Abstract
MicroRNAs are small (∼22 nt) RNAs that regulate gene expression and play important roles in both normal and disease physiology. The use of microarrays for global characterization of microRNA expression is becoming increasingly popular and has the potential to be a widely used and valuable research tool. However, microarray profiling of microRNA expression raises a number of data analytic challenges that must be addressed in order to obtain reliable results. We introduce here a universal reference microRNA reagent set as well as a series of nonhuman spiked-in synthetic microRNA controls, and demonstrate their use for quality control and between-array normalization of microRNA expression data. We also introduce diagnostic plots designed to assess and compare various normalization methods. We anticipate that the reagents and analytic approach presented here will be useful for improving the reliability of microRNA microarray experiments.
INTRODUCTION
Accumulating evidence suggests that many molecular processes are controlled by changes in microRNA abundance. Consequently, the use of microarrays to characterize microRNA expression is becoming an increasingly popular research tool. However, this application raises data analytic challenges that must be addressed in order for the results to be reliable. Foremost among these is between-array data normalization. Substantial differences between the nature of typical microRNA and mRNA expression experiments suggest that common methods of data normalization employed for mRNA expression arrays may not be ideal for microRNA arrays (1). There are relatively few known microRNAs for any species (∼600 for humans), and the proportion of microRNAs abundantly expressed in a given sample tends to be much smaller than mRNAs (reflected in the tissue-specific expression pattern of many microRNAs). These two observations suggest that the usual assumptions for normalization between mRNA arrays, that most mRNAs are not differentially expressed across samples and that the number of such mRNAs is large, are unlikely to hold true for microRNA arrays. Since the number of expressed microRNAs in a given sample tends to be small, the proportion of those that are differentially expressed (among those expressed at all) is much larger than that observed when profiling global mRNA expression.
The overall set of procedures that need to be followed to process raw microarray data is well known from the use of microarrays for many other purposes, such as detection of differential mRNA expression and array competitive genomic hybridization (aCGH). Briefly, one begins with some form of quality assessment of the obtained images, followed by normalization, and finally estimation of differential expression. As an approach for quality assessment, we constructed a synthetic universal microRNA reference comprised of a pool of 480 chemically synthesized microRNAs corresponding to all known human microRNA probes present on the microarray. We used a platform based on locked nucleic acid probes spotted on glass slides and used in two-channel mode, in which the sample was labeled and hybridized in red (Cy5) and the universal reference pool in green (Cy3). The universal microRNA reference pool was optimized to comprise individual microRNAs at concentrations selected to provide relatively uniform level of intensity in the green channel. This approach provides an excellent basis for quality assessment procedures that could potentially detect bad arrays or regions of arrays that are not functioning appropriately. The low number of expressed microRNAs in individual biological samples and the intrinsic between-sample variability makes use of the experimental samples (red channel) for assessing quality difficult.
As an approach to facilitate data normalization, we show how chemically synthesized spike-in microRNAs can be employed. We synthesized a set of 15 nonhuman microRNAs as spike-in controls to be included in the labeling reaction for each experimental sample. Spike-ins corresponded to non-human microRNA probes present on the array and were further empirically confirmed to show no cross-hybridization with human probes on the array. The concentration of each spike-in was empirically optimized to maximize the range of the signal provided by the spike-ins as a group. Signal obtained from these reagents is useful in assessing the performance of various normalization methods, and plays a central role in a novel normalization method we propose here. This approach is applicable to any microRNA microarray where unique, non-cross-hybridizing probes for a different species than the one of interest are present. This is in fact true for most popular microRNA microarray platforms, and we provide specific suggestions for microRNAs that are likely to serve as successful spike-ins on other platforms. Although initial experiments are required to optimize the concentration of spike-ins, this step needs to be performed only once for a given platform.
We provide explicit methods to address the issues of quality assessment, normalization and the detection of differential expression. We demonstrate implementation of our methods using a dataset representing microRNA intensity profiles of two histologic types of ovarian cancer as well as primary cultures of human ovarian surface epithelial cells. We also report qRT-PCR results for selected microRNAs to provide independent assessment of our methods.
We note that Hua et al. (2) have also compared various normalization methods for microRNA microarrays, using correlation with PCR results to quantify performance. Although they report print-tip loess normalization as the method that performs best, we find no statistically significant difference between it and other standard methods compared in their study (median normalization, VSN, etc.), as we show in the Supplementary Material. We have not considered print-tip loess normalization in our analysis as it does not seem applicable to our data; in particular, unlike for Hua et al. (2), a substantial proportion of microRNAs appear to be unexpressed in our samples.
