Abstract
Mitochondrial biogenesis occurs in response to increased cellular ATP demand. The mitochondrial electron transport chain requires molecular oxygen to produce ATP. Thus, increased ATP generation after mitochondrial biogenesis results in increased oxygen demand that must be matched by a corresponding increase in oxygen supply. We found that overexpression of peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α), which increases mitochondrial biogenesis in primary skeletal muscle cells, leads to increased expression of a cohort of genes known to be regulated by the dimeric hypoxia-inducible factor (HIF), a master regulator of the adaptive response to hypoxia. PGC-1α-dependent induction of HIF target genes under physiologic oxygen concentrations is not through transcriptional coactivation of HIF or up-regulation of HIF-1α mRNA but through HIF-1α protein stabilization. It occurs because of intracellular hypoxia as a result of increased oxygen consumption after mitochondrial biogenesis. Thus, we propose that at physiologic oxygen concentrations, PGC-1α is coupled to HIF signaling through the regulation of intracellular oxygen availability, allowing cells and tissues to match increased oxygen demand after mitochondrial biogenesis with increased oxygen supply.
Keywords: hypoxia, metabolism, mitochondria, transcription
In a range of physiologic states where increased tissue workload elevates ATP demand, cells respond by increasing mitochondrial mass. Mitochondrial biogenesis occurs in response to a number of environmental cues where energy requirements are increased, including skeletal muscle exercise (1), brown fat cold exposure (during adaptive thermogenesis) (2), and in the developing myocardium (3, 4). Mitochondria consume pyruvate to generate ATP, through the activity of the citric acid cycle and electron transport chain, the terminal enzyme of which (cytochrome oxidase), requires molecular oxygen. Under physiologic conditions, tissue oxygen levels are generally limiting. Therefore, increased mitochondrial activity results in elevated ATP only when accompanied by a corresponding increase in oxygen supply.
Peroxisome proliferator-activated receptor-γ (PPAR) coactivator 1α (PGC-1α) regulates mitochondrial biogenesis (5) in multiple cell types (6–9) through coactivation of key transcription factors including nuclear respiratory factors (NRF-1 and NRF-2) (5, 10), estrogen-related receptor-α (ERRα) (11, 12), Gabpa/b (5, 7), PPARs (13), and the thyroid hormone receptor (14). These transcription factors support mitochondrial biogenesis through the expression of genes encoding proteins involved in oxidative phosphorylation, fatty acid oxidation, heme biosynthesis, and mitochondrial protein import (15). The coordinated transcription of genes enhanced by PGC-1α supports a net increase in oxidative phosphorylation with an accompanying increase in oxygen demand.
PGC-1 α is enriched in skeletal muscle (7) and is induced in response to exercise (16–18). Although recent studies have identified an important physiological role of PGC-1α in skeletal muscle responses to exercise (19, 20) the underlying mechanisms remain unclear. To investigate these mechanisms, we performed gene expression profiling of primary human skeletal muscle cells overexpressing PGC-1α. We found that PGC-1α overexpression leads to increased expression of a cohort of genes previously shown to be strongly induced in hypoxia in a manner dependent on hypoxia inducible factor (HIF-1), a master regulator of hypoxia-dependent gene expression that consists of a HIF-1α/ARNT dimer (21, 22).
In hypoxia, HIF-1 initiates a program of gene expression that facilitates increased oxygen supply to overcome the initial hypoxic insult. HIF-1 binds to the core hypoxia response element (HRE) motif in target genes (22, 23). HIF-1α, the oxygen-sensitive subunit is constitutively expressed; however, it is continuously targeted for ubiquitination and degradation in a manner dependent on the hydroxylation of 2 proline residues within the oxygen-dependent degradation domain (ODDD) (24–26). The HIF hydroxylases responsible for this reaction require molecular oxygen and ferrous iron (Fe2+) and 2-oxoglutarate for activity. Upon hydroxylation, HIF-1α is targeted by the Von Hipple-Lindau (VHL) protein to ubiquitination and degradation (24, 27, 28). Hydroxylase inhibition in hypoxia leads to a rapid stabilization of HIF-1α with a subsequent increase in HIF-1 target gene expression.
In the current study, we show that PGC-1α-induced mitochondrial biogenesis results in increased oxygen consumption, leading to a decrease in intracellular oxygen availability to HIF-hydroxylases, resulting in stabilization of HIF-1α. We hypothesize that this HIF activation facilitates the elevated generation of ATP after mitochondrial biogenesis by matching the increased oxygen demand with an increase in oxygen supply.
Results
PGC-1α Activates HIF-1 Target Genes in Primary Human Skeletal Muscle Myotubes.
