Abstract
Macquarie Island, a small subantarctic island, is home to rockhopper, royal and king penguins, which are often infested with the globally distributed seabird tick, Ixodes uriae. A flavivirus, an orbivirus, a phlebovirus, and a nairovirus were isolated from these ticks and partial sequences obtained. The flavivirus was nearly identical to Gadgets Gully virus, isolated some 30 year previously, illustrating the remarkable genetic stability of this virus. The nearest relative to the orbivirus (for which we propose the name Sandy Bay virus) was the Scottish Broadhaven virus, and provided only the second available sequences from the Great Island orbivirus serogroup. The phlebovirus (for which we propose the name Catch-me-cave virus) and the previously isolated Precarious Point virus were distinct but related, with both showing homology with the Finnish Uukuniemi virus. These penguin viruses provided the second and third available sequences for the Uukuniemi group of phleboviruses. The nairovirus (for which we propose the name Finch Creek virus) was shown to be related to the North American Tillamook virus, the Asian Hazara virus and Nairobi sheep disease virus. Macquarie Island penguins thus harbour arboviruses from at least four of the seven arbovirus-containing genera, with related viruses often found in the northern hemisphere.
Introduction
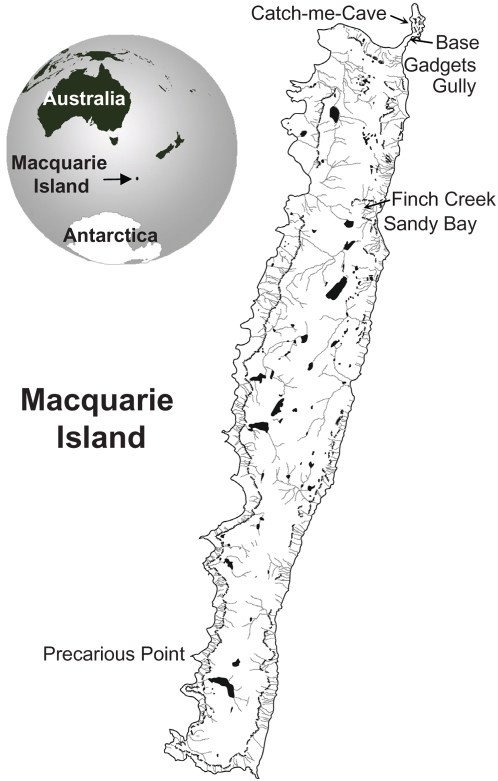
Macquarie Island is a small (34 km long and 5 km wide) subantarctic island, which lies between Australia and Antarctica, 1,466 km SSE of Tasmania and 1,294 km N of the Antarctic continent (54°30′S, 158°55′E) (Fig. 1). The island is the only Southern Ocean biosphere reserve within the Man and the Biosphere Program of the United Nations Educational, Scientific and Cultural Organization (UNESCO). In 1997, Macquarie Island plus the surrounding 12 nautical miles of ocean became a world heritage area for geological and aesthetic reasons. The Australian Antarctic Division currently maintains a base on the island, which is resupplied twice yearly by sea. A number of tourist vessels also call for short visits each southern hemisphere summer. Macquarie Island is home to millions of seabirds, including four species of penguins; King (Aptenodytes patagonicus), rockhopper (Eudyptes chrysocome), royal (Eudyptes schlegeli) and gentoo (Pygoscelis papua) penguins. The only place in the world where royal penguins breed is Macquarie Island. The populations of these species have recovered well since Macquarie Island was declared a sanctuary in 1933 and the rendering down of penguins for oil ceased [1].
Figure 1. Map of Macquarie Island and its location on the globe.
The locations of the Penguin colony sites where ticks were collected and the Antarctic Division base are shown.
Arboviruses are distributed globally and belong mainly to the genera alphavirus (family Togaviridae), flavivirus (family Flaviviridae), nairovirus (family Bunyaviridae), phlebovirus (family Bunyaviridae), orbivirus (family Reoviridae), coltivirus (family Reoviridae) and vesiculovirus (family Rhabdoviridae). Each of these genera contains viruses, which are a significant cause of human and animal diseases globally. Arboviruses of the Antarctic region have not been studied extensively [2] and sequence information is available for only two, the flavivirus, Gadgets Gully [3], [4] and the Southern Elephant Seal alphavirus [5]. Given the rise in Antarctic tourism, the emergence and re-emergence of arboviral disease globally and the general concern for wildlife [6], [7], [8], [9], [10], we undertook to resurvey the tick-borne viruses associated with the different penguin species on Macquarie Island.
Results
Collection of ticks
I. uriae ticks were found under rocks near a King penguin colony at Sandy Bay, near a Royal penguin colony at upper Finch Creek, and inside Catch-me-cave (Fig. 1), a favoured roost of rockhopper penguins. Ticks could often be seen on King penguins, especially on young birds. Despite extensive searching, no ticks were found under rocks around the coastal gentoo penguin colonies near the Australian Antarctic Division base.