MATERIALS AND METHODS
Biological samples
Primary human ovarian surface epithelial cultures were derived from histologically normal oopherectomy specimens and cultured as described in detail in the Supplementary Material. Snap frozen ovarian cancer tissue specimens corresponding to serous and endometrioid histologies were obtained from the Pacific Ovarian Cancer Research Consortium Repository. All clinical samples in this study were collected under Institutional Review Board-approved protocols.
Experimental methods
MicroRNA microarrays
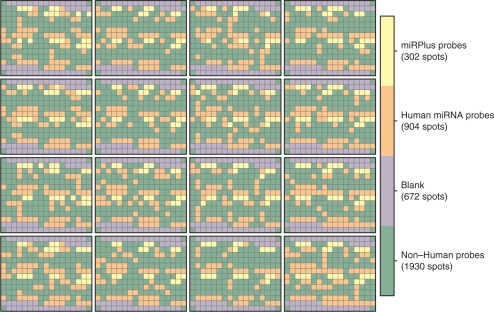
Explicit details regarding the microarrays are given in the Supplementary Material. Briefly, as shown in Figure 1, the arrays had 16 print-tip blocks, each with 238 spots laid out in a 14 × 17 grid. Of these, 672 spots were blank (primarily at block boundaries), 1930 represented nonhuman microRNAs, 904 were human microRNAs and 302 were proprietary probes (miRPlus). Each probe was spotted in duplicate. Two arrays were printed on each slide.
Figure 1.
Layout of microRNA arrays. There were 16 print-tips, and blocks were laid out on a 4 by 4 grid. The majority of spots do not represent human microRNAs. Of the ones that do, each are spotted in duplicate, so there are ∼450 unique microRNAs in total. Included among the nonhuman microRNAs are 15 spike-in controls, also spotted in duplicate.
PCR validation
For some microRNAs a TaqMan microRNA assay (Applied Biosystems) was used for qRT-PCR. Normalization was done with the RNU24 endogenous control assay. Reverse transcription was carried out with the ABI microRNA Reverse Transcription kit using the manufacturer's; recommended protocol. Real-time PCR was performed on an ABI Prism 7900HT Sequence detection system using 2× Universal PCR Master Mix, no AmpErase UNG. A total of seventeen microRNAs (listed in the Supplementary Material) were assessed by qRT-PCR.
Spiked-in synthetic nonhuman microRNAs
Fifteen nonhuman microRNAs that did not show cross-hybridization with multiple human tissue RNA samples and that exhibited sufficient signal intensities were used as spike-ins in varying amounts adjusted to maximize the range of the signal they provided, covering the expected span of intensities of biological samples. These were added to samples for both the red and the green channels. See the Supplementary Material for more details, including the identities of the microRNAs used.
Synthetic human microRNA universal reference pool
A synthetic human microRNA universal reference pool was constructed and was used in the green channel in all arrays (we will sometimes refer to this as the reference channel). Briefly, RNA oligonucleotides were synthesized corresponding to 454 microRNAs, of which 56 failed to provide sufficient intensity, for a variety of reasons, leaving 398 that were used on our arrays. Their concentrations were adjusted to provide approximately uniform intensity across spots.
Quality assessment
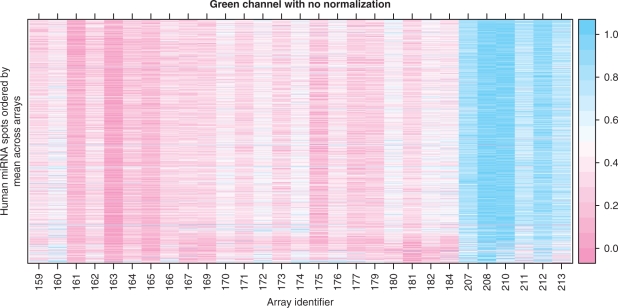
The measured per-spot log2 intensities may be used to assess the quality of the arrays and spots. The values in the red channel are not suitable for quality assessment, but the green reference channel is useful because it should be the same on all arrays, and the Cy3-labeled (green) synthetic universal reference pool oligos should hybridize to most spots that correspond to human microRNAs. We see an example of this in Figure 2, which is much like the heatmap, or false color image, that is widely used to convey gene expression data. Rows represent microRNAs, and the columns represent arrays. The microRNAs are ordered by average intensity across arrays, as there is no particularly natural ordering for them. The false color represents intensity, which is scaled to lie between 0 and 1 for each spot; this enhances comparison across arrays at the cost of comparison across spots, which is not of interest.