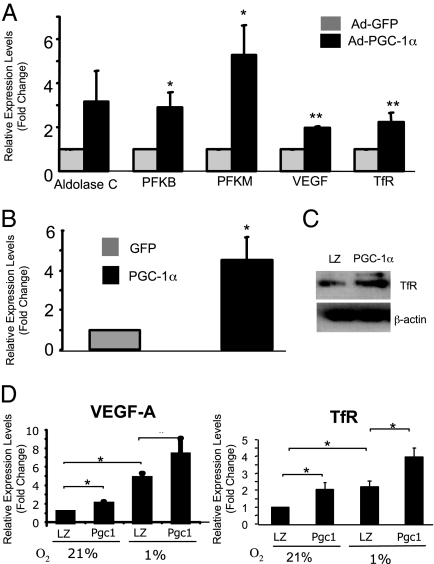
We used gene expression profiling to determine global transcriptional responses in primary differentiated human skeletal muscle myotubes infected with either an adenovirus expressing PGC-1α (Ad-PGC-1α) or a control adenovirus expressing GFP (Ad-GFP). PGC-1α overexpression resulted in elevated PGC-1α mRNA (99.6 ± 36.1-fold; n = 3, P < 0.01) and increased expression of a cohort of known PGC-1α target genes including genes involved in mitochondrial function (Table S1). We also identified a cohort of up-regulated genes previously shown to be regulated by HIF in response to cellular hypoxia. These included genes involved in glycolysis, angiogenesis, and iron transport (Table S2). Real-time PCR was performed to confirm the up-regulation of a selected group of these HIF-1 target genes including the transferrin receptor (TfR; Fig. 1A). The TfR provides the main route for iron uptake, which is a critical cofactor for a number of mitochondrial enzymes. For this reason, we selected the TfR for further investigations into the mechanisms by which PGC-1α regulates gene expression. We confirmed PGC-1α-dependent induction of TfR mRNA levels by real-time PCR in 2 skeletal muscle cell lines. First, differentiated C2C12 mouse skeletal muscle myotubes demonstrated a 4.5-fold increase in TfR mRNA when infected with Ad-PGC-1α (Fig. 1B). Next, we used rat skeletal muscle cells (L6) to generate a cell line stably overexpressing PGC-1α (leading to a 10-fold increase in PGC-1α mRNA) and found that these cells expressed TfR protein at higher levels than control cells (Fig. 1C). Furthermore, both VEGF-A and TfR mRNA were expressed at higher levels in PGC-1α-L6 cells than in control cells and this up-regulation was enhanced in cells cultured at 1% oxygen (Fig. 1D).
Fig. 1.
Identification of HIF target gene expression in PGC-1α-overexpressing cells. (A) RT-PCR of HIF target genes in primary human skeletal muscle cells infected with either Ad-GFP or Ad-PGC-1α. (B) RT-PCR of TfR mRNA in C2C12 myotubes infected with either Ad-GFP or Ad-PGC1α. (C) TfR protein expression in L6 myotubes stably expressing PGC-1α (L6-PGC-1α) compared with control L6-laczeo (LZ) cells. β-Actin was used as a loading control. (D) RT-PCR analysis of VEGF-A and TfR in L6-PGC-1α and L6-laczeo myotubes maintained at 21% or 1% atmospheric oxygen for 24 h. *, P < 0.05; **, P < 0.01; mean ± SEM; n = 3 throughout.
PGC-1α Activates the TfR in a HIF-Dependent Manner.
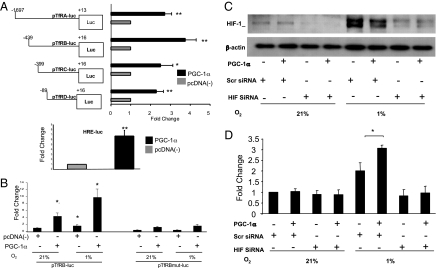
To examine whether PGC-1α induction of TfR mRNA is caused by activation of TfR transcription, we cotransfected a 1.7-kb TfR promoter luciferase reporter construct (pTfRA-luc) with a PGC-1α expression vector (or an empty vector) into C2C12 cells. Cotransfection of pTfRA-luc with PGC-1α resulted in a 2.7-fold increase in reporter activity relative to control (Fig. 2A). To identify the DNA elements that PGC-1α is acting through on the TfR promoter a series of promoter truncations were generated. PGC-1α-dependent activation of the TfR occurred with all of the deletion constructs, including the shortest construct containing the minimal active transcriptional region (refs. 29 and 30; Fig. 2A). Sequence analysis of the TfR promoter region revealed a consensus HRE that has been identified as a functional HIF binding site at position −90 to −83 relative to the transcription start site (31, 32). Having identified this HRE, we next tested whether PGC-1α overexpression increases HRE-luciferase reporter activity. PGC-1α overexpression in C2C12 cells resulted in increased HRE activity compared with controls (Fig. 2A Lower).
Fig. 2.
Role of HIF-1α in the transactivation of the TfR promoter in cells overexpressing PGC-1α. (A) (Upper) A TfR promoter-luciferase truncation reporter series was cotransfected with PGC-1α expression vector or control vector. Luciferase activity was measured. (Lower) HRE-luciferase activity was measured in C2C12 cotransfected with PGC-1α expression vector. (B) Wild-type (pTfRB-luc; Left) or HRE-mutant TfR reporter (pTfRBmut-luc; Right) constructs were cotransfected in C2C12 myoblasts with a PGC-1α expression vector or control vector. Luciferase expression was measured after 24-h exposure to 21% or 1% O2. (C) Immunoblot analysis demonstrates siRNA silencing of HIF1-α in HeLa cells exposed to hypoxia and cotransfected with PGC-1α expression vector or a control vector. (D) HIF-1α siRNA was used to reduce HIF-1α expression in HeLa cells. Cells were cotransfected with TfR luciferase reporter and PGC-1α expression vector or control vector. Luciferase expression was measured after exposure to 21% or 1% O2 for 24 h. *, P < 0.05; **, P < 0.01; mean ± SEM; n = 3 throughout.