Isolation of viruses and transmission electron microscopy
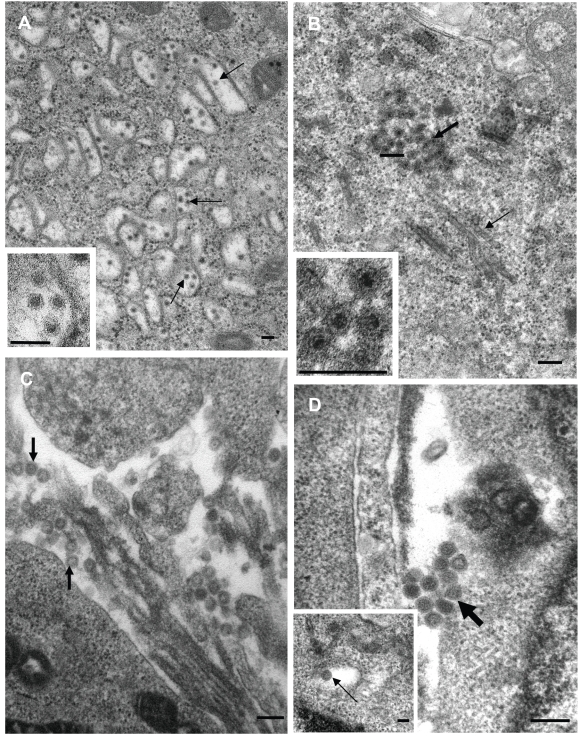
A summary of all the viruses isolated by cell culture is shown in Table 1. Of the 90 ticks processed from the King penguin colony at Sandy Bay, 7 yielded virus. By electron microscopy the virus from 2 ticks (F3/2 and F4 BM) were identified as belonging to the genus Flavivirus (Fig. 2A). These were consistently about 45 nm in diameter, contained an inner electron dense core, were enveloped and were present within cytoplasmic vesicles (Fig. 2A). The viruses isolated from the other 5 ticks from the King penguin colony were identified as belonging to the genus Orbivirus or Coltivirus (Fig. 2B). The viruses were ≈65 nm in diameter, were non-enveloped, isometric in shape and cytoplasmic in location. Semi electron dense material was associated with the viruses and is characteristic of orbivirus inclusion bodies [11]. Cytoplasmic structures resembling virus-specific tubules [12] were also observed (Fig. 2B). Of the 36 ticks processed from the rockhopper colony at Catch-me-cave, 2 yielded virus and both were identified as belonging to the family Bunyaviridae (Fig. 2C). The enveloped viruses were spherical and 80–100 nm in diameter. These viruses were observed budding into smooth surfaced cytoplasmic vesicles (Fig. 2D insert) or associated with the external aspect of the plasma membrane (Fig. 2D). For ticks from the royal penguin colony at Finch Creek, only 1 out of the 43 yielded virus, and this was also identified as belonging to the family Bunyaviridae.
Table 1. Viruses isolated from ticks associated with penguin colonies.
| Virus code | Location | I. Uriae stage sex | Penguins species | CPE day | EM ID | Sequence ID | Name | |
| BHK | Vero | |||||||
| F3/2 | Sandy Bay | Female | King | +++ d4 | ND | Flavi | Flavi | Gadgets Gully |
| F4 BM | Sandy Bay | Female | King | +++ d4 | − d5 | Flavi | Flavi | Gadgets Gully |
| F3/4 | Sandy Bay | Female | King | ++++ d1 | +++ d2 | Orbi/Colti | Orbi | Sandy Baya |
| F1/11 | Sandy Bay | Adult female | King | ND | ++++ d2 | Orbi/Colti | ND | - |
| F1/40 | Sandy Bay | Adult female | King | ++ d2 | - | Orbi/Colti | ND | - |
| F2/21 | Sandy Bay | Adult male | King | ++++ d2 | ND | Orbi/Colti | ND | - |
| F2-22 | Sandy Bay | Adult male | King | ++++ d2 | − d5 | Orbi/Colti | ND | - |
| I2-19 | Catch-me-cave | Adult male | Rockhopper | ND | ++++ d6 | Bunya | Phlebo | Catch-me-cavea |
| I2-7 | Catch-me-cave | Adult male | Rockhopper | ND | ++++ d6 | Bunya | Phlebo | Catch-me-cavea |
| EB-6 | Upper Finch Ck | ND | Royal | − d11 | +/− d13 | Bunya | Nairo | Finch Creeka |
Names proposed for the new virus isolates. CPE-cytopathic effect. ID-identification. Gadgets Gully virus infected cells did not react with the pan-flavi antibody 4G2 (data not shown). (CPE; + <20%, ++ 20–40%, +++ 40–60%, ++++ 60–80%).
Figure 2. Identification of viruses by transmission electron microscopy.
(A) F3/2 from a King penguin colony identified as a flavivirus after infection of BHK cells. Arrows indicate viral particles within cytoplasmic vesicles. Bars = 100 nm. (B) F3/4 from a King penguin colony identified as an orbi- or colti-virus (family Reoviridae) after infection of Vero cells. The thin arrow indicates a virus-specific tubule; the thick arrow aggregation of viruses, the associated electron semi-dense material represents material usually associated with viral assembly bodies. Bars = 100 nm. (C) I2-19 from a rockhopper penguin colony identified as a bunyavirus after infection of Vero cells. Arrows indicate extracellular viruses. Bar = 200 nm. (D) EB6 from a royal penguin colony identified as a bunyavirus after infection of Vero cells. Arrows indicated extracellular viruses. Bar = 200 nm. Inset shows virus within an intracellular vesicle. Bar = 100 nm.
