollen collection: 553 ± 82 sec, N = 5). It might be reasonable to think that ingestion of sugar syrup into the honey crop might provoke more changes in brain peptide abundances compared with placing pollen into the external pollen baskets on the legs, but there were more differences as a result of pollen collection. The longer duration and the more vigorous movements involved in pollen collection may account for the larger peptide changes detected. The peptides that are regulated in association with both nectar and pollen collection perhaps are involved in more general aspects of foraging behavior, e.g., the initiation and termination of the behavioral sequence, or changes in response thresholds to food stimuli as a consequence of food collection (27, 32, 33). Other peptides that responded to just pollen collection may have a more specialized function.
It is noteworthy that 3 of the 8 peptides highlighted in our experiments [APMGFQGMRa (tachykinin), SPSLRLRFa (sNPF), and IYLPLFASRLa (PBAN)] are involved in feeding regulation in solitary insects (34, 35). Perhaps social evolution has modified their function somewhat to be involved in the regulation of one or more aspects of honey bee social foraging. Tachykinins are involved in feeding regulation in insects and affect olfactory and locomotor behavior in Drosophila melanogaster (36–38). Multifunctionality is common for brain peptides (1, 2); perhaps the same peptide can influence the perception and localization of a food source and its actual collection. PBAN also is well known to be a regulator of pheromone biosynthesis in a variety of insects (39), and pheromone signaling is important in the recruitment of honey bee foragers via the famous dance communication system (40). This multifunctionality makes it easier to envision how such neuropeptides could be coopted in social evolution to regulate honey bee foraging behavior (41).
Neuropeptides are often colocalized with traditional neurotransmitters, or function as neuromodulators and neurohormones produced by relatively small numbers of large neurons released into large brain areas or the circulatory system (42, 43). Conversely, the brain also receives peptide signals produced in other organs (36, 44). We can interpret an increase in peptide abundance in our datasets as the inflow of peptides into the brain, either from neurons located elsewhere in the nervous system or from the circulatory system, and a decrease in abundance as either a release of peptides from the brain to other parts of the nervous system or their proteolytic degradation after release. Additional information, perhaps obtained by immunocytochemical analysis, would help in understanding the distribution and spatial dynamics of the 8 peptides that showed the strongest differences in abundance. It also is important to determine how these peptides interact with other neurochemicals such as biogenic amines to regulate foraging behavior (27, 45).
Materials and Methods
Collection of Bees and Preparation of Brain Samples.
Apis mellifera colonies were maintained according to standard methods at the University of Illinois Bee Research Facility (Urbana, IL). These colonies represent a mixture of European races (predominantly A.m. ligustica). Collection procedures in both experiments took <5 sec, thus minimizing possible effects on brain chemistry. Brains (with attached corpora allata-corpora cardiaca complexes) were dissected on dry ice and stored at −80 °C.
Collections for Experiment 1.
Bees were collected from 3 different colonies (with populations of ≈20,000 bees) maintained under typical conditions in the field. Nurses and returning (pollen) foragers were identified according to established procedures (25). They were caught with tweezers and immediately frozen in liquid nitrogen. There were 4 samples per behavioral group, 2 behavioral groups, from 3 colonies (n = 20 brains per sample) for a total of 480 brains.
Collections for Experiment 2.
This experiment also included brains from a total of 480 bees. There were 2 samples per behavioral group, 4 behavioral groups, from 3 colonies (n = 20 brains per sample). We collected enough bees for 40 dissected brains per sample and then split each pool of 40 brains into 2 samples of 20 brains for isotopic labeling (Fig. 1B). Three small colonies (n = 3) were set up in an outdoor flight cage. Each colony was composed of 2,000 adult bees, 1 honeycomb frame containing brood, and 2 honeycomb frames containing honey and pollen. Sugar syrup (2M) and pollen were provided ad libitum at 2 separate feeders for a few hours each day. The feeders were located 50 cm apart from each other. Collections of foragers were performed after a few days of strong foraging activity; at this point, the colony was acclimated to the feeders and little cross-feeder foraging was observed. Bees were either caught shortly after they landed on the feeder table before they started to collect food or just after they finished food collection and were ready to take off. Collections occurred in the morning and afternoon and were pooled to diminish circadian effects. Each bee was caught in a small glass vial and frozen as above. Bees were not age-marked; there are no differences in the age distributions of nectar and pollen forager honey bees, so we relied solely on behavioral identification (25, 46).
