Abstract
Purpose
Three previous studies have tested for an association between high myopia and polymorphisms in the open angle glaucoma gene, myocilin (MYOC), all in subjects of Chinese ethnicity. In two of the studies, a significant association was found while in the third, there was no association. We sought to investigate the association between high myopia and polymorphisms in MYOC in subjects of European ethnicity.
Methods
Subjects were recruited from two sites, Cardiff University in the UK and Duke University in the United States. The Cardiff University cohort was comprised of 164 families with high myopia (604 subjects) plus 112 unrelated, highly myopic cases and 114 emmetropic controls. The Duke University cohort was comprised of 87 families with high myopia (362 subjects) plus 59 unrelated, highly myopic cases. Subject DNA was genotyped with a panel of MYOC single nucleotide polymorphisms (SNPs) including those found previously associated with high myopia. The Cardiff cohort was also genotyped for two flanking microsatellite markers analyzed in prior studies. Association between high myopia and MYOC polymorphisms was assessed using the Unphased program.
Results
Since there was no evidence of heterogeneity in genotype frequencies between families and singleton samples or between cohorts, both subject groups (families and unrelated subjects) from both recruitment sites were analyzed jointly for those SNPs genotyped in common. Two variants showed significant association before correction for multiple testing. These two variants were rs16864720 (p=0.043) and NGA17 (p=0.026). However, there was no significant association after Bonferroni correction. The estimated relative risk (RR) conferred by each of the MYOC variants was low (RR<1.5).
Conclusions
Our results suggest that MYOC polymorphisms have a very low, or possibly negligible, influence on high myopia susceptibility in subjects of European ethnicity.
Introduction
Myopia is a common cause of visual impairment throughout the world, and its prevalence is increasing [1-3]. The World Health Organization has listed myopia among the leading five causes of blindness [4]. Currently, there is no effective treatment to arrest myopia progression [5]. As myopia is highly heritable [6,7], the identification of genetic variants that confer susceptibility to the condition is likely to further our understanding of its pathophysiology and may make it possible to design rational therapies to thwart myopia progression.
Several highly penetrant genetic loci for non-syndromic myopia have been mapped [8]. However, none of the causative mutations has yet been found. Candidate gene association studies have led to the identification of several high myopia susceptibility genes (Table 1) including the myocilin gene (MYOC) on chromosome 1. Nonetheless, replication of these findings is necessary to separate true positives from false positives.
Table 1. High myopia susceptibility genes.
| Gene | Locus | Reference |
|---|---|---|
| Myocilin (MYOC) |
1q23 |
[17] |
| Hepatocyte growth factor (HGF) |
7q21 |
[51] |
| Paired box gene 6 (PAX6) |
11p13 |
[52] |
| Collagen, Type II alpha 1 (COL2A1) |
12q13 |
[42] |
| Lumican (LUM) |
12q21 |
[53] |
| Collagen, Type I alpha 1 (COL1A1) |
17q21 |
[54] |
| Transforming growth induced factor (TGIF) |
18p11 |
[55] |
| Transforming growth factor beta 1 (TGFB1) | 19q13 | [24] |
MYOC is best known for its role in glaucoma. Mutations in MYOC can cause both juvenile-onset and adult-onset open-angle glaucoma [9,10]. MYOC consists of three exons, and it has been shown that an upstream stimulatory factor is critical for its basal promoter activity [11]. Myocilin (also known as trabecular meshwork inducible glucocorticoid response or TIGR), the protein product of MYOC, was discovered during studies examining proteins that could be induced upon long-term treatment of human trabecular meshwork cells (TMC) with glucocorticoids [12]. In the human eye, myocilin is highly expressed in the TMC, sclera, ciliary body, and iris with considerably lower amounts in the retina and optic nerve head. The secreted protein is present in the aqueous humor [11]. Aside from glucocorticoid stimulation, the expression of myocilin in TMC is affected by the transcription protein transforming growth factor β (TGF β), mechanical stretch, basic fibroblast growth factor (bFGF), and oxidative stress [11,13,14]. Experimental studies show that mutant myocilin isoforms found in patients with juvenile-onset glaucoma are not secreted but accumulate in the TMC where they are thought to interfere with cell functions. For example, mutant myocilin disturbs the mitochondrial membrane potential [15]. Despite intensive research efforts, however, the precise role of MYOC mutations in glaucoma is unclear.
In addition to glaucomatous involvement, genetic variants in MYOC have also been implicated in causing susceptibility to high myopia [16,17]. This involvement would be consistent with the increased frequency of myopia in patients with open-angle glaucoma [18-20], the observation (though only in a proportion of studies) that intraocular pressure (IOP) is higher in myopes than in emmetropes [21], and the identification of significant genetic linkage close to the MYOC locus on chromosome 1 in families with myopia from the Beaver Dam Eye Study [22]. It is also noteworthy that some factors that stimulate myocilin expression in TMC have also been implicated in the regulation of postnatal eye growth and myopia, e.g., bFGF, TGF β, and oxidative mitochondrial pathways [23-25].
Association between MYOC polymorphisms and high myopia was first reported in a case-control study of Chinese subjects from Singapore [16]. An initial attempt to replicate this finding using a similar case-control design in Hong Kong Chinese subjects, however, did not support the association [26]. Later, a larger, family based association study also in Chinese subjects from Hong Kong yielded a significant result [17]. In this latter study, association was found with two microsatellite polymorphisms (NGA17 at the promoter region and NGA19 at the 3′ region) and two single nucleotide polymorphisms (SNPs; rs2421853 and rs235858 at the 3′ flanking region). Herein, association between myocilin polymorphisms and high myopia was examined in two independent Caucasian subject groups.
Methods
Subjects
This research followed the principles of the Declaration of Helsinki. Signed, informed consents were obtained from all participants. The number of subjects participating in the study is shown in Table 2.
