Abstract
Humans are exposed to N-nitroso compounds from both endogenous and exogenous sources. Many N-nitroso compounds can be metabolically activated to give diazoacetate, which can result in the carboxymethylation of DNA. The remarkable similarity in p53 mutations found in human gastrointestinal tumors and in shuttle vector studies, where the human p53 gene-containing vector was treated with diazoacetate and propagated in yeast cells, suggests that diazoacetate might be an important etiological agent for human gastrointestinal tumors. The O6-carboxymethyl-2′-deoxyguanosine was previously detected in isolated DNA upon exposure to diazoacetate and in blood samples of healthy human subjects. The corresponding modifications of thymidine and 2′-deoxyadenosine have not been assessed, though significant mutations at A:T base pairs were found in the p53 tumor suppressor gene in human gastrointestinal tumors and in shuttle vector studies. To understand the implications of the carboxymethylation chemistry of thymidine in the observed mutations at A:T base pairs, here we synthesized authentic N3-carboxymethylthymidine (N3-CMdT) and O4-carboxymethylthymidine (O4-CMdT), incorporated them into DNA, and demonstrated, for the first time, that they were the major carboxymethylated derivatives of thymidine formed in calf thymus DNA upon exposure to diazoacetate. The demonstration of the formation of N3-CMdT and O4-CMdT in isolated DNA upon treatment with diazoacetate, together with the preparation of authentic oligodeoxyribonucleotide substrates housing these two lesions, laid the foundation for investigating the replication and repair of these lesions and for understanding their implications in the mutations observed in human gastrointestinal tumors.
INTRODUCTION
Humans are exposed to N-nitroso compounds (NOCs) from diet, tobacco smoke and other environmental sources as well as from endogenous sources, and it was estimated that the endogenous sources account for 45–75% of the total NOC exposure (1). The exposure to endogenous NOCs was found to be significantly associated with the risk of developing non-cardia gastric cancer in a recent large-scale epidemiological study, i.e. European Prospective Investigation into Cancer and Nutrition (EPIC) (2). This study included more than half a million individuals and 314 incident cases of gastric cancer that had occurred after an average of 6.6 years of follow up (2).
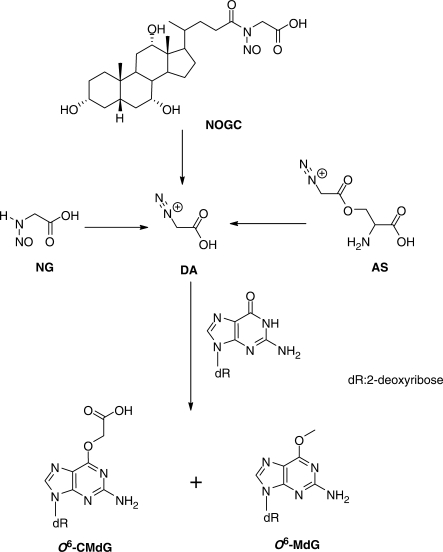
Some endogenously produced NOCs are known to cause damage to DNA and they are both mutagenic and carcinogenic. Earlier studies by Wogan et al. (3) revealed that the treatment of male Fischer rats with nitrosated bile acid conjugates N-nitrosoglycocholic acid (NOGC) and N-nitrosotaurocholic acid (NOTC) could induce significant levels of hepatocarcinoma in 54–70% of the treated animals, and 12–13% of the treated rats developed gastric tumors. Forward mutation assay in Salmonella typhimurium and in TK6 diploid human lymphoblasts showed that NOGC and NOTC are both mutagenic, with NOGC being more effective than NOTC in inducing mutations in TK6 cells (4). In this regard, NOGC could arise from the nitrosation of glycocholic acid under simulated gastric conditions (5). Shuker et al. (6) later demonstrated that the treatment of calf thymus DNA with NOGC can induce the formation of O6-carboxymethyl-2′-deoxyguanosine (O6-CMdG) and, to a much lesser extent, O6-methyl-2′-deoxyguanosine (O6-MdG, Scheme 1). By using immunoslot blot assay or immunoaffinity/HPLC method with antibody recognizing O6-carboxymethylguanine, Shuker et al. (7–9) showed that O6-CMdG can be induced in DNA by N-nitroso glycine (NG, Scheme 1). In addition, O-diazoacetyl-l-serine (also known as azaserine, AS, Scheme 1), a pancreatic carcinogen (10), could induce the carboxymethylation of guanine residues in DNA in pancreatic acinar cells (11).
Scheme 1.
The formation of diazoacetate and its modification of 2′-deoxyguanosine.
The above carboxymethylation was thought to proceed through a common reactive intermediate, diazoacetate (DA, Scheme 1), and it was demonstrated that potassium diazoacetate (KDA) can induce the formation of O6-CMdG and, to a lesser degree (about one-sixteenth of that of O6-CMdG), O6-methyl-2′-deoxyguanosine in DNA (7–9). Furthermore, it was observed that humans consuming red meat have a higher level of nitrosated compounds in feces (12) and an elevated level of O6-CMdG in colonic exfoliated cells than control group consuming vegetarian diet (13). O6-CMdG was also detected at a level of three to eight lesions per 107 nucleosides in the blood DNA of healthy human subjects (9).
Some initial replication and repair studies have been carried out for O6-CMdG. In this context, it was found that the azaserine-mediated cell killing cannot be rescued by the overexpression of O6-methylguanine methyltransferase (MGMT) (14), which is keeping with the observation that O6-CMdG is not subjected to repair by MGMT in vitro (6). On the other hand, lymphoblastoid cells with deficiency in nucleotide excision repair (NER) were significantly more sensitive to azaserine than the control repair-proficient cells, suggesting that O6-CMdG can be repaired by NER factors (14).