Peptide Extraction and Isotope Labeling.
Frozen brain samples were transferred to a glass homogenizer (1 mL; Kontes Glass,) by using a pair of tweezers and homogenized in 400 μL of acidified acetone solution (acetone/water/conc. HCl, 40:6:1, vol/vol/v). The homogenates were centrifuged (12,000 rpm) and the supernatant filtered (10 kDa molecular mass cutoff filter, Millipore) to remove larger proteins. The solvent in the filtrate was removed in vacuo by using a Savant SpeedVac (Savant Instruments). The residue containing the extracted peptides was reconstituted in 25 μL of 5% aqueous acetonitrile (ACN) solution [water/ACN (95:5, vol/vol) containing 0.1% formic acid, 0.01% TFA] and buffered by using 2 μL of 1 M phosphate buffer at pH 8.0. Samples were grouped based on the isotopic chemical labels they would receive, and 5 μL of 1 M NaOH was added to each of the vials.
Covalent modification of the extracted peptides for quantitation was performed by adding 2 μL of light (2 M succinic anhydride in DMSO) or heavy (2 M succinic [2H4] anhydride in DMSO) label. The solutions were vortexed, centrifuged, and incubated for 15 min at room temperature (RT). After incubation, the solutions were readjusted to pH 9 with 1 M NaOH. We repeated the labeling procedure with subsequent pH adjustments (pH 9) 4 times. Any remaining unbound labeling reagent was quenched by using 10 μL of 2.5 M glycine for 1 h at RT, followed by 5 μL of 2 M hydroxylamine solution for 30 min at RT, as described in ref. 47.
Corresponding light and heavy isotope-labeled samples were then combined as in Fig. 1B and desalted by using PepClean C18 spin columns (Pierce Biotechnology) as per manufacturer's protocol and eluted with 70% aqueous ACN solution. The solvent in the eluant was removed on a SpeedVac and the residue reconstituted in 20 μL of solvent A (5% ACN, 0.1% formic acid, and 0.01% TFA in water). Aliquots (15 μL) from each of the samples were used for LC-MS analysis. All chemical reagents were obtained from Sigma-Aldrich unless otherwise mentioned.
Mass Spectrometric Analysis.
Samples were analyzed by using a UPLC system directly coupled to an electrospray ionization quadrupole time-of-flight (ESI-QTOF) mass spectrometer (Waters). In brief, samples were separated on a reversed phase C18 nanoAcquity column (Waters) by using a gradient system containing solvent A (5% aqueous ACN with 0.1% formic acid in water) and solvent B (95% ACN and 0.1% formic acid in water) at 250 nL/min. The 90-min gradient included 3 steps: from 5 to 50% solvent B in 60 min (linear); then up to 90% solvent B in 10 min (linear); followed by 5% solvent B for 20 min (isocratic). Nitrogen was used as the nebulizing gas and argon as the collision gas. Data were acquired in MS scan mode and the detection was set in the m/z range, 400–1,500.
Data Analysis.
Peptide peak identifications were made by using retention time, charge state, and mass accuracy from previous analyses (6). Extracted ion chromatograms were constructed for each of the peptides analyzed and the combined mass spectrum across their elution period was used for quantification. Isotopic labeling imparts a mass difference of 4 Da between the light- and heavy-labeled samples. The singly charged peptides were separated by 4 Da, doubly charged peptides by 2 Da, and triply charged peptides by 1.33 Da in the MS data. For each of the peptides, quantitative information was obtained by calculating the peak intensity ratios of the light and heavy form of a peptide by using the sum of 3 abundant isotopic peaks arising from each of the labels (light and heavy).
Statistical Analyses.
