Abstract
Members of the Nodal family regulate left-right asymmetry during vertebrate organogenesis, but it is unclear how Nodal signaling controls asymmetric morphogenesis at the cellular level. We used high-resolution time-lapse imaging in zebrafish to compare the movements of cardiomyocytes in the presence or absence of Nodal signaling. Loss of Nodal signaling in late-zygotic mutants for the Nodal co-receptor one-eyed pinhead (LZoep) abolished the leftward movement of cardiomyocytes. Global heart rotation was blocked but cardiomyocyte neighbor relationships were maintained as in wild type. Cardiomyocytes in LZoep mutants moved more slowly and less directionally than their wild-type counterparts. The phenotypes observed in the absence of Nodal signaling strongly resemble abnormalities found in BMP signaling mutants. These results indicate that a Nodal-BMP signaling cascade drives left-right heart morphogenesis by regulating the speed and direction of cardiomyocyte movement.
Keywords: Nodal, heart, zebrafish, morphogenesis, left-right
INTRODUCTION
Vertebrate organs such as heart, intestine, spleen and liver have left-right asymmetric morphologies and positions within the body cavity. Left-sided expression of Nodal signals during embryogenesis is thought to initiate organ lateralization (reviewed in Levin, 2005; Tabin, 2005; Raya and Belmonte, 2006; Shiratori and Hamada, 2006). Nodal signals are expressed in the left lateral plate mesoderm (LPM) and activate the asymmetric expression of downstream genes such as lefty2 (lft2) ( reviewed in Hamada et al., 2002; Schier, 2003; Shen, 2007). In the absence of Nodal signaling, organ laterality becomes randomized or disrupted (Levin et al., 1995; Chen et al., 1997; Yan et al., 1999; Bamford et al., 2000; Concha et al., 2000; Liang et al., 2000; Brennan et al., 2002; Long et al., 2003; Duboc et al., 2005), but it is largely unclear how Nodal affects asymmetric organogenesis at the cellular level.
The heart is the first organ to exhibit asymmetry in vertebrates (reviewed in Trinh and Stainier, 2004; Ramsdell, 2005). In the zebrafish, bilateral cardiac precursors form in the anterior lateral plate mesoderm (LPM) and then undergo a series of morphogenetic movements to assemble the heart tube. First, cardiac precursors migrate toward the embryonic midline and meet anteriorly and posteriorly. This process leads to the formation of a cardiac cone that is left-right symmetric (Holtzman et al., 2007). Second, in a process called cardiac jogging, cardiomyocytes are displaced both anteriorly and to the left (Chen et al., 1997; Rohr et al., 2008; Smith et al., 2008). Concomitantly, cells within the right side of the cardiac cone involute, and the entire cardiac cone undergoes a clockwise rotation (Rohr et al., 2008; Smith et al., 2008). These morphogenetic processes are the first manifestations of left-right asymmetry and lead to the positioning of right and left cardiac cone cells to ventral and dorsal parts of the heart tube, respectively. In parallel, central and peripheral cells of the cardiac cone give rise to the ventricle and atrium, respectively (reviewed in Trinh and Stainier, 2004). Finally, the linear heart tube loops rightwards, positioning the ventricle to the right side of the atrium (reviewed in Trinh and Stainier, 2004).
Several signaling pathways have been implicated as regulators of asymmetric heart morphogenesis, including components of the Nodal and BMP signaling pathways (Zhang and Bradley, 1996; Chen et al., 1997; Ramsdell and Yost, 1999; Schilling et al., 1999; Branford et al., 2000; Chang et al., 2000; Breckenridge et al., 2001; Hamada et al., 2002; Piedra and Ros, 2002; Schlange et al., 2002; Schier, 2003; Kishigami et al., 2004; Kishigami and Mishina, 2005; Chocron et al., 2007; Shen, 2007; Mine et al., 2008; Monteiro et al., 2008; Smith et al., 2008). The BMP signaling pathway is activated predominantly within the left side of the zebrafish heart (Chen et al., 1997; Chocron et al., 2007; Smith et al., 2008). When this asymmetry is disrupted by hyperactivation or block of BMP signaling, cardiomyocyte movement slows and meandering increases, and the heart tube fails to jogs leftwards or rotate clockwise (Chen et al., 1997; Schilling et al., 1999; Monteiro et al., 2008; Smith et al., 2008). Moreover, ectopic sources of BMP can direct the direction of cardiac jogging and cardiomyocyte movement (Smith et al., 2008). These results have suggested that asymmetric BMP signaling underlies the asymmetric movement and rotation of the zebrafish heart.
Several loss-of-function studies have revealed the importance of Nodal signaling in the lateralization of the zebrafish heart. The Nodal gene spaw is expressed in the left LPM and activates the Nodal pathway within the left side of the heart, as evidenced by the left-side specific activation of the Nodal downstream gene lft2 (Long et al., 2003). Morpholino-mediated knock-down of spaw affects the leftward but not the anterior displacement of the heart (Long et al., 2003). Loss of Nodal signaling leads to involution in the posterior region instead of the right section of the cardiac cone (Rohr et al., 2008). These morphogenetic abnormalities disrupt cardiac jogging and lead to a linear heart tube positioned at the midline. Heart morphogenesis is similarly disrupted in late-zygotic mutants for the Nodal co-receptor oep (Yan et al., 1999). The late expression of oep in the left and right LPM can be abolished by injecting oep mRNA into maternal-zygotic oep mutants, restoring oep function in mesendoderm induction but not in left-right patterning (Gritsman et al., 1999; Yan et al., 1999). In these LZoep mutants, the Nodal pathway is not activated in the LPM, and heart looping is randomized (Yan et al., 1999). Similarly, loss of FoxH1 (zebrafish schmalspur), a transcriptional mediator of Nodal signaling, disrupts cardiac jogging (Chen et al., 1997; Pogoda et al., 2000; Sirotkin et al., 2000).
These findings have suggested that the Nodal signal spaw activates the Nodal pathway via Oep and FoxH1 within the left side of the developing heart and promotes lateralization by driving asymmetric morphogenesis. Yet it is largely unclear how Nodal signaling affects heart morphogenesis at the single cell level. Interestingly, it has been found that asymmetric BMP expression is dependent on asymmetric Nodal signaling (Chen et al., 1997; Chocron et al., 2007). In light of recent cell tracking studies (Rohr et al., 2008; Smith et al., 2008), we therefore hypothesized that loss of Nodal signaling might lead to similar cellular defects as observed in loss- or gain-of-function BMP mutants. Here we test this idea and use time-lapse in vivo imaging to analyze the movement of cardiomyocytes in wild type and LZoep mutants. We find that disruption of Nodal signaling reduces the speed and directional movement of cardiomyocytes and disrupts the leftward morphogenesis and rotation of the cardiac cone. These results indicate that Nodal generates left-right asymmetry by regulating the speed and direction of cardiomyocyte movement. The striking similarity to BMP pathway mutants suggests that a TGFß signaling cascade involving Nodal and BMP underlies the asymmetric behavior of zebrafish cardiomyocytes.