To investigate the importance of this HRE in PGC-1α-induced TfR activation, a TfR promoter construct containing a TACGT to AATTC mutation in the HIF binding site was generated. Mutation at this site has been shown to inactivate this HRE (32). C2C12 cells were cotransfected with either a wild-type (pTfRB-luc) or mutant (pTfRBmut-luc) reporter construct and a PGC-1α expression vector or empty vector. At 21% O2, cells overexpressing PGC-1α demonstrated a significantly increased activity of the wild-type pTfRB-luc. Hypoxia resulted in a 1.5-fold increase in basal pTfRB-luc reporter activity. Similarly, the hydroxylase inhibitor dimethyloxallyl glycine (1 mM) increased TfR expression by 95 ± 41.2% (n = 3). Furthermore, C2C12 cells overexpressing PGC-1α responded to hypoxia with a 9.7-fold increase in wild-type pTfRB-luc reporter activity. Mutation of the HRE greatly attenuated enhanced luciferase expression by PGC-1α at 21% and 1% O2 (Fig. 2B). These data show that PGC-1α overexpression increases activation of the TfR promoter at 1% O2 in a manner at lease in part dependent on the presence of an intact HRE at positions −90 to −83.
We next investigated the role of HIF-1α, the transcription factor that binds to this site, by using siRNA that effectively depletes HIF-1α when compared with scrambled control siRNA (≈74% decrease in protein expression). In HeLa cells, at 1% O2 HIF is maximally stabilized and PGC-1α overexpression does not enhance this stabilization, furthermore HIF-1α siRNA abolishes HIF-1α expression in all cases (Fig. 2C). HeLa cells were transfected with the pTfRB-luc reporter construct and either a pcDNA-PGC-1α expression vector or an empty pcDNA control vector. Luciferase expression was measured after 24-h exposure to 21% or 1% O2. Unlike C2C12 cells, PGC-1α expression in HeLa cells did not activate luciferase expression at 21% O2 (likely because of different levels of basal oxygen consumption); however, hypoxia induced pTfRB-luc by ≈2-fold in HeLa cells transfected with control siRNA. Overexpression of PGC-1α potentiated this response. Silencing of HIF-1α abolished both hypoxia-induced pTfRB-luc activity and PGC-1α-dependent potentiation of this response (Fig. 2D). These data indicate that PGC-1α overexpression enhances TfR promoter activity at 1% O2 via HIF-1α-dependent mechanisms.
Using both DNA-binding assays and ChIP, we found no evidence that PGC-1α becomes physically associated with the HRE of the TfR promoter. We also discounted the possibility that overexpression of PGC-1α activates HIF-1α mRNA expression by using gene array and real-time PCR. Therefore, PGC-1α regulates HIF-1-dependent gene expression in a manner that is caused by PGC-1α acting as a direct coactivator of HIF-1 or by increasing HIF-1α mRNA expression.
PGC-1α Overexpression Decreases Intracellular O2 Levels.
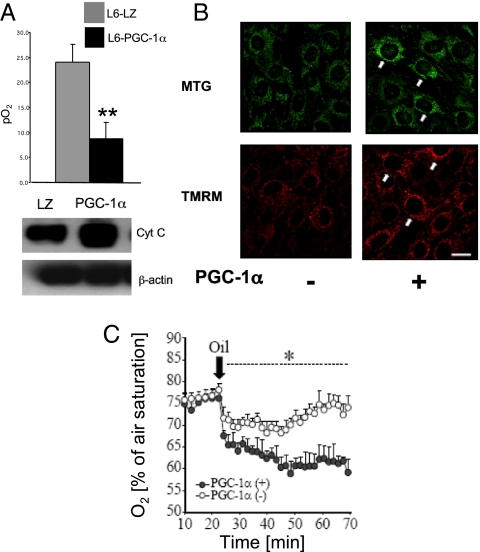
The HIF-1α protein is extremely oxygen labile because of prolyl hydroxylase (PHD)/VHL activity and proteosome-dependent degradation as outlined in the Introduction. Therefore, a possible mechanism by which PGC-1α could activate HIF-1α is via regulation of intracellular oxygen availability through increased mitochondrial biogenesis. It has been shown that inhibition of mitochondrial respiration by nitric oxide and other inhibitors of the respiratory chain results in redistribution of oxygen to nonmitochondrial targets such as PHD enzymes, resulting in destabilization of HIF-1α in hypoxia (33). Conversely, we hypothesized that an increase in mitochondrial respiration subsequent to PGC-1α-induced mitochondrial biogenesis may lead to a decrease in intracellular oxygen availability and the stabilization and accumulation of HIF-1α. To investigate whether PGC-1α expression results in decreased cellular O2 availability, we measured extracellular O2 concentration in L6 cells stably expressing PGC-1α by using fluorescence quenching oxymetry. To mimic tissue oxygen levels, this experiment was carried out at 10% atmospheric oxygen. L6-PGC-1α myotubes demonstrated a marked reduction in extracellular pO2 levels compared with control cells (Fig. 3A Upper), suggesting that these cells are consuming oxygen at a significantly greater rate than control cells. Increased mitochondrial mass in L6-PGC-1α cells was confirmed by immunoblot analysis of cellular levels of cytochrome c (Fig. 3A Lower).
Fig. 3.
Effects of PGC-1α overexpression on intracellular oxygen levels. (A) (Upper) O2 concentration in L6-PGC-1α myotubes was measured by fluorescence quenching oxymetry. (Lower). Increased mitochondrial mass in L6-PGC-1α myotubes was confirmed by immunoblot analysis of cellular levels of cytochrome c. (B) C2C12 cells were transfected with either PGC-1α expression vector or control vector, and mitochondrial mass and activity was imaged with MTG (Upper) and TMRM (Lower), respectively. (Bar: 20 μm.) (C) C2C12 cells were cotransfected with near-infrared (NIR) oxygen probes, and either PGC-1α expression vector or control vector and oxygen consumption was determined. *, P < 0.05; **, P < 0.01; mean ± SEM; n = 3 throughout.