A very recent study by Gottschalg et al. (15) revealed that the passage of KDA-treated, human p53 gene-containing plasmid in yeast cells could result in substantial single-base substitutions. Interestingly, KDA induced almost equal distributions of transition and transversion mutations, which differs markedly from the predominant GC→AT transition mutation (>80%) induced by methylating agent methylnitrosourea (15), suggesting that DNA methylation is not the major contributor to KDA-induced mutations. Moreover, the identities and frequencies of mutations induced by diazoacetate at non-CpG sites are remarkably similar to the mutations at non-CpG sites in p53 gene observed in human stomach and colorectal tumors (15), suggesting that diazoacetate may constitute an important etiological agent for the development of gastrointestinal cancer.
Gottschalg et al. (15) also observed a substantial frequency of single-base substitutions at A:T base pairs in p53 gene from shuttle vector studies and in human gastrointestinal tumors. For instance, the percentages of mutations at A:T base pairs account for 43% and 28% of all the observed mutations while the vector was treated with KDA in PBS and Tris–EDTA, respectively (15). The observation of a significant proportion of mutations at A:T base pairs supports that KDA can also induce promutagenic lesions at adenine and/or thymine. Although KDA has been shown to induce the formation of O6-CMdG, it remains unexplored what types of lesions can be induced at adenine and thymine sites in DNA by KDA and what the mutagenic properties are for the lesions induced at these two nucleobases.
As a first step toward answering these questions, we report here the chemical syntheses of N3- and O4-carboxymethylthymidine, their incorporation into oligodeoxyribonucleotides (ODNs), and the demonstration of their formation in calf thymus DNA upon treatment with diazoacetate.
MATERIALS AND METHODS
Materials
All chemicals, unless otherwise specified, were from Sigma-Aldrich (St. Louis, MO). Common reagents for solid-phase DNA synthesis were obtained from Glen Research Co. (Sterling, VA).
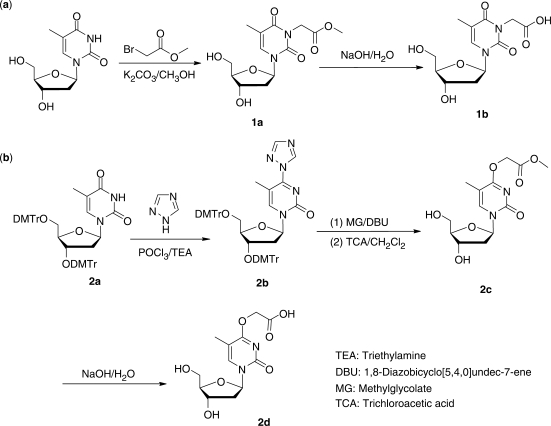
Synthesis of standard N3-carboxymethylthymidine (N3-CMdT, 1b, Scheme 2a)
Scheme 2.
The chemical synthesis of N3-CMdT and O4-CMdT.
In a flask, 242 mg of thymidine and 166 mg of potassium carbonate were dissolved in 50 ml of anhydrous methanol. To the solution was subsequently added 184 mg of methyl bromoacetate. The resulting solution was refluxed for 6 h. Evaporation of solvent under reduced pressure afforded white solid which was then purified by silica gel column chromatography with EtOAc/MeOH (85/15, v/v) as mobile phase to give 152 mg of 1a as colorless viscous liquid (yield 48%). Hydrolysis of 1a (152 mg) with 2.0 ml of aqueous NaOH solution (0.5 M) rendered 1b in sodium salt form (119 mg; yield 84%).
1a. 1H NMR (300 MHz, CDCl3, 25°C, Figure S1): δ 7.49 (s, 1H, H-6), 6.20 (t, 1H, H-1′, J = 6.6 Hz), 4.69 (s, 2H, N3-CH2), 4.54 (m, 1H, H-3′), 3.98 (m, 1H, H-4′), 3.83–3.80 (m, 2H, H-5′ and H-5″), 3.75 (s, 3H, 3-OCH3), 2.35 (m, 2H, H-2′ and H-2″), 1.94 (s, 3H, 5-CH3).
HRMS (ESI): [M + H]+ calcd m/z 315.1192, found 315.1184.
1b. 1H NMR (300 MHz, D2O, 25°C, Figure S2): δ 7.72 (s, 1H, H-6), 6.34 (t, 1H, H-1′, J = 6.6 Hz), 4.50 (m, 3H, N3-CH2 and H-3′), 4.07 (m, 1H, H-4′), 3.93–3.79 (m, 2H, H-5′ and H-5″), 2.41–2.45 (m, 2H, H-2′ and H-2″), 1.96 (s, 3H, 5-CH3).
HRMS (ESI): [M + H]+ calcd m/z 301.1025, found 301.1028.
Synthesis of O4-carboxymethylthymidine (O4-CMdT, 2d, Scheme 1b)
Compound 2a was prepared following the previously published procedures (16). Compound 1,2,4-triazole (414 mg) was suspended in anhydrous acetonitrile (9.0 ml) in an ice bath, and then POCl3 (110 μl) was slowly added with rapid stirring followed by dropwise addition of TEA (840 μl). Compound 2a (261 mg) in ∼2.0 ml acetonitrile was subsequently added, and the solution was continuously stirred for 90 min. The reaction was terminated by adding saturated NaHCO3 solution and extracted with CH2Cl2. The organic layer was washed sequentially with saturated NaHCO3 and brine, dried with Na2SO4, and the solvent was evaporated under reduced pressure. Purification with silica gel column chromatography using ethyl ether as mobile phase rendered 2b as white solid (154 mg; yield 63%).