Figure 2.
False color image of raw intensities in the reference channel. Columns represent arrays and rows represent spots. Only human microRNA spots are shown. The intensities have been location- and scale-transformed within each row (spot) to lie between 0 and 1; this enables comparison across arrays. Other transformations are possible, and give similar results. There are two interesting features in this plot. First, there is a banding pattern of high and low intensity on alternate arrays. This is presumably due to the fact that two arrays are hybridized on each slide. Second, the last six arrays, run on a different day, have consistently higher intensity, suggesting a batch effect. This effect is also seen in Figure 3.
We use this graphic to demonstrate the presence of a batch effect in our experiment. There are two well-defined batches: Arrays 159–184 were labeled on 4/13/07 and hybridized on 4/16/07 (159–182) and 4/17/07 (184). Arrays 207–213 were labeled on 5/5/07 and hybridized on 5/14/07. The arrays from the second batch (207–213) have larger intensity in the green channel. The banding pattern visible in this figure is presumably due to the fact that two arrays were printed on each slide.
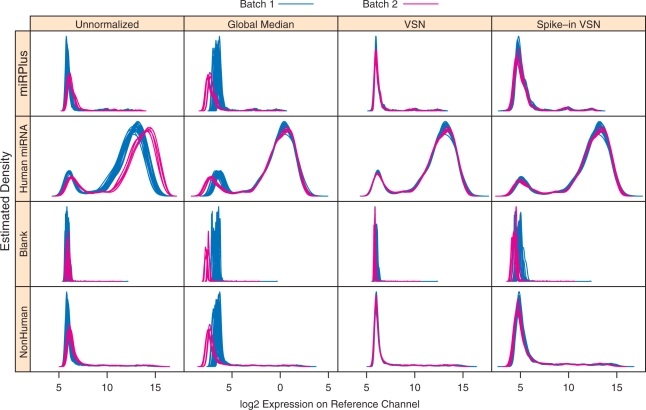
Other plots designed to highlight this batch effect can also serve as diagnostics for assessing different normalization methods. Supplementary Figure S2, which encodes a pairwise distance between all arrays, is an example of such a plot. Another example is Figure 3, which plots the estimated densities for the green channel log2 intensities. The two batches are indicated by different colors in Figure 3, and a visual separation of the two batches is readily apparent in the densities for the unnormalized intensities. The separation is removed by all three normalization methods considered in this article (see the next section), but with some differences in behavior for low intensities. Figure 3 also serves as a diagnostic of array quality; had there been any bad arrays, one or more of the corresponding density estimates are likely to have been aberrant.
Figure 3.
Histograms (kernel density estimates) of log2 intensity in the reference channel. The rows correspond to different types of probes, and the columns represent the different normalization methods used. Colors distinguish the two batches, and show a systematic upward shift in the second batch. All normalization methods appear to rectify the batch effect for high intensity spots, but have different effects on the low intensity spots.
Normalization of measured intensity
Normalization is an essential preprocessing step in the analysis of any microarray experiment. Its primary purpose is to try to ensure that the observed between-array differences are due to biological phenomena, and not due to artifacts that arise due to differences in handling or processing of the samples. Normalization of microRNA microarrays is problematic primarily for two reasons. First, the number of microRNAs measured by the arrays is fairly small, numbering only in the hundreds (Figure 1). Second, even among the measured microRNAs, only a small number are expressed at all in a given tissue, and consequently, most of the spots on the arrays cannot be used for normalization. In other words, the usual normalization assumptions do not apply. A careful analysis requires the development of a reasonable normalization strategy, as well as diagnostics that can be used to assess the performance of potential normalization strategies.
We examined the properties of three different normalization strategies: global median normalization, variance stabilizing normalization and a spike-in based normalization. Global median normalization (3) has been previously used for normalizing microRNA data (4). Variance stabilizing normalization (VSN) (5) is another method widely used for microarray data. In both cases, we only use the human microRNAs in the normalization, as the remainder are unlikely to be expressed in our samples. The third method (spike-in VSN normalization) is described next.