Table 2. Number of subjects in the study.
| Subject group |
Subjects (families) participating |
Subjects (families) analyzed |
||
|---|---|---|---|---|
| Cardiff University | Duke University | Cardiff University | Duke University | |
| Related |
604 (164) |
358 (86) |
551 (142) |
358 (86) |
| Cases |
112 |
56 |
121 |
56 |
| Controls |
114 |
0 |
116 |
0 |
| Total | 830 | 414 | 788 | 414 |
Note that subjects for whom all relatives were excluded were reclassified as cases or controls if they met the necessary refractive criteria.
Cardiff University (UK) cohort
The cohort comprised of 164 families with high myopia (604 subjects) along with an additional set of unrelated individuals comprised of 112 highly myopic cases and 114 “emmetropic” controls (spherical equivalent refractive error in both eyes >−1.00 D and <+1.00 D). Subjective refraction details were obtained from the subjects’ optometrists. DNA was extracted from saline mouthwashes and mailed to our laboratory as previously described [27]. Individuals with known syndromic disorders or a systemic condition that could predispose them to myopia were excluded. All subjects were of Caucasian ethnicity (self-reported “White Europeans”). Ethical approval for the study was granted by the Cardiff University Human Sciences Research Ethics committee (Cardiff, Wales).
Duke University Center for Human Genetics (USA) cohort
The cohort comprised of 86 families with high myopia (358 subjects) along with an additional set of unrelated individuals comprising of 56 highly myopic cases. All subjects underwent a complete ophthalmic examination, and individuals with syndromic conditions that could predispose them to myopia were excluded. Genomic DNA was extracted from venous blood using the AutoPure LS® DNA Extractor and PUREGENE™ reagents (Gentra Systems Inc., Minneapolis, MN). The study was approved by the Institutional Review Board at the Duke University Medical Center (Durham, NC).
Molecular genetics
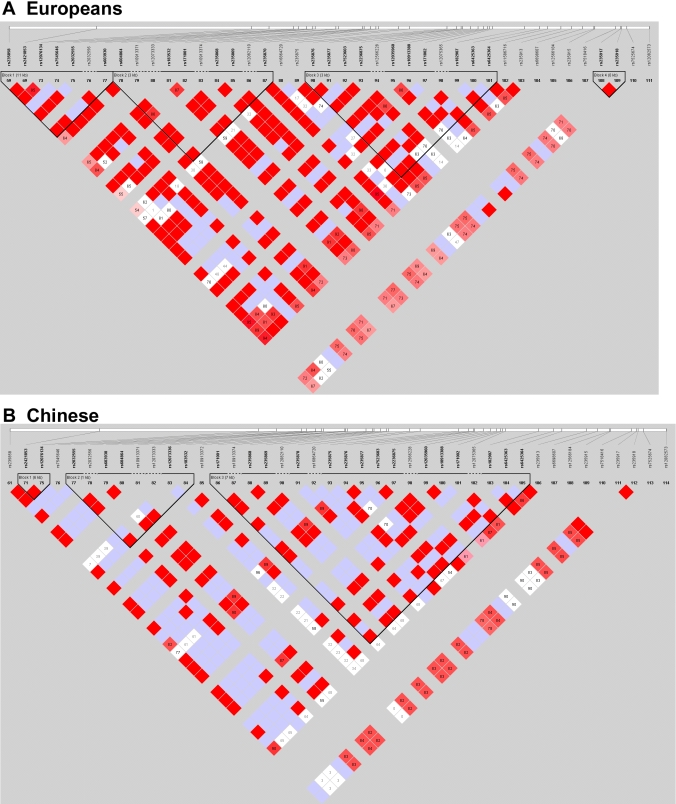
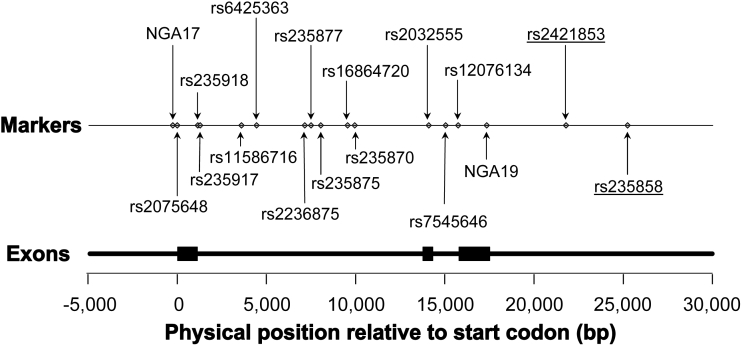
The HapMap database lists 25 SNPs with minor allele frequencies (MAF) greater than 5% in MYOC in subjects of European descent. The linkage disequilibrium (LD) structure of the gene in Europeans is shown in Figure 1A. The positions of the SNPs genotyped in this study are shown in Figure 2.
Figure 1.
The linkage disequilibrium pattern of MYOC SNPs in European and Han Chinese subjects. The figure shows LD patterns in (A) European and (B) Han Chinese subjects in the HapMap database for the region running from SNP rs235858 to SNP rs12082573 on human chromosome 1 (position 142819774 to 142844986 of Genome Build 36.3 of the NCBI Human Reference sequence).
Figure 2.
MYOC polymorphisms genotyped in the present study. The figure shows the positions of the polymorphisms genotyped in the present study relative to the exon structure of the MYOC gene. Exons are depicted as black rectangles, introns as intervening thick black lines. The start codon of the MYOC gene is labelled as position zero.