Compound 2b (450 mg) was dissolved in anhydrous acetonitrile (8.0 ml), and to this solution were added methyl glycolate (240 μl) and DBU (60 μl). The resulting solution was stirred at room temperature overnight and subsequently acidified by adding an equal volume of 3% trichloroacetic acid (TCA) in CH2Cl2. The solvent was removed under reduced pressure, and the resulting residues were subjected to purification with silica gel column chromatography by using EtOAc/MeOH (85/15, v/v) as the mobile phase. Compound 2c (182 mg) was obtained as colorless viscous liquid (yield 58%). Hydrolysis of 2c (150 mg) with 2.0 ml of 0.1 M aqueous NaOH solution rendered 2d in sodium salt form (142 mg; yield 92%).
2b. 1H NMR (300 MHz, DMSO-d6, 25°C, Figure S3): δ 9.35 (s, 1H, triazolyl H), 8.35 (s, 1H, triazolyl H), 8.27 (s, 1H, H-6), 6.80–7.32 (m, 26H, aromatic H of DMTr), 6.22 (t, 1H, H-1′, J = 6.2 Hz), 4.09 (m, 1H, H-3′), 3.82 (m, 1H, H-4′), 3.71 (m, 12H, OCH3 of DMTr), 3.23–3.04 (m, 2H, H-5′ and H-5″), 2.23 (m, 1H, H-2′), 1.92 (m, 1H, H-2″), 2.04 (s, 3H, 5-CH3).
HRMS (ESI): [M + Na]+ calcd m/z 920.3635, found 920.3606.
2c. 1H NMR (300 MHz, D2O, 25°C, Figure S4): δ 7.78 (s, 1H, H-6), 6.32 (t, 1H, H-1′, J = 6.0 Hz), 4.78 (s, 2H, O4-CH2), 4.51 (m, 1H, H-3′), 3.91 (m, 1H, H-4′), 3.87 (m, 2H, H-5′ and H-5″), 3.82 (s, 3H, O4-OCH3), 2.41 (m, 2H, H-2′ and H-2″), 1.96 (s, 3H, 5-CH3).
HRMS (ESI): [M + H]+ calcd m/z 315.1192, found 315.1186.
2d. 1H NMR (300 MHz, D2O, 25°C, Figure S5): δ 7.71 (s, 1H, H-6), 6.34 (t, 1H, H-1′, J = 6.0 Hz), 4.48 (m, 3H, O4-CH2 and H-3′), 4.06 (m, 1H, H-4′), 3.87 (m, 2H, H-5′ and H-5″), 2.43 (m, 2H, H-2′ and H-2″), 1.96 (s, 3H, 5-CH3).
HRMS (ESI): [M + H]+ calcd m/z 301.1035, found 301.1034.
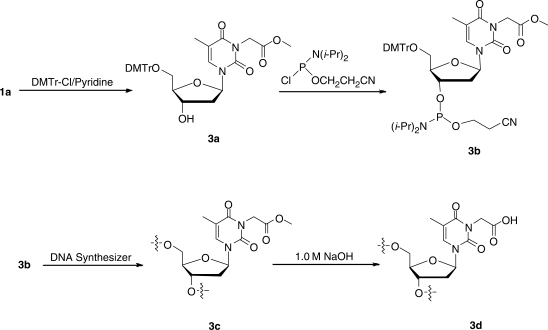
Synthesis of 5′-O-(4,4′-dimethoxytrityl)-N3-methoxycarbonylmethylthymidine (3a, Scheme 3)
Scheme 3.
The incorporation of N3-CMdT into ODNs.
Compound 3a was prepared from 1a following standard procedures (17). Briefly, 450 mg of compound 1a was dissolved in anhydrous pyridine (10 ml) and the solution was cooled in an ice-bath. 4-Dimethylaminopyridine (DMAP, 10 mg) and dimethoxytrityl chloride (DMTr-Cl, 340 mg) were added, and the resulting solution was stirred at room temperature for 90 min. The reaction was then quenched with methanol (0.5 ml), and the solvent was removed under reduced pressure. The product was isolated by silica gel column chromatography using 15% MeOH in EtOAc as mobile phase, and dried under reduced pressure to give a foam (565 mg; yield: 64%).
3a. 1H NMR (300 MHz, DMSO-d6, 25°C, Figure S6): δ 7.65 (s, 1H, H-6), 7.45–6.80 (m, 13H, aromatic H of DMTr), 6.25 (t, 1H, H-1′, J = 6.3 Hz), 5.38 (s, 1H, 3′-OH), 4.58 (s, 2H, N3-CH2), 4.36 (m, 1H, H-3′), 4.04 (m, 1H, H-4′), 3.72 (s, 6H, OCH3 of DMTr), 3.67 (s, 3H, 3-OCH3), 3.23 (m, 2H, H-5′ and H-5″), 2.25 (m, 2H, H-2′ and H-2″), 1.99 (s, 3H, 5-CH3).
HRMS (ESI): [M + Na]+ calcd m/z 639.2318, found 639.2300.
Synthesis of 5′-O-(4,4′-dimethoxytrityl)-N3-methoxycarbonylmethylthymidine-3′-O-[(2-cyanoethyl)-N,N-diisopropylphosphoramidite] (3b)
To a flask, which was suspended in an ice bath and contained a solution of compound 3a (92.4 mg) in dry CH2Cl2 (3.0 ml), was added N,N-diisopropylethylamine (DIEA, 52 μl) followed by dropwise addition of 2-cyanoethyl-N,N-diisopropyl chlorophosphoramidite (60 μl). The mixture was stirred at room temperature for 30 min under an argon atmosphere. The reaction was quenched by cooling the mixture in an ice bath followed by slow addition of CH3OH (0.40 ml). The solution was quickly extracted with EtOAc (8.0 ml). The organic layer was washed with saturated NaHCO3 (4.0 ml) and brine (4.0 ml), and dried with anhydrous Na2SO4. The solvent was evaporated in vacuo to yield 3b in white foam that was used for ODN synthesis (101 mg; yield: 84%).