RESULTS
Distinct expression patterns of Nodal signaling genes during heart lateralization
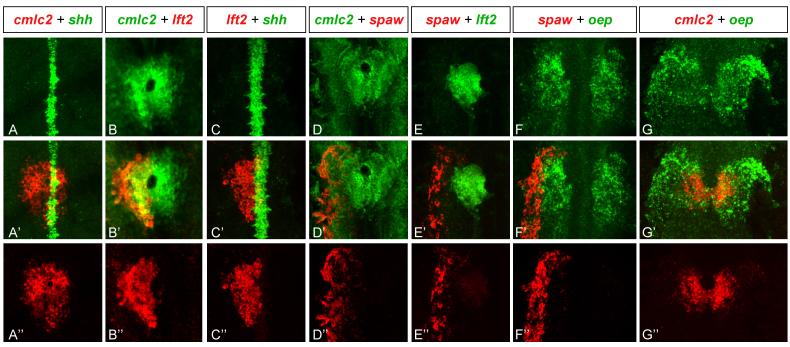
Previous studies have shown that spaw, lft2 and oep are expressed in the LPM in the region of the developing heart (Zhang et al., 1998; Bisgrove et al., 1999; Thisse and Thisse, 1999; Bisgrove et al., 2000; Long et al., 2003), but the exact overlap of expression patterns has not been studied in detail. To gain a higher resolution view of Nodal signaling during heart lateralization, we performed double in situ hybridization analyses of Nodal pathway genes, the heart marker cmlc2 (Yelon et al., 1999) and the midline marker shh (Krauss et al., 1993)(Fig.1). At the 21-somite stage cmlc2 marked the cardiac cone shortly after fusion of the left and right heart fields (Fig. 1A). spaw was expressed in the anterior LPM (Long et al., 2003), adjacent to but not overlapping the left domain of cmlc2 expression (Fig. 1D). In contrast, lft2 expression overlapped the left domain of cmlc2 expression (Bisgrove et al., 1999; Bisgrove et al., 2000; Long et al., 2003) (Fig. 1B and C) and juxtaposed the spaw expression domain (Fig. 1E). oep was expressed in the left and right LPM (Zhang et al., 1998), adjacent to and within the developing heart (Fig. 1E and G), thus defining the maximum territory competent to respond to Nodal signals (Gritsman et al., 1999). Together with previous studies (Zhang et al., 1998; Bisgrove et al., 1999; Thisse and Thisse, 1999; Bisgrove et al., 2000; Long et al., 2003), these results indicate that spaw expression in the left non-cardiac LPM activates the Nodal signaling pathway in adjacent ipsilateral heart progenitors in a oep-dependent manner at the onset of asymmetric morphogenesis.
Figure 1. Expression of Nodal pathway genes in the lateral plate mesoderm.
Confocal images of double fluorescent in situ RNA hybridization of spaw, lft2, and oep with respect to the midline marker shh and the heart marker cmlc2 at the 21-somite stage. Single channel expression (A-G and A”-G”) and overlay (A’-G’). All images are dorsal views of the embryo, anterior to the top and left to the left. Note broad expression of oep in left and right LPM (F and G) and more restricted expression of spaw in left non-cardiac LPM (F’ and F”) and lft2 in left cardiac LPM (B’ and B”). spaw is expressed adjacent to lft2 (E’).
Position-dependent differences in cardiomyocyte movements during heart lateralization
To analyze the cellular basis of heart lateralization, we used high-resolution 4D confocal time-lapse imaging of cardiomyocyte movement using a similar approach as Smith et al. (2008). Transgenic cmlc2:gfp embryos express GFP in all cardiomyocytes throughout cardiac morphogenesis (Huang et al., 2003) (Supplemental Movie 1). Cell tracking and isosurface rendering revealed that the first sign of asymmetry was apparent shortly after cardiac fusion, when anterior and leftward displacement initiated and cardiomyocytes located on the right involuted (Fig. 2A-E; Supplemental Movies 1 and 2) (Rohr et al., 2008; Smith et al., 2008). To gain quantitative insights into heart morphogenesis, we tracked individual cells using Imaris software and measured the direction, displacement, meandering, and speed of cells (Fig. 3; Supplemental Movie 1; Supplemental Fig. 1) (Smith et al., 2008). In contrast to a recent study (Smith et al., 2008), we analyzed heart morphogenesis per se, independent of the movement of the entire heart within the embryo. Using the position of the lumen as a stationary reference point, we tracked the movement of cells within the heart. This allowed us to measure cardiomyocyte movements with respect to each other rather than the potential drift of the embryo during imaging or the displacement of the entire heart within the embryo. As described below and in Supplemental Figure 2, our conclusions largely agree with the recent study by Smith et al. (2008), despite the different means of analysis.
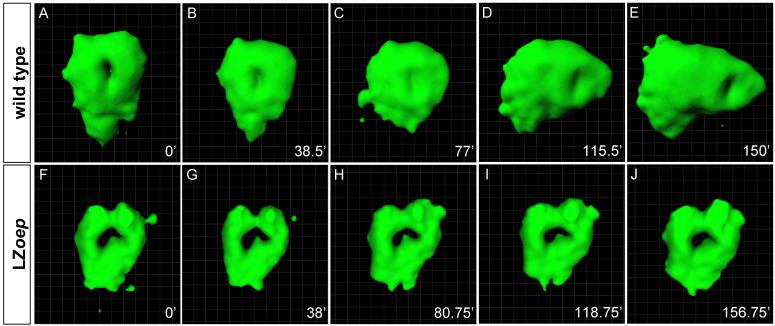
Figure 2. Isosurface rendering of heart morphogenesis in wild type and LZoep mutants.
cmlc2-GFP cells label the cardiac cone and heart tube. Imaris software created an isosurface representing points of constant fluorescent intensity within the 3D volume. Cardiac cone is initially symmetric in both the wild type (A, B) and the LZoep mutant (F, G). Note the leftwards and anterior displacement of the heart in wild type (C-E) but not in LZoep mutants (H-J). For detailed views from different angles see Supplemental Movies 2 (wild type) and 6 (LZoep). Pictures were cropped to keep the heart in the center and do not maintain the positions of the coordinate systems. For movements with respect to coordinate systems, see Supplemental Movies 2 (wild type) and 6 (LZoep).