Spike-in VSN normalization
One way to assess whether or not normalization is needed is to plot the raw log2 intensity values of each spike-in control against their median across all arrays. If no normalization is needed we expect these points to all fall along the diagonal line y = x. This is not quite true (Supplementary Figure S5), but the departures are small, and can be accounted for by an affine transformation. This observation suggests a normalization procedure where all values on an array are transformed so that the spike-ins become approximately aligned. We find such a per-array transformation using VSN, restricting the model fit to the spiked-in spots. Normalized intensities for all microRNAs are then obtained by applying the resulting transformation to all spots of interest on the array. We will henceforth refer to this procedure as ‘spike-in VSN normalization’. One limitation of this approach is that we can only expect reliable results for intensities within the range covered by the spike-ins, which excludes targets that are not expressed. To address this problem, we augmented the list of spike-ins used for the initial VSN fit with 15 randomly chosen probes that correspond to rice (Oryza sativa) microRNA targets and have no known human counterparts. Further details are given in the Supplementary Material.
Assessment
Assessing different normalization schemes is somewhat problematic, and is an issue that is difficult to adequately address due to the lack of gold standard references where the true values of some of the features (in our case microRNAs) are known. The approach taken in (6), for Affymetrix arrays, was to make use of spike-in datasets, where concentrations were known for a small number of mRNAs. While such an experiment has not yet been performed for microRNAs, our use of internal spike-ins provides a reasonable metric for assessing the performance of different normalization schemes; the variability in intensity for a given spike-in, across arrays, should be small, and consequently, normalization methods that tend to reduce that variability should be preferred over those that do not.
However, the reduction of variability alone does not make a good normalization method. One must also ensure that the signal is maintained. For that purpose we used an external method, qRT-PCR, to assess the levels of 17 microRNAs (Supplementary Table S2). These measurements can be used to assess the performance of various normalization methods; we expect the normalized expressions to correlate well with the qRT-PCR measurements, and better normalization schemes to have stronger correlation. In practice, for this dataset, the observed correlations are fairly strong for most microRNAs, both for unnormalized and normalized expressions, with no significant differences between methods. Details can be found in the Supplementary Material.
Differential Expression
Differential expression was assessed using an empirical Bayes approach, as implemented in the software package limma (7), available from the Bioconductor project (8). Comparison was between the serous and endometrioid subtypes of ovarian cancer.
RESULTS AND DISCUSSION
Diagnostics
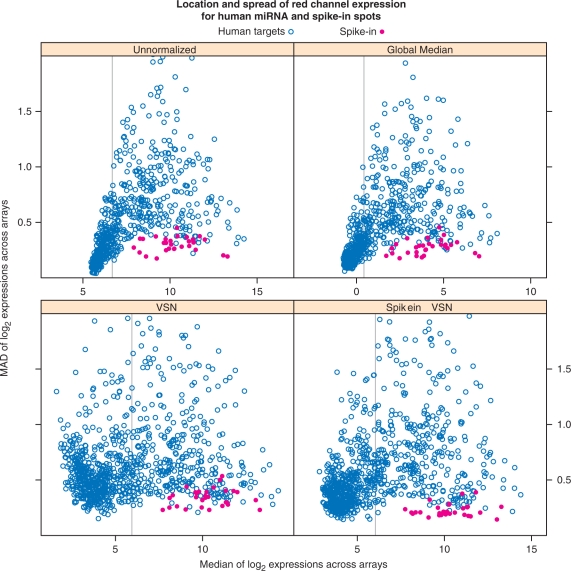
As noted previously, Figure 3 and other plots designed to emphasize potential batch effects can be used to assess success of normalization. Another useful diagnostic can be derived from the spike-ins. By design, the observed intensities for the spike-ins should cover the range of expressed microRNAs, and each spike-in control should have essentially the same intensity in every array. Thus, after normalization, these spots should have low variability across arrays in the red channel. In Figure 4 we plot, for each human probe and spike-in, a measure of spread (MAD) across arrays against a measure of location (median), before and after normalization. For successful normalization, we expect the spike-ins to have lower variability than regular probes with similar median intensities.
Figure 4.
Spread-location plot for probes on the red channel. A measure of spread (MAD) across arrays is plotted against a measure of location (median), before and after normalization. Only the spots corresponding to human microRNAs are shown, along with the spike-ins. For successful normalization, we expect the spike-ins to have low variability. By this criterion, all methods are successful; indeed, this holds even for the unnormalized intensities. There is a clear relationship between location and spread in the unnormalized data. The VSN-based methods make the spreads more homogeneous, as expected. The vertical grey line represents the 60th percentile of the median values; we only retain spots with median higher than this value (i.e., the top 40% spots) for further analysis.
Normalization
Our proposed diagnostics, namely, Figures 3, 4, and Supplementary Figure S2, suggest that all normalization methods perform adequately in our study. Here, we only show results of subsequent analysis using spike-in VSN normalization; results for other methods are given in the Supplementary Material.