3b. 1H NMR (300 MHz, DMSO-d6, 25°C, Figure S7): δ 7.70 (s, 0.5H, H-6, diastereomer 1), 7.65 (s, 0.5H, H-6, diastereomer 2), 7.39–6.82 (m, 13H, aromatic H of DMTr), 6.43 (m, 1H, H-1′), 4.71 (s, 2H, N3-CH2), 4.64 (m, 1H, H-3′), 4.15 (m, 1H, H-4′), 3.80 (s, 6H, OCH3 of DMTr), 3.76 (s, 3H, 3-OCH3), 3.56–3.34 (m, 5H, 2H from CH2CN, 2H from 2CH and 2H from H-5′&H-5″), 2.59 (m, 2H, OCH2), 2.39 (m, 2H, H-2′ and H-2″), 1.46 (s, 3H, 5-CH3), 1.15-1.04 (m, 12H, CH3 from isopropyl). 31P NMR: δ 149.9, 149.8.
HRMS (ESI): [M + Na]+ calcd m/z 839.3397, found 839.3379.
ODN synthesis
ODNs (sequences shown in the text) were synthesized on a Beckman Oligo 1000S DNA synthesizer (Fullerton, CA) at 1 μmol scale. The synthesized phosphoramidite building block was dissolved in anhydrous acetonitrile at a concentration of 0.07 M. Standard phosphoramidite building blocks (Glen Research Inc., Sterling, VA) of dA, dC, dG and dT were employed, and a standard ODN assembly protocol was used without any modification.
Synthesis of the ODNs containing N3-CMdT (3d) started with 3b as phosphoramidite building block, after synthesis (Scheme 3), the ODNs containing N3-methoxycarbonylmethylthymidine were cleaved from the controlled-pore glass (CPG) support with 1.0 M NaOH (3.0 ml) at room temperature for 2 days. Under such conditions, the nucleobase protecting groups were removed and the ester was hydrolyzed to the corresponding carboxylic acid derivative to yield the ODN containing 3c.
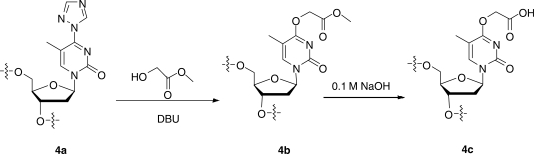
An O4-triazolyl derivative of standard dT phosphoramidite building block was used for synthesis of the ODNs containing O4-CMdT (4c, Scheme 4). After the ODN assembly, the CPG support bearing 4a was first suspended in anhydrous acetonitrile (3.0 ml). To the solution were then added 50 equivalents of methyl glycolate and 15 equivalents of DBU, and the suspension was stirred at room temperature for 24 h, which offered the CPG support bearing 4b. The solvent was removed by filtration. The 4b-containing ODN was hydrolyzed with 0.1 M NaOH (3.0 ml) at room temperature for 3 days to afford 4c, and in the mean time 4c was cleaved from the CPG support and deprotected. The beads were then removed, and the solution was neutralized with 0.1 M HCl to pH 7.0 and lyophilized to dryness.
Scheme 4.
The incorporation of O4-CMdT into ODNs.
KDA treatment of calf thymus DNA
KDA was synthesized via alkaline hydrolysis of ethyl diazoacetate (18). The concentration of KDA was estimated by assuming that the hydrolysis reaction was quantitative. Calf thymus DNA (∼1.0 mg/ml) was treated with freshly prepared KDA (8 or 20 mM) in PBS buffer (pH 7.3) at 37°C for 8 h. The resulting DNA was isolated by ethanol precipitation, and the recovered pellets were taken up into doubly distilled water.
Enzymatic digestion of KDA-treated calf thymus DNA
One unit of nuclease P1, 0.01 unit of calf spleen phosphodiesterase, and 2.5 μl of buffer solution, which contained sodium acetate (300 mM, pH 5.0) and zinc acetate (10 mM), were added to a 20 μl solution of KDA-treated calf thymus DNA. The digestion was continued at 37°C for 6 h. To the digestion mixture were then added 10 units of alkaline phosphatase, 0.05 unit of snake venom phosphodiesterase, and 5 μl of 0.5 M Tris–HCl (pH 8.9). The digestion was continued at 37°C for 6 h, and the enzymes were removed by chloroform extraction. The resulting aqueous solution was lyophilized to dryness and redissolved in ultra-pure water for LC/MS analysis.
Mass spectrometry (MS) and NMR
Electrospray ionization-MS (ESI-MS) and tandem MS (MS/MS) experiments were carried out on an LCQ Deca XP ion-trap mass spectrometer (Thermo Fisher Scientific, San Jose, CA). A mixture of acetonitrile and water (50:50, v/v) was used as solvent for electrospray. The spray voltage was 3.0 kV, and the temperature of the heated capillary was maintained at 275°C. High-resolution mass spectra (HRMS) were acquired on an Agilent 6510 Q-TOF LC/MS instrument (Agilent Technologies, Palo Alto, CA) equipped with an electrospray ionization (ESI) source. 1H NMR spectra were recorded at 300 MHz on a Varian Inova 300 NMR spectrometer (Varian Inc., Palo Alto, CA), and 31P NMR spectra were acquired at 80 MHz on the same instrument.
2D NMR spectra were recorded in D2O using a Varian Unity (Varian, Inc., Palo Alto, CA) spectrometer operating at 500 MHz at 25°C. A triple resonance 5 mm 1H13C15N probe was used. Resonance assignments were made based on 1H–1H COSY and 1H–13C HMBC experiments. The HMBC spectra were acquired using sweep widths of 5006.3 Hz and 30 177.3 Hz for 1H and 13C, respectively. The first delay was set to match a 140 Hz coupling constant, and the second delay was set to match a long-range coupling constant of 8 Hz.