The same approaches were used for both experiments. Raw peptide ratios were normalized, log2-transformed, and centered. Log2 ratios were analyzed by using a linear mixed-effects model, accounting for effects of behavior (nurse/forager; and pollen/nectar foraging) and run (combined run, colony, and isotope label effects). Differential abundance was tested by using F test statistics. Adjustment for multiple testing was implemented by using the FDR criterion (48). A peptide was considered differentially abundant across behavioral groups when the FDR was P < 0.1, which approximately corresponds to raw unadjusted P values of P < 0.05. FDRs were calculated across the entire set of peptides. In Experiment 2, for those peptides with significant differences in abundance, Student's t tests were used to identify the behavioral groups that were significantly different from zero (P value < 0.05). In addition, for the subset of peptides that showed significant differences in the above tests, canonical discriminant analysis was used to determine whether the differences in brain peptide abundances were sufficient to classify individual brain samples to one of the 4 behavioral groups. Data preprocessing and analysis was performed by using the SAS statistical package (SAS Inc.).
Supplementary Material
Acknowledgments.
We thank P. Yau and B. Imai at the University of Illinois at Urbana–Champaign (UIUC) Protein Sciences Facility and L. Fricker for useful discussions and technical help, M. Kolodokin for help with the experiments, and members of the Robinson and Sweedler labs for comments that improved the manuscript. This work was supported by National Institute on Drug Abuse Award P30 DA 018310 (UIUC Neuroproteomics Center), National Institute of General Medical Sciences Award GM073644 (to G.E.R.), and the Illinois Sociogenomics Initiative (G.E.R.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813021106/DCSupplemental.
References
- 1.Strand FL. Neuropeptides: Regulators of Physiological Processes. Cambridge, MA: MIT Press; 1999. [Google Scholar]
- 2.Li L, Sweedler JV. Peptides in our brain: Mass spectrometric-based measurement approaches and challenges. Annu Rev Anal Chem. 2008;1:451–483. doi: 10.1146/annurev.anchem.1.031207.113053. [DOI] [PubMed] [Google Scholar]
- 3.Amare A, et al. Bridging neuropeptidomics and genomics with bioinformatics: Prediction of mammalian neuropeptide prohormone processing. J Proteome Res. 2006;5:1162–1167. doi: 10.1021/pr0504541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Southey BR, Sweedler JV, Rodriguez-Zas SL. Prediction of neuropeptide cleavage sites in insects. Bioinformatics. 2008;24:815–825. doi: 10.1093/bioinformatics/btn044. [DOI] [PubMed] [Google Scholar]
- 5.Clynen E, et al. Peptidomics of the locust corpora allata: Identification of novel pyrokinins (-FXPRLamides) Peptides. 2003;24:1493–1500. doi: 10.1016/j.peptides.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Hummon AB, et al. From the genome to the proteome: uncovering peptides in the Apis brain. Science. 2006;314:647–649. doi: 10.1126/science.1124128. [DOI] [PubMed] [Google Scholar]
- 7.Hewes RS, Taghert PH. Neuropeptides and neuropeptide receptors in the Drosophila melanogaster genome. Genome Res. 2001;11:1126–1142. doi: 10.1101/gr.169901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Svensson M, Skold K, Svenningsson P, Andren PE. Peptidomics-based discovery of novel neuropeptides. J Proteome Res. 2003;2:213–219. doi: 10.1021/pr020010u. [DOI] [PubMed] [Google Scholar]
- 9.Dowell JA, Heyden WV, Li L. Rat neuropeptidomics by LC-MS/MS and MALDI-FTMS: Enhanced dissection and extraction techniques coupled with 2D RP-RP HPLC. J Proteome Res. 2006;5:3368–3375. doi: 10.1021/pr0603452. [DOI] [PubMed] [Google Scholar]
- 10.Predel R, Neupert S. Social behavior and the evolution of neuropeptide genes: Lessons from the honeybee genome. BioEssays. 2007;29:416–421. doi: 10.1002/bies.20571. [DOI] [PubMed] [Google Scholar]