Differential expression
We are interested in microRNAs that have expression roughly in the range of the spike-ins. However, Figure 4 suggests that the majority of the spots have median intensity below this range, and are presumably not expressed. We thus retain for further analysis only those spots corresponding to the top 40% median intensity values. This nonspecific filtering reduces problems associated with multiple testing. Note that the filtering step is by nature somewhat arbitrary, and may need adjustments depending on the purpose of the study; for example, our method may not identify potentially interesting spots that are expressed only in a few samples.
Table 1 presents the list of the top ten differentially expressed microRNAs between the serous and endometrioid subtypes of ovarian cancer. When available, the table is augmented by the result of qRT-PCR (in the form of a log2 fold change and P-value). The results for other normalization methods, as well as for unnormalized intensities, are provided in the Supplementary Material. The results are largely invariant to the normalization method used; in particular, the same microRNAs appear as the most significantly differentially expressed, with largely similar adjusted P-value.
Table 1.
The top 10 hits using Spike-in VSN normalization
| logFC. array | t | adj. P-value | logFC. PCR | P-value PCR | |
|---|---|---|---|---|---|
| hsa-miR-422b | 2.15 | 6.430 | 0.000 | 1.878 | 0.000 |
| hsa-miR-146b | 1.53 | 5.712 | 0.000 | 1.842 | 0.005 |
| hsa-miR-625 | 1.24 | 5.289 | 0.001 | ND | ND |
| hsa-miR-155 | 1.41 | 4.167 | 0.013 | 3.139 | 0.012 |
| hsa-miR-200a | −1.18 | −3.829 | 0.025 | −0.943 | 0.058 |
| hsa-miR-200b | −1.23 | −3.441 | 0.050 | −0.914 | 0.054 |
| hsa-miR-193a | 0.94 | 3.400 | 0.050 | ND | ND |
| hsa-miR-99a | 1.42 | 3.379 | 0.050 | ND | ND |
| hsa-miR-148a | −1.01 | −3.216 | 0.056 | ND | ND |
| hsa-miR-23b | 1.07 | 3.189 | 0.056 | ND | ND |
ND, Not done.
External validation
It should be noted that most of the diagnostic plots we propose are based on the premise that successful normalization methods should reduce variability (across arrays) at spots that correspond to microRNAs that should have constant expression. A good method should additionally ensure that the true signals of interest are also reported accurately. This can be checked by comparing the normalized intensities to expression data obtained using an independent method. The supplementary file qrtpcr.pdf plots average ΔCt values obtained using qRT-PCR against raw and normalized intensities from microarrays for the same set of tissue samples for 17 microRNAs. The agreement can be summarized by correlations, which were fairly strong for most microRNAs, both for unnormalized and normalized intensities. Analysis of variance, described in the Supplementary Material, suggests that the amount of correlation did not differ significantly between normalization methods, especially if we consider the variability inherent in qRT-PCR.
Reagent availability and applicability to other platforms
Many of our proposed methods depend on a synthetic universal reference pool and spike-in controls (both available from author M. Tewari upon request). The choice and concentrations of the spike-in controls are specific to the microarray platform used, and will need to be tuned for each array platform. This is possible as long as the relevant platform contains a sufficient number of probes that do not cross-hybridize to human microRNAs. One approach to predict cross-hybridization is to compute similarity with known human microRNA sequences, and although further experiments are required to confirm suitability, an initial set of candidate microRNAs can be obtained using such a score. We used a similarity measure that is essentially the length of the longest common substring, with penalties for mismatches and gaps, to obtain explicit lists of candidate microRNAs for popular commercial platforms, given in Table 2–6. Computations are based on mature sequences in miRBase release 12.0 (9) and available information on the contents of each platform. Candidates are sorted by similarity, but are arbitrarily limited to 20 after removing those too similar to candidates already on the list, to ensure that the selected spike-ins do not cross-hybridize with each other. Further details are given in the Supplementary Material.
Table 2.