LC/MS
Coupled liquid chromatography and ESI tandem MS experiments were performed on an Agilent 1100 capillary HPLC pump (Agilent Technologies) and an LTQ linear ion trap mass spectrometer (Thermo Fisher Scientific, San Jose, CA). A 0.5 × 150 mm Zorbax SB-C18 column (5 μm in particle size, Agilent Technologies) was used, and a 0.1% formic acid in water (Solution A) and a 0.1% formic acid in methanol (Solution B) were employed as mobile phases. The ESI spray voltage was 3.0 kV, and the temperature of the heated capillary was maintained at 275°C.
HPLC purification
The purification of ODNs was performed on a Beckman HPLC System (32 Karat software version 3.0, pump module 125) with a UV detector (module 126) monitoring at 260 nm. A 4.6 × 250 mm Apollo C18 column (5 μm in particle size and 300 Å in pore size, Alltech Associate Inc., Deerfield, IL) was used, and a 50 mM triethylammonium acetate buffer (TEAA, pH 6.6, Solution A) and a mixture of 50 mM TEAA and acetonitrile (70/30, v/v, Solution B) were employed as mobile phases.
RESULTS
Chemical synthesis of N3- and O4-carboxymethylthymidine
To understand the implications of carboxymethylation of thymidine in the diazoacetate-induced mutations observed at A:T base pairs in human p53 tumor suppressor gene, we set out to assess the carboxyalkylation chemistry of thymidine. To this end, we first synthesized the authentic N3- and O4-carboxymethylthymidine (N3- and O4-CMdT).
We introduced the methyl ester derivative of the carboxymethyl functionality to the N3 position of thymidine by alkylation with methyl bromoacetate, and treated the resulting compound with 0.5 M NaOH to give the desired N3-CMdT (Scheme 2a).
The chemical synthesis of O4-CMdT was inspired by the previous synthesis of ODNs bearing an O4-methylthymidine (19) and O6-CMdG (20) (Scheme 2b). We incorporated the 1,2,4-triazolyl moiety to the C4 carbon atom of 3′,5′-O-di-DMTr-protected thymidine, which, upon treatment with methyl glycolate in the presence of DBU, renders the corresponding methyl ester of the di-DMTr-protected O4-CMdT. The DMTr groups in the latter derivative were removed by using 3% TCA in CH2Cl2 (Scheme 2b, details shown in Materials and methods section), and the resulting product was incubated with 0.1 M NaOH to give the corresponding carboxylic acid derivative (Scheme 2).
The structures of the two synthesized carboxymethylated thymidine derivatives were supported by 1H NMR and 2D heteronuclear multi-bond correlation (HMBC) measurements. In particular, the two methylene protons in the carboxymethyl functionality of N3-CMdT exhibit strong correlation with the C2 and C4 of the thymine ring in the 2D HMBC spectrum, whereas the two corresponding methylene protons in O4-CMdT show correlation with the C4, but not the C2, of the thymine ring (Figures S8 and S9). These distinct spectral features support the sites of carboxymethylation in these two thymidine lesions.
Formation of O4-CMdT and N3-CMdT in calf thymus DNA upon treatment with KDA
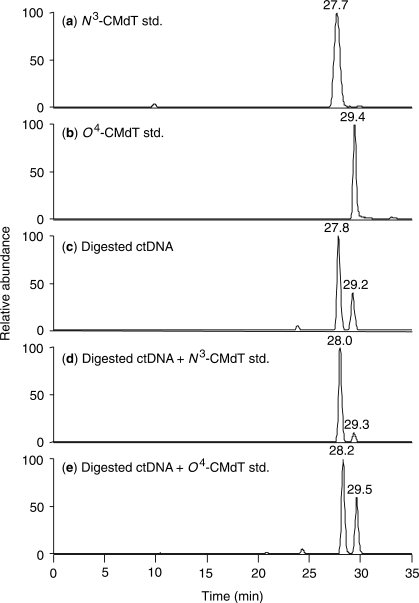
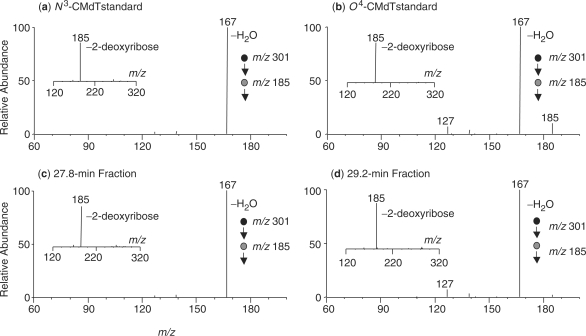
The availability of the standard O4-CMdT and N3-CMdT enabled us to assess whether these two lesions can be induced in calf thymus DNA. To this end, we treated calf thymus DNA with potassium diazoacetate, removed the salts from the reaction mixture by ethanol precipitation, digested the DNA with enzymes, and subjected the resulting nucleoside mixture to LC-MS/MS analysis, where we monitored selectively the fragmentation of the [M + H]+ ion (m/z 301) of carboxymethylated thymidines. The selected-ion chromatogram (SIC) for monitoring the formation of the protonated carboxymethylthymine (i.e. m/z 185) revealed two peaks with elution time at 27.8 and 29.2 min, respectively (Figure 1c). The product-ion spectra averaged from these two peaks showed the formation of a predominant fragment ion of m/z 185 (insets in Figure 2c–d), which is attributed to form from the elimination of a 2-deoxyribose component. Further fragmentation of this ion (i.e. MS3 experiment) leads to the facile loss of a water molecule to give the ion of m/z 167 (Figure 2c–d).
Figure 1.
LC-MS/MS for monitoring the formation of N3-CMdT and O4-CMdT in calf thymus DNA upon treatment with diazoacetate. Shown are the SICs for monitoring the m/z 301→185 transition, corresponding to the loss of a 2-deoxyribose, from the LC-MS/MS analyses with the injection of: (a) N3-CMdT standard; (b) O4-CMdT standard; (c) enzymatic digestion mixture of calf thymus (ct) DNA treated with 20 mM potassium diazoacetate (KDA, and the LC-MS/MS results from the reaction with 8 mM KDA are shown in Figure S7); (d) the sample in (c) with the addition of standard N3-CMdT; and (e) the sample in (c) with the addition of standard O4-CMdT.
