Abstract
Damaged DNA-binding protein (DDB), consisting of DDB1 and DDB2 subunits recognizes a wide spectrum of DNA lesions. DDB is dispensable for in vitro nucleotide excision repair (NER) reaction, but stimulates this reaction especially for cyclobutane pyrimidine dimer (CPD). Here we show that DDB directly interacts with XPA, one of core NER factors, mainly through DDB2 subunit and the amino-acid residues between 185 and 226 in XPA are important for the interaction. Interestingly, the point mutation causing the substitution from Arg-207 to Gly, which was previously identified in a XP-A revertant cell-line XP129, diminished the interaction with DDB in vitro and in vivo. In a defined system containing R207G mutant XPA and other core NER factors, DDB failed to stimulate the excision of CPD, although the mutant XPA was competent for the basal NER reaction. Moreover, in vivo experiments revealed that the mutant XPA is recruited to damaged DNA sites with much less efficiency compared with wild-type XPA and fails to support the enhancement of CPD repair by ectopic expression of DDB2 in SV40-transformed human cells. These results suggest that the physical interaction between DDB and XPA plays an important role in the DDB-mediated NER reaction.
INTRODUCTION
Nucleotide excision repair (NER) is the major mechanism for removing helix-distorting DNA lesions induced by sunlight and chemical mutagens (1–4). Defects in the NER pathway give rise to xeroderma pigmentosum (XP), an autosomal recessive disease characterized by photosensitivity, pigment changes and a predisposition to skin cancer and in some cases neurological abnormalities (1,5). In human cells, the early process of NER from damage recognition to dual incision is accomplished by six core NER factors, XP complementation group A (XPA), RPA, XPC-RAD23B, TFIIH, XPF-ERCC1 and XPG in vitro (6,7).
Damaged DNA-binding protein (DDB) is a heterodimeric complex comprising DDB1 and DDB2 subunits and binds to a wide spectrum of DNA lesions including a UV-induced (6–4) photoproduct (6-4PP) and cyclobutane pyrimidine dimer (CPD) (8–11). However, DDB is dispensable for in vitro NER reaction in the reconstituted systems with purified proteins (6,12). The DDB2 gene is responsible for XP complementation group E (13) and the cells derived from XP-E patients show a lack of DDB activity (14–16) and a partial deficiency in NER (17,18). DNA lesions induced in transcriptionally inactive DNA regions or the non-transcribed strand of expressed genes are repaired by a global genome repair (GGR) sub-pathway of NER, whereas those in the transcribed strand of expressed genes are repaired by a transcription-coupled repair (TCR) sub-pathway. DDB2-deficient cells exhibit normal TCR activity but significantly or slightly reduced GGR activity for CPD or 6-4PP, respectively (17,18). The accessory roles of DDB in GGR have been suggested from different aspects.
We found using an in vitro excision repair assay that DDB greatly stimulates the excision of CPD by cell-free extracts (CFEs) (19) or purified NER factors (20), although the excision of 6-4PP was rather inhibited under the same conditions and weakly stimulated by a less amount of DDB. Interestingly, the DDB-mediated stimulation of CPD excision was further enhanced by the addition of XPA and/or RPA to CFEs (19). Moreover, in an electrophoretic mobility shift assay, DDB was found to strikingly elevate the binding of XPA to DNA substrates containing a single CPD and to make a ternary complex with XPA and the DNA substrate. These results suggest that there is some link between DDB and XPA on damaged DNA sites, although there was an argument about the stimulatory role of DDB in NER reaction (21).
Local UV irradiation experiments have revealed a significant role of DDB in recruiting core NER factors to damaged DNA in vivo. DDB rapidly accumulates at damaged DNA sites in the absence of core NER factors (20,22), suggesting that DDB can bind to UV lesions before other core NER factors. Moreover, the ectopic overexpression of DDB2 in SV40-transformed human cell lines, which exhibit a low endogenous DDB2 level due to p53 inactivation by SV40 large T antigen, enhances the recruitment of XPC (22,23). A more detailed study using XP-A cells stably expressing CPD- or 6-4PP-specific photolyase showed that DDB activates the recruitment of XPC to CPD rather than 6-4PP (24).
A recent finding that DDB is a component of E3 ubiquitin ligase complex together with Cullin4A and Roc1 has given a new insight into the accessory role of DDB in NER (25). The E3 ligase complex was demonstrated to polyubiquitinate XPC and DDB2, leading to the alteration of DNA binding properties of XPC-RAD23B and DDB, respectively (26). The polyubiquitination appears to be required for cell-free NER reaction when DDB is bound to 6-4PP. Furthermore, it has been shown that XPC is modified by SUMO-1 and ubiquitin following UV irradiation and these modifications require DDB2 and XPA (27).
DDB has been also suggested to function in recognizing the lesions in the context of chromatin and/or remodeling chromatin to facilitate repair (28). DDB2 subunit shares homology with chromatin reorganization proteins (29) and DDB heterodimer has been shown to interact with histone acetyltransferases, CBP/p300 and GCN5 (30,31). Moreover, the E3 ubiquitin ligase complex containing DDB was reported to ubiquitinate histone H2A, H3 and H4 (32,33). DDB seems to be involved in several reactions at different levels during NER, but those still remain to be fully understood.
In this study, we have focused on the stimulatory role of DDB in an early step of NER and on a molecular link between DDB and core NER factors. We have found that DDB2 physically interacts with XPA in vitro as well as in vivo, and identified a DDB2-interactive domain in XPA. Functional analyses of a binding-defective XPA mutant indicate that the interaction between DDB2 and XPA is required for the stimulatory role of DDB in NER reaction especially for CPD.
MATERIALS AND METHODS
Plasmid constructs
The pGEX18/XPA construct for expressing glutathione S-transferase (GST)-XPA fusion protein and its deletion derivatives have been described (34). The mammalian expression plasmids for myc-XPA were constructed by subcloning the XPA cDNA from pRSET/XPA into the pCMV-myc vector (Clontech) (pCMV-myc/XPA). To express the fusion protein of maltose-binding protein (MBP) and DDB2, pMAL/DDB2 was generated by subcloning the DDB2 cDNA from pTB13 (35) into the pMAL-c2 vector (New England Biolabs). The DDB2 cDNA was also subcloned into the p3xFLAG-CMV-10 vector (Sigma) to express DDB2 tagged with three consecutive Flag epitopes in mammalian cells (p3xFLAG-CMV/DDB2). For establishing a human cell line that conditionally expresses 3xFlag-DDB2 in the presence of doxycycline, pTRE2/3xF-DDB2 was generated by subcloning 3xFlag sequences generated by PCR and the DDB2 cDNA into the pTRE2 vector (Clontech).
