Abstract
The assembly of large recombinant DNA encoding a whole biochemical pathway or genome represents a significant challenge. Here, we report a new method, DNA assembler, which allows the assembly of an entire biochemical pathway in a single step via in vivo homologous recombination in Saccharomyces cerevisiae. We show that DNA assembler can rapidly assemble a functional d-xylose utilization pathway (∼9 kb DNA consisting of three genes), a functional zeaxanthin biosynthesis pathway (∼11 kb DNA consisting of five genes) and a functional combined d-xylose utilization and zeaxanthin biosynthesis pathway (∼19 kb consisting of eight genes) with high efficiencies (70–100%) either on a plasmid or on a yeast chromosome. As this new method only requires simple DNA preparation and one-step yeast transformation, it represents a powerful tool in the construction of biochemical pathways for synthetic biology, metabolic engineering and functional genomics studies.
INTRODUCTION
Methods that enable design and rapid construction of biochemical pathways will be invaluable in pathway engineering, metabolic engineering, combinatorial biology and synthetic biology. In pathway and metabolic engineering, complete biosynthetic pathways are often transferred from native hosts into heterologous organisms in order to obtain products of interest at high yields (1–5). Consequently, gene expression needs to be balanced, promoter strength needs to be tuned and the endogenous regulatory network needs to be modified (6–8). In combinatorial biology, genes are rearranged in order to generate new small molecules (9); whereas, in synthetic biology, complex combinations of genetic elements are used to create new circuits with desired properties (10). In all these studies, the conventional multi-step, sequential-cloning method, including primer design, PCR amplification, restriction digestion, in vitro ligation and transformation, is typically involved and multiple plasmids are often required (2,4). This method is not only time consuming and inefficient but also relies on unique restriction sites that become limited for large recombinant DNA molecules.
To address these limitations, a new cloning method, sequence and ligation-independent cloning (SLIC), was recently reported (11). Using SLIC, up to 10 DNA fragments including nine PCR-generated fragments of sizes ranging between 274 and 980 bp and a 3.1-kb vector were assembled into a recombinant DNA molecule using in vitro homologous recombination. However, the success rate was only 17% (7 of 42 Escherichia coli transformants were proven to be correct), and the size of the recombinant DNA was only approximately 8 kb. More recently, a protocol called the ‘domino method’ was developed to construct a number of DNA pieces, known as ‘domino clones’, in a vector through in vivo homologous recombination in Bacillus subtilis (12). Using two selection markers alternatively, a 16.3-kb mouse mitochondrial genome and a 134.5-kb rice chloroplast genome were successfully assembled. However, each ‘domino clone’ needs to be prepared in the vector first, and the whole assembly process is carried out in a sequential manner, thus requiring a great amount of time and labor.
Here, we developed a new method, called ‘DNA assembler’, that enables design and rapid construction of large biochemical pathways in one-step fashion by exploitation of in vivo homologous recombination mechanism in S. cerevisiae. Thanks to its high efficiency and ease to work with, in vivo homologous recombination in yeast has been widely used for gene cloning, plasmid construction and library creation (13–20). For example, Oldenburg and coworkers incorporated a linker of 30–40 bp, homologous to a linearized yeast plasmid, into primers to PCR-amplify the target DNA. Co-transformation of the PCR products and the plasmid into yeast resulted in homologous recombination at a position directed by the PCR primers (18). Raymond and coworkers (13,19) used synthetic, double-stranded ‘recombination linkers’ to subclone a DNA fragment, and counter-selection was later used to improve the efficiency of plasmid construction (19,20). Manivasakam et al. and Langle-Rouault et al. further extended homologous recombination into genome modification and determined the length of sequence homology required for targeted gene integration and deletion (21,22). In addition, Larionov and coworkers (23–27) developed a transformation-associated recombination cloning method in yeast, which was used to isolate large chromosomal fragments or entire gene clusters from complex genomes and to assemble synthetic DNA arrays of long tandem repeats up to 120 kb (28). Most recently, Gibson and coworkers described the construction of the entire Mycoplasma genitalium genome from 101 fragments. The assembly process was partitioned to several rounds and in vitro recombination followed by transformation into E. coli was used to assemble the synthetic cassettes. In the final step, the complete synthetic genome was assembled by transformation-associated recombination cloning in yeast when the size of the synthetic molecule became too big to be stable in E. coli (29). Despite these impressive applications, yeast in vivo homologous recombination has not been used to assemble multiple-gene biochemical pathways in a one-step fashion either on a plasmid or on a chromosome.
As proof of concept, we used DNA assembler to rapidly assemble three distinct functional biochemical pathways with sizes ranging from 9 to 19 kb on both a plasmid and a chromosome. This method is highly efficient and circumvents the potential problems associated with the conventional cloning method, representing a versatile approach for the construction of biochemical pathways for synthetic biology, metabolic engineering and functional genomics studies.
MATERIALS AND METHODS
Microorganisms, plasmids, reagents and media
S. cerevisiae YSG50 (MATα, ade2-1, ade3Δ22, ura3-1, his3-11,15, trp1-1, leu2-3,112 and can1-100) was used as the host for DNA assembly and integration. The pRS426 and pRS416 plasmid (New England Biolabs, Beverly, MA) were modified by incorporating the hisG and partial δ sequence (δ2) that flank the multiple cloning site and serve as the vectors for assembly of various pathways (Supplementary Figure 1). The resulting pRS426m and pRS416m were linearized by BamHI. The δ1-hisG-ura3-hisG fragment was cut from pdδUB (30) with BamHI and XhoI. Plasmid containing the cDNA (psXKS1) encoding d-xylulokinase (XKS) was a gift from T.W. Jeffries, University of Wisconsin, Madison. pCAR-ΔCrtX was kindly provided by E.T. Wurtzel at the City University of New York, which contains CrtE, B, I, Y and Z from Erwinia uredovora for zeaxanthin biosynthesis. The restriction enzymes and Phusion DNA polymerase were purchased from the New England Biolabs. Yeast YPAD medium-containing 1% yeast extract, 2% peptone and 2% dextrose supplied with 0.01% adenine hemisulphate was used to grow S. cerevisiae YSG50 strain. Synthetic complete drop-out medium lacking uracil (SC-Ura) was used to select transformants or integrants containing the assembled biochemical pathways of interest.