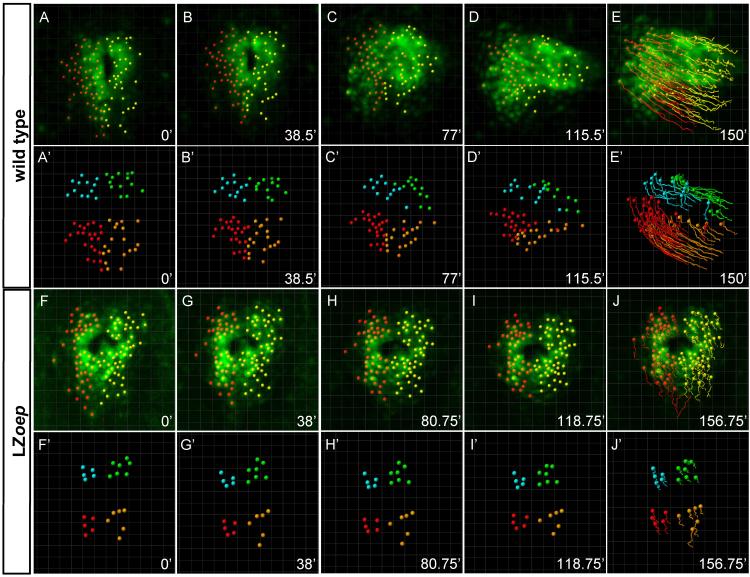
Figure 3. Tracking of single cells in wild type and LZoep mutants.
Confocal 3D-images showing individual cmlc2-GFP cells that were manually tracked starting at the onset of cardiac cone formation. First (A-E) and third (F-J) rows: green cmlc2-GFP expression; red: cells within the left side of cardiac cone; yellow: cells within the right side of cardiac cone. For detailed views from different angles see Supplemental Movies 1 (wild type) and 5 (LZoep). Second (A’-E’) and fourth (F’-J’) rows: tracking of quadrants. Quadrants were defined by grouping left-anterior cells (blue), left-posterior cells (red), right anterior cells (green) and right-posterior cells (orange). For detailed views from different angles see Supplemental Movies 3 (wild type) and 7 (LZoep). Pictures were cropped to keep the heart in the center and do not maintain the position of the coordinate systems. For movements with respect to coordinate systems, see Supplemental Movies 3 (wild type) and 7 (LZoep).
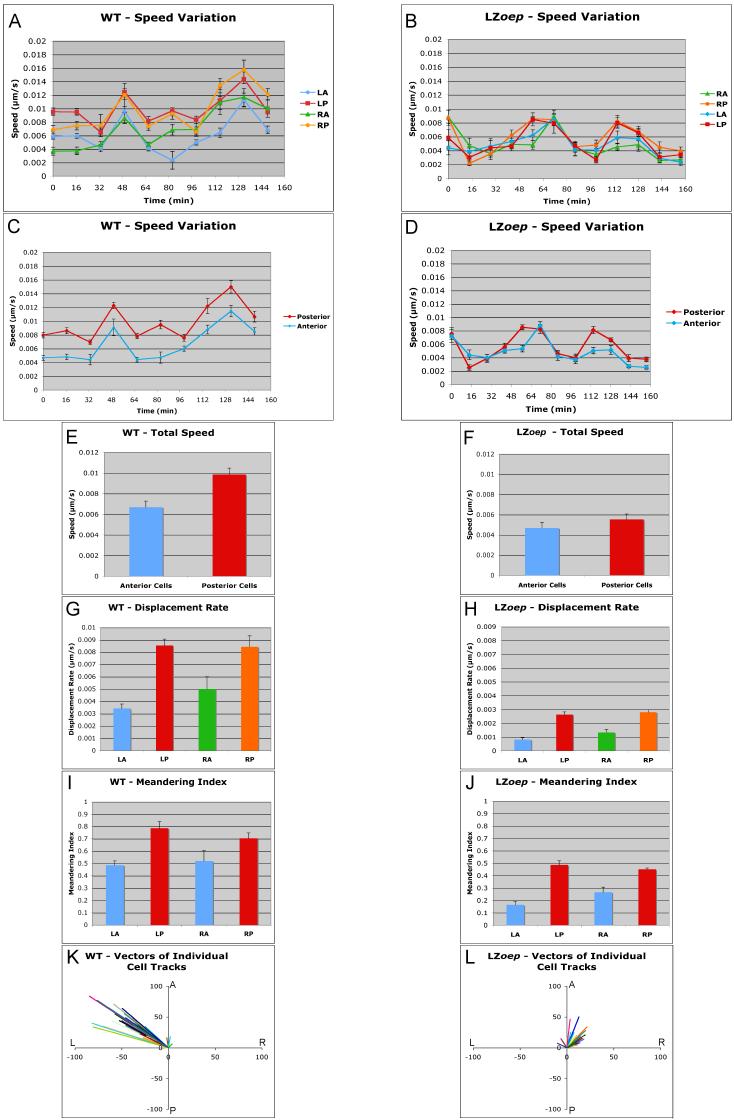
To quantitate heart morphogenesis, we first compared four territories: left anterior (LA), right anterior (RA), left posterior (LP) and right posterior (RP) (Suppl. Movie 3; Figs. 3, 4 and 5). Analysis of each cell track provided measurements of the speed, displacement and meandering of a cell (Smith et al., 2008). Speed was defined as distance moved/time interval, displacement rate as distance from first to last location/entire time interval, and meandering index as total distance traveled/displacement (Suppl. Fig. 1) (Smith et al., 2008). Comparison of posterior and anterior cells revealed that posterior cells moved faster than anterior cells, had higher displacement rates, and a higher meandering index (Fig. 4A, C, E, G, and I). These results indicate that posterior cells move faster and more directionally than anterior cells (Smith et al., 2008). To determine the direction of movement, we performed vector analyses that included displacement length and angle with respect to the anterior-posterior and left-right axes. These measurements showed that the large majority of individual cardiomyocytes moved leftwards and anteriorly (Fig. 4K), consistent with the overall morphogenetic change of the cardiac cone (Rohr et al., 2008; Smith et al., 2008).