We next confirmed increased mitochondrial mass in C2C12 cells overexpressing PGC-1α by independent quantitative fluorescence imaging of the mitochondrial markers tetramethylrhodamine methyl ester (TMRM) and mitotracker green (MTG). In each case cells overexpressing PGC-1α demonstrated increases in fluorescence, thus confirming mitochondrial biogenesis in PGC-1α-overexpressing cells (Fig. 3B). In separate experiments, C2C12 cells were cotransfected with an intracellular oxygen probe (34) and either a PGC-1α expression vector or an empty vector and intracellular oxygen levels were measured under oil (to prevent oxygen diffusion). Cells overexpressing PGC-1α had decreased intracellular oxygen levels, reflecting increased oxygen consumption compared with control cells (Fig. 3C). Similar results were obtained for the rat pheochromocytoma cell line PC-12 for which this protocol was originally developed (Fig. S1). In summary, these data suggest that PGC-1α overexpression leads to mitochondrial biogenesis and subsequently decreased intracellular oxygen levels in skeletal muscle cells.
PGC-1α Overexpression Results in HIF-1α Stabilization and Decreased PHD Activity.
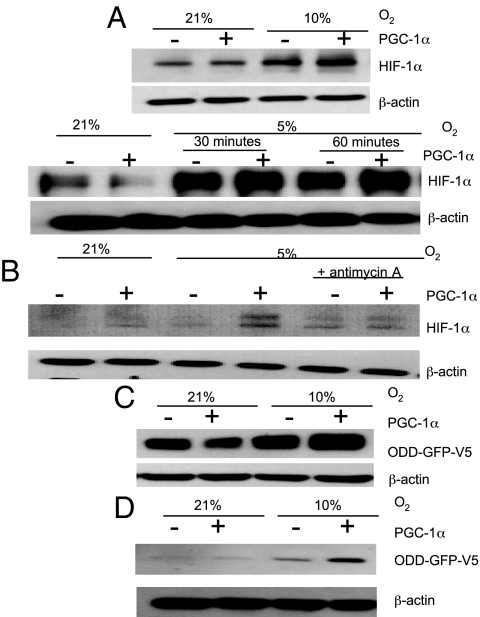
The data presented thus far led us to hypothesize that PGC-1α-dependent increases in mitochondrial oxygen consumption results in a decrease in intracellular oxygen availability for nonrespiratory dioxygenases such as HIF hydroxylases. To test this hypothesis, HeLa cells expressing PGC-1α were exposed to either 21%, 10%, or 5% atmospheric O2 for 30 min. Immunoblot analysis revealed no increase in HIF-1α levels in HeLa cells overexpressing PGC-1α at 21% O2 where oxygen availability is not likely to be a limiting factor. However, at 10% and 5% atmospheric O2 (which represent intermediate levels of tissue oxygen where HIF is not maximally activated; see Fig. S2), PGC-1α overexpression resulted in increased HIF-1α levels when compared with control cells (Fig. 4A). Importantly, inhibition of mitochondrial respiration with antimycin A reversed this effect (Fig. 4B). These data suggest that when oxygen is limited PGC-1α-induced increases in oxygen consumption results in HIF-1α stabilization.
Fig. 4.
Effect of PGC-1α overexpression on HIF-1α stabilization and PHD activity. (A) Immunoblot analysis of HIF-1α in HeLa cells transfected with either PGC-1α expression vector or control vector at either 21% and 10% atmospheric O2 (Upper) or 21% and 5% atmospheric O2 (Lower). (B) HeLa cells were exposed to 21% and 5% oxygen with or without PGC-1α overepression and antimycin A (1 μg/mL), and HIF-1α levels were determined by Western blot analysis. (C) Immunoblot analysis of ODD-GFP-V5 expression in HeLa cells transfected with either PGC-1α expression vector or control vector at either 21% or 10% atmospheric O2. (D) Immunoblot analysis of ODD-GFP-V5 expression in C2C12 cells transfected with either PGC-1α expression vector or control vector in 21% or 10% O2. n = 3 throughout.
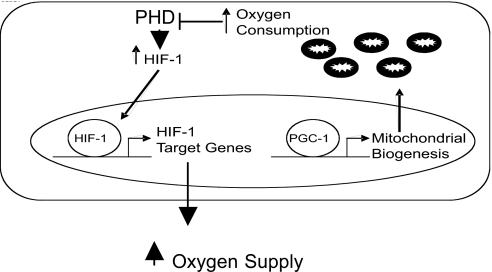
To investigate whether the increased HIF-1α in PGC-1α-overexpressing cells is a result of inhibition of the oxygen-dependent HIF hydroxylases, we used a construct containing amino acid residues 521–652 of HIF-1, which comprises the ODD of HIF-1α, fused to a V5-tagged GFP (ODD-GFP-V5). The ODD of HIF-1α confers hydroxylase-dependent oxygen sensitivity to the V5-tagged GFP protein (33). HeLa cells were transfected with an ODD-GFP-V5 expression vector and either a PGC-1α expression vector or an empty vector. Cells were then exposed to either 21% or 10% atmospheric O2 for 30 min. Immunoblot analysis of ODD-GFP-V5 expression demonstrated that, like endogenous HIF-1α levels, ODD-GFP-V5 expression is increased with PGC-1α at 10% oxygen, indicating that PGC-1α-induced mitochondrial biogenesis results in the inhibition of HIF hydroxylases when oxygen is limited (Fig. 4C). We carried out similar experiments in C2C12 cells and found the same results (Fig. 4D). Interestingly, PGC-1α expression in C2C12 cells was sufficient to stabilize ODD-GFP-V5 protein levels at 21% O2. However, a further increase in ODD-GFP-V5 protein was seen in cells overexpressing PGC-1α exposed to 10% O2 for 30 min. Importantly, PGC-1α overexpression did not significantly alter PHD mRNA expression levels in primary human skeletal muscle cells (PHD1 fold change = 0.94 ± 0.22, P = 0.18; PHD2 fold change = 1.00 ± 0.08, P = 0.48; PHD3 fold change = 1.01 ± 0.17, P = 0.47) or protein levels in HeLa or C2C12 cells (Fig. S3). Furthermore, hypoxia did not alter PGC-1α expression levels, indicating that these potentially alternative mechanisms are not mediating the effects of PGC-1α overexpression on HIF or the ODD-GFP construct (Fig. S3). These data suggest that PGC-1α overexpression results in HIF-1α stabilization, which is caused by inhibition of hydroxylation of the ODD of HIF-1α. Taken together, the results of this study indicate that PGC-1α-induced increases in oxygen consumption at physiologic oxygen levels results in decreased intracellular oxygen availability with the subsequent stabilization/transactivation of HIF-1α and resultant expression of HIF-1-dependent target genes (Fig. 5).