DNA manipulation
To prepare individual gene expression cassettes, yeast promoters (ADH1p, 1500 bp; PGK1p, 750 bp; PYK1p, 1000 bp, TEF1p, 412 bp; truncated HXT7p, 395 bp; TEF2p, 560 bp; FBA1p, 820 bp; and PDC1p, 800 bp) and terminators (ADH1t, 327 bp; CYC1t, 250 bp; ADH2t, 400 bp; PGIt, 400 bp; TPI1t, 400 bp; FBA1t, 400 bp; ENO2t, 400 bp; and TDH2t, 400 bp) were PCR-amplified individually from the genomic DNA isolated from S. cerevisiae YSG50 using the Wizard Genomic DNA isolation kit from Promega (Madison, WI). The structural genes XR and XDH were amplified from Neurospora crassa cDNA, and the structural genes including XKS, CrtE, CrtB, CrtI, CrtY and CrtZ were amplified from the corresponding plasmids. HisG and δ2 were amplified from pdδUB. All the primers are listed in Supplementary Table 1. Each individual gene expression cassette was assembled by overlap extension PCR (OE-PCR) (31). Following electrophoresis, the OE-PCR product was individually gel-purified from a 0.7% agarose gel using Qiagen Gel Purification kit (Valencia, CA). Each individual gene cassette (2.1–3.7 kb, 300 ng) was mixed with the linearized pRS426m (6.3 kb, 500 ng) or pRS416m (5.5 kb, 500 ng) and precipitated with ethanol. The resulting DNA pellet was air-dried and resuspended in 4 μl Milli-Q double deionized water for transformation into S. cerevisiae. To perform one-step integration targeting the δ sites on yeast chromosomes, DNA mixture of individual cassettes together with the fragment, δ1-hisG-ura3-hisG, digested from pdδUB with BamHI and XhoI, was used to transform S. cerevisiae.
Yeast transformation
Yeast transformation was performed by electroporation. YPAD medium (50 ml) was inoculated with a 0.5 ml overnight S. cerevisiae YSG50 culture and shaken at 30°C and 250 rpm for 4–5 h until OD600 reached 0.8–1.0. Yeast cells were harvested by centrifugation at 4°C and 4000 rpm for 5 min. The supernatant was discarded and the cell pellet was washed with 50 ml ice-cold Milli-Q double-deionized water, followed by another wash with cold 1 M sorbitol and finally resuspended in 250 μl cold sorbitol. An aliquot of 50 μl of yeast cells together with 4 μl DNA mixture was electroporated in a 0.2 cm cuvette at 1.5 kV. The transformed cells were immediately mixed with 1 ml room temperature YPAD medium and shaken at 30°C for 1 h. Following that, the cells were harvested by centrifugation, washed with room temperature 1 M sorbitol several times to remove the remaining medium and finally resuspended in 1 ml sorbitol. Aliquots of 30–50 μl were spread on SC-Ura plates for the selection of the transformants, and the plates were incubated at 30°C for 2–4 days until colonies appeared. Colonies were randomly picked to SC-Ura liquid medium and grown for 1 day, after which yeast plasmid was isolated using Zymoprep II Yeast plasmid Miniprep kit (Zymo Research, CA), and genomic DNA was prepared using Wizard Genomic DNA isolation kit from Promega (Madison, WI).
PCR analysis of the assembled pathways
For each pathway, confirmation of the correct DNA assembly was performed by PCR amplification of each gene cassette from the isolated yeast plasmids and the genomic DNAs. Generally, the PCRs were carried out in 20 μl reaction mixture consisting of 10 μl FailSafe™ PCR 2× PreMix G from Epicentre Biotechnologies (Madison, WI), 0.5 pmol of each primer, 1 μl yeast plasmid or genomic DNA and 0.4 U Phusion DNA polymerase for 35 cycles on a PTC-200 Thermal Cycler (MJ Research, Watertown, MA). Each cycle consisted of 10 s at 98°C, 30 s at 58°C and 3 min at 72°C, with a final extension of 10 min.
Physical characterization of recombinant clones
Yeast plasmids were transformed to E. coli strain BW25141 and selected on Luria Broth (LB) agar plates supplemented with 100 μg/ml ampicillin. Colonies were inoculated into 3 ml LB medium supplemented with 100 μg/ml ampicillin, and plasmids were isolated from the liquid culture using the plasmid miniprep kit from Qiagen (Valencia, CA). Plasmids isolated from E. coli were then subjected to restriction digestion by BamHI and EcoRI, after which the reaction mixtures were loaded to 1% agarose gels to check the correct restriction digestion pattern by DNA electrophoresis.
Functional analysis of the assembled pathways
The recombinant S. cerevisiae carrying the d-xylose utilization pathway was analyzed by monitoring the cell growth in SC-Ura liquid medium supplemented with 2% d-xylose (SC-Ura+xylose) as the sole carbon source. Colonies were picked into 2 ml SC-Ura liquid medium supplemented with 2% glucose and grown for 1–2 days. Cells were spun down and washed with 1 ml SC-Ura+xylose medium three times to remove the remaining glucose and finally resuspended in 1 ml SC-Ura+xylose. The resuspended cells were normalized to an OD600 of 0.1 in 3 ml of fresh SC-Ura+xylose medium. Cells were grown at 30°C for 192 h and OD600 was measured every 24 h.