Figure 4. Quantitative analysis of anterior and posterior cell movements in wild type and LZoep mutants.
cmlc2-GFP cells in left-anterior (LA), left-posterior (LP), right-anterior (RA), and right-posterior (RP) quadrants were tracked and different parameters were measured using Imaris Software. Speed variation indicates speed of a cell during a given time interval. Total speed, displacement rate, and meandering index were determined as described in Experimental Procedures and Supplemental Figure 1. Note the higher speed (A, C and E), displacement rate (G) and meandering index (I) of posterior cells as compared to anterior cells in wild type. These differences are reduced in LZoep mutants. Cardiomyocytes in LZoep mutants have lower speed (B, D and F), displacement rate (H) and meandering index (J) as compared to wild type. Differences between posterior and anterior cells are attenuated in LZoep mutants. Vectors of individual cells illustrate the angle and length of x/y displacement of cells (A: anterior; P: posterior; L: left; R: right). Note the leftwards and anterior displacement of wild type cells (K). LZoep mutant cells displace less and mostly anteriorly (L). Statistics: E) p<0.05 ; F) p>0.05; G) LA vs LP, p<0.05; RA vs RP, p<0.05; LA vs RA, p>0.05; LP vs RP, p>0.05; H) LA vs LP, p<0.05; RA vs RP, p<0.05; LA vs RA, p>0.05; LP vs RP, p>0.05; I) LA vs LP, p<0.05; RA vs RP, p<0.05; LA vs RA, p>0.05; LP vs RP, p>0.05; J) LA vs LP, p<0.05; RA vs RP, p<0.05; LA vs RA, p>0.05; LP vs RP, p>0.05.
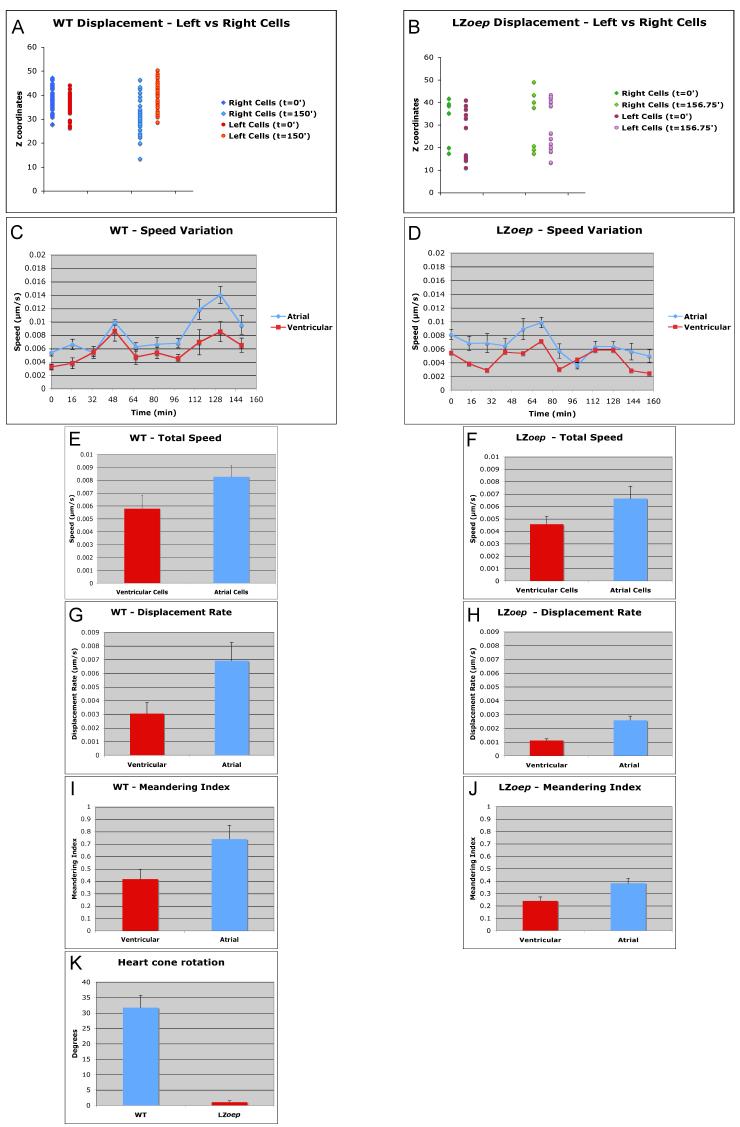
Figure 5. Quantitative analysis of left, right, ventricular and atrial cell movements in wild type and LZoep mutants.
Left and right cmlc2-GFP cells were tracked and Z-axis (dorsal-ventral) displacement of cells was measured. Note the asymmetric, ventral displacement of cells on the right in wild type (A) but not in LZoep mutants (B). Ventricular cells (located at the apex of the cardiac cone) and atrial cells (located at the base of the cardiac cone) were tracked using Imaris. Speed variation indicates speed of a cell during a given time interval. Total speed, displacement rate, and meandering index were determined as described in Experimental Procedures and Supplemental Figure 1. Note the higher speed (C and E), displacement rate (G) and meandering index (I) of atrial cells as compared to ventricular cells in wild-type. These differences are reduced in LZoep mutants. LZoep mutant cells have lower speed (D and F), displacement rate (H) and meandering index (J) compared to wild type. (K) Degree of clockwise rotation of cardiac cone. Note the lack of rotation in LZoep mutants. Statistics: E-K p<0.05.
To determine if there are differences between cardiomyocytes located on the left or right side, we compared speed, displacement rate and meandering index. Consistent with the findings by Smith et al. (2008), we did not find significant differences between left and right cells using these measures (Fig. 4A, G, I). However, extending previous studies (Rohr et al., 2008; Smith et al., 2008), we quantified the dorsal-ventral displacement of cells and found that cells on the right, but not the left, were displaced ventrally (Fig. 5A). This observation is consistent with the observed involution of cells located in the right heart cone (Rohr et al., 2008) and reveals that the first signs of asymmetric morphogenesis are the leftwards displacement of all cardiomycytes and the ventral displacement of cardiomyocytes located in the right heart cone.
The large number of tracked cells also allowed us to study the movement of cells located in the inner and outer regions of the cardiac cone. Consistent with gene expression patterns for atrial and ventricular markers (Yelon et al., 1999), we found that the atrial precursors were located in the outer region of the cone and had a higher speed, displacement rate and meandering index than the ventricular cells, which were located in the inner region of the cone (Fig. 5C, E. G, and I).
To quantify the global changes in heart morphogenesis, we measured the initial and final position of cells placed in regions LA, LP, RA and RP. This analysis confirmed the previously observed ∼30° clockwise rotation of the cardiac cone. (Fig. 5K) (Smith et al., 2008). The clockwise rotation and the leftwards and anterior movement of individual cardiomyocytes reflected the overall morphogenesis of the heart. This result indicated that the coherent movement of individual cardiomyocytes underlies overall heart morphogenesis. Coherence could be generated if neighbor relationships between individual cardiomyocytes did not change extensively (Rohr et al., 2008; Smith et al., 2008). Tracking of groups of cells confirmed that neighbor relationships were maintained during heart lateralization (Suppl. Movie 4). These results confirm and extend previous studies (Rohr et al., 2008; Smith et al., 2008) and indicate that differential but coherent cell displacement and direction underlie asymmetric heart morphogenesis in wild type.