Figure 2.
Tandem mass spectra supporting the formation of N3-CMdT and O4-CMdT in calf thymus DNA. Shown are the MS3 results, which monitors the further fragmentation of the protonated carboxymethylthymine (m/z 185), for: (a) standard N3-CMdT; (b) standard O4-CMdT; (c) the 27.8 min fraction shown in Figure 1c; and (d) the 29.2 min fraction shown in Figure 1c. Depicted in the insets are the corresponding MS/MS for the [M + H]+ ions of the modified nucleosides. The ion of m/z 185 found in MS/MS is attributed the elimination of a 2-deoxyribose.
We also subjected standard O4-CMdT and N3-CMdT to LC-MS/MS analysis under the same experimental conditions. It turned out that these two modified nucleosides share the same retention times (Figure 1a–c) and the identical product-ion spectra (both MS/MS and MS3, Figure 2) as the two components present in the enzymatic digestion mixture of KDA-treated calf thymus DNA. Moreover, doping the nucleoside mixture arising from the KDA-treated calf thymus DNA with N3-CMdT leads to increase in signal for the first peak (Figure 1d), similar doping with O4-CMdT results in a rise in signal intensity for the later-eluting peak (Figure 1e), lending further support that both O4-CMdT and N3-CMdT could be induced in calf thymus DNA upon KDA treatment.
Incorporation of O4-CMdT and N3-CMdT into ODNs
To understand the implications of O4-CMdT and N3-CMdT in the KDA-induced mutations observed at T:A base pairs, it is important to obtain ODN substrates housing a site-specifically inserted O4-CMdT and N3-CMdT for future replication and repair studies. Therefore, we developed synthetic methods for the insertion of these two lesions into ODNs at a unique site. In this respect, the phosphoramidite building block of the methyl ester derivative of N3-CMdT was prepared from the above-described modified nucleoside following standard procedures (Scheme 3) (17). After solid-phase ODN assembly, we cleaved the ODN from the resin and removed the nucleobase protecting groups by treatment with 1.0 M NaOH, which also converted the methyl ester derivative to the corresponding carboxylic acid (20).
A similar nucleoside conversion strategy as described above for the synthesis of O4-CMdT was employed for the synthesis of ODNs housing a site-specifically inserted O4-CMdT (Scheme 4). The 1,2,4-triazolyl-protected thymidine was incorporated into ODNs by using standard phosphoramidite chemistry, and the triazolyl functionality on thymidine was displaced with methyl glycolate while the ODN was still on the resin. The resin was subsequently treated with 0.1 M NaOH to convert the ester derivative to the corresponding carboxylic acid; such treatment also resulted in the cleavage of the ODN from the solid support and the removal of the nucleobase protecting groups (Scheme 4).
It is worth noting that alkaline conditions were employed for the above conversion from methyl ester to the corresponding carboxylic acid and for the nucleobase deprotection, it is important to assess the stability of the lesion-carrying ODNs under these basic conditions. Our LC-MS/MS analyses showed that while the dodecameric ODN containing N3-CMdT is stable under the alkaline conditions (1.0 M NaOH for 24 h), the corresponding O4-CMdT-bearing ODN is not very stable under strong alkaline conditions. Thus, we treated the ODN-carrying solid support with 0.1 M NaOH for 36 h to facilitate the above ester-to-acid conversion and the nucleobase deprotection. In this context, our LC-MS analysis showed that approximately 13% of the ODN has lost the carboxymethyl group to produce the corresponding unmodified thymidine-containing ODN. The thymidine and O4-CMdT-bearing ODNs, however, can be resolved by HPLC.
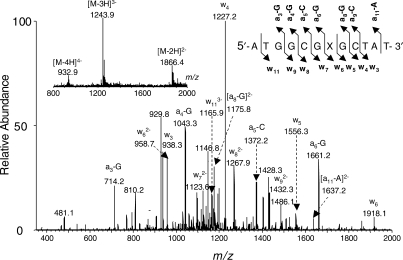
The ODNs obtained from the above-described procedures were characterized by ESI-MS and MS/MS to confirm their sequence and the location of carboxymethylation. Taking the characterizations of the d(ATGGCGXGCTAT) (where ‘X’ represents O4-CMdT) as an example, we found that the measured m/z values for the multiply deprotonated ions are consistent with their calculated ones (Figure 3, inset). In addition, MS/MS of the [M – 3H]3– ion (m/z 1243.9, Figure 3) gives a series of sequence ions, allowing us to establish unambiguously that there is an O4-CMdT at the seventh position in this ODN counting from the 5′ terminus. Upon collisional activation in a mass spectrometer, an ODN undergoes cleavages at the N-glycosidic bond and the 3′ C–O bond of the same nucleoside to form [an – Base] and its complementary wn ions, which carry the 5′ and 3′ termini of the ODN, respectively (21). Because of the low proton affinity of thymine, the N-glycosidic bond in thymidine is usually not susceptible to cleavage and, as a result, chain cleavage is often not observed at the 3′ C–O bond of this nucleoside (22). In Figure 1, we observed [a3 – G], [a4 – G], [a5 – C], [a6 – G], [a8 – G], [a9 – C], [a11 – A], w3, w4, w5, w6, w7, w8, w9 and w11 ions. The observed masses of w6, w7, w8, w9 and w11 ions are 58 Da higher in masses than the calculated masses of the corresponding fragments for the unmodified ODN, whereas the masses of the w3, w4, w5 ions are the same the calculated masses for the corresponding fragments from the unmodified ODN, supporting the presence of an O4-CMdT at the seventh position of this ODN. The same conclusion can be drawn based on the observed masses of the [a – Base] series fragment ions. Likewise, we characterized the synthesized N3-CMdT-containing ODN by ESI-MS and MS/MS analyses (spectra not shown).