Candidate spike-in controls for Exiqon human, mouse and rat array (v 11.0)
| microRNA | Mature sequence | Similarity score |
|---|---|---|
| mmu-miR-701 | UUAGCCGCUGAAAUAGAUGGA | 7 |
| kshv-miR-K12-2 | AACUGUAGUCCGGGUCGAUCUG | 7 |
| hcmv-miR-US25-2-3p | AUCCACUUGGAGAGCUCCCGCGG | 8 |
| mghv-miR-M1-3 | GAGGUGAGCAGGAGUUGCGCUU | 8 |
| mmu-miR-693-3p | GCAGCUUUCAGAUGUGGCUGUAA | 8 |
| mcmv-miR-m108-2-3p | GUGACUCGAGACGAGUGACCGGU | 8 |
| mmu-miR-714 | CGACGAGGGCCGGUCGGUCGC | 8 |
| mmu-miR-681 | CAGCCUCGCUGGCAGGCAGCU | 8 |
| rno-miR-742 | GAAAGCCACCAUGUUGGGUAAA | 8 |
| mmu-miR-1195 | UGAGUUCGAGGCCAGCCUGCUCA | 8 |
| mmu-miR-700 | CACGCGGGAACCGAGUCCACC | 8 |
| mmu-miR-804 | UGUGAGUUGUUCCUCACCUGGA | 8 |
| mmu-miR-540-3p | AGGUCAGAGGUCGAUCCUGG | 8 |
| mmu-miR-770-3p | CGUGGGCCUGACGUGGAGCUGG | 8 |
| ebv-miR-BART17-5p | UAAGAGGACGCAGGCAUACAAG | 8 |
| mcmv-miR-M87-1 | AGGCAGCCGUCGGCAGCGGCAGC | 8 |
| ebv-miR-BART1-3p | UAGCACCGCUAUCCACUAUGUC | 8 |
| ebv-miR-BART6-3p | CGGGGAUCGGACUAGCCUUAGA | 8 |
| ebv-miR-BART19-5p | ACAUUCCCCGCAAACAUGACAUG | 8 |
| mcmv-miR-m01-1 | AGAGGAGAAUAACGUCGAACGG | 8 |
The similarity score represents the maximum sequence similarity of the given candidate spike-in miRNA to any known human miRNA in miRBase release 12.0.
Table 3.
Candidate spike-in controls for Exiqon 9.2 all species array
| microRNA | Mature sequence | Similarity score |
|---|---|---|
| ath-miR401 | CGAAACUGGUGUCGACCGACA | 7 |
| osa-miR169n | UAGCCAAGAAUGACUUGCCUA | 7 |
| kshv-miR-K12-2 | AACUGUAGUCCGGGUCGAUCUG | 7 |
| ppt-miR535a | UGACAACGAGAGAGAGCACGC | 7 |
| mmu-miR-701 | UUAGCCGCUGAAAUAGAUGGA | 7 |
| ppt-miR1218 | CCUUAGAGUCGUAGGCCUCUG | 7 |
| bta-miR-425-3p | AUCGGGAAUGUCGUGUCCGCCC | 7 |
| sme-miR-754 | GUUGCUUGGGGUUAUUACUA | 7 |
| ath-miR829.2 | CAAAUUAAAGCUUCAAGGUAG | 7 |
| ppt-miR894 | CGUUUCACGUCGGGUUCACC | 7 |
| rrv-miR-rR1-5 | CCGGAACCCAAAGACACGUGCCCG | 7 |
| mdv2-miR-M30 | CAACACUCCCUCGGACGCAGCA | 7 |
| cel-miR-36 | UCACCGGGUGAAAAUUCGCAUG | 8 |
| cel-miR-46 | UGUCAUGGAGUCGCUCUCUUCA | 8 |
| cel-miR-49 | AAGCACCACGAGAAGCUGCAGA | 8 |
| cel-miR-54 | UACCCGUAAUCUUCAUAAUCCGAG | 8 |
| cel-miR-58 | UGAGAUCGUUCAGUACGGCAAU | 8 |
| cel-miR-64 | UAUGACACUGAAGCGUUACCGAA | 8 |
| cel-miR-75 | UUAAAGCUACCAACCGGCUUCA | 8 |
| cel-miR-77 | UUCAUCAGGCCAUAGCUGUCCA | 8 |
The similarity score represents the maximum sequence similarity of the given candidate spike-in miRNA to any known human miRNA in miRBase release 12.0.
Table 4.