- 11.Schoofs L, Baggerman G. Peptidomics in Drosophila melanogaster. Brief Funct Genomic Proteomic. 2003;2:114–120. doi: 10.1093/bfgp/2.2.114. [DOI] [PubMed] [Google Scholar]
- 12.Schoofs L, Vanden Broeck J, De Loof A. The myotropic peptides of Locusta migratoria: Structures, distribution, functions and receptors. Insect Biochem Mol Biol. 1993;23:859–881. doi: 10.1016/0965-1748(93)90104-z. [DOI] [PubMed] [Google Scholar]
- 13.Svensson M, et al. Neuropeptidomics: MS applied to the discovery of novel peptides from the brain. Anal Chem. 2007;79:15–16. 18–21. doi: 10.1021/ac071856q. [DOI] [PubMed] [Google Scholar]
- 14.Hatcher NG, et al. Mass spectrometry-based discovery of circadian peptides. Proc Natl Acad Sci USA. 2008;105:12527–12532. doi: 10.1073/pnas.0804340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fricker LD, Lim J, Pan H, Che FY. Peptidomics: Identification and quantification of endogenous peptides in neuroendocrine tissues. Mass Spectrom Rev. 2006;25:327–344. doi: 10.1002/mas.20079. [DOI] [PubMed] [Google Scholar]
- 16.Che FY, Fricker LD. Quantitation of neuropeptides in Cpe(fat)/Cpe(fat) mice using differential isotopic tags and mass spectrometry. Anal Chem. 2002;74:3190–3198. doi: 10.1021/ac015681a. [DOI] [PubMed] [Google Scholar]
- 17.Robinson G. Regulation of division of labor in insect societies. Annu Rev Entomol. 1992;37:637–665. doi: 10.1146/annurev.en.37.010192.003225. [DOI] [PubMed] [Google Scholar]
- 18.Lindauer M. Ein Beitrag zur Frage der Arbeitsteilung im Bienenstaat (A contribution to the question of division of labor in honey bee colonies) Z Vergl Physiol. 1952;34:299–345. [Google Scholar]
- 19.Dreller C, Page RE, Fondrk MK. Regulation of pollen foraging in honeybee colonies: Effects of young brood, stored pollen, and empty space. Behav Ecol Sociobiol. 1999;45:227–233. [Google Scholar]
- 20.Dreller C, Tarpy DR. Perception of the pollen need by foragers in a honeybee colony. Anim Behav. 2000;59:91–96. doi: 10.1006/anbe.1999.1303. [DOI] [PubMed] [Google Scholar]
- 21.Estoup A, Solignac M, Cornuet J. Precise assessment of the number of patrilines and of genetic relatedness in honeybee colonies. Proc Biol Sci. 1994;258:1–7. [Google Scholar]
- 22.Page RE, Robinson GE, Fondrk MK, Nasr M. Effects of worker genotypic diversity on honey bee colony development and behavior (Apis mellifera L.) Behav Ecol Sociobiol. 1995;36:387–396. [Google Scholar]
- 23.Winston M, Katz S. Foraging differences between cross-fostered honeybee workers (Apis mellifera) of European and Africanized races. Behav Ecol Sociobiol. 1982;10:125–129. [Google Scholar]
- 24.Seeley TD. The Wisdom of the Hive. Cambridge, MA: Harvard Univ Press; 1995. [Google Scholar]
- 25.Winston M. The Biology of the Honey Bee. Cambridge, MA: Harvard Univ Press; 1987. [Google Scholar]
- 26.Dyer FC, Dickinson JA. Development of sun compensation by honeybees: How partially experienced bees estimate the sun's course. Proc Natl Acad Sci USA. 1994;91:4471–4474. doi: 10.1073/pnas.91.10.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barron AB, Maleszka R, Vander Meer RK, Robinson GE. Octopamine modulates honey bee dance behavior. Proc Natl Acad Sci USA. 2007;104:1703–1707. doi: 10.1073/pnas.0610506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ament SA, Corona M, Pollock HS, Robinson GE. Insulin signaling is involved in the regulation of worker division of labor in honey bee colonies. Proc Natl Acad Sci USA. 2008;105:4226–4231. doi: 10.1073/pnas.0800630105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeuchi H, Yasuda A, Yasuda-Kamatani Y, Kubo T, Nakajima T. Identification of a tachykinin-related neuropeptide from the honeybee brain using direct MALDI-TOF MS and its gene expression in worker, queen and drone heads. Insect Mol Biol. 2003;12:291–298. doi: 10.1046/j.1365-2583.2003.00414.x. [DOI] [PubMed] [Google Scholar]