Figure 3.
The product-ion spectrum of the ESI-produced [M – 3H]3– ions of d(ATGGCGXGCTAT), where ‘X’ represents O4-CMdT. Illustrated in the insets is the negative-ion ESI-MS for the modified ODN.
DISCUSSION
Humans are exposed to NOCs from both endogenous and exogenous sources (1), and a recent epidemiological study found a significant correlation between the human exposure toward endogenous NOCs and the risk of developing non-cardia gastric cancer (2). Endogenously induced NOCs can be metabolically activated to give diazoacetate, which can result in the alkylation of DNA (1). An earlier study by Zurlo et al. (11) showed that the exposure of pancreatic acinar cells to O-diazoacetyl-l-serine, which can be metabolically activated to give diazoacetate (Scheme 1), could induce the formation of 7-carboxymethylguanine in DNA. It was also observed that the treatment of calf thymus DNA with potassium diazoacetate or NOCs could result in the formation of O6-CMdG (6–9). In addition, O6-CMdG was detected at a level of three to eight lesions per 107 nucleosides in the blood DNA of healthy human subjects (9), and humans consuming red meat have elevated level of O6-CMdG in colonic exfoliated cells than control group consuming vegetarian diet (13). Moreover, the propagation of diazoacetate-treated, human p53 gene-containing shuttle vector in yeast cells gave similar mutation pattern at non-CpG sites as that found in human stomach and colon cancers (15). Together, these previous studies established a strong link between human exposure to N-nitroso compounds and the carboxymethylation of DNA and suggested that the exposure to N-nitroso compounds may constitute an important etiological factor in human cancer development.
Aside from the above two dG adducts, the diazoacetate-mediated carboxymethylation chemistry of other nucleosides in DNA has not been assessed. Shuttle vector studies revealed that diazoacetate treatment can also lead to significant mutations at T:A base pairs (15), suggesting that diazoacetate can also lead to the modification of thymine and/or adenine in DNA. Here, we synthesized authentic N3-CMdT and O4-CMdT, and demonstrated, by using LC-MS/MS, that indeed these two lesions could be induced and they constitute the main thymidine products formed in calf thymus DNA upon exposure to diazoacetate. The methods for the syntheses of N3-CMdT and O4-CMdT can be readily adapted for the preparation of the corresponding stable isotope-labeled derivatives, which facilitates the future LC-MS/MS quantification of these lesions formed in vitro and in vivo by using the isotope dilution technique. In this context, it is worth noting LC-MS/MS also revealed the carboxymethylation of 2′-deoxyadenosine in calf thymus DNA. The chemical nature of the carboxymethylated adenine is currently under investigation.
The formation of N3-CMdT and O4-CMdT in calf thymus DNA calls for the examination of the replication and repair of these lesions. To carry out such studies, we need to obtain ODN substrates housing a site-specifically incorporated N3-CMdT and O4-CMdT. Thus, we also developed facile synthetic methods for the site-specific incorporation of these two lesions into ODNs. In this respect, the methods facilitated the preparation of 12-mer ODNs in reasonably good yield. After HPLC separation and desalting, we were able to obtain ∼100 nmol of the lesion-bearing ODNs from a 1 µmol scale synthesis. Thus, the synthetic strategy facilitates the availability of sufficient ODN substrates for future NMR structural studies. The lesion-bearing substrates can also be employed to construct, by enzymatic ligation, longer lesion-bearing ODNs for in vitro DNA repair studies (23) and damage-containing shuttle vectors for in vivo replication and repair studies (24,25).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the National Institutes of Health (Grant No R01 CA101864). Funding for open access charge: National Institutes of Health (R01 CA101864).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors want to thank Dr Dan Borchardt for assistance with the HMBC experiments.
REFERENCES
- 1.Tricker AR. N-nitroso compounds and man: sources of exposure, endogenous formation and occurrence in body fluids. Eur. J. Cancer Prev. 1997;6:226–268. [PubMed] [Google Scholar]
- 2.Jakszyn P, Bingham S, Pera G, Agudo A, Luben R, Welch A, Boeing H, Del Giudice G, Palli D, Saieva C, et al. Endogenous versus exogenous exposure to N-nitroso compounds and gastric cancer risk in the European Prospective Investigation into Cancer and Nutrition (EPIC-EURGAST) study. Carcinogenesis. 2006;27:1497–1501. doi: 10.1093/carcin/bgl019. [DOI] [PubMed] [Google Scholar]
- 3.Busby WF, Jr, Shuker DE, Charnley G, Newberne PM, Tannenbaum SR, Wogan GN. Carcinogenicity in rats of the nitrosated bile acid conjugates N-nitrosoglycocholic acid and N-nitrosotaurocholic acid. Cancer Res. 1985;45:1367–1371. [PubMed] [Google Scholar]
- 4.Puju S, Shuker DE, Bishop WW, Falchuk KR, Tannenbaum SR, Thilly WG. Mutagenicity of N-nitroso bile acid conjugates in Salmonella typhimurium and diploid human lymphoblasts. Cancer Res. 1982;42:2601–2604. [PubMed] [Google Scholar]