Zeaxanthin was extracted with acetone from S. cerevisiae bearing either the five-gene pathway, the eight-gene pathway, or the control with an empty vector. Briefly, cells from 250 ml culture were collected by centrifugation, resuspended with 5 ml acetone and passed through French press several times at 10 000 psi. Supernatants were collected after centrifugation and evaporated to dryness. After resuspension in 0.5–1 ml methanol, 100 μl of sample was loaded onto the Agilent ZORBAX SB-C18 column and analyzed at 450 nm on an Agilent 1100 series HPLC (Agilent Technologies, Palo Alto, CA). The solvent program with a 0.5 ml/min flow rate was as follows: 0–3 min, 60% CH3OH; 3–15 min, linear gradient from 60% CH3OH to 100% CH3OH; 15–17 min, 100% CH3OH; 17–20 min, linear gradient from 100% CH3OH to 60% CH3OH. Authentic zeaxanthin from Sigma (St Louis, MO) was used as a standard.
RESULTS
Design of the DNA assembler method
As proof of concept, we sought to design and construct various multiple-gene biosynthetic pathways. As shown in Scheme 1, for each individual gene in a pathway, an expression cassette including a promoter, a structural gene and a terminator is PCR-amplified and assembled using OE-PCR (31). The 5′-end of the first gene expression cassette is designed to overlap with a vector (or a helper fragment carrying a selection marker and a sequence sharing homology with a targeted locus of a chromosome for integration), while the 3′-end is designed to overlap with the second cassette. Each successive cassette is designed to overlap with the two flanking ones, and the 3′-end of the last cassette overlaps with the vector (or the targeted locus of the chromosome for integration). All overlaps are designed to have >40 bp for efficient in vivo homologous recombination. The resulting multiple expression cassettes are co-transformed into S. cerevisiae with the linearized vector (or the helper fragment) through electroporation, which allows the entire pathway to be either assembled into a vector or integrated onto the chromosome.
Scheme 1.
A one-step method for assembly and integration of a biochemical pathway using in vivo homologous recombination in S. cerevisiae. (a) One-step assembly into a vector. (b) One-step integration to a δ site on a S. cerevisiae chromosome. n represents the number of DNA fragments.
Construction of a three-gene d-xylose utilization pathway
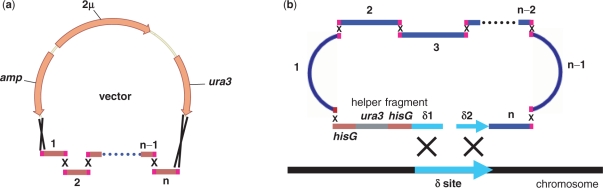
We first applied the DNA assembler method to construct the known d-xylose utilization pathway consisting of xylose reductase, xylitol dehydrogenase from N. crassa (32) and XKS from Pichia stipitis (33). The vector pRS426m, which was created by incorporating a hisG sequence and a δ2 sequence in the pRS426, a 2 μm multiple copy vector (unpublished data), was used and linearized with BamHI (Supplementary Figure 1). Promoters and terminators were PCR-amplified from the genomic DNA of S. cerevisiae. Individual cassettes, hisG-ADH1p-XR-ADH1t, PGK1p-XDH-CYC1t and PYK1p-XKS-ADH2t-δ2, were assembled by OE-PCR, purified by agrose gel electrophoresis and mixed with the linearized vector. After precipitation in ethanol, the mixture was re-dissolved in ddH2O and co-transformed into S. cerevisiae by electroporation and spread onto plates containing synthetic complete drop-out medium lacking uracil (SC-Ura). In parallel, a 4.7-kb integration fragment δ1-hisG-ura3-hisG, carrying a ura3 selection marker and δ sequence that is homologous to the partial sequence of the δ sites on the chromosome of S. cerevisiae, was generated from the δ-integration vector pdδUB (30) via BamHI and XhoI digestion and then co-transformed with the hisG-ADH1p-XR-ADH1t, PGK1p-XDH-CYC1t and PYK1p-XKS-ADH2t-δ2 cassettes into S. cerevisiae. The two repeated hisG sequences in the δ1-hisG-ura3-hisG fragment could be used for excising ura3 from the chromosome and thus recycling the marker using 5-fluoroorotic acid counter selection (30). Typically, 100–150 colonies appeared after 2–3 days of incubation at 30°C and then were randomly picked for plasmid or genomic DNA isolation. In order to confirm the correct assembly and integration, a comprehensive set of PCR was performed. As shown in Figure 1a, three pairs of primers annealing to the corresponding two overlap regions flanking each cassette were used as verification primers (Supplementary Table 2). A correct assembly (both plasmid based and chromosome based) would generate all three expected PCR products (Figure 1b). In order to determine the assembly efficiency, 10 colonies were randomly picked and checked by PCR. For both plasmid-based DNA assembly and chromosome based DNA assembly, high efficiencies (80–100%) were obtained for all three different lengths of overlaps and use of longer overlaps (125 bp and 270–430 bp) between the adjacent cassettes seems not to be necessary (Table 1).
Figure 1.
Construction of the d-xylose utilization pathway in Saccharomyces cerevisiae. (a) The assembled d-xylose utilization pathway with its corresponding verification primers; (b) PCR analysis of the assembled d-xylose utilization pathway on a plasmid [d-xylose utilization (p), using pRS426m as a backbone] and on a chromosome [d-xylose utilization (c), integrated to the δ site(s)]. M represents 1 kb DNA ladder from Invitrogen; (c) Growth of S. cerevisiae strains carrying d-xylose utilization pathway using d-xylose as the sole carbon source. Cells carrying the empty vector pRS426 were used as the negative control.
Table 1.
The efficiency of one-step DNA assembly (using pRS426m as a backbone) and integration of three-, five- and eight-gene pathways for d-xylose utilization, zeaxanthin biosynthesis and the combined d-xylose utilization and zeaxanthin biosynthesis, respectively, with different lengths of overlap
| Length of the overlap (bp) |
|||
|---|---|---|---|
| ∼50 | ∼125 | 270–430 | |
| Three-gene pathway | |||
| Plasmid-based (%) | 100 | 90 | 80 |
| Chromosome-based (%) | 80 | 80 | 100 |
| Five-gene pathway | |||
| Plasmid-based (%) | 80 | 60 | 80 |
| Chromosome-based (%) | 80 | 70 | 80 |
| Eight-gene pathway | |||
| Plasmid-based (%) | 20 | 50 | 40 |
| Chromosome-based (%) | 10 | 60 | 70 |
For each pathway, 10 clones were randomly picked and analyzed.