Loss of Nodal signaling decreases the speed and increases the meandering of cardiomyocytes
To determine how and when lack of Nodal signaling affects heart morphogenesis, we analyzed LZoep embryos (Suppl. Movies 5-8). These mutants lack spaw and lft2 expression in the left LPM, indicating the loss of Nodal signaling, but form normal mesendoderm and are viable (Gritsman et al., 1999; Yan et al., 1999). To analyze overall heart morphogenesis, we used isosurface rendering. Cardiomyocytes formed the cardiac cone in LZoep mutants, but in striking contrast to wild type, the heart failed to tilt leftwards and displayed reduced anterior displacement (Fig. 2F-J; Suppl. Movie 6). Consequently, the heart in LZoep mutants remained positioned at the midline. As an additional measure of asymmetry, we analyzed cardiac rotation. Strikingly, clockwise rotation was abolished in LZoep mutants (Fig. 5K). These results support the model that absence of Nodal signaling disrupts the earliest steps of cardiac lateralization.
To quantitatively compare wild type and LZoep mutants, we used cell tracking analyses (Figs. 3, 4 and 5; Suppl. Movies 5 and 7). Strikingly, cardiomyocytes in LZoep mutants moved more slowly and meandered more (Fig. 4B, D, F, and J). These defects resulted in a strongly reduced displacement rate (Fig. 4J). To determine the direction of movement, we performed vector analysis. Cardiomyocytes in LZoep mutants did not move leftwards and displaced anteriorly much less than wild type (Fig. 4L). These results reveal that cardiomyocyte movement in LZoep mutants is slower, less directional, and largely symmetric compared to wild type.
To determine if different regions in the LZoep mutant heart have distinct characteristics as in wild type, we performed detailed tracking analyses. Strikingly, differences between anterior and posterior, between left and right, and between atrial and ventricular cells were strongly reduced or absent (Fig. 4 and 5). For example, right cells were not displaced ventrally (Fig. 5B) and posterior cells were not significantly faster than anterior cells (Fig. 4F). These results suggest that the loss of asymmetric behavior of cardiomyocytes underlies the loss of cardiac cone asymmetry in LZoep mutants.
To determine if cardiomyocytes in LZoep mutants lose their coherence, we analyzed neighbor relationships (Suppl. Movie 8). Despite the abnormal direction and speed of cardiomycytes, cells continued to move as cohorts, as in wild type. These results indicate that Nodal signaling is not required for the integrity of the heart tube but regulates the speed and directionality of cardiomyocyte movement.
DISCUSSION
The cellular mechanisms underlying left-right asymmetric organogenesis are largely elusive. A deeper understanding rests on detailed analyses of cell movements, shapes and rearrangements during this process in wild type (Horne-Badovinac et al., 2003; Yashiro et al., 2007; Davis et al., 2008; Kurpios et al., 2008; Rohr et al., 2008; Smith et al., 2008). For instance, cell shape analyses in the amniote dorsal mesentery have suggested a structural basis for the asymmetric tilting and rotation of the gut tube (Davis et al., 2008; Kurpios et al., 2008) Columnar epithelization and mesenchymal condensation on the left side lead the mesentery to take on a trapezoidal shape that leads to a leftward tilt of the gut. In the zebrafish heart, cell movement analyses have revealed that cardiomyocytes move anteriorly and leftwards as a cohort while the heart cone rotates clockwise (Chen et al., 1997; Rohr et al., 2008; Smith et al., 2008). Our studies confirm these results and reveal two additional quantitative differences. First, cells on the right are displaced more ventrally than their left-side neighbors. This displacement reflects the involution of cells in the right heart cone (Rohr et al., 2008). Second, atrial cells move faster and more directionally than ventricular cells and therefore have higher displacement rates. These results highlight the potential of single cell tracking to dissect the cellular processes underlying asymmetric morphogenesis during left-right development.
Components of the Nodal signaling pathway are expressed asymmetrically during left-right heart morphogenesis (Hamada et al., 2002; Schier, 2003; Shen, 2007). Our double in situ hybridization analysis confirms and extends previous studies and shows that spaw and lft2 are expressed in adjacent regions of the left LPM, with only lft2 being activated in heart progenitors (Zhang et al., 1998; Bisgrove et al., 1999; Thisse and Thisse, 1999; Bisgrove et al., 2000; Long et al., 2003). The largely non-overlapping expression domains of spaw and lft2 are unusual for nodal and lefty genes. For example, during mesendoderm induction, the Nodal signals cyclops and squint are co-expressed with lefty1 and lefty2 in the blastula margin (Schier and Talbot, 2005). It is conceivable that lft2 can only be activated in the heart because other LPM cells lack components of the Nodal signaling pathway (Gritsman et al., 1999). However, our finding that the Nodal co-receptor oep is expressed bilaterally in all LPM cells suggests that the restricted expression of oep is unlikely to restrict Nodal signaling to the heart. It is possible that other components of the Nodal signaling pathway are absent in non-cardiac cells. Blocking BMP signaling during mid-somitogenesis blocks induction of lft2 (Chocron et al., 2007), indicating that other signaling pathways or transcription factors might also restrict lft2 expression to the left heart field.
Nodal signaling is required for normal left-right morphogenesis (Hamada et al., 2002; Schier, 2003; Shen, 2007). Previous studies have shown that loss of Nodal signaling in zebrafish results in abnormal heart morphogenesis, resulting in randomized heart looping and loss of initial leftwards jogging (Chen et al., 1997; Yan et al., 1999; Long et al., 2003; Rohr et al., 2008). For example, cardiac cells in spaw morphants do not displace leftwards and cells on the right side of the heart do not involute (Long et al., 2003; Rohr et al., 2008). Our quantitative analysis of LZoep mutants shows that loss of Nodal signaling affects the direction of movement of individual cardiomyocytes during early heart development. Anterior movement is reduced and leftwards migration is abolished. In addition, our results show that loss of Nodal signaling also affects the speed of cardiomyocyte movement. Cardiomyocytes move more slowly and meander more, resulting in strongly reduced displacement. The differences between anterior/posterior, left/right and atrial/ventricular cells are reduced or absent. Taken together, these results suggest that Nodal signaling provides a directional cue and promotes the speed of heart cells.
It is unclear whether specific cell shape changes underlie the differential movement and rotation of anterior/posterior, left/right and atrial/ventricular cardiomyocytes. Studies of cell movements during zebrafish gastrulation and germ cell migration have shown that changes in cell polarity and protrusions are associated with abnormal movement (Solnica-Krezel, 2005; Raz and Reichman-Fried, 2006; Rohde and Heisenberg, 2007). Cardiomyocytes display small extensions restricted to the basal surface and chamber-specific cell shapes (Rohr et al., 2008). Future studies will address whether these properties are regulated by Nodal signaling.