After 2–4 weekly passages the flaviviruses and orbi/coltiviruses produced overt CPE in BHK cells within 2–4 days, and the rockhopper-associated bunyavirus produced overt CPE in Vero cells within 6 days. The royal penguin-associated bunyavirus showed only mild CPE after 12–14 days in Vero cells (Table 1).
The flavivirus from ticks associated with King penguins
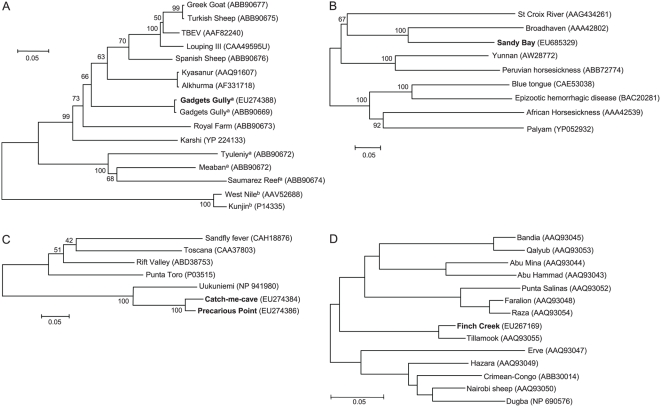
The two flavivirus isolates from separate ticks collected at the Sandy Bay King Penguin colony (Table 1) were sequenced using primers based on Saumarez reef NS5 and E proteins [13]. The NS5 sequence (EU274387) showed a 96% sequence identity with nucleic acids 9307–9662 of the published Gadgets Gully NS5 gene (DQ235145.1), with no amino acid changes. The E protein sequence showed a 94% sequence identity with nucleic acids 1241–1659 of the published Gadgets Gully E gene (DQ235145) and resulted in only one conservative (I454 to V) substitution in the envelope protein (ABB90669). Phylogenetic analysis is shown in Fig. 3A and the closest relatives to Gadgets Gully virus are described in Table 2.
Figure 3. Dendograms showing the relationship between viruses.
The scale shows the distance in terms of proportion of amino acids difference. The percent bootstrap support levels for each node are shown. (A) Gadgets Gully virus, genus Flavivirus. The dendogram was constructed from a 176 amino acid sequence of the E protein and recapitulates the dendogram shown previously for the complete polyprotein sequences [4]. Gadgets Gully in bold - virus collected in 2002. Gadgets Gully in normal text - virus collected in 1972. aSeabird viruses. b Mosquito borne viruses. (B) Sandy Bay virus, genus Orbivirus. The dendogram was constructed from a 320 amino acid sequence of the VP5 outer capsid protein. Broadhaven, Nugget and possibly St Croix River virus [15] are tick born. (C) Precarious Point and Catch-me-cave virus, genus Phlebovirus. The dendogram was constructed from a 59 amino acid sequence of the N protein. Only a partial sequence was available for Precarious Point. Uukuniemi, Precarious Point and Catch-me-cave viruses are tick borne. (D) Finch Creek virus, genus Nairovirus. The dendogram was constructed from a 120 amino acid sequence from RNA polymerase and recapitulates the segregation previously reported between nairoviruses transmitted by soft ticks (Bandia to Raza) and hard ticks (Taggert to Dugba) [16].
Table 2. Penguin virus genera and their closest relatives.
| Virus name | Genus Family | Penguin species | Nearest relativesa | Known hosts | Location | Amino acid divergence %b |
| Gadgets Gully | Flavivirus Flaviviridae | King (Royalc) | Tick borne flaviviruses [4], excluding seabird viruses. | Mammals, humans | Northern hemisphere, Africa | Polyproteind |
| 28–31% | ||||||
| VP5 | ||||||
| Sandy Baye | Orbivirus Reoviridae | King | 1. Broadhaven [42] | Sea birds | Scotland | 40 |
| 2. Yunnan [15] | Micef | China | 57.2 | |||
| 3. Peruvian horse sickness | Horse | S. America | 62.8 | |||
| N protein | ||||||
| Catch-me-cave | Phlebovirus Bunyaviridae | Rock-hopper | 1. Precarious Point | Penguin | Macquarie | 5.1 |
| 2. Uukuniemi [24], [25] | Birds/mammals | Finland | 32.7 | |||
| 3. Rift valley fever | Ruminants | Africa/M. East | 60.9 | |||
| N proteing | ||||||
| Precarious Point (Uukuniemi serogroup) | Phlebovirus Bunyaviridae | Royalc | 1. Catch-me-cave | Penguin | Macquarie | 5.1 |
| 2. Uukuniemi | Birds/mammals | Finland | 24.6 | |||
| 3. Rift valley fever | Ruminants | Africa/M. East | 57.4 | |||
| RNA pol | ||||||
| Finch Creeke | Nairovirus Bunyaviridae | Royal | 1. Tillamook [16] | Unknown | N. America | 4.6 |
| 2. Hazara [43] | Rodents | Asia | 23.1 | |||
| 3. Nairobi sheep disease [44] | Goats/Sheep | Africa/Asia | 24.6 |
All viruses listed are tick borne except Rift valley fever virus. Underlined viruses or virus families are or contain, respectively, known pathogens of humans or animals.