Preparation of repair factors
Various GST-XPA fusion proteins or MBP-DDB2 were expressed in Escherichia coli DR153 or BL21(DE3)plys, respectively, under the optimal conditions and the cells were resuspended in buffer-A [1 M Tris–HCl (pH 7.4), 5 M NaCl, 10% sucrose] and quickly frozen. After thawing on ice, the cell suspension was sonicated and centrifuged at 35 000 rpm at 4°C for 30 min, and aliquots of the supernatant were stored at –80°C until their use. Wild-type or R207G mutant (His)6-XPA was expressed and purified as described previously (19). Other core NER factors for the reconstituted NER reaction were prepared as described (20). Flag-DDB1 was co-overexpressed with Flag-DDB2 in a baculovirus/insect cell system and DDB heterodimer was purified by sequential column steps of SP-sepharose, heparin-sepharose (GE Healthcare) and anti-FLAG M2 affinity gels (Sigma) as described previously (20). It should be noted that the heparin-sepharose purification step is critical for higher DDB activity of CPD binding and NER stimulation. Flag-DDB1 or Flag-DDB2 alone was also expressed in insect cells and partially purified by SP-sepharose and anti-FLAG M2 affinity gels (19). Biotin-labeled ERCC1 was prepared with a TNT quick coupled transcription/translation system using Transcend tRNA according to the manufacture's instruction (Promega).
Pull-down assay
The protein–protein interactions between DDB subunits and XPA or its deletion mutants were analyzed using a pull-down assay. Appropriate amounts of lysates from E. coli overexpressing each factor fused with GST or MBP were incubated with 50 µl of glutathione–sepharose 4B beads (GE Healthcare) or amylose beads (New England Biolabs), respectively, in 500 µl of buffer-B [50 mM Tris–HCl (pH 7.4), 1 mM EDTA, 0.1 M KCl, 20% glycerol, 1 mM dithiothreitol (DTT)] at 4°C overnight with gentle rocking. The beads were washed with the same buffer thrice and equilibrated in IP buffer [20 mM Tris–HCl (pH 7.4), 0.1 M KCl, 4 mM MgCl2, 0.5 mM EDTA. 0.1% NP-40, 1 mM DTT]. The GST- or MBP-fusion proteins bound to each beads were incubated with other factors at 4°C for 1 h with gentle rocking. After extensive washing with IP buffer, the bound proteins were eluted in SDS-sample buffer (Bio-Rad) by boiling for 5 min and analyzed by SDS–PAGE followed by western blotting using specific antibodies.
In vitro excision repair assay
The substrate for an in vitro excision repair assay was a 136-bp duplex-DNA containing a 6-4PP or CPD in the center and 32P-label at fourth phosphodiester bond 5′ to the lesion (19,36). CFEs were prepared from HeLa S3 and XP12ROSV cell lines according to the method of Manley et al. (37). Three femtomoles of substrates were incubated at 30°C for 45 min with 50 µg of CFEs in 25 µl of reaction buffer [32 mM Hepes–KOH (pH 7.9), 64 mM KCl, 6.44 mM MgCl2, 0.16 mM of DTT, 2 mM ATP, 4% glycerol]. In a reconstituted system, 6 fmol of substrates were incubated with six core NER factors (60 ng of XPA, 150 ng of RPA, 20 ng of XPC-RAD23B, 150 ng of TFIIH, 10 ng of XPG and 20 ng of XPF-ERCC1) in the presence or absence of 50 ng of DDB in 25 µl of reaction buffer. DNAs were extracted with phenol:chloroform and separated on 8% denaturing polyacrylamide gels. The excision products were visualized by autoradiography and quantitated with a Fuji Bas 2000 Bio-Imaging Analyzer.
Cell lines and culture
Tet-on U2OS/3xF-DDB2 cells were established by cotransfecting pTRE2/3xF-DDB2 and pTK-Hyg (Clontech) into Tet-on U2OS cells (Clontech) according to the manufacturer's instructions. The XP2YOSV (XP-F) cell line was provided by Dr Takashi Yagi (Osaka Prefecture University) and its derivative cell line stably expressing FLAG-tagged DDB2 (XP2YOSV/F-DDB2) was generated as described previously (20). The XP-A revertant cell line XP129 was transfected with p3xFlag-CMV/DDB2 using the Effectene transfection reagent (Qiagen) to generate a stable cell line expressing 3xF-DDB2 (XP129/3xF-DDB2). Those cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and gentamicin in a 5% CO2 incubator at 37°C.
Transient overexpression and immunoprecipitation
For transient expression, typically, 2 µg of pCMV-myc/XPA or pCMV-myc/XPA(R207G) were transfected into Tet-on U2OS/3xF-DDB2 cells in a 90 mm plastic dish by Effectene transfection reagent. After incubation for 40 h in the presence or absence of doxycycline, cells were lyzed in 650 µl of NP40 buffer [10 mM Tris–HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 1% NP40, 1 mM DTT]. The lysates were incubated for 1.5 h with anti-FLAG M2 agarose (Sigma) or anti-myc antibody (Clontech) followed by protein A/G plus agarose (Oncogene Science). The beads were collected by centrifugation and washed with NP40 buffer thrice. Proteins retained on the beads were boiled at 100°C for 5 min and analyzed by western blotting using antibodies against the corresponding epitope tag.
Micropore UV irradiation and immunostaining
XP2YOSV or XP2YOSV/F-DDB2 cells were transfected with 50 ng of pCMV-myc/XPA (wild-type or R207G mutant) using the Fugene HD transfecting reagent (Roche). In some experiments, 50 ng of p3xFlag-CMV/DDB2 were co-transfected into XP2YOSV cells. Two days after the transfection, cells were briefly rinsed twice with Dulbecco's phosphate-buffered saline (−) and irradiated with 20 J/m2 of UV-C light (254 nm) through an isopore polycarbonate membrane filter (Millipore Corp., pore size 5 µm in diameter). Following 30 min incubation, cells were fixed with 4% formaldehyde (Wako) at room temperature for 15 min and permeabilized with 0.2% Triton X-100 in 10 mM phosphate-buffered saline on ice for 5 min. After blocking the non-specific antibody-binding sites with 20% fetal bovine serum in 10 mM phosphate-buffered saline, cells were sequentially stained with anti-myc polyclonal antibody (Santa Cruz Biotechnology) and Alexa Fluor 488 goat anti-rabbit IgG (H+L) conjugate (Invitrogen) to visualize the localization of myc-XPA. For the co-detection of UV-induced CPD, the stained cells were refixed with 2% formaldehyde and treated with 2 M HCl for 10 min at 37°C to denature the DNAs. The cells were incubated with anti-CPD monoclonal antibody (TDM-2) (38) and subsequently with Alexa Fluor 594 goat anti-mouse IgG conjugate (Invitrogen). To obtain fluorescence images, a Leica DMIRBE microscope equipped with a cooled CCD camera (CoolSNAP HQ, Photometrics) was used.