Cardiff University (UK) cohort
Tagging SNPs were selected using the Haploview program [28] conditional on LD (r2) being less than 0.8 and MAF being greater than 5% (Table 3). Genotyping was performed for 12 SNPs within and in the vicinity of MYOC, including the significant SNPs from the Tang et al. [17] study, and for two microsatellites in the untranslated regions of the gene (NGA17 at the 5' end and NGA19 at the 3' end). SNP genotyping was performed by Kbiosciences Ltd., Hoddesdon, Hertfordshire, UK. Microsatellite genotyping was performed using conventional methods [27]. Briefly, the polymerase chain reaction (PCR) mixture contained 1X HotStar PCR buffer (Qiagen Ltd., Crawley, West Sussex, UK), 1.5 mM MgCl2, 200 µM each dNTP, 0.3 µM of fluorescently-labeled forward primer, 0.3 µM of reverse primer, 0.1 U HotStar Taq polymerase (Qiagen Ltd), and ~20 ng genomic DNA. Amplification was achieved using PCR (35 cycles; denaturation at 94 °C for 1 min, annealing at 56 °C for 1 min, and extension at 72 °C for 1 min) after a preliminary step of 15 min at 95 °C to activate the enzyme. The primers are shown in Table 4. Amplicons were sized using an ABI Prism 310 Genetic Analyzer® (Applied Biosystems, Warrington, Cheshire, UK), run on program D with Genotyper® software (Applied Biosystems) used to call the alleles.
Table 3. Allele frequencies of MYOC SNPs.
| SNP name | SNP allele |
Cardiff University Cohort |
Duke University Cohort |
Tang et al. [17] |
||||
|---|---|---|---|---|---|---|---|---|
| Family Founders | Cases | Controls | Family Founders | Cases | Controls | Family Founders | ||
|
rs235877 |
C |
0.685 |
0.655 |
0.670 |
- |
- |
- |
- |
| T |
0.315 |
0.345 |
0.330 |
- |
- |
- |
- |
|
|
rs235870 |
A |
0.560 |
0.556 |
0.551 |
- |
- |
- |
- |
| T |
0.440 |
0.444 |
0.449 |
- |
- |
- |
- |
|
|
rs2236875 |
G |
0.920 |
0.940 |
0.930 |
- |
- |
- |
- |
| T |
0.080 |
0.060 |
0.070 |
- |
- |
- |
- |
|
|
rs235918 |
A |
0.353 |
0.366 |
0.347 |
- |
- |
- |
- |
| T |
0.647 |
0.634 |
0.653 |
- |
- |
- |
- |
|
|
rs11586716 |
C |
0.264 |
0.239 |
0.269 |
- |
- |
- |
- |
| T |
0.736 |
0.761 |
0.731 |
- |
- |
- |
- |
|
|
rs2075648 |
C |
0.869 |
0.866 |
0.836 |
- |
- |
- |
- |
| T |
0.131 |
0.134 |
0.164 |
- |
- |
- |
- |
|
|
rs16864720 |
A |
0.131 |
0.118 |
0.115 |
0.121 |
0.116 |
- |
- |
| G |
0.869 |
0.882 |
0.885 |
0.879 |
0.884 |
- |
- |
|
|
rs7545646 |
C |
0.087 |
0.074 |
0.078 |
0.100 |
0.116 |
- |
- |
| T |
0.913 |
0.926 |
0.922 |
0.900 |
0.884 |
- |
- |
|
|
rs12076134 |
G |
0.210 |
0.202 |
0.232 |
0.272 |
0.232 |
- |
- |
| T |
0.790 |
0.798 |
0.768 |
0.728 |
0.768 |
- |
- |
|
|
rs235858 |
A |
0.584 |
0.590 |
0.573 |
0.639 |
0.607 |
- |
0.600 |
| G |
0.416 |
0.410 |
0.427 |
0.361 |
0.393 |
- |
0.400 |
|
|
rs2421853 |
A |
0.232 |
0.217 |
0.245 |
0.300 |
0.277 |
- |
0.270 |
| G |
0.768 |
0.783 |
0.755 |
0.700 |
0.723 |
- |
0.730 |
|
|
rs6425363 |
C |
- |
- |
- |
0.886 |
0.900 |
- |
- |
| T |
- |
- |
- |
0.114 |
0.100 |
- |
- |
|
|
rs235917 |
A |
- |
- |
- |
0.284 |
0.277 |
- |
- |
| G |
- |
- |
- |
0.716 |
0.723 |
- |
- |
|
|
rs235875 |
C |
- |
- |
- |
0.792 |
0.815 |
- |
- |
| T |
- |
- |
- |
0.208 |
0.185 |
- |
- |
|
|
rs2032555 |
C |
- |
- |
- |
0.239 |
0.277 |
- |
- |
| T | - | - | - | 0.761 | 0.723 | - | - | |
The values shown in the table are the SNP allele frequencies for the subjects in the Cardiff University and Duke University cohorts, who were all of Caucasian ethnicity, and for subjects in the cohort of Tang et al. [17], who were all of Chinese ethnicity.
Table 4. MYOC microsatellite primer sequences.
| Primer name | Primer sequence |
|---|---|
| NGA17 forward |
GCACAGTGCAGGTTCTCAA |
| NGA17 reverse |
CCAACCATCAGGTAATTCCTT |
| NGA19 forward |
CCGAGCTCCAGAGAGGTTTA |
| NGA19 reverse | CCTCAAAACCAGGCACAA |
Duke University Center for Human Genetics (USA) cohort
Tagging SNPs were selected using SNPSelector conditional on LD (r2) being less than 0.8 and MAF being greater than 5% in the CEU HapMap population. Genotyping was performed for nine SNPs including the significant SNPs from the Tang et al. [17] study using TaqMan® (Applied Biosystems) allelic discrimination assays (Table 3).