The function of the assembled d-xylose utilization pathway was analyzed by growing the strains containing the d-xylose utilization pathway in SC-Ura medium supplemented with d-xylose instead of d-glucose. Note that, in contrast to S. cerevisiae, which is unable to ferment d-xylose directly, S. cerevisiae expressing XR, XDH and XKS can use d-xylose as the sole carbon source (34,35). As shown in Figure 1c, only the strains carrying the whole d-xylose utilization pathway either assembled into the vector or integrated onto the chromosome were able to grow on d-xylose, while the S. cerevisiae containing the empty pRS426 did not grow.
Construction of a five-gene zeaxanthin biosynthetic pathway
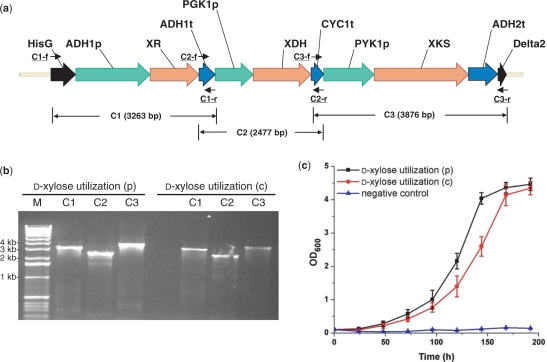
We then extended the DNA assembler method to construct the five-gene zeaxanthin biosynthetic pathway from E. uredovora (36–38). The individual cassettes, hisG-TEF1p-CrtE-PGIt, HXT7p-CrtB-TPI1t, TEF2p-CrtI-FBA1t, FBA1p-CrtY-ENO2t and PDC1p-CrtZ-TDH2t-δ2 were prepared by OE-PCR. The total size of the five-gene pathway is 10.6 kb, and the entire pathway was assembled into the pRS426m or the S. cerevisiae chromosome in a one-step fashion within a week. Typically, 50–80 colonies appeared after 2–3 days of incubation at 30°C and then were randomly picked for plasmid or genomic DNA isolation. To confirm the correct assembly, five PCRs were performed using the verification primers (Figure 2a and Supplementary Table 2 online). Various lengths of overlap regions were tested, and 10 colonies were picked from each group to determine the assembly efficiency. The correct assembly exhibited all five expected bands obtained from PCR on a DNA agarose gel (Figure 2b). The efficiency of one-step assembly of this five-gene pathway is summarized in Table 1. Similar to the efficiency of assembling three-gene d-xylose utilization pathway, the efficiency is high (60–80%) and longer overlap regions between adjacent gene expression cassettes had little effect on the efficiency.
Figure 2.
Construction of the zeaxanthin biosynthesis pathway in Saccharomyces cerevisiae. (a) The assembled zeaxanthin biosynthesis pathway with its corresponding verification primers; (b) PCR analysis of the assembled zeaxanthin biosynthesis pathway on a plasmid [zeaxanthin biosynthesis (p), using pRS426m as a backbone] and on a chromosome [zeaxanthin biosynthesis (c), integrated to the δ site(s)]. M represents 1 kb DNA ladder from Invitrogen; (c) HPLC analysis of the cell extracts from S. cerevisiae carrying the zeaxanthin biosynthesis pathway. Cells carrying the empty vector pRS426 were used as the negative control.
S. cerevisiae strains containing the zeaxanthin biosynthetic pathway (plasmid based and chromosome based) were grown in SC-Ura medium at 30°C for 72 h for production of zeaxanthin. Following the experimental procedures reported elsewhere (38,39), the cells were collected by centrifuge and subjected to acetone extraction several times before evaporation to dryness. The extracts were then re-dissolved in methanol and analyzed by HPLC. When monitored at 450 nm, a peak appeared at 19.9 min, consistent with the elution time of authentic zeaxanthin, whereas no such peak was observed for the cell extracts from the strains carrying the empty pRS426 vector (Figure 2c).
Construction of a combined eight-gene d-xylose utilization and zeaxanthin biosynthetic pathway
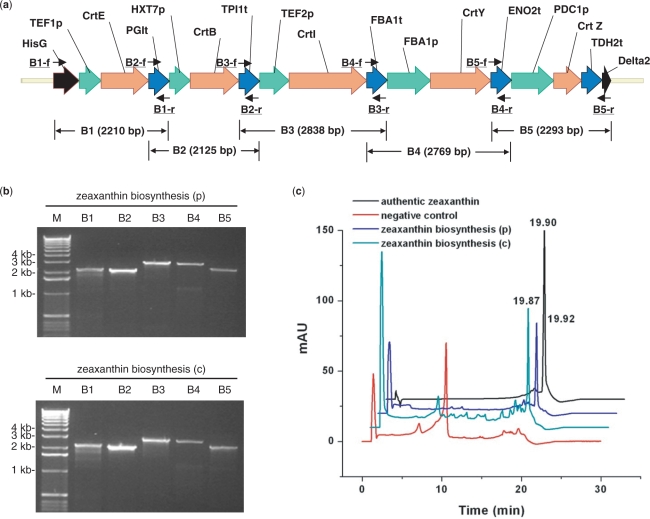
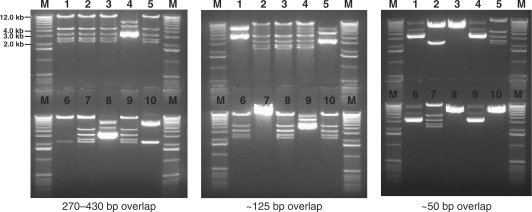
To further demonstrate the capacity and versatility of DNA assembler on constructing even longer biochemical pathways, the two above-mentioned pathways, the d-xylose utilization pathway and the zeaxanthin biosynthesis pathway, were combined and assembled onto one vector or integrated onto a δ site of the chromosome as a single piece of DNA. Eight gene cassettes including hisG-ADH1p-XR-ADH1t, PGK1p-XDH-CYC1t, PYK1p-XKS-ADH2t, TEF1p-CrtE-PGIt, HXT7p-CrtB-TPI1t, TEF2p-CrtI-FBA1t, FBA1p-CrtY-ENO2t and PDC1p-CrtZ-TDH2t-δ2 were constructed by OE-PCR and co-transformed to S. cerevisiae with the linearized vector or the δ1-hisG-ura3-hisG fragment. Typically, 30–50 colonies appeared after 2–3 days of incubation at 30°C and then were randomly picked for plasmid or genomic DNA isolation. Eight PCRs were performed to confirm the correct assembly (Figure 3a and b, and Supplementary Table 2). The total size of the combined eight-gene pathway is around 19 kb (or 23.7 kb when the 4.7 kb δ1-hisG-Ura3-hisG helper fragment is used). The efficiency of DNA assembly decreased dramatically with a shorter overlap (50 bp), as indicated in Table 1 (10–20%). However, for longer overlaps, 125 bp and 270–430 bp, a much higher efficiency (40–70%) was obtained. Thus, it seems that unlike the short pathways, the dependence of the efficiency of DNA assembly on the length of the overlap becomes more significant with the increase of the number of the DNA fragments co-transformed into S. cerevisiae.