In vivo repair assay
Human cells cultured in 90 mm plastic dishes were irradiated with 10 J/m2 of UV-C from germicidal lamps (Toshiba, GL-10) and incubated for various periods. Genomic DNAs were purified with the DNeasy kit (Qiagen), and the amounts of CPD were measured by an enzyme-linked immunosorbent assay with TDM-2 antibody (38).
RESULTS
Physical interaction between DDB and XPA
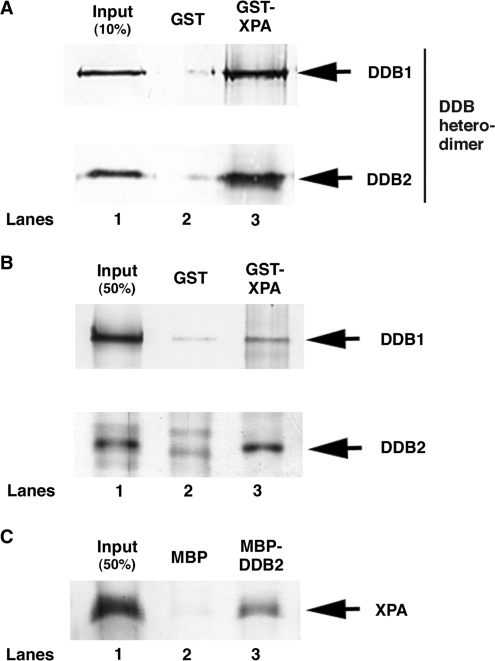
As we previously found that DDB markedly elevates the binding of XPA to damaged DNA and makes a ternary complex with XPA and damaged DNA (19), we tested whether DDB physically interacts with XPA using a pull-down assay with recombinant GST-XPA and DDB heterodimer. As shown in Figure 1A, DDB specifically bound to GST-XPA but not GST. To determine which subunit of DDB is responsible for XPA binding, a similar experiment was conducted with individual subunits and revealed that DDB2 binds to XPA, although DDB1 also binds to XPA with less efficiency (Figure 1B). In a reciprocal pull-down experiment, MBP-DDB2 specifically pulled down His-tagged XPA (Figure 1C), leading us to conclude that DDB physically interacts with XPA, mainly through DDB2 subunit.
Figure 1.
DDB directly binds to XPA through DDB2 subunit. Purified DDB heterodimer (A) or each DDB subunit (B) was incubated with GST alone (lane 2) or GST-XPA (lane 3) coupled to glutathione–sepharose 4B beads. The bound proteins were separated on a SDS–polyacrylamide gel and analyzed by western blotting with anti-Flag antibody for detecting DDB1 and DDB2. (C) Purified (His)6-XPA protein was incubated with MBP alone (lane 2) or MBP-DDB2 (lane 3) coupled to amylose beads. The bound proteins were analyzed by western blotting with anti-His antibody.
Identification of a DDB2-interactive domain in XPA
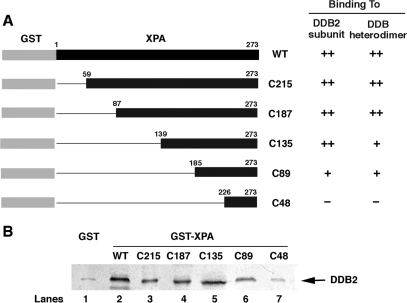
In order to determine a DDB2-interactive domain in XPA, a series of N-terminal deletion mutants of XPA fused with GST (Figure 2A) were prepared and used for pull-down experiments with the lysates from SF21 insect cells overexpressing Flag-tagged DDB2. A typical result from repeated experiments is shown in Figure 2B. N-terminal deletion up to 184 amino-acids from XPA did not significantly affect its binding ability to DDB2, but additional 41 amino-acid deletion greatly diminished the interaction. The similar binding properties were obtained with purified DDB heterodimer (data not shown, see summary in Figure 2A), consistent with the notion that XPA binds to DDB heterodimer by interacting with DDB2. These results indicate that the amino-acid domain between 185 and 225 is required for DDB2 interaction.
Figure 2.
Domain mapping of XPA responsible for the binding to DDB2. (A) Schematic diagram of various XPA deletion mutants and summary of pull-down experiments with these mutants. (B) The lysates from insect cells overproducing DDB2 were incubated with GST alone (lane 1) or various GST-XPA derivatives (lanes 2–7) coupled to glutathione–sepharose 4B beads. The bound proteins were analyzed by western blotting with anti-Flag antibody.
R207G mutation in XPA affects its interaction with DDB
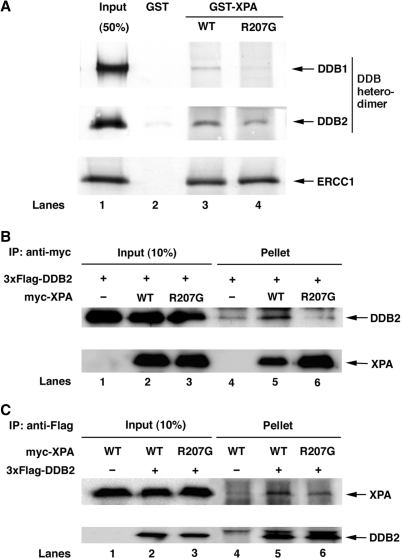
We searched various XP-A patients for naturally occurring missense mutations within the DDB2-interactive domain, but we could not find such mutations. However, we noticed that the XP129 UV-resistant revertant cell line obtained by mutagenizing XP12ROSV cells with methyl methane sulfonate contains a missense mutation in this domain, which causes Arg-207 to Gly substitution (39,40). The repair kinetics data of this cell line (39) is reminiscent of that of XP-E and Chinese hamster V79 cells lacking DDB2, characterized by a specific deficiency in GGR of CPD (17,41). We prepared R207G-type mutant XPA protein fused with GST and tested for the binding to DDB heterodimer (Figure 3A). As we expected, the binding ability of GST-XPA(R207G) to DDB was significantly impaired, whereas the mutant XPA showed normal binding to ERCC1 that interacts with XPA through a different domain (residues 75–114) (42).