Fig. 5.
Schematic representation of HIF target gene expression by PGC-1α. PGC-1α expression results in mitochondrial biogenesis and increased oxygen consumption. The resultant drop in intracellular pO2 levels leads to decreased HIF hydroxylase activity and HIF stabilization with resultant gene expression that facilitates an increase in oxygen supply.
Discussion
In a number of physiologic conditions, increased cellular ATP demand is facilitated by mitochondrial biogenesis. A central regulator of the expression of mitochondrial genes during biogenesis is PGC-1α (5). Because mitochondria require oxygen to produce ATP, an increase in oxygen consumption occurs to support increased oxidative metabolism after mitochondrial biogenesis. In the current study, we demonstrate that PGC-1α expression in primary human skeletal muscle cells activates the expression of HIF-1 target genes and provide evidence that it is the increased consumption of oxygen caused by PGC-1α-dependent mitochondrial biogenesis that leads to HIF-1α stabilization in skeletal muscle cells. A consequence of HIF-1α stabilization in vivo is elevated tissue oxygen supply that would facilitate the increased level of oxidative phosphorylation occurring as a result of mitochondrial biogenesis. We show that physiologically relevant increases in O2 consumption can lead to HIF-1α stabilization and activity.
A number of studies have demonstrated that inhibition of mitochondrial function in cells exposed to low oxygen levels (1–3%) reverses hypoxia-induced HIF-1 activity (35–38), suggesting an important role of mitochondria in oxygen sensing. Hagen et al. (33) demonstrated that inhibition of mitochondrial respiration during hypoxia results in a destabilization of HIF-1α. Furthermore, that study showed that mitochondrial inhibition in hypoxia results in a redistribution of oxygen from the mitochondria to the oxygen-dependent prolyl hydroxylases located elsewhere in the cell. Our current data provide evidence that the converse is also true, i.e., that activation of mitochondrial activity, through the physiological activator PGC-1α, results in stabilization of HIF-1α. A critical issue is that increasing mitochondrial activity will only impact nonmitochondrial oxygen-dependent enzymes when oxygen is limiting but not unavailable. When a system is anoxic, differences in mitochondrial activity will not impact on oxygen availability because there is no oxygen to redistribute to mitochondria. However, in the physiologic range, where oxygen is limiting and the constraints on nonmitochondrial oxygen availability are determined by mitochondrial respiration, an increase in respiratory activity will impact cellular oxygen availability to nonmitochondrial dioxygenases.
Interestingly, a recent study (39) has demonstrated that PGC-1α can activate VEGF expression by binding and coactivating ERRα at conserved sites on the VEGF promoter in a manner independent of HIF-1α. Stabilization of HIF-1α in primary skeletal muscle cells infected with a PGC-1α adenovirus was not detected in these experiments. However, HIF-1α activity was examined at near anoxic conditions (0.2% oxygen). For the reason stated above, one would not expect to see a HIF-dependent role at these extremely low oxygen concentrations. In addition to demonstrating this HIF-independent role of PGC-1α, we provide evidence for a HIF-dependent role for PGC-1α in the regulation of genes involved in increasing tissue oxygen supply at higher oxygen concentrations.
In vivo, the activation of HIF-1 would result in enhanced oxygen delivery to sustain augmented mitochondrial activity in PGC-1α-expressing cells. HIF-1 plays a well-established role in the induction of factors involved in angiogenesis and erythropoesis, resulting in increased vessel formation, heme synthesis, and red blood cell production. The net effect of these actions will result in optimal blood flow and oxygen supply to tissues with limiting oxygen supply. Here, we provide evidence that in skeletal muscle HIF-1 may be coupled to mitochondrial biogenesis through activation of TfR and increased cellular uptake of iron in response to increased oxygen demand, while at the same time facilitating oxygen delivery through VEGF expression that may be both PGC-1α dependent and independent.
Increased oxidative phosphorylation occurring as a result of mitochondrial biogenesis is not without its consequences for the cell. Oxidative phosphorylation results in the generation of mitochondrial reactive oxygen species (ROS). One mechanism in which this may be counteracted is by HIF regulation of mitochondrial activity. HIF-1 activates the expression of COX4–2 subunit and a protease involved in COX4–1 degradation in hypoxia, resulting in increased efficiency in electron transfer to molecular oxygen, minimizing ROS production (40). HIF also increases PDK1 expression, a negative regulator of the citric acid cycle, which in turn will lead to a decrease in ROS production (41, 42). Thus, it is possible that a second function of PGC-1α-dependent HIF-1 activation is the control of mitochondrial ROS production after mitochondrial biogenesis, complementing PGC-1α activation of genes encoding ROS-detoxifying enzymes (43). Furthermore, a recently described role for HIF as a possible negative regulator of mitochondrial biogenesis (44) may represent a negative feedback loop to prevent excessive mitochondrial accumulation.