Figure 3.
Construction of the combined d-xylose utilization and zeaxanthin biosynthesis pathway in Saccharomyces cerevisiae. (a) The assembled combined d-xylose utilization and zeaxanthin biosynthesis pathway with its corresponding verification primers; (b) PCR analysis of the assembled combined d-xylose utilization and zeaxanthin biosynthesis pathway on a plasmid [combined pathway (p), using pRS426m as a backbone] and on a chromosome [combined pathway (c), integrated to the δ site(s)]. M represents 1 kb DNA ladder from Invitrogen; (c) Growth of S. cerevisiae strains carrying the combined pathway using d-xylose as the sole carbon source; (d) HPLC analysis of the cell extracts from S. cerevisiae carrying the combined pathway. Cells carrying the empty vector pRS426 were used as the negative control.
S. cerevisiae strains containing the combined d-xylose utilization and zeaxanthin biosynthesis pathway were grown in SC-Ura medium supplemented with 2% d-xylose as the sole carbon source for d-xylose utilization. As shown in Figure 3c, S. cerevisiae bearing the combined pathway either on the plasmid or integrated to the chromosome were able to grow, in contrast to the strain carrying an empty pRS426 vector. In addition, as shown by HPLC analysis (Figure 3d), zeaxanthin was detected from the strains carrying the combined pathway, suggesting the presence of a functional zeaxanthin biosynthesis pathway.
The use of single-copy plasmid as a vehicle for DNA assembly
It was noticed that sometimes there was heterogeneity in the plasmids even though single yeast colonies were analyzed (data not shown). We reasoned that the DNA fragments and the vector could be assembled in more than one fashion and the assembled plasmids could be maintained within the cell, because pRS426 is a multiple-copy plasmid.
To address this issue, we sought to use a single-copy vector, pRS416m, containing a centromere and an autonomously replicating sequence for DNA assembly. The eight-gene pathway with various lengths of overlaps was chosen as a model system, and the same experiments were performed using the linearized pRS416m as the backbone. Yeast plasmids were isolated and transformed into E. coli strain BW25141, after which multiple colonies were picked from each plate. The plasmids isolated from the clones on the same E. coli transformation plate all showed the same restriction digestion pattern on the agarose gel (data not shown), indicating a single form of plasmid was maintained in each yeast transformant. The E. coli plasmids were then subjected to restriction digestion by BamHI and EcoRI. As shown in Figure 4, 6 of 10 clones exhibited the expected restriction digestion pattern (five bands: 2233, 2551, 3218, 4454 and 11 386 bp) for both ∼125 bp and 270–430-bp overlaps, and 2 of 10 clones exhibited the expected restriction digestion pattern for ∼50 bp overlap, resulting in an assembly efficiency similar to that when pRS426m was used.
Figure 4.
Physical characterization of the recombinant clones using pRS416m as a backbone through restriction digestion. Plasmids corresponding to the clones from the eight-gene pathway with ∼50 bp, ∼125 bp and 270–430 bp overlaps were digested by BamHI and EcoRI. The correct clones should exhibit five bands with sizes of 2233, 2551, 3218, 4454 and 11 386 bp.
DISCUSSION
Unlike the conventional cloning approach that relies on site-specific digestion and ligation, homologous recombination aligns complementary sequences and enables the exchange between homologous fragments. Homologous recombination is much more efficient in yeast than in bacteria and other higher eukaryotes. Therefore, it has been exploited as one of the most important tools in gene cloning, site-specific mutagenesis, plasmid construction and target gene disruption and deletion (13,18–22). In this report, we further extended its application to assemble biochemical pathways in a single step.
The novelty of this DNA assembler method lies on the following aspects: (a) it is the first demonstration of yeast in vivo homologous recombination in assembling multiple-gene biochemical pathways in a single step, which distinguishes it from routine single-gene plasmid construction; (b) compared to the two related methods including the SLIC method and the domino method, DNA assembler is the most rapid and efficient approach to construct large recombinant DNA molecules; (c) unlike the SLIC method and the domino method, DNA assembler can be used to construct custom-designed DNA molecules not only on a plasmid but also on a chromosome. The latter enables the exogenous DNA stably maintained in the cell and has the potential to assemble a very large biochemical pathway or even a whole genome.
Similar to E. coli, because of its well-characterized physiology and genetics and ample genetic tools, S. cerevisiae has been widely used as a platform organism for heterologous expression of biochemical pathways in the fields of pathway engineering and metabolic engineering (1–5). In contrast to the conventional techniques for pathway cloning, DNA assembler avoids repeated cycles of multiple-step cloning, does not rely on the restriction digestion and in vitro ligation and takes only 1–2 weeks to assemble multiple DNA fragments into S. cerevisiae either on a plasmid or on a chromosome. It simply takes the advantage of the pre-existing homologous recombination machinery in yeast and thus does not require any exogenous recombinase.