Figure 3.
R207G mutation reduces XPA binding to DDB. (A) Purified DDB heterodimer or in vitro translated ERCC1 was incubated with GST alone (lane 2), GST-XPA (lane 3) or GST-XPA(R207G) (lane 4) coupled to glutathione–sepharose 4B beads. The bound proteins were analyzed by western blotting with either anti-Flag followed by anti-mouse IgG conjugated with alkaline phosphatase (for DDB1 and DDB2) or streptavidin conjugated with alkaline phosphatase (for ERCC1). (B) and (C) myc-tagged XPA protein, wild-type or R207G mutant, was expressed in Tet-on U2OS/3xF-DDB2 cells in the presence of doxycycline and cell lysates were prepared after 40-h incubation. One or 0.3 mg of the lysates were incubated for 1.5 h with anti-FLAG M2 agarose (B) or anti-myc antibody followed by protein A/G plus agarose (C), respectively. Proteins retained on the beads were analyzed by western blotting using anti-Flag and anti-myc antibodies.
To examine whether the effect of this mutation is observed in vivo as well, we ectopically expressed myc-tagged XPA (wild-type or R207G mutant) in Tet-on U2OS/3xF-DDB2 cells in the presence of doxycycline and conducted the immunoprecipitation with anti-myc or anti-Flag antibody. DDB2 was specifically coprecipitated with wild-type XPA, indicating their physical association in vivo, whereas R207G mutant XPA coprecipitated DDB2 only marginally as seen in control lane (Figure 3B). A reciprocal experiment exhibited the consistent result that mutant XPA was less efficiently (10.0%) coprecipitated with DDB2 compared to wild-type XPA (Figure 3C). These results indicate that R207G mutation affects the ability of XPA to interact with DDB in vitro as well as in vivo.
R207G mutant XPA fails to support the stimulation of CPD excision by DDB in vitro
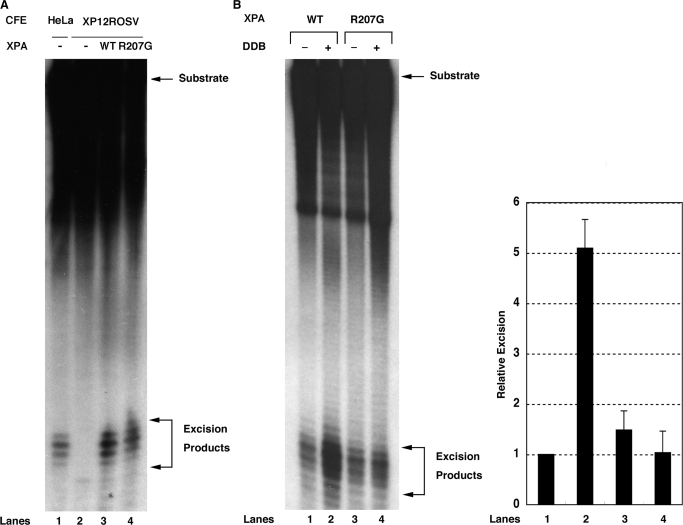
Having given that R207G mutant XPA shows a defect in interacting with DDB but not ERCC1, we wished to know whether the mutant XPA is able to support the excision reaction in vitro. First, we conducted an in vitro complementation assay with the substrate containing a 6-4PP, in which (His)6-XPA or (His)6-XPA(R207G) was mixed with XP-A CFEs showing no excision activity (Figure 4A, lane 2). R207G mutant XPA gave a comparable signal of 6-4PP excision to wild-type XPA (Figure 4A, lanes 3 and 4), indicating no or little effects of this mutation on complementing the repair defect of XP-A CFEs. We next examined the excision of CPD in a reconstituted system containing either wild-type or R207G mutant (His)6-XPA and other five NER factors. The substrate containing a CPD was incubated with the purified proteins in the presence or absence of recombinant DDB. As shown in Figure 4B, under DDB-free conditions, R207G mutant XPA was competent for excising CPD in vitro with other NER components (lane 1 versus lane 3). However, the addition of DDB to the reaction containing R207G mutant XPA exhibited no stimulation (lane 4), whereas DDB did stimulate the excision of CPD ∼5-fold in the presence of wild-type XPA (lane 2) as reported previously (20). These results strongly suggest that the interaction between DDB and XPA is required for the DDB-mediated stimulation of CPD excision in vitro.
Figure 4.
Effect of R207G mutation on XPA activity in NER reaction in vitro. (A) Three femtomoles of internally-labeled 136-bp substrates containing a 6-4PP were incubated with 50 µg of XP-A CFEs in the absence (lane 2) or presence of 60 ng of wild-type (lane 3) or R207G mutant XPA (lane 4) for 45 min. As a control, the excision reaction was also conducted with 50 µg of HeLa CFEs (lane 1). DNAs were extracted, separated on an 8% sequencing gel and detected by autoradiography. The signals were quantified by a Fuji Bas 2000 Bio-imaging analyzer and each excision efficiency was determined as 0.66% (lane 1), 0.017% (lane 2), 2.2% (lane 3) and 2.9% (lane 4). (B) Six femtomoles of internally-labeled 136-bp substrates containing a CPD were incubated with 60 ng of wild-type (lanes 1 and 2) or R207G mutant XPA (lanes 3 and 4) and other core NER factors (150 ng of RPA, 20 ng of XPC-RAD23B, 150 ng of TFIIH, 10 ng of XPG and 20 ng of XPF-ERCC1) in the presence (lanes 2 and 4) or absence (lanes 1 and 3) of DDB heterodimer (50 ng) for 4 h. DNAs were analyzed as described in (A) and each excision efficiency was 0.98% (lane 1), 5.6% (lane 2), 1.2% (lane 3) and 0.74% (lane 4). A right panel shows quantitative data from triplicate experiments and percent excision was expressed as relative excision to control reaction (wild-type XPA, no DDB). Error bars indicate the SD.