PGC-1α plays an important role in the skeletal muscle response to exercise. PGC-1α expression is robustly induced by exercise (16, 17), resulting in mitochondrial biogenesis, a switch from glycolytic fibers to increased mitochondrial-rich oxidative fibers and increased glycogen deposition (19, 20). This PGC-1α-induced mitochondrial biogenesis prepares the muscle for the next bout of exercise, with an increased oxidative phosphorylation capacity, resulting in greater capacity for endurance training. HIF-1 α protein levels also increase markedly in skeletal muscle in response to a single bout of exercise (45), and the effect of PGC-1α on HIF-1 that we describe here may prolong the activity of HIF-1 allowing for a sustained adaptive response to exercise. Finally, skeletal muscle-specific HIF-1α knock out mice demonstrate that the increased expression of HIF target genes normally observed in response to exercise is HIF-1 dependent, suggesting a role for HIF in the adaptation of skeletal muscle in response to exercise (46). We propose that PGC-1α activation in skeletal muscle cells leads to increased oxygen consumption secondary to increased mitochondrial mass (Fig. 5). The resultant drop in intracellular pO2 leads to HIF activation that in turn increases iron delivery and tissue perfusion by increasing the expression of the TfR and angiogenic and vasoactive factors. Thus, increased perfusion will facilitate the increased oxygen requirements of the mitochondrial biogenesis that initiated the initial drop in oxygen concentration. In parallel, increased HIF-1 target gene expression may protect cells undergoing mitochondrial biogenesis from oxidative damage by decreasing mitochondrial respiration. In summary, this study along with other recent developments place the interaction of PGC-1α and HIF-1α as central in the skeletal muscle response to increases in oxygen consumption during exercise.
Materials and Methods
Tissue Culture.
HeLa cells were cultured in MEM. C2C12 myoblasts were cultured in DMEM with 10% FBS. L6-PGC-1α myoblasts were grown in α-MEM containing hygromycin (12.5 μg/mL). L6-LZ cells were grown in α-MEM zeocin (25 μg/mL). For myotube formation, C2C12 L6 myoblasts were cultured in DMEM or α-MEM containing 2% horse serum (Lonza) for 96 h. Primary human skeletal muscle myoblasts were grown in skeletal muscle cell growth medium and differentiated in skeletal muscle cell differentiation medium (Promocell) for 13 days. For hypoxia studies, cell culture dishes were maintained in a humidified hypoxia workstation (Coy Laboratories) at the indicated oxygen concentration with 5% CO2 and the balance N2. Extracellular pO2 measurements were made by fluorescence quenching oxymetry (oxylite-2000; Oxford Optronix).
Adenoviral Infections.
Recombinant Ad-GFP alone or PGC-1α plus eGFP (Ad-Pgc1α) were gifts from B. Lavan (Metabolex, Hayward, CA). Cells were infected at a multiplicity of infection sufficient to infect >95% of cells based on GFP fluorescence. Primary human skeletal muscle and C2C12 cells were analyzed after 96 and 72 h, respectively.
Stable Transfection of L6 Cells.
L6-Glut4myc cells (a generous gift from A. Klip, University of Toronto, Toronto) were converted to stable Flp-In host cells (L6-lacZeo) by transfection with pFRT/lacZeo2 (Invitrogen). L6-lacZeo cells were subsequently cotransfected with pOG44 and pEF5/FRT/V5-Dest containing full-length human PGC-1α with a stop codon to prevent V5 tagging of PGC-1α, at a 9:1 ratio. Stably-transfected L6-PGC-1α cells were selected for hygromycin resistance.
RNA Isolation and RT-PCR.
Total RNA was extracted by using RNeasy mini elute columns (Qiagen). One microgram of total RNA was reverse-transcribed by using random primers (Invitrogen) and SuperScript II (Invitrogen). RT-PCR was performed on an Applied Biosystems 7900HT fast real-time PCR system using primers (Table S3). Results were normalized to 18s expression.
Microarray Analysis.
One microgram of total RNA was labeled according to the Gene Chip Whole Transcript Assay as provided by manufacturer (Affymetrix) and hybridized to Human Exon 1.0ST Arrays (Affymetrix) overnight before scanning in an Affymetrix GCS3000 7G scanner.
Whole-Cell Protein Extract Preparation and Immunoblot Analysis.
Whole-cell extracts were prepared in whole-cell lysis buffer [150 mM NaCL/25 mM Tris·HCl, pH 8.0./1 mM EDTA/1% Nonidet P-40 and 1× protease inhibitor mixture (Sigma)]. Protein content was quantified and normalized by using the Bradford method (Bio-Rad) and separated by electrophoresis on SDS/PAGE gels. TfR mAb (Zymed), HIF-1α mAb (BD Transduction Laboratories), V5 mAb (Invitrogen), monoclonal N-terminal GFP antibody (Sigma), monoclonal cytochrome c antibody (BD Biosciences,) and monoclonal β-actin (Sigma) were used. For mitochondrial inhibition, HeLa cells were treated with 1 μg/mL with Antimycin A for 60 min.
Luciferase Reporter Assays and Expression Constructs.