Statistics
High myopia was examined as a dichotomous trait. Subjects with a spherical equivalent refractive error of less than −6.00 D (averaged between eyes) were classified as affected [17]. All other subjects were classified as unaffected. The Pedstats package [29] was used to carry out an exact test for Hardy–Weinberg equilibrium (HWE) on unrelated subjects and to check for Mendelian consistency in pedigrees. Association analyses were performed on family data only and jointly on pedigree and case-control subject data to maximize the power of association testing between MYOC polymorphisms and high myopia [30]. Tests were performed using the Unphased program [31], which in addition to family based assays, is able to jointly examine pedigrees and case/control samples. The recruited pedigrees from both centers included families with either one or both parents missing. However, this missingness was accounted for by Unphased, which has been shown to be free from bias in such circumstances [31]. A Bonferroni correction was applied to account for multiple testing. Importantly, the association test results for SNPs genotyped in both the Cardiff University and Duke University cohorts are only reported for combined analyses. The implications of this approach with respect to potential population stratification between subjects from the UK and USA are discussed below.
Results
Subjects and genotyping
The combined study population included a total of 1251 subjects (Table 2). Forty-nine subjects were excluded due to genotyping failure. The genotyping failure rate of each polymorphism is shown in Table 5. This left 293 unrelated and 909 related individuals available for association analyses: 788 subjects in the UK cohort (142 families, 121 cases, and116 controls) and 414 subjects in the USA cohort (86 families and 56 cases). Subjects for whom all relatives failed to pass our genotyping quality control threshold were classified as cases or controls if they met the necessary refractive criteria.
Table 5. Tests of association between MYOC polymorphisms and high myopia.
| Polymorphism | Failed genotypes (%) | HWE p value | Unphased p value (corrected p value) | Unphased relative risk (95% CI) |
|---|---|---|---|---|
| Duke University Cohort | ||||
|
rs6425363 |
1.5 |
1.00 |
0.57 |
1.15 (0.71–1.86) |
|
rs235917 |
4.4 |
0.55 |
0.49 |
1.13 (0.79–1.59) |
|
rs235875 |
2.7 |
0.20 |
0.36 |
1.20 (0.81–1.75) |
|
rs2032555 |
3.5 |
0.01 |
Not tested due to HWE status |
|
| Cardiff University Cohort | ||||
|
rs235877 |
12.0 |
0.09 |
0.57 |
1.07 (0.84–1.37) |
|
rs235870 |
9.0 |
0.27 |
0.53 |
0.93 (0.74–1.17) |
|
rs2236875 |
10.0 |
0.01 |
Not tested due to HWE status |
|
|
rs235918 |
8.0 |
0.19 |
0.53 |
1.07 (0.86–1.34) |
|
rs11586716 |
8.6 |
0.13 |
0.38 |
0.73 (0.84–1.44) |
|
rs2075648 |
9.8 |
0.07 |
0.59 |
0.91 (0.64–1.28) |
| NGA17 |
0.1 |
0.08 |
0.03 (0.39) |
0.70 (0.55–0.92) |
| NGA19 |
0.2 |
0.49 |
0.97 |
1.02 (0.82–1.26) |
| Combined Cohorts | ||||
|
rs16864720 |
7.9 |
0.85 |
0.04 (0.65) |
1.30 (1.004–1.73) |
|
rs7545646 |
12.0 |
0.05 |
0.06 |
1.30 (0.98–1.8) |
|
rs12076134 |
9.4 |
0.81 |
0.09 |
1.20 (0.97–1.48) |
|
rs235858
* |
13.0 |
0.86 |
0.87 |
1.02 (0.84–1.22) |
| rs2421853 * | 13.0 | 0.18 | 0.25 | 1.13 (0.91–1.39) |
The results shown in the table are for the analysis of the full set of subjects (i.e. families, cases and controls). For each marker studied, the table gives (column 1) the genotyping error rate, (column 2) the p-value for a contingency test examining whether the marker allele frequencies are in Hardy-Weinberg equilibrium (HWE), (column 3) the uncorrected, and in brackets the Bonferroni-corrected, p-values for an Unphased analysis examining whether the marker allele frequencies are associated with high myopia affectation status, and (column 4) the relative risk of high myopia calculated by Unphased for subjects carrying the second allele relative to carrying the first (reference) allele, along with the 95% confidence interval of the relative risk estimate. The 2 SNPs marked with an asterisk were found to be significantly associated with high myopia in the study of Tang et al. [17].
Genotyping of the two microsatellite markers, NGA17 and NGA19, revealed four alleles each. For each marker, there were three common alleles and one rare allele. The observed allele frequencies of the microsatellite polymorphisms are shown in Table 6. Since the sample size was modest, the rare allele of each microsatellite marker was combined with the allele next in size to it (allele 1 with allele 2 for both markers). Genotyping for SNP marker rs235875 failed.
Table 6. Allele frequencies of MYOC microsatellites.
| Microsatellite allele |
Cardiff University Cohort |
Tang et al. [17] |
||
|---|---|---|---|---|
| Family founders | Cases | Controls | Family founders | |
| NGA17 alleles | ||||
| 12 repeats |
0.000 |
0.033 |
0.028 |
- |
| 13 repeats |
0.597 |
0.637 |
0.550 |
0.501 |
| 14 repeats |
0.184 |
0.156 |
0.170 |
0.184 |
| 15 repeats |
0.219 |
0.174 |
0.252 |
0.312 |
| 16 repeats |
- |
- |
- |
0.003 |
| NGA19 alleles | ||||
| 11 repeats |
- |
- |
- |
0.0015 |
| 12 repeats |
0.000 |
0.014 |
0.000 |
- |
| 13 repeats |
0.342 |
0.344 |
0.400 |
0.218 |
| 14 repeats |
0.039 |
0.047 |
0.004 |
0.008 |
| 15 repeats |
0.619 |
0.595 |
0.596 |
0.711 |
| 16 repeats |
- |
- |
- |
0.060 |
| 17 repeats | - | - | - | 0.0015 |
The values shown in the table are the microsatellite marker allele frequencies for the subjects in the Cardiff University cohort, who were all of Caucasian ethnicity, and for subjects in the cohort of Tang et al. [17], who were all of Chinese ethnicity.