In addition, unlike the recently reported in vitro SLIC recombination method that requires T4 DNA polymerase and RecA recombinase to treat the DNA substrates before transformation (11), DNA assembler only requires simple DNA preparation via PCR and one-step yeast transformation and yet yields a much higher assembly efficiency than the SLIC method. As shown in our report, for a shorter pathway (3–5 genes, ∼10 kb), a very high efficiency of 80–100% was obtained with a ∼50-bp overlap, while for a longer pathway (8 genes, ∼19 kb), a relatively high efficiency of 40–70% was achieved with a slightly longer overlap (∼125–430 bp). In addition, for a long pathway, increasing the ratio between the inserts and the backbone could further improve the assembly efficiency. For example, the eight-gene pathway was reassembled with a ∼50-bp overlap. The amount of the inserts was doubled while the amount of the linearized vector was maintained (i.e. 600 ng of each insert was combined with 500 ng of the linearized vector). As a result, more colonies on the SC-Ura plates were observed, and an assembly efficiency of as high as 70% was obtained, compared with the efficiency of 20% when lower amount of the inserts was used. This indicates that DNA assembler could efficiently assemble a large DNA molecule even with a short overlap by manipulating the ratio between the inserts and the vector backbone.
It is possible that deletions may be caused by a repeat or repeats in the DNA fragments. To address this concern, we examined the existence of repeats in pRS426m-xylose-zeaxanthin. We found that short repeats are very common in such a 24.6-kb DNA molecule. As shown in Supplementary Table 3, 7 sequences with a length of 14 bp appeared twice, which were widely distributed among promoters, terminators and structural genes. In addition, 29 sequences (13 bp), 76 sequences (12 bp), 197 sequences (11 bp) and 527 sequences (10 bp) were found to occur at least twice in the same molecule. These abundant repeats up to 14 bp did not appear to be problematic in our experiment considering that assembling efficiencies of 40–70%, 50–60% and 10–20% were obtained with 270–430-bp, ∼125-bp and ∼50-bp overlaps, respectively. Previous homologous recombination-based gene-cloning experiments by other groups indicated the length of overlap required for efficient recombination between two DNA fragments: 40-bp overlaps yielded an efficiency of greater than 90% (18,19,40). The efficiency was slightly reduced using 30-bp overlaps (∼80%) and rapidly dropped to 3.4% when 20-bp overlaps were used (40). Similarly, in terms of deletions, Manivasakam and coworkers (22) studied the length of a repeat required for targeted deletion. A repeat of a 45 bp, a 30 bp and a 25 bp resulted in efficiencies of 84%, 54% and 4%, respectively (22). In addition, Langle-Rouault and Jacobs reported that selection of marker excision failed using 25-bp direct repeats (21). All these studies suggest that the efficiency of recombination-based deletions is fairly low when repeats of shorter than 25 bp are involved. It should also be noted that in all these mentioned experiments, a selectable marker was used to specifically select the deletion event, and the efficiency was calculated based on the percentage of the correct deletions among all the transformants. Therefore, short repeats (<25 bp) in a long DNA molecule should not cause too much trouble, especially because such a deletion event is not specifically selected, making the chance even much lower. When the repeats become longer (>25 bp, but not too long), the recombination between the repeats will likely compete with the recombination between the overlaps at the ends of each fragment in the assembling process. In this case, a longer overlap (>100 bp) will benefit the recombination between the ends. The deletion between repeats could happen, but the chance will be several orders of magnitude lower than the above efficiency considering that no selection is used in such a deletion process. Of special note, we recently developed an efficient method for gene deletions in yeast, and we studied the length of the repeats required to yield decent deletion efficiencies (41). Our results showed that 25-bp repeats are not sufficient to generate deletions. It should be noted that when the repeats become very long (>100 bp), the deletion event may occur with a higher efficiency. However, such long repeats are not common in a typical biochemical pathway. If long repeats exist in structural genes, silent mutations may be incorporated during the fragment preparation process in order to reduce the length of the repeated regions.
In our studies, S. cerevisiae YSG50 was used as a host for DNA assembly and integration. This strain carries a ura3-1 point mutation on its genome, which may be converted to wild type by the ura3 gene in the pRS626m vector (or in the helper fragment). To address this concern, we also tested a strain HZ848 with a complete deletion of ura3 (the strain HZ848 was obtained by deleting the ura3 gene in YSG50). The same amount of DNA mixture (eight DNA fragments plus the digested vector) was transformed to YSG50 and HZ848, and in parallel, the same amount of the linearized vector was transformed to both strains as controls.
Similar numbers of colonies were obtained in the two samples (42 for YSG50 versus 38 for HZ848), whereas only two colonies appeared in both controls. That the same number of colonies appeared in both controls suggested that the vector had not been completely digested. Although it is possible that the chromosomal gene ura3-1 may be converted to wild type, it should occur at a very low frequency (<10−6). In comparison, the frequency of DNA assembly was estimated to be 5.7 × 10−6 (∼40 colonies appearing on the Sc-Ura plates from 7 × 106 cells), which is relatively high. Ten clones from both of YSG50 transformants and HZ88 transformants were selected for further analysis. Of 10, 5 YSG50 transformants and 4 of 10 HZ848 transformants were correct, indicating that for a biochemical pathway consisting of up to eight genes, there is no advantage to use a strain with a complete deletion of ura3. However, when the assembly efficiency drops to a similar level with that of the reversion rate of ura3-1 back to wild type, using a strain with ura3 completely deleted might become advantageous. Such scenario may occur when a much longer biochemical pathway is assembled.