R207G mutation in XPA affects its recruitment to locally UV-damaged subnuclear regions in vivo
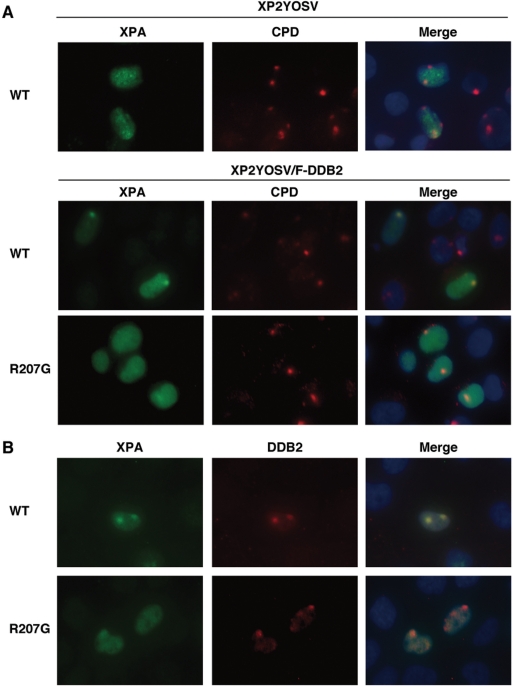
We next asked whether R207G mutation also affects the recruitment of XPA to damaged DNA sites in vivo following local UV irradiation. For this experiment, we used a SV40-transformed XP-F cell line lacking 5′ endonuclease of dual incision, in which the accumulation of XPA at locally damaged DNA sites can be observed more clearly and persistently than in NER-proficient cells (43, our unpublished data). XP2YOSV or its derivative cell line stably expressing Flag-tagged DDB2 (XP2YOSV/F-DDB2) was transiently transfected with a plasmid expressing myc-XPA (wild-type or R207G mutant) and exposed to UV light through a 5 µm isopore membrane filter. The expression levels of wild-type and R207G mutant XPA were verified to be comparable by immunoblotting (Supplementary Figure 1S). As shown in Figure 5A and Supplementary Figure 2S, the accumulation of myc-XPA at locally UV-damaged regions (indicated by CPD staining) was more significant in XP2YOSV/F-DDB2 cells compared with its parental XP2YOSV cells, indicating that DDB enhances the recruitment of XPA to damaged DNA sites. On the other hand, myc-XPA(R207G) exhibited poor accumulation even in the presence of F-DDB2. To verify the co-localization of XPA and DDB at damaged DNA sites, myc-XPA and 3xFlag-DDB2 were transiently co-expressed in XP2YOSV cells and detected by each epitope-tag-specific antibody following micropore UV irradiation (Figure 5B). The cells harboring 3xFlag-DDB2 signals showed clear co-localization of myc-XPA, whereas myc-XPA(R207G) mutant exhibited only marginal accumulation at 3xFlag-DDB2 sites (Supplementary Figure 2S). These results suggest that the DDB2–XPA interaction is also required for the recruitment of XPA to damaged DNA sites in vivo.
Figure 5.
Effect of R207G mutation on XPA accumulation at damaged DNA sites after local UV irradiation. (A) myc-tagged XPA, wild-type or R207G mutant, was transiently expressed in XP2YOSV (upper panel) or XP2YOSV/F-DDB2 cells (middle and lower panels). Cells were irradiated with 20 J/m2 of UV light through an isopore polycarbonate membrane filter. After 30 min incubation, cells were fixed and co-stained with anti-myc antibody (XPA) and TDM-2 antibody (CPD). (B) myc-tagged XPA, wild-type or R207G mutant, and 3xFlag-tagged DDB2 were transiently co-expressed in XP2YOSV cells. Following micropore UV irradiation, cells were fixed and co-stained with anti-myc antibody (XPA) and anti-Flag antibody (DDB2).
R207G mutant XPA fails to support the enhancement of CPD repair by ectopic DDB2 expression in SV40-transformed human cells
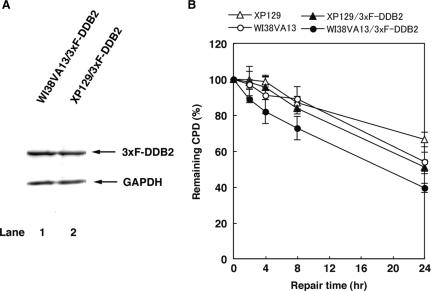
To further ascertain our model, we transfected WI38VA13 and XP129 cell lines with p3xFlag-CMV/DDB2 and generated stable cell lines expressing 3xF-DDB2, WI38VA13/3xF-DDB2 and XP129/3xF-DDB2, respectively, since SV40-immortalized human cell lines show reduced GGR activity of CPD (44). We compared CPD repair in WI38V13 (wild-type XPA) and XP129 (R207G mutant XPA) cells with or without stable expression of 3xF-DDB2 (Figure 6). The repair kinetics of CPD is significantly slower in WI38VA13 compared to primary diploid human fibroblasts (45), and indistinguishable from that in XP129, consistent with the observation by Kobayashi et al. (46). Interestingly, the stable expression of DDB2 conferred the enhanced CPD repair in WI38VA13 but not XP129 cells, suggesting that R207G mutant XPA does not support the enhancement of CPD repair by ectopic expression of DDB2 in SV40-transformed human cells.
Figure 6.
Effect of R207G mutation on the enhancement of CPD repair by ectopic expression of DDB2 in SV40-transformed human cells. (A) Cell lysates were prepared from WI38VA13/3xF-DDB2 and XP129/3xF-DDB2 cells and analyzed by immunoblotting with anti-Flag and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies. (B) WI38VA13/3xF-DDB2, XP129/3xF-DDB2 and their parental cells were irradiated with 10 J/m2 of UV-C light and incubated for the indicated periods. Genomic DNAs were purified and used for the determination of CPD content by an enzyme-linked immunosorbent assay. Each point represents the mean of three independent experiments and error bars indicate the SD.
DISCUSSION
The protein–protein interaction studies on core NER factors have greatly contributed to the understanding of basal NER reaction (47). However, only a few studies on the interaction between DDB and core NER factors have been reported (26,48), in contrast to numerous reports of DDB-interactive proteins involved in other cellular processes (49). The first core NER factor shown to interact with DDB is RPA, based on immunoprecipitation using partially purified DDB fraction or nuclear extracts (48). RPA is a eukaryotic single-stranded DNA-binding protein essential for chromosomal DNA replication as well as NER and shows a preferential binding to damaged DNA compared with native DNA (50,51). The interaction between DDB and RPA appears to enhance damaged DNA-binding activity of both proteins (48). The authors (48) and we (19) also observed a ternary complex of DDB, RPA and damaged DNA in an electrophoretic mobility shift assay. In addition, RPA is known to interact with XPA physically and functionally and this interaction again enhances both binding activities to damaged DNA (50,52). Together with our previous observation of a higher complex containing DDB, RPA, XPA and damaged DNA (19), DDB might also play a role in recruiting RPA along with XPA to damaged DNA sites.