Based on amino acid identity.
Based on complete polyprotein sequence [4].
Laboratory based experimental infection only [15].
Based on partial sequence.
No significant neutralising activity was found in sera from 6 Antarctic division staff to our Gadgets Gully isolate or to Saumarez Reef virus (data not shown).
The orbivirus from ticks associated with a King penguin colony
The EM results of the 5 viruses isolated from ticks associated with the King penguin colony at Sandy Bay suggested they were all orbi- or coltiviruses (Fig. 2). PCR primers based on conserved regions of VP3 from Broadhaven, Yunnan and St Croix River viruses failed to generate products (data not shown). Using the method of Potgieter et al. 2002 [14] to isolate and amplify viral dsRNA, several viral gene segments were cloned and sequence was obtained from the VP5 (EU685329), VP4 (Cap) (EU685332, EU685333), NS2 (ViP) (EU685331) and NS3 (EU685330) genes. BLAST searches illustrated that sequence for this or closely related viruses have not previously been reported; we thus propose the name Sandy Bay virus for this virus. Phylogenetic analysis using the VP5 protein sequence is shown in Fig. 3B and the nearest relatives are described in Table 2. NS3 sequence is also available for Broadhaven virus and showed a 85% amino acid sequence identity with Sandy Bay virus NS3 sequence (ACD38336), with Peruvian Horse sickness and Yunnan viruses again the next closest relatives (Supplemental Fig. S1). Sandy Bay virus also showed homology with Yunnan virus VP4 and NS2 (ViP) sequences (EU685333 and EU685331, respectively) with amino acid sequence identities of 40% and 32%, respectively (data not shown). Similar relationships to those shown in Fig. 3B were evident when VP4 sequences were used (data not shown). No sequence is available for Broadhaven virus VP4 and NS2 (ViP). The relationship between the close relatives of Sandy Bay virus and the rest of the viruses in the family Reoviridae have recently been described elsewhere [15].
The phlebovirus from ticks associated with rockhopper penguins
Using our own (see Materials and methods) or published primers [16] for the nairovirus L polymerase gene, we were unable to obtain PCR products from the bunyavirus virus isolated from ticks associated with the rockhopper colony at Catch-me-cave. Using a primer, containing a 3′-terminus based on the conserved RNA segment termini of Uukuniemi virus, clones containing N (EU274386) and NS-s gene (EU274385) sequences of the S RNA segment of Precarious Point virus [3] were obtained. Using primers based on the Precarious Point virus sequence, N and NS-s gene sequences were obtained from the rockhopper penguin-associated virus (EU274384). The more conserved N protein sequence was used for phylogenetic analysis (Fig. 3C). The 5.1 % sequence divergence between the two viruses suggested the rockhopper penguin-associated virus was distinct from Precarious Point virus and we propose the name Catch-me-cave virus for this virus. The closest relatives for these viruses are described in Table 2.
A nairovirus from ticks associated with the royal penguin colony
Sequence for the royal penguin-associated bunyavirus was obtained using newly designed degenerate PCR primers based on the conserved regions of nairovirus RNA polymerase genes [16]. The same or similar sequences have not previously been reported; we thus propose the name Finch Creek virus for this viral isolate. Phylogenetic analysis is shown in Fig. 3D and the closest relatives are described in Table 2.
Discussion
The current study provides electron microscopic and sequence data for five arboviruses from four virus genera associated with Macquarie Island penguins and I. uriae ticks (summarized in Table 2). Together with the Southern Elephant Seal alphavirus [5], this island thus contains arboviruses from five of the seven genera of viruses known to contain arboviruses.
The nucleotide sequence similarity between Gadgets Gully virus isolated in the 1970's [3] and the isolate described herein suggest that this flavivirus has maintained remarkable genetic stability over ≈30 years, an observation consistent with the finding that flaviviruses have high fidelity RNA polymerases [17], [18]. Gadgets Gully virus has been referred to as a mammalian tick-borne flavivirus [4]. However, it has now been independently isolated twice from the seabird tick I. uriae associated with penguin colonies on Macquarie Island, suggesting that the enzootic host for Gadgets Gully virus is a seabird. Nevertheless, as shown previously [4], this virus does not group with the other seabird flaviviruses (Fig. 3A).
An orbivirus and a nairovirus have previously been isolated from I. uriae ticks associated with Royal Penguin colonies on Macquarie Island [19], [20]; Nugget virus (Kemerovo or Great Island virus serogroup) and Taggert virus (Sakhalin serogroup), respectively. As Broadhaven and Tillamook virus belong to the same serogroups [16], [21], [22], Sandy Bay and Finch Creek virus may be related to Nugget and Taggert virus, respectively. However, neither the viruses nor sequences from these latter viruses are available.
The observation that each virus was only associated with ticks from one penguin species might suggest that each virus is host specific. An alternative explanation is that fluctuating but nevertheless widespread immunity in these populations limits cycling of most of the viruses at any given time in any given bird population. As National Parks and Wildlife service has not allowed the trauma of blood collection from these recovering populations of nesting penguins, we were unable to resolve this issue. As arboviruses from these genera often have broad vertebrate host ranges [23], [24], [25], [26], [27], [28], and other seabird colonies have shown widespread immunity to multiple viruses from these genera [29], we would currently favour the latter explanation.