The TfR promoter (pTfRA-Luc) in a promoterless luciferase reporter plasmid, pGL2-basic, and a pGL2-basic vector containing a 455-bp fragment of the TfR promotor (pTfRB-Luc) was kindly provided by Gaetano Cairo (Istituto Patologia Generale, Milan). The pTfR-Luc gene truncation series was generated by PCR amplification from the pTfRB-Luc construct, The following 5′ primers were used: 5′-AACAGATTGGACCTAGCACTG-3′ (pTfRC-luc), 5′- TACGTGCCTCAGGAAGTG-3′ (pTfRD-luc). The same 3′ primer was used for both constructs: 5′-CTGATATCCCGACGCTCT. Site-directed mutagenesis was performed by using the Quik Change Kit (Stratagene) complementary oligonucleotides 5′-CAGAGCACCTCGCGAGCGaattcGCCTCAG G A AGTGACGC 5′-GCGTCACTTCTTCCTGAGGCGaattcGCTCGCGAGGTGCTCTG. PGC-1α was subcloned into pDONR207 by using the gateway BP clonase in vitro recombination reaction (Invitrogen) after PCR amplification from a human skeletal muscle cDNA library, using forward primer 5′-GGGG-attB1-ATGGCGTGGGACATGTGCAACCAG-3′ and reverse primer 5′-GGGG-attB2-TTACCTGCGCAAGCTTCTCTGAGC-3′ and shuttled into pcDNA3.1/Nv5-DEST expression vector by using the gateway LR in vitro recombination reaction (Invitrogen). pcDNA3-ODD-GFP-V5 has been described (33). For RNA interference experiments, siRNA against HIF-1α and a nonspecific control siRNA (Dharmacon) were added at a final concentration of 50 pmol/mL. At 80% confluence, cells were transfected with reporter plasmids (0.5 μg), expression vectors (0.5 μg), and control β-galactosidase vector (0.1 μg) per well of a 24-well plate by using fugen-6 (Roche). Expression of reporter gene activity was normalized to β-galactosidase levels by using a β-galactosidase enzyme assay system (Promega).
Oxygen Probe Analysis.
At 50% confluence, C2C12 cells were transfected with expression vectors (0.1 μg per well of 96-well plate) with Lipofectamine 2000. On day 1 of differentiation, cells were transfected with 1.2 μM MitoXpress O2 probe (Luxcell Biosciences)/6 μM Endo-Porter (Gene Tools) as described (34). Cells were washed and analyzed on a fluorescent plate reader in 100 μL of RPMI medium 1640 without serum or phenol red. To minimize contribution of glycolysis to ATP production, C2C12 cells were preincubated for 2 h in serum-free RPMI medium 1640 containing 10 mM galactose and 1 mM pyruvate. To prevent oxygen diffusion, experimental wells were covered in mineral oil.
Live Cell Monitoring of Mitochondrial Membrane Potential.
C2C12 cells were transfected at 50% confluence on glass-bottom dishes. On day 2 of differentiation, cells were loaded with either 50 nM MTG or 20 nM TMRM for 30 min at 37 °C (in the dark) in serum-free DMEM. The cells were washed twice and monitored in serum-free DMEM by using an Olympus FU1000 laser scanning confocal microscope. MTG and TMRM were maintained during a whole experiment at 10 nM and 20 nm, respectively. MTG and TMRM were excited at 488 nM and 543 nm with an argon laser (5% maximum power) or a helium neon laser (1.5%), respectively, and the emission was collected through a 500- to 550-nm filter (MTG) or 550- to 600-nm filter (TMRM) with photomultiplier adjusted to 555 mV. Fluorescence and differential interference contrast (DIC) images were collected through an oil-immersion 60× objective in 2 (for PC12) or 5 (C2C12) planes. Images were analyzed by using FV1000 Viewer software (Olympus) and Adobe Photoshop.
Statistical Analysis.
All experiments were carried out with a minimum of n = 3. Intergroup comparisons were made by Student's t test with P < 0.05 considered statistically significant.
Supplementary Material
Acknowledgments.
We thank A. Murphy for assistance with the microarray analysis, Gaetano Cairo for the full-length TfR luciferase construct, and B. Lavan for recombinant adenoviruses. This work was supported by Science Foundation Ireland.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808801106/DCSupplemental.