Recently, Sugasawa et al. (26) have reported that DDB physically interacts with XPC based on immunoprecipitation using cell lysates as well as purified proteins. The interaction between DDB and XPC is implicated in the ubiquitination of XPC by the E3 ligase complex with Cullin4A, Roc1 and DDB, and the XPC polyubiquitination alters its DNA-binding properties. In the literature, XPC is also known to interact with XPA (53). Furthermore, other group has recently shown that XPC is modified by SUMO-1 and ubiquitin in untransformed cells exposed to UV light and these modifications require not only DDB2 but also XPA (27). Taken all together, it is plausible that DDB bound to damaged DNA sites may function in the recruitment of XPC-RAD23B, XPA and RPA and further in the modification of XPC at damaged DNA sites.
A recently developed micropore UV irradiation method has contributed to our understanding of sequential assembly of NER factors at damaged DNA sites in the nucleus (43,54). We originally found that DDB rapidly accumulates at locally UV-damaged subnuclear regions in the absence of XPC or XPA (20). Other groups reported similar observations and further showed that the recruitment of XPC to UV-damaged sites, especially CPD, requires functional DDB2 (as a DDB complex) (22–24). In this study, we showed that the ectopic expression of DDB2 in SV40-transformed XP-F cells enhances the recruitment of XPA to UV-damaged sites (Figure 5). Using the same local UV irradiation, Volker et al. (43) demonstrated that XPC is required for the recruitment of XPA to damaged-DNA sites in primary diploid human fibroblasts containing a normal level of DDB2. DDB may enhance the recruitment of XPA to DNA lesions in vivo by two possible mechanisms, directly through its interaction with XPA and indirectly through efficient XPC loading onto DNA lesions.
XPA is known to interact with all of other five core NER factors (34,42,50,52,53,55,56), and their interaction domains as well as DNA-binding domain in XPA have been mapped. The DDB2-interactive domain (residues 185–225) determined in this study partially overlaps with a DNA-binding domain (residues 98–219) reported previously (57,58) and is located between an RPA1-binding domain (residues 98–187) (52,59) and a C-terminal TFIIH-binding domain (residues 226–273) (34). We further demonstrated that R207G mutation in the DDB2-interactive domain results in the reduced binding to DDB2 or DDB heterodimer. Importantly, this amino-acid substitution caused the attenuated XPA recruitment to locally UV-damaged subnuclear regions (Figure 5 and Supplementary Figure 2S) and impaired enhancement of CPD repair by DDB in vitro (Figure 4) as well as in vivo (Figure 6), clearly indicating that the interaction between DDB and XPA plays an important role in the DDB-mediated NER reaction for CPD.
The R207G mutation was found in the XP129 revertant cell line, which had been isolated from a SV40-transfomed XP12ROSV cell line following the repetitious treatment of methyl methane sulfonate and reported to have a unique DNA repair phenotype of removing 6-4PP normally but not CPD (39,40). On the other hand, Kobayashi et al. (46) examined the effects of this mutation on NER activity and UV sensitivity using various XP12ROSV transfectants stably expressing wild-type or R207G mutant XPA. Based on the observation that the rate of CPD repair in the transfectant expressing R207G mutant XPA was almost normal, the authors concluded that the R207G mutation might not be responsible for the selective GGR defect for CPD repair in XP129 cells. The apparent discrepancy between the two studies seems explainable by the findings that SV40 transformation reduces GGR activity for CPD significantly (but not completely) due to dysfunction of p53 by the large T antigen (44) and this repair deficiency can be reversed by ectopic expression of DDB2 (20). Our data shown in Figure 6 strongly support this possible explanation.
In conclusion, this study has found that DDB2 physically interacts with XPA and this interaction is required for the stimulatory effect of DDB on CPD excision in vitro, the efficient recruitment of XPA to damaged sites in vivo and the enhancement of CPD repair by ectopic expression of DDB2 in SV40-transformed human cells. These findings reveal a new link between DDB and the core NER factors, and indicate that XPA interacts with not only five core NER factors but also an accessory factor DDB. The critical role of XPA as a scaffold might explain why XP-A patients or XPA-deficient cells exhibit the most severe phenotypes among seven complementation groups of NER-defective XP.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by grants 12143202 (T.M.) and 16710031 (M.W.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and National Institutes of Health grant R01 NS052781 (J.E.C.). Funding for open access charges: Kanazawa University.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr J. T. Reardon, Dr C. P. Selby and Dr A. Sancar (University of North Carolina at Chapel Hill) for various constructs for the repair factors and TFIIH used in the reconstitution experiment. We are grateful to Dr T. Yagi (Osaka Prefecture University) for the XP2YOSV cell line. We also thank Dr S. Linn (University of California, Berkeley) and Dr H. Morioka (Kumamoto University) for the cDNAs of DDB subunits and oligonucleotides containing a DNA lesion, respectively.