There is no currently indication that the penguin arboviruses pose a significant threat to the recovering penguin populations on Macquarie Island. To our knowledge there are currently only two reports of disease in penguins caused by tick-borne arboviruses from the genera described herein; (i) an uncharacterised arbovirus from I. uriae ticks from Macquarie Island (AUST-MI-411) was associated with disease and mortality in experimentally infected little blue penguins (Eudyptula minor) [9], and (ii) Humboldt penguins (Spheniscus Humboldti) and Black-footed penguins (S. demersus) have been fatally affected by West Nile virus [10]. Nevertheless, it is tempting to speculate that the gentoo penguins (unlike the other penguin species) move their nesting sites every year to avoid tick infestation [30] and perhaps also the associated arboviral infections. Interestingly, they are unique amongst Macquarie Island penguins in having no flavivirus antibodies [31].
The considerable sequence divergence of the viruses described herein from pathogenic viruses (Table 2) gives no indication that these penguin viruses pose an imminent threat to the health of humans or live stock. We and others [32] also failed to find evidence of human infection with Gadgets Gully or Saumerez reef virus. However, a serosurvey in the Australian Great Barrier Reef found that 4% of humans and birds were seropositive for Gadgets Gully [33]. Although no disease was reported [33], this study illustrated that Gadgets Gully virus can infect humans and has a range that includes tropical regions of Australia. In addition, tick borne arboviruses from seabirds have been reported to infect humans in Europe [34], and two seabird Sakhalin serogroup nairoviruses, Soldado and Avalon, have been reported to cause febrile illness and pruitus [35], and polyradiculoneuritis [36] in humans, respectively.
The rich arboviral diversity circulating in Macquarie Island penguins may be supported by the ideal transmission conditions associated with the crowded penguin nesting colonies. The geographic isolation of the island is probably not a barrier to the introduction of arboviruses as the extraordinary annual long distance migration of several seabird species may provide regular transcontinental transport of ticks and viruses. For instance, both sooty shearwaters [37] and Arctic Terns [38] visit Macquarie Island and migrate to all the continents of the northern hemisphere.
Materials and Methods
Tick collection
During the 6 day resupply of the Australian Antarctic Division base on Macquarie Island (12-16/3/02) by the RSV Aurora Australis, I. uriae ticks were collected from under rocks very close to three penguin colonies. An approach distance of no less than 5 m was maintained from the roosting birds at all times. Ticks were transported back to the base at ambient temperature, placed into Nunc vials, kept in a −70°C freezer at the base and on the return voyage, and were then couriered on dry ice from Tasmania to Queensland Institute of Medical Research, where they were stored at −70°C.
Virus isolation
Individual ticks were placed into a plastic Petri dish containing 500 ul cold medium (RPMI 1640 supplemented with 5% FCS and 100 ug/ml streptomycin and 100 IU/ml penicillin) and were extensively chopped with two scalpel blades. The suspension was vigorously pipetted and then placed into an Eppendorf tube, which was spun at 8000 g for 10 mins at 4°C. Individual supernatants were added to BHK-21 (ATCC CCL-10) and/or Vero (ATCC CCL-8) cells at 1/10 and 1/100 dilutions. Cells were maintained as described [5]. The cells (105 cells per 24 well) were cultured overnight before addition of tick extract, and the medium changed to RPMI 1640 supplemented with 2.5% FCS and antibiotics. When wells showed signs of cytopathic effects (CPE) ≈100 ul of supernatant was removed and placed onto fresh cells. After 3–5 passages viral stocks were prepared using cell grown in T75 flasks and were frozen in aliquots at −70°C.
Precarious Point virus (isolate CS0123) [3] was recovered from long term storage and had been passaged twice in suckling mouse brain and twice in BHK21 cells.
Electron microscopy
Vero or BHK cells were infected with virus. When CPE was visible, the cells were fixed (1 h) in 2.5% (v/v) phosphate buffered (pH 7.2, 300 mOsmol/Kg) glutaraldehyde, rinsed in the same buffer (3×30 min) and then transferred to the CSIRO Australian Animal Health Laboratory where the cells were processed into ultrathin sections and examined by transmission electron microscopy as described previously [5].
Virus sequencing
RNA was extracted from BHK or Vero cells (for the orbivirus) 2–3 days after infection (MOI≈1) using TRIzol reagent (Invitrogen). cDNA synthesis was performed using random hexamers, and Superscript III (Invitrogen) as per manufacturer's instructions. PCR amplification used 2 µl of cDNA, 200 nM primers and 2.5 U Pfu Turbo polymerase (Stratagene) as per manufacturer's instructions. Cycling conditions were 1 cycle of 95°C/1 min, 35 cycles of 95°C/30 sec, annealing at 40–55°C/30 sec and 68°C/5 mins. PCR products were gel purified using QIAquick Gel Extraction Kit (Qiagen). The purified PCR product was directly sequenced using Big Dye 3.1 sequencing mix (Applied Biosystems Inc).