Statistical analysis
Tests for HWE showed that two SNPs, rs2236875 and rs2032555, were not in equilibrium in the unrelated subjects (Table 5). Therefore, these two markers were dropped from further analyses. Thus, association tests were performed for the remaining 15 variants, which were 13 SNPs and two microsatellites.
There was no significant heterogeneity in genotype frequencies between families and singleton samples either within or between cohorts (Table 3 and Table 6). Therefore, families and unrelated subjects were analyzed jointly [31]. Likewise, subjects recruited at Duke University and Cardiff University were analyzed jointly for those SNPs genotyped in common (i.e., ignoring potential population stratification issues). The association test results are shown in Table 5. Prior to correction for multiple testing, two variants showed significant association, rs16864720 (p=0.043) and NGA17 (p=0.026). However, neither association retained statistical significance after Bonferroni correction (Table 5). Evaluation of relative risk highlighted the same two polymorphisms, rs16864720 and NGA17, with 95% confidence intervals that did not include 1.0 (Table 5). The relative risk conferred by each of these variants, however, was low (RR<1.5). When the analysis was restricted to the family data alone, there was also no significant association between MYOC and high myopia (Table 7) in concordance with the joint analysis.
Table 7. Test of association between MYOC and myopia: family data only.
| Polymorphism | Failed genotypes (%) | HWE p value | Unphased p value (corrected p value) | Unphased relative risk (95% CI) |
|---|---|---|---|---|
| Duke University Cohort | ||||
|
rs6425363 |
6.30 |
1.00 |
0.59 |
1.18 (0.64–2.20) |
|
rs235917 |
6.60 |
0.40 |
0.72 |
0.66 (0.60–1.75) |
|
rs235875 |
5.30 |
1.00 |
0.30 |
1.30 (0.78–2.16) |
| Cardiff University Cohort | ||||
|
rs235877 |
13.50 |
0.22 |
0.76 |
1.06 (0.74–1.50) |
|
rs235870 |
10.15 |
0.10 |
0.58 |
0.91 (0.64–1.28) |
|
rs235918 |
8.80 |
0.10 |
0.16 |
0.78 (0.55–1.10) |
|
rs11586716 |
9.20 |
0.23 |
0.62 |
1.10 (0.77–1.54) |
|
rs2075648 |
10.85 |
0.31 |
0.77 |
0.92 (0.55–1.57) |
| NGA17 |
0.20 |
0.33 |
0.28 |
0.77 (0.56–1.07) |
| NGA19 |
0.40 |
0.08 |
0.37 |
1.04 (0.79–1.38) |
| Combined Cohorts | ||||
|
rs16864720 |
5.50 |
0.80 |
0.017 (0.289) |
1.56 (1.07–2.27) |
|
rs7545646 |
9.40 |
0.33 |
0.017 (0.289) |
1.62 (1.07–2.46) |
|
rs12076134 |
6.70 |
0.87 |
0.12 |
1.25 (0.94–1.67) |
|
rs235858
* |
11.00 |
0.33 |
0.69 |
1.05 (0.81–1.36) |
| rs2421853 * | 10.80 | 0.88 | 0.36 | 1.15 (0.85–1.54) |
The results shown in the table are for the analysis of the families only (i.e. cases and controls were excluded from the analysis). For each marker studied, the table gives (column 1) the genotyping error rate, (column 2) the p-value for a contingency test examining whether the marker allele frequencies are in Hardy-Weinberg equilibrium (HWE), (column 3) the uncorrected, and in brackets the Bonferroni-corrected, p-values for an Unphased analysis examining whether the marker allele frequencies are associated with high myopia affectation status, and (column 4) the relative risk of high myopia calculated by Unphased for subjects carrying the second allele relative to carrying the first (reference) allele, along with the 95% confidence interval of the relative risk estimate. The 2 SNPs marked with an asterisk were found to be significantly associated with high myopia in the study of Tang et al. [17].
Discussion
A joint analysis of subjects from the UK and USA was performed for those SNPs that were genotyped in both groups of subjects. This pooling of subjects could potentially have given rise to a “false positive” or “false negative” association due to population stratification. However, population stratification can only give rise to a significant association between a disease phenotype and a marker genotype if the prevalence of the disease differs between the two subject groups and if the allele frequency of the marker of interest differs between the two subject groups. For high myopia, exact figures on the prevalences in Caucasian subjects from the UK and USA are lacking, but estimates suggest these rates are similar [32-34]. Furthermore, the MYOC polymorphisms studied here had statistically similar allele frequencies in the UK and USA subjects (Table 3 and Table 6).
In contrast to previously published significant association between MYOC and high myopia in subjects of Chinese ethnicity [16,17], this study suggests that there is no such relationship in subjects of Caucasian ethnicity. The ethnic difference of the respective study populations is an appealing explanation for these discrepant findings. Different populations may exhibit differences in allele frequencies and linkage disequilibrium patterns at specific loci (Figure 1 and Table 6). Thus, the role of MYOC in high myopia in Chinese subjects may be dissimilar to that in Caucasians.
An alternative explanation could be the power of association analyses. The estimated relative risk of the genetic variants examined here was less than 1.5, which suggests that the power of this study would be approximately 75% [35]. On the other hand, Tang et al. [17] investigated a smaller sample size (557 individuals in 162 nuclear families) and reported a relative risk greater than 1.5 for two significant SNPs (rs235858 and rs2421853). To gain 80% power, a family based association study of a variant with relative risk greater than 1.5 and allele frequency of 0.5 would need approximately 200 subjects under an additive model and approximately 1100 subjects under a dominant model [36].
A final potential reason for our failure to detect an association between MYOC polymorphisms and high myopia is that MYOC may not in fact be a high myopia susceptibility gene (i.e., the significant associations reported previously [16,17] could have been false positive findings). Several studies have suggested that many of the other high myopia genetic association results that have been published are likely to be false positives [37-48]. Moreover, candidate gene based association studies for other disorders have also yielded numerous false positive findings over the years [49].