References
- 1.Holloszy JO, Coyle EF. Adaptations of skeletal muscle to endurance exercise and their metabolic consequences. J Appl Physiol. 1984;56:831–838. doi: 10.1152/jappl.1984.56.4.831. [DOI] [PubMed] [Google Scholar]
- 2.Nicholls DG, Locke RM. Thermogenic mechanisms in brown fat. Physiol Rev. 1984;64:1–64. doi: 10.1152/physrev.1984.64.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Attardi G, Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- 4.Mayor F, Cuezva JM. Hormonal and metabolic changes in the perinatal period. Biol Neonate. 1985;48:185–196. doi: 10.1159/000242171. [DOI] [PubMed] [Google Scholar]
- 5.Wu Z, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 6.Lehman J, et al. Peroxisome proliferator-activated receptor γ coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin J, et al. Transcriptional coactivator PGC-1α drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 8.St-Pierre J, et al. Bioenergetic analysis of peroxisome proliferator-activated receptor γ coactivators 1α and 1β (PGC-1α and PGC-1β) in muscle cells. J Biol Chem. 2003;278:26597–26603. doi: 10.1074/jbc.M301850200. [DOI] [PubMed] [Google Scholar]
- 9.Uldry M, et al. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 10.Gleyzer N, Vercauteren K, Scarpulla RC. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol Cell Biol. 2005;25:1354–1366. doi: 10.1128/MCB.25.4.1354-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huss JM, Kopp RP, Kelly DP. Peroxisome proliferator-activated receptor coactivator-1α (PGC-1α) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-α and -γ. Identification of novel leucine-rich interaction motif within PGC-1α. J Biol Chem. 2002;277:40265–40274. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- 12.Schreiber SN, Knutti D, Brogli K, Uhlmann T, Kralli A. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor α (ERRα) J Biol Chem. 2003;278:9013–9018. doi: 10.1074/jbc.M212923200. [DOI] [PubMed] [Google Scholar]
- 13.Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor α in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puigserver P, et al. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 15.Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 16.Baar K, et al. Adaptations of skeletal muscle to exercise: Rapid increase in the transcriptional coactivator PGC-1. FASEB J. 2002;16:1879–1886. doi: 10.1096/fj.02-0367com. [DOI] [PubMed] [Google Scholar]
- 17.Terada S, et al. Effects of low-intensity prolonged exercise on PGC-1 mRNA expression in rat epitrochlearis muscle. Biochem Biophys Res Commun. 2002;296:350–354. doi: 10.1016/s0006-291x(02)00881-1. [DOI] [PubMed] [Google Scholar]
- 18.Pilegaard H, Saltin B, Neufer PD. Exercise induces transient transcriptional activation of the PGC-1α gene in human skeletal muscle. J Physiol (London) 2003;546:851–858. doi: 10.1113/jphysiol.2002.034850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wende AR, et al. A role for the transcriptional coactivator PGC-1α in muscle refueling. J Biol Chem. 2007;282:36642–36651. doi: 10.1074/jbc.M707006200. [DOI] [PubMed] [Google Scholar]
- 20.Handschin C, et al. Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1α muscle-specific knockout animals. J Biol Chem. 2007;282:30014–30021. doi: 10.1074/jbc.M704817200. [DOI] [PubMed] [Google Scholar]
- 21.Semenza GL, Wang GL. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 1992;12:5447–5454. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic helix–loop–helix–PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang GL, Semenza GL. Characterization of hypoxia-inducible factor 1 and regulation of DNA binding activity by hypoxia. J Biol Chem. 1993;268:21513–21518. [PubMed] [Google Scholar]
- 24.Ivan M, et al. HIFα targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 25.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 26.Epstein AC, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 27.Jaakkola P, et al. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 28.Maxwell PH, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 29.Ouyang Q, Bommakanti M, Miskimins WK. A mitogen-responsive promoter region that is synergistically activated through multiple signaling pathways. Mol Cell Biol. 1993;13:1796–1804. doi: 10.1128/mcb.13.3.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Owen D, Kuhn LC. Noncoding 3′ sequences of the transferrin receptor gene are required for mRNA regulation by iron. EMBO J. 1987;6:1287–1293. doi: 10.1002/j.1460-2075.1987.tb02366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lok CN, Ponka P. Identification of a hypoxia response element in the transferrin receptor gene. J Biol Chem. 1999;274:24147–24152. doi: 10.1074/jbc.274.34.24147. [DOI] [PubMed] [Google Scholar]
- 32.Tacchini L, Bianchi L, Bernelli-Zazzera A, Cairo G. Transferrin receptor induction by hypoxia. HIF-1-mediated transcriptional activation and cell-specific posttranscriptional regulation. J Biol Chem. 1999;274:24142–24146. doi: 10.1074/jbc.274.34.24142. [DOI] [PubMed] [Google Scholar]
- 33.Hagen T, Taylor CT, Lam F, Moncada S. Redistribution of intracellular oxygen in hypoxia by nitric oxide: Effect on HIF1α. Science. 2003;302:1975–1978. doi: 10.1126/science.1088805. [DOI] [PubMed] [Google Scholar]
- 34.Zhdanov AV, Ward MW, Prehn JH, Papkovsky DB. Dynamics of intracellular oxygen in PC12 cells upon stimulation of neurotransmission. J Biol Chem. 2008;283:5650–5661. doi: 10.1074/jbc.M706439200. [DOI] [PubMed] [Google Scholar]
- 35.Brunelle JK, et al. Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metab. 2005;1:409–414. doi: 10.1016/j.cmet.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 36.Guzy RD, et al. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005;1:401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 37.Mansfield KD, et al. Mitochondrial dysfunction resulting from loss of cytochrome c impairs cellular oxygen sensing and hypoxic HIF-α activation. Cell Metab. 2005;1:393–399. doi: 10.1016/j.cmet.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon MC. Mitochondrial reactive oxygen species are required for hypoxic HIFα stabilization. Adv Exp Med Biol. 2006;588:165–170. doi: 10.1007/978-0-387-34817-9_15. [DOI] [PubMed] [Google Scholar]
- 39.Arany Z, et al. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1α. Nature. 2008;451:1008–1012. doi: 10.1038/nature06613. [DOI] [PubMed] [Google Scholar]
- 40.Fukuda R, et al. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129:111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 41.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 42.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively down-regulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 43.St-Pierre J, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 44.Mason SD, et al. HIF-1α in endurance training: Suppression of oxidative metabolism. Am J Physiol. 2007;293:R2059–R2069. doi: 10.1152/ajpregu.00335.2007. [DOI] [PubMed] [Google Scholar]
- 45.Ameln H, et al. Physiological activation of hypoxia inducible factor-1 in human skeletal muscle. FASEB J. 2005;19:1009–1011. doi: 10.1096/fj.04-2304fje. [DOI] [PubMed] [Google Scholar]
- 46.Mason SD, et al. Loss of skeletal muscle HIF-1α results in altered exercise endurance. PLoS Biol. 2004;2:e288. doi: 10.1371/journal.pbio.0020288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.