The forward and reverse primers and annealing temperature used for the flaviviruses were 5′ GGATGGGGCAACCATTGTGG 3′, 5′ TCGTGCTGGCTTCCTGTTGG 3′ [13] and 50°C, for the E protein, and 5′ CCTACCACGCCAAGGTGGTCAG 3′, 5′ AGCARAAYGGGACCTCTTCCC 3′ and 55°C for NS5. The primer sequences for nairovirus L polymerase gene were forward 5′ GTIAGRAGYAARGTIRTITAIGA 3′ and reverse 5′ GCYTTIGGIGCYARIACIGCA 3′ or reverse 5′ GTRAARTCICCIGAIATRCA 3′ (annealing temperatures were 48 and 40°C, respectively). The primer sequences for the phlebovirus were based on sequence from Precarious Point virus and were forward 5′ GGCAGATGATGGACAGTGG 3′ and reverse 5′ GTCTGAGGAAGGCAAGAAGG 3′ (annealing temperature 55°C).
Precarious point virus RNA was amplified by RT-PCR using a single primer (5′-GCCGGAGCTCTGCAGAATTCACACAAAGAC-3′) in which the 3′-terminal 10 nucleotides matched the conserved sequence at the RNA segment termini of Uukuniemi virus. cDNA was synthesised using 0.5 µg total RNA, primer and Superscript II reverse transcriptase (Invitrogen) and amplified by PCR using the same primer, Taq DNA polymerase (Promega) and 2.5 mM MgCl2. PCR conditions were 95°C/2 min, 10 cycles of 95°C/30 sec, 25°C/30 sec, 72°C/3 min, 25 cycles of 95°C/30 sec, 60°C/30 sec, 72°C/3 min and final extension at 72°C/10 min. DNA products were gel purified and cloned into pGEM-T vector and DNA from colonies containing inserts sequenced.
Sequence from the orbivirus was obtained by preparing dsRNA from infected cells as described [14]. Purified viral dsRNA (≈500 ng) was ligated to the PC3 primer [14] using T4 RNA ligase 1 (New England Biolabs). The primer tagged dsRNA was purified using QIAprep Spin Miniprep Kit (QIAGEN) and reverse transcribed using the PC2 primer [14] and Superscript(III) (Invitrogen). The complementary cDNA strands were annealed by heating to 95°C and cooling slowly to 72°C. PCR amplification was undertaken using Pfu Turbo polymerase (Stratagene) and the PC2 primer . PCR conditions were an initial step of 72°C/5 mins and 94°C/2 mins, followed by 40 cycles 94°C/15 sec, 58°C/30 sec and 72°C/2 min, and finally 72°C/5 min. PCR products were gel purified, cut with BamH1 and cloned into pEGFP-N1 using T4 DNA polymerase (Roche). Viral fragments were sequenced using pEGFP-N1 primers 5′ CAGAGCTGGTTTAGTGAACCGTC 3′ and 5′ CCGTCCAGCTCGACCAG 3′.
Sequence analysis
Predicted amino acid sequences were determined using the Translate Tool ExPASy (http:au.expasay.org). The amino acid sequences were then used to query the non-redundant protein sequence database at NCBI using the blastp program (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Alignments of related sequences were created using the ClustalW program [39] incorporated into the MEGA3 program [40], and then adjusted manually if necessary (the alignments are available on request). Distances between sequences were proportion of amino acid differences (using complete deletion). Phylogenetic reconstructions were done using the neighbour-joining method [41] as implemented in MEGA3. Bootstrap support levels at nodes were calculated after 500 replications.
Supporting Information
(0.40 MB EPS)
Acknowledgments
We wish to thank all the staff at the Australian Antarctic Division, the National Parks and Wildlife Service, and the crew of the RSV Aurora Australis. Special thanks go to Kerri Clark for help with virus isolation, and Kate Kiefer and Tasmanian National Parks ranger Chris Hall for their help with tick collection on Macquarie Island. We would also like to thank Dr M-L Johnson (University of Queensland) for identification of the ticks.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Funding was provided by the Australian Antarctic Science Advisory Committee (Project 1334) and the Australian Centre for International and Tropical Health. Australian Antarctic Division, NH&MRC Australia. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wienecke B, Robertson G. Foraging areas of king penguins from Macquarie Island in relation to a marine protected area. Environ Manage. 2002;29:662–672. doi: 10.1007/s00267-0015-1. [DOI] [PubMed] [Google Scholar]
- 2.Pearce DA, Wilson WH. Viruses in the Antarctic ecosystem. Antarctic Science. 2003;15:319–331. [Google Scholar]