Candidate spike-in controls for NCode Multi-Species miRNA Microarray Kit V2
| microRNA | Mature sequence | Similarity score |
|---|---|---|
| mmu-miR-701 | UUAGCCGCUGAAAUAGAUGGA | 7 |
| cel-miR-36 | UCACCGGGUGAAAAUUCGCAUG | 8 |
| cel-miR-46 | UGUCAUGGAGUCGCUCUCUUCA | 8 |
| cel-miR-49 | AAGCACCACGAGAAGCUGCAGA | 8 |
| cel-miR-54 | UACCCGUAAUCUUCAUAAUCCGAG | 8 |
| cel-miR-58 | UGAGAUCGUUCAGUACGGCAAU | 8 |
| cel-miR-64 | UAUGACACUGAAGCGUUACCGAA | 8 |
| cel-miR-75 | UUAAAGCUACCAACCGGCUUCA | 8 |
| cel-miR-77 | UUCAUCAGGCCAUAGCUGUCCA | 8 |
| cel-miR-80 | UGAGAUCAUUAGUUGAAAGCCGA | 8 |
| dme-miR-3 | UCACUGGGCAAAGUGUGUCUCA | 8 |
| cel-miR-242 | UUGCGUAGGCCUUUGCUUCGA | 8 |
| cel-miR-248 | AUACACGUGCACGGAUAACGCUCA | 8 |
| cel-miR-252 | AUAAGUAGUAGUGCCGCAGGUAA | 8 |
| cel-miR-259 | AAAUCUCAUCCUAAUCUGGUAGCA | 8 |
| cel-miR-261 | UAGCUUUUUAGUUUUCACG | 8 |
| cel-miR-271 | UCGCCGGGUGGGAAAGCAUU | 8 |
| cel-miR-273 | UGCCCGUACUGUGUCGGCUG | 8 |
| dme-miR-275 | UCAGGUACCUGAAGUAGCGCGCG | 8 |
| dme-miR-276a | UAGGAACUUCAUACCGUGCUCU | 8 |
The similarity score represents the maximum sequence similarity of the given candidate spike-in miRNA to any known human miRNA in miRBase release 12.0.
Table 5.
Candidate spike-in controls for Agilent Human miRNA Microarray, Version 1
| microRNA | Mature sequence | Similarity score |
|---|---|---|
| kshv-miR-K12-2 | AACUGUAGUCCGGGUCGAUCUG | 7 |
| ebv-miR-BART6-3p | CGGGGAUCGGACUAGCCUUAGA | 8 |
| ebv-miR-BART17-5p | UAAGAGGACGCAGGCAUACAAG | 8 |
| ebv-miR-BART10 | UACAUAACCAUGGAGUUGGCUGU | 8 |
| hcmv-miR-US25-2-3p | AUCCACUUGGAGAGCUCCCGCGG | 8 |
| ebv-miR-BART1-3p | UAGCACCGCUAUCCACUAUGUC | 8 |
| kshv-miR-K12-4-5p | AGCUAAACCGCAGUACUCUAGG | 8 |
| kshv-miR-K12-8 | UAGGCGCGACUGAGAGAGCACG | 8 |
| kshv-miR-K12-10a | UAGUGUUGUCCCCCCGAGUGGC | 8 |
| ebv-miR-BART6-5p | UAAGGUUGGUCCAAUCCAUAGG | 8 |
| kshv-miR-K12-9 | CUGGGUAUACGCAGCUGCGUAA | 8 |
| hcmv-miR-US5-1 | UGACAAGCCUGACGAGAGCGU | 8 |
| kshv-miR-K12-4-3p | UAGAAUACUGAGGCCUAGCUGA | 8 |
| ebv-miR-BART11-5p | UCAGACAGUUUGGUGCGCUAGUUG | 8 |
| hsv1-miR-H1 | UGGAAGGACGGGAAGUGGAAG | 9 |
| ebv-miR-BART11-3p | ACGCACACCAGGCUGACUGCC | 9 |
| hcmv-miR-UL70-3p | GGGGAUGGGCUGGCGCGCGG | 9 |
| hcmv-miR-US4 | CGACAUGGACGUGCAGGGGGAU | 9 |
| hcmv-miR-US25-1 | AACCGCUCAGUGGCUCGGACC | 9 |
| ebv-miR-BART12 | UCCUGUGGUGUUUGGUGUGGUU | 9 |
The similarity score represents the maximum sequence similarity of the given candidate spike-in miRNA to any known human miRNA in miRBase release 12.0.
Table 6.