- 5.Shuker DEG, Tannenbaum SR, Wishnok JS. N-nitroso bile acid conjugates. 1. Synthesis, chemical reactivity and mutagenic activity. J. Org. Chem. 1981;46:2092–2096. [Google Scholar]
- 6.Shuker DE, Margison GP. Nitrosated glycine derivatives as a potential source of O6-methylguanine in DNA. Cancer Res. 1997;57:366–369. [PubMed] [Google Scholar]
- 7.Harrison KL, Jukes R, Cooper DP, Shuker DE. Detection of concomitant formation of O6-carboxymethyl- and O6-methyl-2′-deoxyguanosine in DNA exposed to nitrosated glycine derivatives using a combined immunoaffinity/HPLC method. Chem. Res. Toxicol. 1999;12:106–111. doi: 10.1021/tx980057n. [DOI] [PubMed] [Google Scholar]
- 8.Harrison KL, Fairhurst N, Challis BC, Shuker DE. Synthesis, characterization, and immunochemical detection of O6-(carboxymethyl)-2'-deoxyguanosine: a DNA adduct formed by nitrosated glycine derivatives. Chem. Res. Toxicol. 1997;10:652–659. doi: 10.1021/tx960203u. [DOI] [PubMed] [Google Scholar]
- 9.Cupid BC, Zeng Z, Singh R, Shuker DE. Detection of O6-carboxymethyl-2'-deoxyguanosine in DNA following reaction of nitric oxide with glycine and in human blood DNA using a quantitative immunoslot blot assay. Chem. Res. Toxicol. 2004;17:294–300. doi: 10.1021/tx0340706. [DOI] [PubMed] [Google Scholar]
- 10.Longnecker DS, Curphey TJ. Adenocarcinoma of the pancreas in azaserine-treated rats. Cancer Res. 1975;35:2249–2258. [PubMed] [Google Scholar]
- 11.Zurlo J, Curphey TJ, Hiley R, Longnecker DS. Identification of 7-carboxymethylguanine in DNA from pancreatic acinar cells exposed to azaserine. Cancer Res. 1982;42:1286–1288. [PubMed] [Google Scholar]
- 12.Bingham SA, Pignatelli B, Pollock JR, Ellul A, Malaveille C, Gross G, Runswick S, Cummings JH, O’Neill IK. Does increased endogenous formation of N-nitroso compounds in the human colon explain the association between red meat and colon cancer? Carcinogenesis. 1996;17:515–523. doi: 10.1093/carcin/17.3.515. [DOI] [PubMed] [Google Scholar]
- 13.Lewin MH, Bailey N, Bandaletova T, Bowman R, Cross AJ, Pollock J, Shuker DE, Bingham SA. Red meat enhances the colonic formation of the DNA adduct O6-carboxymethyl guanine: implications for colorectal cancer risk. Cancer Res. 2006;66:1859–1865. doi: 10.1158/0008-5472.CAN-05-2237. [DOI] [PubMed] [Google Scholar]
- 14.O’Driscoll M, Macpherson P, Xu YZ, Karran P. The cytotoxicity of DNA carboxymethylation and methylation by the model carboxymethylating agent azaserine in human cells. Carcinogenesis. 1999;20:1855–1862. doi: 10.1093/carcin/20.9.1855. [DOI] [PubMed] [Google Scholar]
- 15.Gottschalg E, Scott GB, Burns PA, Shuker DE. Potassium diazoacetate-induced p53 mutations in vitro in relation to formation of O6-carboxymethyl- and O6-methyl-2'-deoxyguanosine DNA adducts: relevance for gastrointestinal cancer. Carcinogenesis. 2007;28:356–362. doi: 10.1093/carcin/bgl150. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Wang Y. Synthesis and thermodynamic studies of oligodeoxyribonucleotides containing tandem lesions of thymidine glycol and 8-oxo-2’-deoxyguanosine. Chem. Res. Toxicol. 2006;19:837–843. doi: 10.1021/tx060032l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gait MJ. Oligonucleotide Synthesis: A Practical Approach. Oxford, England: IRL Press Limited; 1984. [Google Scholar]
- 18.Anderson D, Hambly RJ, Yu TW, Thomasoni F, Shuker DE. The effect of potassium diazoacetate on human peripheral lymphocytes, human adenocarcinoma colon Caco-2 cells, and rat primary colon cells in the comet assay. Teratog. Carcinog. Mutagen. 1999;19:137–146. doi: 10.1002/(sici)1520-6866(1999)19:2<137::aid-tcm6>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 19.Xu YZ, Swann PF. A simple method for the solid phase synthesis of oligodeoxynucleotides containing O4-alkylthymine. Nucleic Acids Res. 1990;18:4061–4065. doi: 10.1093/nar/18.14.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Y-Z. Synthesis and characterization of DNA containing O6-carboxymethylguanine. Tetrahedron. 2000;56:6075–6081. [Google Scholar]
- 21.McLuckey SA, Van Berkel GJ, Glish GL. Tandem mass spectrometry of small, multiply charged oligonucleotides. J. Am. Soc. Mass Spectrom. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
- 22.Wang Z, Wan KX, Ramanathan R, Taylor JS, Gross ML. Structure and fragmentation mechanisms of isomeric T-rich oligodeoxynucleotides: a comparison of four tandem mass spectrometric methods. J. Am. Soc. Mass Spectrom. 1998;9:683–691. doi: 10.1016/S1044-0305(98)00178-0. [DOI] [PubMed] [Google Scholar]
- 23.Gu C, Zhang Q, Yang Z, Wang Y, Zou Y, Wang Y. Recognition and incision of oxidative intrastrand cross-link lesions by UvrABC nuclease. Biochemistry. 2006;45:10739–10746. doi: 10.1021/bi060423z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delaney JC, Essigmann JM. Assays for determining lesion bypass efficiency and mutagenicity of site-specific DNA lesions in vivo. Methods Enzymol. 2006;408:1–15. doi: 10.1016/S0076-6879(06)08001-3. [DOI] [PubMed] [Google Scholar]
- 25.Yuan B, Cao H, Jiang Y, Hong H, Wang Y. Efficient and accurate bypass of N2-(1-carboxyethyl)-2′-deoxyguanosine by DinB DNA polymerase in vitro and in vivo. Proc. Natl Acad. Sci. USA. 2008;105:8679–8684. doi: 10.1073/pnas.0711546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.