It should be noted that DNA assembler has the potential to assemble much longer biochemical pathways in S. cerevisiae. Conceptually, a long biochemical pathway can be split into several segments (8–10 genes each) that will be sequentially integrated into the yeast chromosome by using a recyclable selection marker such as Ura. Once a segment is integrated, 5-fluoroorotic acid can be used to remove the integrated Ura selection marker from the chromosome (42), so that another segment can be integrated subsequently. Through recycling the selection marker, horizontally moving any biochemical pathways from their native hosts to S. cerevisiae will be no longer limited by the size of the pathways. For example, a biochemical pathway composed of 30–50 genes (around 100–200 kb) could be integrated to the chromosome of S. cerevisiae within several weeks. Of course, when a biochemical pathway is very long, PCR-introduced mutations may become a concern even though high-fidelity DNA polymerases are used. Note that this concern is also common to other reported DNA assembly methods using high-fidelity polymerases such as the Phusion polymerase (with a very low error rate, 4.4 × 10−7) should greatly reduce the mutation frequency. Fortunately, with the development of new powerful DNA sequencing technologies, validation of the correctly assembled long biochemical pathway by DNA sequencing is no longer a burden in terms of time and cost. For example, the recently developed 454 sequencing technology (Roche, Branford, CT) supports the sequencing of samples from a wide variety of starting materials including genomic DNA, PCR products, BACs and cDNA (http://www.454.com/). And it costs less than $15K to sequence the entire genome of a microorganism of typical size (∼6 Mb) within a few days.
In principle, this method may be even used to assemble a DNA molecule as large as an entire chromosome or genome. As a matter of fact, Gibson and coworkers’ success in the construction of the entire M. genitalium genome from 101 fragments (29) indicates that it is possible to assemble a DNA molecule as large as a genome from short DNA fragments using DNA assembler. Also, in addition to assembling biochemical pathways and genomes, DNA assembler has many other applications such as library creation in combinatorial biology and construction of complicated DNA molecules in the field of synthetic biology.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by National Institutes of Health (GM077596). Funding for open access charge: National Institutes of Health (GM077596).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank N. U. Nair, R. P. Sullivan, M. McLachlan and T. Johannes for valuable discussion. We thank F. H. Arnold, P. Orlean, T. W. Jeffries, E. T. Wurtzel and N. A. Da Silva for providing plasmids and yeast strains.
REFERENCES
- 1.Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 2.Dejong JM, Liu Y, Bollon AP, Long RM, Jennewein S, Williams D, Croteau RB. Genetic engineering of taxol biosynthetic genes in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2006;93:212–224. doi: 10.1002/bit.20694. [DOI] [PubMed] [Google Scholar]
- 3.Szczebara FM, Chandelier C, Villeret C, Masurel A, Bourot S, Duport C, Blanchard S, Groisillier A, Testet E, Costaglioli P, et al. Total biosynthesis of hydrocortisone from a simple carbon source in yeast. Nat. Biotechnol. 2003;21:143–149. doi: 10.1038/nbt775. [DOI] [PubMed] [Google Scholar]
- 4.Yan Y, Kohli A, Koffas MA. Biosynthesis of natural flavanones in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2005;71:5610–5613. doi: 10.1128/AEM.71.9.5610-5613.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang MC, Keasling JD. Production of isoprenoid pharmaceuticals by engineered microbes. Nat. Chem. Biol. 2006;2:674–681. doi: 10.1038/nchembio836. [DOI] [PubMed] [Google Scholar]
- 6.Pitera DJ, Paddon CJ, Newman JD, Keasling JD. Balancing a heterologous mevalonate pathway for improved isoprenoid production in Escherichia coli. Metab. Eng. 2007;9:193–207. doi: 10.1016/j.ymben.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 7.Alper H, Fischer C, Nevoigt E, Stephanopoulos G. Tuning genetic control through promoter engineering. Proc. Natl Acad. Sci. USA. 2005;102:12678–12683. doi: 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hjersted JL, Henson MA, Mahadevan R. Genome-scale analysis of Saccharomyces cerevisiae metabolism and ethanol production in fed-batch culture. Biotechnol. Bioeng. 2007;97:1190–1204. doi: 10.1002/bit.21332. [DOI] [PubMed] [Google Scholar]
- 9.Menzella HG, Reid R, Carney JR, Chandran SS, Reisinger SJ, Patel KG, Hopwood DA, Santi DV. Combinatorial polyketide biosynthesis by de novo design and rearrangement of modular polyketide synthase genes. Nat. Biotechnol. 2005;23:1171–1176. doi: 10.1038/nbt1128. [DOI] [PubMed] [Google Scholar]
- 10.Keasling JD. Synthetic biology for synthetic chemistry. ACS Chem. Biol. 2008;3:64–76. doi: 10.1021/cb7002434. [DOI] [PubMed] [Google Scholar]
- 11.Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods. 2007;4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- 12.Itaya M, Fujita K, Kuroki A, Tsuge K. Bottom-up genome assembly using the Bacillus subtilis genome vector. Nat. Methods. 2008;5:41–43. doi: 10.1038/nmeth1143. [DOI] [PubMed] [Google Scholar]
- 13.Raymond CK, Pownder TA, Sexson SL. General method for plasmid construction using homologous recombination. Biotechniques. 1999;26:134–141. doi: 10.2144/99261rr02. [DOI] [PubMed] [Google Scholar]
- 14.Schaerer-Brodbeck C, Barberis A. Coupling homologous recombination with growth selection in yeast: a tool for construction of random DNA sequence libraries. Biotechniques. 2004;37:202–206. doi: 10.2144/04372BM05. [DOI] [PubMed] [Google Scholar]
- 15.Gunyuzlu PL, Hollis GF, Toyn JH. Plasmid construction by linker-assisted homologous recombination in yeast. Biotechniques. 2001;31:1246–1250. doi: 10.2144/01316bm03. [DOI] [PubMed] [Google Scholar]