The cellular and morphogenetic phenotypes in LZoep mutants become apparent at the same time, making it difficult to resolve cause-and-effect relationships. Slower and more meandering movements of individual cells might lead to a block of the overall asymmetric morphogenesis of the heart. Alternatively, overall abnormal morphogenesis and lack of directionality might restrict the speed of individual cells. Detailed analysis of additional mutants might be able to separate the processes we quantified here. For instance, mutants that do not slow down cardiomyocytes might still lead to left-right defects.
How does Nodal signaling drive asymmetric morphogenesis? The activation of the Nodal signaling pathway in the left heart cone suggests that Nodal regulates heart morphogenesis directly. This contrasts with the rotation of the gut, which is driven by shape changes in the associated dorsal mesentery, and the asymmetric maintenance of the sixth branchial arch artery, which is regulated by differential haemodynamics caused by asymmetric development of the heart outflow tract (Yashiro et al., 2007; Davis et al., 2008; Kurpios et al., 2008). It is unclear how activation of Nodal signaling within the left side of the cardiac cone changes cell behavior throughout the heart. We favor a model wherein Nodal signaling acts quite locally in lft2-expressing cells and secondary interactions within the heart lead to the complex morphogenetic behaviors. These interactions could involve relay signals (lft2-expressing cells signal to neighboring cells) or mechano-physical interactions (shape changes in lft2-expressing cells induce shape changes in neighboring cells). It is thus conceivable that a small group of cells (e.g. the lft2-expressing cells) act as organizers to drive asymmetric morphogenesis. Mosaic analysis could address this question.
Interestingly, morphogenesis in the absence of Nodal signaling is not completely blocked. Anterior movement still occurs and heart jogging and heart looping eventually take place in the absence of Nodal signaling, resulting in heterotaxic but viable animals (Yan et al., 1999). These observations suggest that there are Nodal-independent morphogenetic events that might be driven autonomously within the heart or indirectly by surrounding tissue. For example, it is conceivable that the elongation and straightening of the embryo (Kimmel et al., 1995) contributes to asymmetric morphogenesis as the heart interacts with surrounding tissues.
Recent studies have revealed that asymmetric BMP signaling regulates asymmetric heart morphogenesis in zebrafish (Chen et al., 1997; Schilling et al., 1999; Chocron et al., 2007; Monteiro et al., 2008; Smith et al., 2008). Loss of BMP signaling reduces the speed, displacement rate and meandering index of heart cells and abolishes leftward displacement and rotation (Smith et al., 2008). These phenotypes are strikingly similar to the LZoep phenotypes we describe here. Moreover, ectopic sources of BMP can attract cardiomyocytes (Smith et al., 2008). Nodal signaling is required for the asymmetric expression of BMP4 in the heart cone (Chen et al., 1997; Chocron et al., 2007). It is therefore conceivable that the effects of asymmetric Nodal signaling are mediated via asymmetric BMP signaling. Moreover, BMP signaling and Nodal signaling also act further upstream during left-right development (Zhang and Bradley, 1996; Chen et al., 1997; Ramsdell and Yost, 1999; Schilling et al., 1999; Branford et al., 2000; Chang et al., 2000; Breckenridge et al., 2001; Hamada et al., 2002; Piedra and Ros, 2002; Schlange et al., 2002; Schier, 2003; Kishigami et al., 2004; Kishigami and Mishina, 2005; Chocron et al., 2007; Shen, 2007; Mine et al., 2008; Monteiro et al., 2008). Hence, a cascade of asymmetrically activated TGFß signaling pathways appears to drive asymmetric heart morphogenesis.
EXPERIMENTAL PROCEDURES
Fish lines
Tg(cmlc2:gfp) and LZoep lines were described previously (Yan et al., 1999; Huang et al., 2003).
In situ hybridization
Double fluorescent in situ hybridization probes were labeled with fluorescein (Roche) and digoxygenin (Roche). Detection was based on fluorescein or Cy3 tyramide deposition (Perkin Elmer; (Schoenebeck et al., 2007)). Embryos were mounted in benzylbenzoate/benzylalcohol for photography.
Imaging and data analysis
At the 15-somite stage embryos were manually dechorionated and then mounted at the 18-somite stage in glass-bottom culture dishes using 0.8% low-melt agarose in E3 embryo medium. Imaging was performed on a Zeiss LSM 5 Pascal confocal microscope with LSM software. The microscope stage was heated to 30°C to keep embryos at 28.5°C during the time-lapse recordings. Stacks were acquired every 5.5 minutes for wild type and 4.75 minutes for LZoep mutants. Z-stacks were rendered in 3D and analyzed with Imaris software (Bitplane). 3D singe cell trackings and measurements (x/y/z coordinates) were performed manually using Imaris. The base of the lumen was chosen as reference point for the x/y/z coordinate system to analyze heart morphogenesis per se and to avoid confounding effects caused by the potential drift of the embryo during imaging or the movement of the heart within the embryo. Each measurement was obtained from the cells depicted in Fig.3A’ and Fig3.F’ and in Supplemental Movies 3 and 7. The time window for quantitative measurements was from the onset of cardiac cone formation until the full leftward displacement of the cardiac cone in wild type (t=150.0 minutes). Since the same morphological landmark does not develop in LZoep mutants and to directly compare LZoep cardiomyocyte movement to wild type cardiomyocyte movement, the time interval for analysis of LZoep mutants was 156.75 minutes from the formation of the cardiac cone. Cell positions were defined by x/y/z coordinates at each time point. Time intervals between each tracked time point were 16.5 minutes in wild type and 14.25 minutes in LZoep mutants. The following parameters were measured (Smith et al., 2008): speed = track length/time interval; total speed = total track length/total time (i.e. sum of the average distance traveled by the cells within each time interval divided by the total track time); displacement rate = total displacement length/total time (i.e. position of each cell at the end of the track - position at t=0 divided by the total track time); meandering index = total displacement length/total track length; displacement of left vs. right cells: the z-axis coordinates of each cell were analyzed at t=0 and at the end of the track. Rotation angle was obtained by drawing an imaginary square connecting cells in each quadrant of the heart cone at the beginning and at the end of the time-lapse (Smith et al., 2008). Statistical analysis and graphics were obtained in Excel (Microsoft).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Steven Zimmerman for fish care and members of the Schier and Yelon labs for helpful discussions. Supported by NIH and Gulbenkian.
Grants: NIH R01 GM56211 (A.F.S); Gulbenkian PhD Program SFRH/BD/11801/2003 (M.I.M.C.-B.)