- 3.St George TD, Doherty RL, Carley JG, Filippich C, Brescia A, et al. The isolation of arboviruses including a new flavivirus and a new Bunyavirus from Ixodes (Ceratixodes) uriae (Ixodoidea: Ixodidae) collected at Macquarie Island, Australia, 1975–1979. Am J Trop Med Hyg. 1985;34:406–412. doi: 10.4269/ajtmh.1985.34.406. [DOI] [PubMed] [Google Scholar]
- 4.Grard G, Moureau G, Charrel RN, Lemasson JJ, Gonzalez JP, et al. Genetic characterization of tick-borne flaviviruses: new insights into evolution, pathogenetic determinants and taxonomy. Virology. 2007;361:80–92. doi: 10.1016/j.virol.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 5.La Linn M, Gardner J, Warrilow D, Darnell GA, McMahon CR, et al. Arbovirus of marine mammals: a new alphavirus isolated from the elephant seal louse, Lepidophthirus macrorhini. J Virol. 2001;75:4103–4109. doi: 10.1128/JVI.75.9.4103-4109.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gould EA, Higgs S, Buckley A, Gritsun TS. Potential arbovirus emergence and implications for the United Kingdom. Emerg Infect Dis. 2006;12:549–555. doi: 10.3201/eid1204.051010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daszak P, Cunningham AA, Hyatt AD. Emerging infectious diseases of wildlife–threats to biodiversity and human health. Science. 2000;287:443–449. doi: 10.1126/science.287.5452.443. [DOI] [PubMed] [Google Scholar]
- 8.Tuttle AD, Andreadis TG, Frasca S, Jr, Dunn JL. Eastern equine encephalitis in a flock of African penguins maintained at an aquarium. J Am Vet Med Assoc. 2005;226:2059–2062, 2003. doi: 10.2460/javma.2005.226.2059. [DOI] [PubMed] [Google Scholar]
- 9.Morgan IR, Westbury HA, Campbell J. Viral infections of little blue penguins (Eudyptula minor) along the southern coast of Australia. J Wildl Dis. 1985;21:193–198. doi: 10.7589/0090-3558-21.3.193. [DOI] [PubMed] [Google Scholar]
- 10.Komar N. West Nile virus: epidemiology and ecology in North America. Adv Virus Res. 2003;61:185–234. doi: 10.1016/s0065-3527(03)61005-5. [DOI] [PubMed] [Google Scholar]
- 11.Hyatt AD, Wise T. Comparison of immunogold methodologies for the detection of low copy number viral antigens in bluetongue virus (BTV)-infected cells. Micron. 1994;25:597–605. doi: 10.1016/0968-4328(94)90022-1. [DOI] [PubMed] [Google Scholar]
- 12.Eaton BT, Hyatt AD, White JR. Association of bluetongue virus with the cytoskeleton. Virology. 1987;157:107–116. doi: 10.1016/0042-6822(87)90319-9. [DOI] [PubMed] [Google Scholar]
- 13.Marin MS, Zanotto PM, Gritsun TS, Gould EA. Phylogeny of TYU, SRE, and CFA virus: different evolutionary rates in the genus Flavivirus. Virology. 1995;206:1133–1139. doi: 10.1006/viro.1995.1038. [DOI] [PubMed] [Google Scholar]
- 14.Potgieter AC, Steele AD, van Dijk AA. Cloning of complete genome sets of six dsRNA viruses using an improved cloning method for large dsRNA genes. J Gen Virol. 2002;83:2215–2223. doi: 10.1099/0022-1317-83-9-2215. [DOI] [PubMed] [Google Scholar]
- 15.Attoui H, Mohd Jaafar F, Belhouchet M, Aldrovandi N, Tao S, et al. Yunnan orbivirus, a new orbivirus species isolated from Culex tritaeniorhynchus mosquitoes in China. J Gen Virol. 2005;86:3409–3417. doi: 10.1099/vir.0.81258-0. [DOI] [PubMed] [Google Scholar]
- 16.Honig JE, Osborne JC, Nichol ST. The high genetic variation of viruses of the genus Nairovirus reflects the diversity of their predominant tick hosts. Virology. 2004;318:10–16. doi: 10.1016/j.virol.2003.09.021. [DOI] [PubMed] [Google Scholar]
- 17.Pugachev KV, Guirakhoo F, Ocran SW, Mitchell F, Parsons M, et al. High fidelity of yellow fever virus RNA polymerase. J Virol. 2004;78:1032–1038. doi: 10.1128/JVI.78.2.1032-1038.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pijlman GP, Suhrbier A, Khromykh AA. Kunjin virus replicons: an RNA-based, non-cytopathic viral vector system for protein production, vaccine and gene therapy applications. Expert Opin Biol Ther. 2006;6:135–145. doi: 10.1517/14712598.6.2.135. [DOI] [PubMed] [Google Scholar]
- 19.Gorman BM, Taylor J, Morton HC, Melzer AJ, Young PR. Characterization of Nugget virus, a serotype of the Kemerovo group of orbiviruses. Aust J Exp Biol Med Sci. 1984;62:101–115. doi: 10.1038/icb.1984.10. [DOI] [PubMed] [Google Scholar]
- 20.Doherty RL, Carley JG, Murray MD, Main AJ, Jr, Kay BH, et al. Isolation of arboviruses (Kemerovo group, Sakhalin group) from Ixodes uriae collected at Macquarie Island, Southern ocean. Am J Trop Med Hyg. 1975;24:521–526. doi: 10.4269/ajtmh.1975.24.521. [DOI] [PubMed] [Google Scholar]