- 30.Takeuchi H, et al. Prepro-tachykinin gene expression in the brain of the honeybee Apis mellifera. Cell Tissue Res. 2004;316:281–293. doi: 10.1007/s00441-004-0865-y. [DOI] [PubMed] [Google Scholar]
- 31.Hunt GJ, et al. Behavioral genomics of honeybee foraging and nest defense. Naturwissenschaften. 2007;94:247–267. doi: 10.1007/s00114-006-0183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pankiw T, Waddington KD, Page RE., Jr Modulation of sucrose response thresholds in honey bees (Apis mellifera L.): Influence of genotype, feeding, and foraging experience. J Comp Physiol A. 2001;187:293–301. doi: 10.1007/s003590100201. [DOI] [PubMed] [Google Scholar]
- 33.Waddington KD, Nelson CM, Page RE. Effects of pollen quality and genotype on the dance of foraging honey bees. Anim Behav. 1998;56:35–39. doi: 10.1006/anbe.1998.0736. [DOI] [PubMed] [Google Scholar]
- 34.Melcher C, Bader R, Walther S, Simakov O, Pankratz MJ. Neuromedin U and its putative Drosophila homolog hugin. PLoS Biol. 2006;4:e68. doi: 10.1371/journal.pbio.0040068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melcher C, Pankratz MJ. Candidate gustatory interneurons modulating feeding behavior in the Drosophila brain. PLoS Biol. 2005;3:e305. doi: 10.1371/journal.pbio.0030305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winther AM, Nassel DR. Intestinal peptides as circulating hormones: Release of tachykinin-related peptide from the locust and cockroach midgut. J Exp Biol. 2001;204:1269–1280. doi: 10.1242/jeb.204.7.1269. [DOI] [PubMed] [Google Scholar]
- 37.Pascual N, Maestro JL, Chiva C, Andreu D, Belles X. Identification of a tachykinin-related peptide with orexigenic properties in the German cockroach. Peptides. 2008;29:386–392. doi: 10.1016/j.peptides.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 38.Winther AM, Acebes A, Ferrus A. Tachykinin-related peptides modulate odor perception and locomotor activity in Drosophila. Mol Cell Neurosci. 2006;31:399–406. doi: 10.1016/j.mcn.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 39.Rafaeli A. Pheromone biosynthesis activating neuropeptide (PBAN): Regulatory role and mode of action. Gen Comp Endocrinol. 2008 Apr 18; doi: 10.1016/j.ygcen.2008.04.004. doi: 10.1016/j.ygcen.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 40.Thom C, Gilley DC, Hooper J, Esch HE. The scent of the waggle dance. PLoS Biol. 2007;5:e228. doi: 10.1371/journal.pbio.0050228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toth AL, Robinson GE. Evo-devo and the evolution of social behavior. Trends Genet. 2007;23:334–341. doi: 10.1016/j.tig.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 42.Nassel DR. Neuropeptides in the nervous system of Drosophila and other insects: Multiple roles as neuromodulators and neurohormones. Prog Neurobiol. 2002;68:1–84. doi: 10.1016/s0301-0082(02)00057-6. [DOI] [PubMed] [Google Scholar]
- 43.Nassel DR, Homberg U. Neuropeptides in interneurons of the insect brain. Cell Tissue Res. 2006;326:1–24. doi: 10.1007/s00441-006-0210-8. [DOI] [PubMed] [Google Scholar]
- 44.Wicher D. Metabolic regulation and behavior: How hunger produces arousal—an insect study. Endocr Metab Immune Disord Drug Targets. 2007;7:304–310. doi: 10.2174/187153007782794290. [DOI] [PubMed] [Google Scholar]
- 45.Giray T, Galindo-Cardona A, Oskay D. Octopamine influences honey bee foraging preference. J Insect Physiol. 2007;53:691–698. doi: 10.1016/j.jinsphys.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fewell JH, Bertram SM. Evidence for genetic variation in worker task performance by African and European honey bees. Behav Ecol Socobiol. 2002;52:318–325. [Google Scholar]
- 47.Che FY, Vathy I, Fricker LD. Quantitative peptidomics in mice: Effect of cocaine treatment. J Mol Neurosci. 2006;28:265–275. doi: 10.1385/JMN:28:3:265. [DOI] [PubMed] [Google Scholar]
- 48.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.