REFERENCES
- Bamford RN, Roessler E, Burdine RD, Saplakoglu U, dela Cruz J, Splitt M, Goodship JA, Towbin J, Bowers P, Ferrero GB, Marino B, Schier AF, Shen MM, Muenke M, Casey B. Loss-of-function mutations in the EGF-CFC gene CFC1 are associated with human left-right laterality defects. Nat Genet. 2000;26:365–369. doi: 10.1038/81695. [DOI] [PubMed] [Google Scholar]
- Bisgrove BW, Essner JJ, Yost HJ. Regulation of midline development by antagonism of lefty and nodal signaling. Development. 1999;126:3253–3262. doi: 10.1242/dev.126.14.3253. [DOI] [PubMed] [Google Scholar]
- Bisgrove BW, Essner JJ, Yost HJ. Multiple pathways in the midline regulate concordant brain, heart and gut left-right asymmetry. Development. 2000;127:3567–3579. doi: 10.1242/dev.127.16.3567. [DOI] [PubMed] [Google Scholar]
- Branford WW, Essner JJ, Yost HJ. Regulation of gut and heart left-right asymmetry by context-dependent interactions between xenopus lefty and BMP4 signaling. Dev Biol. 2000;223:291–306. doi: 10.1006/dbio.2000.9739. [DOI] [PubMed] [Google Scholar]
- Breckenridge RA, Mohun TJ, Amaya E. A role for BMP signalling in heart looping morphogenesis in Xenopus. Dev Biol. 2001;232:191–203. doi: 10.1006/dbio.2001.0164. [DOI] [PubMed] [Google Scholar]
- Brennan J, Norris DP, Robertson EJ. Nodal activity in the node governs left-right asymmetry. Genes Dev. 2002;16:2339–2344. doi: 10.1101/gad.1016202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H, Zwijsen A, Vogel H, Huylebroeck D, Matzuk MM. Smad5 is essential for left-right asymmetry in mice. Dev Biol. 2000;219:71–78. doi: 10.1006/dbio.1999.9594. [DOI] [PubMed] [Google Scholar]
- Chen JN, van Eeden FJ, Warren KS, Chin A, Nusslein-Volhard C, Haffter P, Fishman MC. Left-right pattern of cardiac BMP4 may drive asymmetry of the heart in zebrafish. Development. 1997;124:4373–4382. doi: 10.1242/dev.124.21.4373. [DOI] [PubMed] [Google Scholar]
- Chocron S, Verhoeven MC, Rentzsch F, Hammerschmidt M, Bakkers J. Zebrafish Bmp4 regulates left-right asymmetry at two distinct developmental time points. Dev Biol. 2007;305:577–588. doi: 10.1016/j.ydbio.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Concha ML, Burdine RD, Russell C, Schier AF, Wilson SW. A nodal signaling pathway regulates the laterality of neuroanatomical asymmetries in the zebrafish forebrain. Neuron. 2000;28:399–409. doi: 10.1016/s0896-6273(00)00120-3. [DOI] [PubMed] [Google Scholar]
- Davis NM, Kurpios NA, Sun X, Gros J, Martin JF, Tabin CJ. The chirality of gut rotation derives from left-right asymmetric changes in the architecture of the dorsal mesentery. Dev Cell. 2008;15:134–145. doi: 10.1016/j.devcel.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboc V, Rottinger E, Lapraz F, Besnardeau L, Lepage T. Left-right asymmetry in the sea urchin embryo is regulated by nodal signaling on the right side. Dev Cell. 2005;9:147–158. doi: 10.1016/j.devcel.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Gritsman K, Zhang J, Cheng S, Heckscher E, Talbot WS, Schier AF. The EGF-CFC protein one-eyed pinhead is essential for nodal signaling. Cell. 1999;97:121–132. doi: 10.1016/s0092-8674(00)80720-5. [DOI] [PubMed] [Google Scholar]
- Hamada H, Meno C, Watanabe D, Saijoh Y. Establishment of vertebrate left-right asymmetry. Nat Rev Genet. 2002;3:103–113. doi: 10.1038/nrg732. [DOI] [PubMed] [Google Scholar]
- Holtzman NG, Schoenebeck JJ, Tsai HJ, Yelon D. Endocardium is necessary for cardiomyocyte movement during heart tube assembly. Development. 2007;134:2379–2386. doi: 10.1242/dev.02857. [DOI] [PubMed] [Google Scholar]
- Horne-Badovinac S, Rebagliati M, Stainier DY. A cellular framework for gut-looping morphogenesis in zebrafish. Science. 2003;302:662–665. doi: 10.1126/science.1085397. [DOI] [PubMed] [Google Scholar]
- Huang CJ, Tu CT, Hsiao CD, Hsieh FJ, Tsai HJ. Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev Dyn. 2003;228:30–40. doi: 10.1002/dvdy.10356. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kishigami S, Mishina Y. BMP signaling and early embryonic patterning. Cytokine Growth Factor Rev. 2005;16:265–278. doi: 10.1016/j.cytogfr.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Kishigami S, Yoshikawa S, Castranio T, Okazaki K, Furuta Y, Mishina Y. BMP signaling through ACVRI is required for left-right patterning in the early mouse embryo. Dev Biol. 2004;276:185–193. doi: 10.1016/j.ydbio.2004.08.042. [DOI] [PubMed] [Google Scholar]
- Krauss S, Concordet JP, Ingham PW. A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell. 1993;75:1431–1444. doi: 10.1016/0092-8674(93)90628-4. [DOI] [PubMed] [Google Scholar]
- Kurpios NA, Ibanes M, Davis NM, Lui W, Katz T, Martin JF, Belmonte JC, Tabin CJ. The direction of gut looping is established by changes in the extracellular matrix and in cell:cell adhesion. Proc Natl Acad Sci U S A. 2008;105:8499–8506. doi: 10.1073/pnas.0803578105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M. Left-right asymmetry in embryonic development: a comprehensive review. Mech Dev. 2005;122:3–25. doi: 10.1016/j.mod.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Levin M, Johnson RL, Stern CD, Kuehn M, Tabin C. A molecular pathway determining left-right asymmetry in chick embryogenesis. Cell. 1995;82:803–814. doi: 10.1016/0092-8674(95)90477-8. [DOI] [PubMed] [Google Scholar]
- Liang JO, Etheridge A, Hantsoo L, Rubinstein AL, Nowak SJ, Belmonte JC Izpisua, Halpern ME. Asymmetric nodal signaling in the zebrafish diencephalon positions the pineal organ. Development. 2000;127:5101–5112. doi: 10.1242/dev.127.23.5101. [DOI] [PubMed] [Google Scholar]
- Long S, Ahmad N, Rebagliati M. The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development. 2003;130:2303–2316. doi: 10.1242/dev.00436. [DOI] [PubMed] [Google Scholar]
- Mine N, Anderson RM, Klingensmith J. BMP antagonism is required in both the node and lateral plate mesoderm for mammalian left-right axis establishment. Development. 2008;135:2425–2434. doi: 10.1242/dev.018986. [DOI] [PubMed] [Google Scholar]