Candidate spike-in controls for Miltenyi Biotec miRNA microarray
| microRNA | Mature sequence | Similarity score |
|---|---|---|
| kshv-miR-K12-2 | AACUGUAGUCCGGGUCGAUCUG | 7 |
| mmu-miR-350 | UUCACAAAGCCCAUACACUUUC | 8 |
| kshv-miR-K12-10a | UAGUGUUGUCCCCCCGAGUGGC | 8 |
| kshv-miR-K12-4-3p | UAGAAUACUGAGGCCUAGCUGA | 8 |
| kshv-miR-K12-4-5p | AGCUAAACCGCAGUACUCUAGG | 8 |
| kshv-miR-K12-8 | UAGGCGCGACUGAGAGAGCACG | 8 |
| kshv-miR-K12-9 | CUGGGUAUACGCAGCUGCGUAA | 8 |
| mmu-miR-879 | AGAGGCUUAUAGCUCUAAGCC | 8 |
| mmu-miR-883b-3p | UAACUGCAACAUCUCUCAGUAU | 8 |
| mmu-miR-883b-5p | UACUGAGAAUGGGUAGCAGUCA | 8 |
| mmu-miR-341 | UCGGUCGAUCGGUCGGUCGGU | 8 |
| ebv-miR-BART1-3p | UAGCACCGCUAUCCACUAUGUC | 8 |
| rno-miR-871 | UAUUCAGAUUGGUGCCGGUCACA | 8 |
| ebv-miR-BART10 | UACAUAACCAUGGAGUUGGCUGU | 8 |
| ebv-miR-BART17-5p | UAAGAGGACGCAGGCAUACAAG | 8 |
| ebv-miR-BART18-3p | UAUCGGAAGUUUGGGCUUCGUC | 8 |
| ebv-miR-BART19-5p | ACAUUCCCCGCAAACAUGACAUG | 8 |
| ebv-miR-BART6-3p | CGGGGAUCGGACUAGCCUUAGA | 8 |
| ebv-miR-BART6-5p | UAAGGUUGGUCCAAUCCAUAGG | 8 |
| hcmv-miR-US25-2-3p | AUCCACUUGGAGAGCUCCCGCGG | 8 |
The similarity score represents the maximum sequence similarity of the given candidate spike-in miRNA to any known human miRNA in miRBase release 12.0.
CONCLUSION
In this paper, we have discussed the need for normalization in microRNA microarray experiments, noted the inadequacy of traditional normalization approaches, and proposed an alternative normalization method, along with diagnostic plots designed to assess and compare various normalization methods. Although our discussion takes place in the context of a specific experiment, its primary messages are more generally relevant. The need to consider alternative normalization techniques arises from the fact that the basic presumptions that underlie normalization methods used for mRNA microarrays do not hold for microRNA microarrays. Our proposed diagnostic plots for comparing normalization methods are applicable to other array designs, although some of the diagnostics benefit from the use of spiked-in spots. Although spike-in controls are not commonly used in mRNA microarrays, they are potentially more valuable tools for quality assessment in microRNA arrays, where traditional tools designed for mRNA arrays are inadequate. The use of spike-ins is also a prerequisite for our proposed normalization method. The other important feature, a universal reference on the green channel, provides a case where the truth is essentially known, thus allowing us to critically assess our methods; Figure 3 gives one example of such use. Although the high quality of array data in this study did not rigorously test our methods, the underlying procedures are sound and generally useful.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Pacific Ovarian Cancer Research Consortium (POCRC)/Ovarian SPORE (Developmental Research Project grant, P50 CA83636 to M.T.); the FHCRC Molecular Diagnostics Program (Pilot grant to M.T.); the POCRC/Ovarian SPORE (P50 CA83636 to R.G.); the National Human Genome Research Institute (5P41 HG004059 to R.G.); and the Human Frontier Science Program (RGP0022/2005 to W.H. and R.G.). Funding for open access charge: POCRC/Ovarian SPORE (P50 CA83636).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ms Carolyn Slater and Lisa Vanderveer for technical assistance.
REFERENCES
- 1.Davison TS, Johnson CD, Andruss BF. Analyzing micro-RNA expression using microarrays. Methods Enzymol. 2006;411:14–34. doi: 10.1016/S0076-6879(06)11002-2. [DOI] [PubMed] [Google Scholar]
- 2.Hau Y-J, Tu K, Tang Z-Y, Li Y-X, Xiao H-S. Genomics. 2008. Comparison of normalization methods with microRNA microarray. doi: 10.1016/j.ygeno.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Simon RM. New York: Springer; 2003. Design and Analysis of DNA Microarray Investigations. [Google Scholar]
- 4.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu C-G, Alder H, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 5.Huber W, von Heydebreck A, Sueltmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl 1):S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 6.Cope LM, Irizarry RA, Jaffe H, Wu Z, Speed TP. A benchmark for affymetrix genechip expression measures. Bioinformatics. 2004;20:323–331. doi: 10.1093/bioinformatics/btg410. [DOI] [PubMed] [Google Scholar]
- 7.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry ZR, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 8.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, El-lis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffiths-Jones S, Saini HK, Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.