REFERENCES
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenger T. DNA Repair and Mutagenesis. 2nd edn. Washington, DC: American Society for Microbiology; 2006. pp. 227–371. [Google Scholar]
- 2.Sancar A. DNA excision repair. Annu. Rev. Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 3.Wood RD. DNA repair in eukaryotes. Annu. Rev. Biochem. 1996;65:135–167. doi: 10.1146/annurev.bi.65.070196.001031. [DOI] [PubMed] [Google Scholar]
- 4.de Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 5.Bootsma D, Kramer KH, Cleaver JE, Hoeijmakers JHJ. Scriver CR, Beaudet AL, Sly WS, Valle E. The MetabolicBasis of Inherited Disease. Vol. 1. New York: McGraw-Hill Book Co; 2001. pp. 677–703. [Google Scholar]
- 6.Mu D, Park CH, Matsunaga T, Hsu DS, Reardon JT, Sancar A. Reconstitution of human DNA repair excision nuclease in a highly defined system. J. Biol. Chem. 1995;270:2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- 7.Araujo SJ, Tirode F, Coin F, Pospiech H, Syvaoja JE, Stucki M, Hubscher U, Egly JM, Wood RD. Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK. Genes Dev. 2000;14:349–359. [PMC free article] [PubMed] [Google Scholar]
- 8.Tang J, Chu G. Xeroderma pigmentosum complementation group E an d UV-damaged DNA-binding protein. DNA Repair. 2002;1:601–616. doi: 10.1016/s1568-7864(02)00052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keeney S, Chang GJ, Linn S. Characterization of a human DNA damage binding protein implicated in xeroderma pigmentosum E. J. Biol. Chem. 1993;268:21293–21300. [PubMed] [Google Scholar]
- 10.Reardon JT, Nichols AF, Keeney S, Smith CA, Taylor JS, Linn S, Sancar A. Comparative analysis of binding of human damaged DNA-binding protein (XPE) and Escherichia coli damage recognition protein (UvrA) to the major ultraviolet photoproducts: T[c,s]T, T[t,s]T, T[6-4]T, and T[Dewar]T. J. Biol. Chem. 1993;268:21301–21308. [PubMed] [Google Scholar]
- 11.Fujiwara Y, Masutani C, Mizukoshi T, Kondo J, Hanaoka F, Iwai S. Characterization of DNA recognition by the human UV-damaged DNA-binding protein. J. Biol. Chem. 1999;274:20027–20033. doi: 10.1074/jbc.274.28.20027. [DOI] [PubMed] [Google Scholar]
- 12.Aboussekhra A, Biggerstaff M, Shivji MK, Vilpo JA, Moncollin V, Podust VN, Protic M, Hubscher U, Egly JM, Wood RD. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 13.Nichols AF, Ong P, Linn S. Mutations specific to the xeroderma pigmentosum group E Ddb- phenotype. J. Biol. Chem. 1996;271:24317–24320. doi: 10.1074/jbc.271.40.24317. [DOI] [PubMed] [Google Scholar]
- 14.Chu G, Chang E. Xeroderma pigmentosum group E cells lack a nuclear factor that binds to damaged DNA. Science. 1988;242:564–567. doi: 10.1126/science.3175673. [DOI] [PubMed] [Google Scholar]
- 15.Kataoka H, Fujiwara Y. UV damage-specific DNA-binding protein in xeroderma pigmentosum complementation group E. Biochem. Biophys. Res. Commun. 1991;175:1139–1143. doi: 10.1016/0006-291x(91)91684-5. [DOI] [PubMed] [Google Scholar]
- 16.Keeney S, Wein H, Linn S. Biochemical heterogeneity in xeroderma pigmentosum complementation group E. Mutat. Res. 1992;273:49–56. doi: 10.1016/0921-8777(92)90049-9. [DOI] [PubMed] [Google Scholar]
- 17.Hwang BJ, Ford JM, Hanawalt PC, Chu G. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc. Natl Acad. Sci. USA. 1999;96:424–428. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang JY, Hwang BJ, Ford JM, Hanawalt PC, Chu G. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol. Cell. 2000;5:737–744. doi: 10.1016/s1097-2765(00)80252-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wakasugi M, Shimizu M, Morioka H, Linn S, Nikaido O, Matsunaga T. Damaged DNA-binding protein DDB stimulates the excision of cyclobutane pyrimidine dimers in vitro in concert with XPA and replication protein A. J. Biol. Chem. 2001;276:15434–15440. doi: 10.1074/jbc.M011177200. [DOI] [PubMed] [Google Scholar]
- 20.Wakasugi M, Kawashima A, Morioka H, Linn S, Sancar A, Mori T, Nikaido O, Matsunaga T. DDB accumulates at DNA damage sites immediately after UV irradiation and directly stimulates nucleotide excision repair. J. Biol. Chem. 2002;277:1637–1640. doi: 10.1074/jbc.C100610200. [DOI] [PubMed] [Google Scholar]
- 21.Reardon JT, Sancar A. Recognition and repair of the cyclobutane thymine dimer, a major cause of skin cancers, by the human excision nuclease. Genes Dev. 2003;17:2539–2551. doi: 10.1101/gad.1131003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fitch ME, Cross IV, Ford JM. p53 responsive nucleotide excision repair gene products p48 and XPC, but not p53, localize to sites of UV-irradiation-induced DNA damage, in vivo. Carcinogenesis. 2003;24:843–850. doi: 10.1093/carcin/bgg031. [DOI] [PubMed] [Google Scholar]
- 23.Wang QE, Zhu Q, Wani G, Chen J, Wani AA. UV radiation-induced XPC translocation within chromatin is mediated by damaged-DNA binding protein, DDB2. Carcinogenesis. 2004;25:1033–1043. doi: 10.1093/carcin/bgh085. [DOI] [PubMed] [Google Scholar]
- 24.Fitch ME, Nakajima S, Yasui A, Ford JM. In vivo recruitment of XPC to UV-induced cyclobutane pyrimidine dimers by the DDB2 gene product. J. Biol. Chem. 2003;278:46906–46910. doi: 10.1074/jbc.M307254200. [DOI] [PubMed] [Google Scholar]
- 25.Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, Kisselev AF, Tanaka K, Nakatani Y. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 2003;113:357–367. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- 26.Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, Mori T, Iwai S, Tanaka K, Tanaka K, et al. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005;121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 27.Wang QE, Zhu Q, Wani G, El-Mahdy MA, Li J, Wani AA. DNA repair factor XPC is modified by SUMO-1 and ubiquitin following UV irradiation. Nucleic Acids Res. 2005;33:4023–4034. doi: 10.1093/nar/gki684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rapic Otrin V, Kuraoka I, Nardo T, McLenigan M, Eker AP, Stefanini M, Levine AS, Wood RD. Relationship of the xeroderma pigmentosum group E DNA repair defect to the chromatin and DNA binding proteins UV-DDB and replication protein A. Mol. Cell. Biol. 1998;18:3182–3190. doi: 10.1128/mcb.18.6.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang BJ, Toering S, Francke U, Chu G. p48 Activates a UV-damaged-DNA binding factor and is defective in xeroderma pigmentosum group E cells that lack binding activity. Mol. Cell. Biol. 1998;18:4391–4399. doi: 10.1128/mcb.18.7.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Datta A, Bagchi S, Nag A, Shiyanov P, Adami GR, Yoon T, Raychaudhuri P. The p48 subunit of the damaged-DNA binding protein DDB associates with the CBP/p300 family of histone acetyltransferase. Mutat. Res. 2001;486:89–97. doi: 10.1016/s0921-8777(01)00082-9. [DOI] [PubMed] [Google Scholar]