- Monteiro R, van Dinther M, Bakkers J, Wilkinson R, Patient R, ten Dijke P, Mummery C. Two novel type II receptors mediate BMP signalling and are required to establish left-right asymmetry in zebrafish. Dev Biol. 2008;315:55–71. doi: 10.1016/j.ydbio.2007.11.038. [DOI] [PubMed] [Google Scholar]
- Piedra ME, Ros MA. BMP signaling positively regulates Nodal expression during left right specification in the chick embryo. Development. 2002;129:3431–3440. doi: 10.1242/dev.129.14.3431. [DOI] [PubMed] [Google Scholar]
- Pogoda HM, Solnica-Krezel L, Driever W, Meyer D. The zebrafish forkhead transcription factor FoxH1/Fast1 is a modulator of nodal signaling required for organizer formation. Curr Biol. 2000;10:1041–1049. doi: 10.1016/s0960-9822(00)00669-2. [DOI] [PubMed] [Google Scholar]
- Ramsdell AF. Left-right asymmetry and congenital cardiac defects: getting to the heart of the matter in vertebrate left-right axis determination. Dev Biol. 2005;288:1–20. doi: 10.1016/j.ydbio.2005.07.038. [DOI] [PubMed] [Google Scholar]
- Ramsdell AF, Yost HJ. Cardiac looping and the vertebrate left-right axis: antagonism of left-sided Vg1 activity by a right-sided ALK2-dependent BMP pathway. Development. 1999;126:5195–5205. doi: 10.1242/dev.126.23.5195. [DOI] [PubMed] [Google Scholar]
- Raya A, Belmonte JC. Left-right asymmetry in the vertebrate embryo: from early information to higher-level integration. Nat Rev Genet. 2006;7:283–293. doi: 10.1038/nrg1830. [DOI] [PubMed] [Google Scholar]
- Raz E, Reichman-Fried M. Attraction rules: germ cell migration in zebrafish. Curr Opin Genet Dev. 2006;16:355–359. doi: 10.1016/j.gde.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Rohde LA, Heisenberg CP. Zebrafish gastrulation: cell movements, signals, and mechanisms. Int Rev Cytol. 2007;261:159–192. doi: 10.1016/S0074-7696(07)61004-3. [DOI] [PubMed] [Google Scholar]
- Rohr S, Otten C, Abdelilah-Seyfried S. Asymmetric involution of the myocardial field drives heart tube formation in zebrafish. Circ Res. 2008;102:e12–19. doi: 10.1161/CIRCRESAHA.107.165241. [DOI] [PubMed] [Google Scholar]
- Schier AF. Nodal signaling in vertebrate development. Annu Rev Cell Dev Biol. 2003;19:589–621. doi: 10.1146/annurev.cellbio.19.041603.094522. [DOI] [PubMed] [Google Scholar]
- Schier AF, Talbot WS. Molecular genetics of axis formation in zebrafish. Annu Rev Genet. 2005;39:561–613. doi: 10.1146/annurev.genet.37.110801.143752. [DOI] [PubMed] [Google Scholar]
- Schilling TF, Concordet JP, Ingham PW. Regulation of left-right asymmetries in the zebrafish by Shh and BMP4. Dev Biol. 1999;210:277–287. doi: 10.1006/dbio.1999.9214. [DOI] [PubMed] [Google Scholar]
- Schlange T, Arnold HH, Brand T. BMP2 is a positive regulator of Nodal signaling during left-right axis formation in the chicken embryo. Development. 2002;129:3421–3429. doi: 10.1242/dev.129.14.3421. [DOI] [PubMed] [Google Scholar]
- Schoenebeck JJ, Keegan BR, Yelon D. Vessel and blood specification override cardiac potential in anterior mesoderm. Dev Cell. 2007;13:254–267. doi: 10.1016/j.devcel.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MM. Nodal signaling: developmental roles and regulation. Development. 2007;134:1023–1034. doi: 10.1242/dev.000166. [DOI] [PubMed] [Google Scholar]
- Shiratori H, Hamada H. The left-right axis in the mouse: from origin to morphology. Development. 2006;133:2095–2104. doi: 10.1242/dev.02384. [DOI] [PubMed] [Google Scholar]
- Sirotkin HI, Gates MA, Kelly PD, Schier AF, Talbot WS. Fast1 is required for the development of dorsal axial structures in zebrafish. Curr Biol. 2000;10:1051–1054. doi: 10.1016/s0960-9822(00)00679-5. [DOI] [PubMed] [Google Scholar]
- Smith KA, Chocron S, von der Hardt S, de Pater E, Soufan A, Bussmann J, Schulte-Merker S, Hammerschmidt M, Bakkers J. Rotation and asymmetric development of the zebrafish heart requires directed migration of cardiac progenitor cells. Dev Cell. 2008;14:287–297. doi: 10.1016/j.devcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Solnica-Krezel L. Conserved patterns of cell movements during vertebrate gastrulation. Curr Biol. 2005;15:R213–228. doi: 10.1016/j.cub.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Tabin C. Do we know anything about how left-right asymmetry is first established in the vertebrate embryo? J Mol Histol. 2005;36:317–323. doi: 10.1007/s10735-005-9000-y. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B. Antivin, a novel and divergent member of the TGFbeta superfamily, negatively regulates mesoderm induction. Development. 1999;126:229–240. doi: 10.1242/dev.126.2.229. [DOI] [PubMed] [Google Scholar]
- Trinh LA, Stainier DY. Cardiac development. Methods Cell Biol. 2004;76:455–473. doi: 10.1016/s0091-679x(04)76020-3. [DOI] [PubMed] [Google Scholar]
- Yan YT, Gritsman K, Ding J, Burdine RD, Corrales JD, Price SM, Talbot WS, Schier AF, Shen MM. Conserved requirement for EGF-CFC genes in vertebrate left-right axis formation. Genes Dev. 1999;13:2527–2537. doi: 10.1101/gad.13.19.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashiro K, Shiratori H, Hamada H. Haemodynamics determined by a genetic programme govern asymmetric development of the aortic arch. Nature. 2007;450:285–288. doi: 10.1038/nature06254. [DOI] [PubMed] [Google Scholar]
- Yelon D, Horne SA, Stainier DY. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev Biol. 1999;214:23–37. doi: 10.1006/dbio.1999.9406. [DOI] [PubMed] [Google Scholar]
- Zhang H, Bradley A. Mice deficient for BMP2 are nonviable and have defects in amnion/chorion and cardiac development. Development. 1996;122:2977–2986. doi: 10.1242/dev.122.10.2977. [DOI] [PubMed] [Google Scholar]
- Zhang J, Talbot WS, Schier AF. Positional cloning identifies zebrafish one-eyed pinhead as a permissive EGF-related ligand required during gastrulation. Cell. 1998;92:241–251. doi: 10.1016/s0092-8674(00)80918-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.