The fact that MYOC polymorphisms are implicated in both myopia and glaucoma is intriguing, especially in light of the higher-than-chance co-occurrence of myopia and glaucoma seen in many studies [18-20]. Nonetheless, the high expression of myocilin in the TMC [50] is easier to reconcile with the role of MYOC polymorphisms in glaucoma than in myopia. Furthermore, the current evidence suggests that those MYOC gene variants that confer an increased risk of open angle glaucoma are different from those that increase susceptibility to myopia. In this respect, the association of MYOC variants with both conditions may be coincidental.
In conclusion, this study found no evidence to support a significant association between MYOC polymorphisms and high myopia in Caucasian subjects from the UK and USA.
Acknowledgments
We are grateful to all of the subjects who participated in this research project. We thank Hywel Williams, George Kirov, and Michael J. Owen for assistance and advice with microsatellite genotyping. This work was supported by the College of Optometrists (grant NdB/TM/Gug/4/6/1), Cardiff University (PhD studentship award), the National Institute of Health (grant RO1 EY014685), and Research to Prevent Blindness, Inc.
References
- 1.Fredrick DR. Myopia: shortsightedness is becoming more common. BMJ. 2002;324:1195–200. doi: 10.1136/bmj.324.7347.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bar Dayan Y, Levin A, Morad Y, Grotto I, Ben-David R, Goldberg A, Onn E, Avni I, Levi Y, Benyamini OG. The changing prevalence of myopia in young adults: a 13-year series of population-based prevalence surveys. Invest Ophthalmol Vis Sci. 2005;46:2760–5. doi: 10.1167/iovs.04-0260. [DOI] [PubMed] [Google Scholar]
- 3.Lee KE, Klein BE, Klein R, Wong TY. Changes in refraction over 10 years in an adult population: The Beaver Dam Eye Study. Invest Ophthalmol Vis Sci. 2002;43:2566–71. [PubMed] [Google Scholar]
- 4.McCarty CA, Taylor HR. Myopia and vision 2020. Am J Ophthalmol. 2000;129:525–7. doi: 10.1016/s0002-9394(99)00444-4. [DOI] [PubMed] [Google Scholar]
- 5.Saw SM, Shih-Yen EC, Koh A, Tan D. Interventions to retard myopia progression in children: an evidence-based update. Ophthalmology. 2002;109:415–21. doi: 10.1016/s0161-6420(01)00972-1. [DOI] [PubMed] [Google Scholar]
- 6.Liang CL, Yen E, Su JY, Liu C, Chang TY, Park N, Wu MJ, Lee S, Flynn JT, Juo SH. Impact of family history of high myopia on level and onset of myopia. Invest Ophthalmol Vis Sci. 2004;45:3446–52. doi: 10.1167/iovs.03-1058. [DOI] [PubMed] [Google Scholar]
- 7.Peet JA, Cotch MF, Wojciechowski R, Bailey-Wilson JE, Stambolian D. Heritability and familial aggregation of refractive error in the Old Order Amish. Invest Ophthalmol Vis Sci. 2007;48:4002–6. doi: 10.1167/iovs.06-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young TL, Metlapally R, Shay AE. Complex trait genetics of refractive error. Arch Ophthalmol. 2007;125:38–48. doi: 10.1001/archopht.125.1.38. [DOI] [PubMed] [Google Scholar]
- 9.Fingert JH, Stone EM, Sheffield VC, Alward WLM. Myocilin glaucoma. Surv Ophthalmol. 2002;47:547–61. doi: 10.1016/s0039-6257(02)00353-3. [DOI] [PubMed] [Google Scholar]
- 10.Stone EM, Fingert JH, Alward WL, Nguyen TD, Polansky JR, Sunden SL, Nishimura D, Clark AF, Nystuen A, Nichols BE, Mackey DA, Ritch R, Kalenak JW, Craven ER, Sheffield VC. Identification of a gene that causes primary open angle glaucoma. Science. 1997;275:668–70. doi: 10.1126/science.275.5300.668. [DOI] [PubMed] [Google Scholar]
- 11.Tamm ER. Myocilin and glaucoma: facts and ideas. Prog Retin Eye Res. 2002;21:395–428. doi: 10.1016/s1350-9462(02)00010-1. [DOI] [PubMed] [Google Scholar]
- 12.Polansky JR, Fauss DJ, Chen P, Chen H, Lutjen-Drecoll E, Johnson D, Kurtz RM, Ma ZD, Bloom E, Nguyen TD. Cellular pharmacology and molecular biology of the trabecular meshwork inducible glucocorticoid response gene product. Ophthalmologica. 1997;211:126–39. doi: 10.1159/000310780. [DOI] [PubMed] [Google Scholar]
- 13.Polansky JR, Fauss DJ, Zimmerman CC. Regulation of TIGR/MYOC gene expression in human trabecular meshwork cells. Eye. 2000;14:503–14. doi: 10.1038/eye.2000.137. [DOI] [PubMed] [Google Scholar]
- 14.Tamm ER, Russell P, Epstein DL, Johnson DH, Piatigorsky J. Modulation of myocilin/TIGR expression in human trabecular meshwork. Invest Ophthalmol Vis Sci. 1999;40:2577–82. [PubMed] [Google Scholar]
- 15.Wang L, Zhuo YH, Liu BQ, Huang SS, Hou F, Ge J. Pro370Leu mutant myocilin disturbs the endoplasm reticulum stress response and mitochondrial membrane potential in human trabecular meshwork cells. Mol Vis. 2007;13:618–25. [PMC free article] [PubMed] [Google Scholar]