- 16.Jansen G, Wu C, Schade B, Thomas DY, Whiteway M. Drag&Drop cloning in yeast. Gene. 2005;344:43–51. doi: 10.1016/j.gene.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 17.Ma H, Kunes S, Schatz PJ, Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- 18.Oldenburg KR, Vo KT, Michaelis S, Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 1997;25:451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raymond CK, Sims EH, Olson MV. Linker-mediated recombinational subcloning of large DNA fragments using yeast. Genome Res. 2002;12:190–197. doi: 10.1101/gr.205201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson PR, Haj-Ahmad Y. Counter-selection facilitated plasmid construction by homologous recombination in Saccharomyces cerevisiae. Biotechniques. 2003;35:692–698. doi: 10.2144/03354bm05. [DOI] [PubMed] [Google Scholar]
- 21.Langle-Rouault F, Jacobs E. A method for performing precise alterations in the yeast genome using a recyclable selectable marker. Nucleic Acids Res. 1995;23:3079–3081. doi: 10.1093/nar/23.15.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manivasakam P, Weber SC, Mcelver J, Schiestl RH. Micro-homology mediated PCR targeting in Saccharomyces cerevisiae. Nucleic Acids Res. 1995;23:2799–2800. doi: 10.1093/nar/23.14.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cocchia M, Kouprina N, Kim SJ, Larionov V, Schlessinger D, Nagaraja R. Recovery and potential utility of YACs as circular YACs/BACs. Nucleic Acids Res. 2000;28:e81. doi: 10.1093/nar/28.17.e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noskov VN, Koriabine M, Solomon G, Randolph M, Barrett JC, Leem SH, Stubbs L, Kouprina N, Larionov V. Defining the minimal length of sequence homology required for selective gene isolation by TAR cloning. Nucleic Acids Res. 2001;29:E32. doi: 10.1093/nar/29.6.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noskov V, Kouprina N, Leem SH, Koriabine M, Barrett JC, Larionov V. A genetic system for direct selection of gene-positive clones during recombinational cloning in yeast. Nucleic Acids Res. 2002;30:e8. doi: 10.1093/nar/30.2.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noskov VN, Kouprina N, Leem SH, Ouspenski I, Barrett JC, Larionov V. A general cloning system to selectively isolate any eukaryotic or prokaryotic genomic region in yeast. BMC Genomics. 2003;4:16. doi: 10.1186/1471-2164-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kouprina N, Larionov V. Innovation—TAR cloning: insights into gene function, long-range haplotypes and genome structure and evolution. Nat. Rev. Genet. 2006;7:805–812. doi: 10.1038/nrg1943. [DOI] [PubMed] [Google Scholar]
- 28.Ebersole T, Okamoto Y, Noskov VN, Kouprina N, Kim JH, Leem SH, Barrett JC, Masumoto H, Larionov V. Rapid generation of long synthetic tandem repeats and its application for analysis in human artificial chromosome formation. Nucleic Acids Res. 2005;33:e130. doi: 10.1093/nar/gni129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 30.Lee FW, Da Silva NA. Sequential delta-integration for the regulated insertion of cloned genes in Saccharomyces cerevisiae. Biotechnol. Prog. 1997;13:368–373. doi: 10.1021/bp970055d. [DOI] [PubMed] [Google Scholar]
- 31.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes—gene-splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 32.Woodyer R, Simurdiak M, van der Donk WA, Zhao HM. Heterologous expression, purification, and characterization of a highly active xylose reductase from Neurospora crassa. Appl Environ. Microbiol. 2005;71:1642–1647. doi: 10.1128/AEM.71.3.1642-1647.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin YS, Jones S, Shi NQ, Jeffries TW. Molecular cloning of XYL3 (D-xylulokinase) from Pichia stipitis and characterization of its physiological function. Appl. Environ. Microbiol. 2002;68:1232–1239. doi: 10.1128/AEM.68.3.1232-1239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ho NW, Chen Z, Brainard AP. Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl. Environ. Microbiol. 1998;64:1852–1859. doi: 10.1128/aem.64.5.1852-1859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karhumaa K, Wiedemann B, Hahn-Hagerdal B, Boles E, Gorwa-Grauslund MF. Co-utilization of L-arabinose and D-xylose by laboratory and industrial Saccharomyces cerevisiae strains. Microb. Cell Fact. 2006;5:18. doi: 10.1186/1475-2859-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chemler JA, Yan Y, Koffas MA. Biosynthesis of isoprenoids, polyunsaturated fatty acids and flavonoids in Saccharomyces cerevisiae. Microb. Cell Fact. 2006;5:20. doi: 10.1186/1475-2859-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Misawa N, Shimada H. Metabolic engineering for the production of carotenoids in non-carotenogenic bacteria and yeasts. J. Biotechnol. 1997;59:169–181. doi: 10.1016/s0168-1656(97)00154-5. [DOI] [PubMed] [Google Scholar]
- 38.Misawa N, Nakagawa M, Kobayashi K, Yamano S, Izawa Y, Nakamura K, Harashima K. Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J. Bacteriol. 1990;172:6704–6712. doi: 10.1128/jb.172.12.6704-6712.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tao L, Schenzle A, Odom JM, Cheng Q. Novel carotenoid oxidase involved in biosynthesis of 4,4′-diapolycopene dialdehyde. Appl. Environ. Microbiol. 2005;71:3294–3301. doi: 10.1128/AEM.71.6.3294-3301.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hua SB, Qiu MS, Chan E, Zhu L, Luo Y. Minimum length of sequence homology required for in vivo cloning by homologous recombination in yeast. Plasmid. 1997;38:91–96. doi: 10.1006/plas.1997.1305. [DOI] [PubMed] [Google Scholar]
- 41.Nair UN, Zhao HM. Mutagenic inverted repeat assisted genome engineering (MIRAGE) Nucleic Acids Res. 2009 doi: 10.1093/nar/gkn943. doi:10.1093/nar/gkn943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goldstein AL, Pan X, McCusker JH. Heterologous URA3MX cassettes for gene replacement in Saccharomyces cerevisiae. Yeast. 1999;15:507–511. doi: 10.1002/(SICI)1097-0061(199904)15:6<507::AID-YEA369>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.