- 21.Moss SR, Ayres CM, Nuttall PA. The Great Island subgroup of tick-borne orbiviruses represents a single gene pool. J Gen Virol. 1988;69(Pt 11):2721–2727. doi: 10.1099/0022-1317-69-11-2721. [DOI] [PubMed] [Google Scholar]
- 22.Lvov DK, Timofeeva AA, Gromashevski VL, Tsyrkin Yu M, Sazonov AA, et al. The ecology of Sakhalin virus in the north of the Far East of the U.S.S.R. J Hyg Epidemiol Microbiol Immunol. 1974;18:87–95. [PubMed] [Google Scholar]
- 23.Briese T, Bernard KA. West Nile virus–an old virus learning new tricks? J Neurovirol. 2005;11:469–475. doi: 10.1080/13550280500187617. [DOI] [PubMed] [Google Scholar]
- 24.Saikku P, Brummer-Korvenkontio M. Tick-borne viruses in Finland. Med Biol. 1975;53:317–320. [PubMed] [Google Scholar]
- 25.Traavik T, Mehl R. Tick-borne viruses in Norway. Med Biol. 1975;53:621–624. [PubMed] [Google Scholar]
- 26.Yunker CE. Tick-borne viruses associated with seabirds in North America and related islands. Med Biol. 1975;53:302–311. [PubMed] [Google Scholar]
- 27.Soldan SS, Gonzalez-Scarano F. Emerging infectious diseases: the Bunyaviridae. J Neurovirol. 2005;11:412–423. doi: 10.1080/13550280591002496. [DOI] [PubMed] [Google Scholar]
- 28.Erasmus BJ. The epizootiology of bluetongue: the African situation. Aust Vet J. 1975;51:196–198. doi: 10.1111/j.1751-0813.1975.tb00054.x. [DOI] [PubMed] [Google Scholar]
- 29.Nuttall PA, Kelly TC, Carey D, Moss SR, Harrap KA. Mixed infections with tick-borne viruses in a seabird colony in Eire. Arch Virol. 1984;79:35–44. doi: 10.1007/BF01314301. [DOI] [PubMed] [Google Scholar]
- 30.Gauthier-Clerc M, Clerquin Y, Handrich Y. Hyperinfestation by Ticks Ixodes uriae: a possible cause of death in adult King penguins, a long lived seabird. Colonial Waterbirds. 1998;21:229–233. [Google Scholar]
- 31.Morgan IR, Westbury HA, Caple IW, Campbell J. A survey of virus infection in sub-antarctic penguins on Macquarie Island, Southern Ocean. Aust Vet J. 1981;57:333–335. doi: 10.1111/j.1751-0813.1981.tb05839.x. [DOI] [PubMed] [Google Scholar]
- 32.Hawkes RA, Boughton CR, Naim HM, Wild J, Chapman B. Arbovirus infections of humans in New South Wales. Seroepidemiology of the flavivirus group of togaviruses. Med J Aust. 1985;143:555–561. [PubMed] [Google Scholar]
- 33.Humphery-Smith I, Cybinski DH, Byrnes KA, St George TD. Seroepidemiology of arboviruses among seabirds and island residents of the Great Barrier Reef and Coral Sea. Epidemiol Infect. 1991;107:435–440. doi: 10.1017/s0950268800049086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chastel C, Guiguen C, Le Lay G, Monnat JY, Quillien MC, et al. [Can arboviruses from seabird colonies in Brittany infect man?]. Rev Epidemiol Sante Publique. 1983;31:445–457. [PubMed] [Google Scholar]
- 35.Chastel C, Bailly-Choumara H, Bach-Hamba D, Le Lay G, Legrand MC, et al. [Tick-transmitted arbovirus in Maghreb]. Bull Soc Pathol Exot. 1995;88:81–85. [PubMed] [Google Scholar]
- 36.Chastel C, Le Lay G, Quillien MC, Simitzis AM. [Polyradiculoneuritis after tick bites and the Avalon virus]. Presse Med. 1986;15:441. [PubMed] [Google Scholar]
- 37.Shaffer SA, Tremblay Y, Weimerskirch H, Scott D, Thompson DR, et al. Migratory shearwaters integrate oceanic resources across the Pacific Ocean in an endless summer. Proc Natl Acad Sci U S A. 2006;103:12799–12802. doi: 10.1073/pnas.0603715103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reed KD, Meece JK, Henkel JS, Shukla SK. Birds, migration and emerging zoonoses: west nile virus, lyme disease, influenza A and enteropathogens. Clin Med Res. 2003;1:5–12. doi: 10.3121/cmr.1.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 41.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 42.Nunn MA, Barton TR, Wanless S, Hails RS, Harris MP, et al. Tick-borne Great Island Virus: (I) Identification of seabird host and evidence for co-feeding and viraemic transmission. Parasitology. 2006;132:233–240. doi: 10.1017/S0031182005008930. [DOI] [PubMed] [Google Scholar]
- 43.Foulke RS, Rosato RR, French GR. Structural polypeptides of Hazara virus. J Gen Virol. 1981;53:169–172. doi: 10.1099/0022-1317-53-1-169. [DOI] [PubMed] [Google Scholar]
- 44.Marczinke BI, Nichol ST. Nairobi sheep disease virus, an important tick-borne pathogen of sheep and goats in Africa, is also present in Asia. Virology. 2002;303:146–151. doi: 10.1006/viro.2002.1514. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(0.40 MB EPS)