- 16.Wu H, Yu XH, Yap EP. Allelic association between trabecular meshwork-induced glucocorticoid response (TIGR) gene and severe myopia. Invest Ophthalmol Vis Sci. 1999;40:S600. [Google Scholar]
- 17.Tang WC, Yip SP, Lo KK, Ng PW, Choi PS, Lee SY, Yap MKH. Linkage and association of myocilin (MYOC) polymorphisms with high myopia in a Chinese population. Mol Vis. 2007;13:534–44. [PMC free article] [PubMed] [Google Scholar]
- 18.Mitchell P, Hourihan F, Sandbach J, Wang JJ. The relationship between glaucoma and myopia - The Blue Mountains Eye Study. Ophthalmology. 1999;106:2010–5. doi: 10.1016/s0161-6420(99)90416-5. [DOI] [PubMed] [Google Scholar]
- 19.Wong TY, Klein BE, Klein R, Knudtson M, Lee KE. Refractive errors, intraocular pressure, and glaucoma in a white population. Ophthalmology. 2003;110:211–7. doi: 10.1016/s0161-6420(02)01260-5. [DOI] [PubMed] [Google Scholar]
- 20.Xu L, Wang Y, Wang S, Wang Y, Jonas JB. High myopia and glaucoma susceptibility: The Beijing Eye Study. Ophthalmology. 2006;114:216–20. doi: 10.1016/j.ophtha.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 21.Abdalla MI, Hamdi M. Applanation ocular tension in myopia and emmetropia. Br J Ophthalmol. 1970;54:122–5. doi: 10.1136/bjo.54.2.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein AP, Duggal P, Lee KE, Klein R, Bailey-Wilson JE, Klein BE. Confirmation of linkage to ocular refraction on chromosome 22q and identification of a novel linkage region on 1q. Arch Ophthalmol. 2007;125:80–5. doi: 10.1001/archopht.125.1.80. [DOI] [PubMed] [Google Scholar]
- 23.Rohrer B, Tao J, Stell WK. Basic fibroblast growth factor, its high- and low-affinity receptors, and their relationship to form-deprivation myopia in the chick. Neuroscience. 1997;79:775–87. doi: 10.1016/s0306-4522(97)00042-0. [DOI] [PubMed] [Google Scholar]
- 24.Lin HJ, Wan L, Tsai Y, Tsai YY, Fan SS, Tsai CH, Tsai FJ. The TGFbeta1 gene codon 10 polymorphism contributes to the genetic predisposition to high myopia. Mol Vis. 2006;12:698–703. [PubMed] [Google Scholar]
- 25.Andrew T, Maniatis N, Carbonaro F, Liew SH, Lau W, Spector TD, Hammond CJ. Identification and replication of three novel myopia common susceptibility gene loci on chromosome 3q26 using linkage and linkage disequilibrium mapping. PLoS Genet. 2008;4:e1000220. doi: 10.1371/journal.pgen.1000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leung YF, Tam POS, Baum L, Lam DSC, Pang CCP. TIGR/MYOC proximal promoter GT-repeat polymorphism is not associated with myopia. Hum Mutat. 2000;16:533. doi: 10.1002/1098-1004(200012)16:6<533::AID-HUMU21>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 27.Farbrother JE, Kirov G, Owen MJ, Pong-Wong R, Haley CS, Guggenheim JA. Linkage analysis of the genetic loci for high myopia on chromosomes 18p, 12q and 17q in 51 UK families. Invest Ophthalmol Vis Sci. 2004;45:2879–85. doi: 10.1167/iovs.03-1156. [DOI] [PubMed] [Google Scholar]
- 28.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 29.Wigginton JE, Abecasis GR. PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics. 2005;21:3445–7. doi: 10.1093/bioinformatics/bti529. [DOI] [PubMed] [Google Scholar]
- 30.Nagelkerke NJD, Hoebee B, Teunis P, Kimman TG. Combining the transmission disequilibrium test and case-control methodology using generalized logistic regression. Eur J Hum Genet. 2004;12:964–70. doi: 10.1038/sj.ejhg.5201255. [DOI] [PubMed] [Google Scholar]
- 31.Dudbridge F. Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Hum Hered. 2008;66:87–98. doi: 10.1159/000119108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vitale S, Ellwein L, Cotch MF, Ferris FL, 3rd, Sperduto R. Prevalence of refractive error in the United States, 1999–2004. Arch Ophthalmol. 2008;126:1111–9. doi: 10.1001/archopht.126.8.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katz J, Tielsch JM, Sommer A. Prevalence and risk factors for refractive errors in an adult inner city population. Invest Ophthalmol Vis Sci. 1997;38:334–40. [PubMed] [Google Scholar]
- 34.Farbrother JE, Kirov G, Owen MJ, Guggenheim JA. Family aggregation of high myopia: Estimation of the sibling recurrence risk ratio. Invest Ophthalmol Vis Sci. 2004;45:2873–8. doi: 10.1167/iovs.03-1155. [DOI] [PubMed] [Google Scholar]
- 35.Epstein MP, Veal CD, Trembath RC, Barker JN, Li C, Satten GA. Genetic association analysis using data from triads and unrelated subjects. Am J Hum Genet. 2005;76:592–608. doi: 10.1086/429225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knapp M. A note on power approximations for the transmission/disequilibrium test. Am J Hum Genet. 1999;64:1177–85. doi: 10.1086/302334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Zhang QJ, Xiao XS, Li JZ, Zhang FS, Li SQ, Li W, Li T, Jia XY, Guo L, Guo XM. The SNPs analysis of encoding sequence of interacting factor gene in Chinese population. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2003;20:454–6. [PubMed] [Google Scholar]
- 38.Scavello GS, Paluru PC, Ganter WR, Young TL. Sequence variants in the Transforming Growth beta-Induced Factor (TGIF) gene are not associated with high myopia. Invest Ophthalmol Vis Sci. 2004;45:2091–7. doi: 10.1167/iovs.03-0933. [DOI] [PubMed] [Google Scholar]
- 39.Paluru PC, Scavello GS, Ganter WR, Young TL. Exclusion of lumican and fibromodulin as candidate genes in MYP3 linked high grade myopia. Mol Vis. 2004;10:917–22. [PubMed] [Google Scholar]
- 40.Tang WC, Yip SP, Yap MKH, Lo KK, Ng PW, Lee SY, Choi PS. Testing for association between COL2A1 and myopia susceptibility in Hong Kong Chinese population. ARVO Annual Meeting; 2004 April 25-29; Fort Lauderdale (FL). [Google Scholar]
- 41.Hasumi Y, Inoko H, Mano S, Ota M, Okada E, Kulski JK, Nishizaki R, Mok J, Oka A, Kumagai N, Nishida T, Ohno S, Mizuki N. Analysis of single nucleotide polymorphisms at 13 loci within the transforming growth factor-induced factor gene shows no association with high myopia in Japanese subjects. Immunogenetics. 2006;58:947–53. doi: 10.1007/s00251-006-0155-9. [DOI] [PubMed] [Google Scholar]
- 42.Mutti DO, Cooper ME, O'Brien S, Jones LA, Marazita ML, Murray JC, Zadnik K. Candidate gene and locus analysis of myopia. Mol Vis. 2007;13:1012–9. [PMC free article] [PubMed] [Google Scholar]
- 43.Simpson CL, Hysi P, Bhattacharya SS, Hammond CJ, Webster A, Peckham CS, Sham PC, Rahi JS. The Roles of PAX6 and SOX2 in Myopia: Lessons from the 1958 British Birth Cohort. Invest Ophthalmol Vis Sci. 2007;48:4421–5. doi: 10.1167/iovs.07-0231. [DOI] [PubMed] [Google Scholar]
- 44.Liang CL, Hung KS, Tsai YY, Chang W, Wang HS, Juo SH. Systematic assessment of the tagging polymorphisms of the COL1A1 gene for high myopia. J Hum Genet. 2007;52:374–7. doi: 10.1007/s10038-007-0117-6. [DOI] [PubMed] [Google Scholar]
- 45.Wang P, Li S, Xiao X, Jia X, Jiao X, Guo X, Zhang Q. High myopia is not associated with the SNPs in the TGIF, Lumican, TGFB1, and HGF genes. Invest Ophthalmol Vis Sci. 2008 doi: 10.1167/iovs.08-2537. [DOI] [PubMed] [Google Scholar]
- 46.Zayats T, Guggenheim JA, Hammond CJ, Young TL. Comment on 'A PAX6 gene polymorphism is associated with genetic predisposition to extreme myopia'. Eye. 2008;22:598–9. doi: 10.1038/sj.eye.6703096. [DOI] [PubMed] [Google Scholar]
- 47.Pertile KK, Schache M, Islam FM, Chen CY, Dirani M, Mitchell P, Baird PN. Assessment of TGIF as a candidate gene for myopia. Invest Ophthalmol Vis Sci. 2008;49:49–54. doi: 10.1167/iovs.07-0896. [DOI] [PubMed] [Google Scholar]
- 48.Nakanishi H, Yamada R, Gotoh N, Hayashi H, Otani A, Tsujikawa A, Yamashiro K, Shimada N, Ohno-Matsui K, Mochizuki M, Saito M, Saito K, Iida T, Matsuda F, Yoshimura N. Absence of Association between COL1A1 Polymorphisms and High Myopia in the Japanese Population. Invest Ophthalmol Vis Sci. 2008 doi: 10.1167/iovs.08-2425. [DOI] [PubMed] [Google Scholar]
- 49.Colhoun HM, McKeigue PM, Davey Smith G. Problems of reporting genetic associations with complex outcomes. Lancet. 2003;361:865–72. doi: 10.1016/s0140-6736(03)12715-8. [DOI] [PubMed] [Google Scholar]
- 50.Wentz-Hunter K, Shen X, Yue B. Distribution of myocilin, a glaucoma gene product, in human corneal fibroblasts. Mol Vis. 2003;9:308–14. [PubMed] [Google Scholar]
- 51.Han W, Yap MK, Wang J, Yip SP. Family-based association analysis of hepatocyte growth factor (HGF) gene polymorphisms in high myopia. Invest Ophthalmol Vis Sci. 2006;47:2291–9. doi: 10.1167/iovs.05-1344. [DOI] [PubMed] [Google Scholar]
- 52.Tsai YY, Chiang CC, Lin HJ, Lin JM, Wan L, Tsai FJA. PAX6 gene polymorphism is associated with genetic predisposition to extreme myopia. Eye. 2008;22:576–81. doi: 10.1038/sj.eye.6702982. [DOI] [PubMed] [Google Scholar]
- 53.Wang IJ, Chiang TH, Shih YF, Hsiao CK, Lu SC, Hou YC, Lin LL. The association of single nucleotide polymorphisms in the 5'-regulatory region of the lumican gene with susceptibility to high myopia in Taiwan. Mol Vis. 2006;12:852–7. [PubMed] [Google Scholar]
- 54.Inamori Y, Ota M, Inoko H, Okada E, Nishizaki R, Shiota T, Mok J, Oka A, Ohno S, Mizuki N. The COL1A1 gene and high myopia susceptibility in Japanese. Hum Genet. 2007;122:151–7. doi: 10.1007/s00439-007-0388-1. [DOI] [PubMed] [Google Scholar]
- 55.Lam DS, Lee WS, Leung YF, Tam PO, Fan DS, Fan BJ, Pang CP. TGFbeta-Induced Factor: A candidate gene for high myopia. Invest Ophthalmol Vis Sci. 2003;44:1012–5. doi: 10.1167/iovs.02-0058. [DOI] [PubMed] [Google Scholar]