- 31.Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, Roeder RG. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 2001;21:6782–6795. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kapetanaki MG, Guerrero-Santoro J, Bisi DC, Hsieh CL, Rapic-Otrin V, Levine AS. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc. Natl Acad. Sci. USA. 2006;103:2588–2593. doi: 10.1073/pnas.0511160103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H, Zhai L, Xu J, Joo HY, Jackson S, Erdjument-Bromage H, Tempst P, Xiong Y, Zhang Y. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell. 2006;22:383–394. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 34.Park CH, Mu D, Reardon JT, Sancar A. The general transcription-repair factor TFIIH is recruited to the excision repair complex by the XPA protein independent of the TFIIE transcription factor. J. Biol. Chem. 1995;270:4896–4902. doi: 10.1074/jbc.270.9.4896. [DOI] [PubMed] [Google Scholar]
- 35.Dualan R, Brody T, Keeney S, Nichols AF, Admon A, Linn S. Chromosomal localization and cDNA cloning of the genes (DDB1 and DDB2) for the p127 and p48 subunits of a human damage-specific DNA binding protein. Genomics. 1995;29:62–69. doi: 10.1006/geno.1995.1215. [DOI] [PubMed] [Google Scholar]
- 36.Mu D, Tursun M, Duckett DR, Drummond JT, Modrich P, Sancar A. Recognition and repair of compound DNA lesions (base damage and mismatch) by human mismatch repair and excision repair systems. Mol. Cell. Biol. 1997;17:760–769. doi: 10.1128/mcb.17.2.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manley JL, Fire A, Cano A, Sharp PA, Gefter ML. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc. Natl Acad. Sci. USA. 1980;77:3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mori T, Nakane M, Hattori T, Matsunaga T, Ihara M, Nikaido O. Simultaneous establishment of monoclonal antibodies specific for either cyclobutane pyrimidine dimer or (6-4)photoproduct from the same mouse immunized with ultraviolet-irradiated DNA. Photochem. Photobiol. 1991;54:225–232. doi: 10.1111/j.1751-1097.1991.tb02010.x. [DOI] [PubMed] [Google Scholar]
- 39.Cleaver JE, Cortes F, Lutze LH, Morgan WF, Player AN, Mitchell DL. Unique DNA repair properties of a xeroderma pigmentosum revertant. Mol. Cell. Biol. 1987;7:3353–3357. doi: 10.1128/mcb.7.9.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cleaver JE, McDowell M, Jones C, Wood R, Karentz D. Mutation and expression of the XPA gene in revertants and hybrids of a xeroderma pigmentosum cell line. Somat. Cell. Mol. Genet. 1994;20:327–337. doi: 10.1007/BF02254721. [DOI] [PubMed] [Google Scholar]
- 41.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 42.Li L, Elledge SJ, Peterson CA, Bales ES, Legerski RJ. Specific association between the human DNA repair proteins XPA and ERCC1. Proc. Natl Acad. Sci. USA. 1994;91:5012–5016. doi: 10.1073/pnas.91.11.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 44.Bowman KK, Sicard DM, Ford JM, Hanawalt PC. Reduced global genomic repair of ultraviolet light-induced cyclobutane pyrimidine dimers in simian virus 40-transformed human cells. Mol. Carcinog. 2000;29:17–24. [PubMed] [Google Scholar]
- 45.Matsumoto M, Yaginuma K, Igarashi A, Imura M, Hasegawa M, Iwabuchi K, Date T, Mori T, Ishizaki K, Yamashita K, et al. Perturbed gap-filling synthesis in nucleotide excision repair causes histone H2AX phosphorylation in human quiescent cells. J. Cell. Sci. 2007;120:1104–1112. doi: 10.1242/jcs.03391. [DOI] [PubMed] [Google Scholar]
- 46.Kobayashi T, Takeuchi S, Saijo M, Nakatsu Y, Morioka H, Otsuka E, Wakasugi M, Nikaido O, Tanaka K. Mutational analysis of a function of xeroderma pigmentosum group A (XPA) protein in strand-specific DNA repair. Nucleic Acids Res. 1998;26:4662–4668. doi: 10.1093/nar/26.20.4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Araujo SJ, Wood RD. Protein complexes in nucleotide excision repair. Mutat. Res. 1999;435:23–33. doi: 10.1016/s0921-8777(99)00042-7. [DOI] [PubMed] [Google Scholar]
- 48.Otrin VR, McLenigan M, Takao M, Levine AS, Protic M. Translocation of a UV-damaged DNA binding protein into a tight association with chromatin after treatment of mammalian cells with UV light. J. Cell. Sci. 1997;110(Pt 10):1159–1168. doi: 10.1242/jcs.110.10.1159. [DOI] [PubMed] [Google Scholar]
- 49.Wittschieben BB, Wood RD. DDB complexities. DNA Repair. 2003;2:1065–1069. doi: 10.1016/s1568-7864(03)00113-7. [DOI] [PubMed] [Google Scholar]
- 50.He Z, Henricksen LA, Wold MS, Ingles CJ. RPA involvement in the damage-recognition and incision steps of nucleotide excision repair. Nature. 1995;374:566–569. doi: 10.1038/374566a0. [DOI] [PubMed] [Google Scholar]
- 51.Burns JL, Guzder SN, Sung P, Prakash S, Prakash L. An affinity of human replication protein A for ultraviolet-damaged DNA. J. Biol. Chem. 1996;271:11607–11610. doi: 10.1074/jbc.271.20.11607. [DOI] [PubMed] [Google Scholar]
- 52.Li L, Lu X, Peterson CA, Legerski RJ. An interaction between the DNA repair factor XPA and replication protein A appears essential for nucleotide excision repair. Mol. Cell. Biol. 1995;15:5396–5402. doi: 10.1128/mcb.15.10.5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.You JS, Wang M, Lee SH. Biochemical analysis of the damage recognition process in nucleotide excision repair. J. Biol. Chem. 2003;278:7476–7485. doi: 10.1074/jbc.M210603200. [DOI] [PubMed] [Google Scholar]
- 54.Katsumi S, Kobayashi N, Imoto K, Nakagawa A, Yamashina Y, Muramatsu T, Shirai T, Miyagawa S, Sugiura S, Hanaoka F, et al. In situ visualization of ultraviolet-light-induced DNA damage repair in locally irradiated human fibroblasts. J. Invest. Dermatol. 2001;117:1156–1161. doi: 10.1046/j.0022-202x.2001.01540.x. [DOI] [PubMed] [Google Scholar]
- 55.Nocentini S, Coin F, Saijo M, Tanaka K, Egly JM. DNA damage recognition by XPA protein promotes efficient recruitment of transcription factor II H. J. Biol. Chem. 1997;272:22991–22994. doi: 10.1074/jbc.272.37.22991. [DOI] [PubMed] [Google Scholar]
- 56.Park CH, Sancar A. Formation of a ternary complex by human XPA, ERCC1, and ERCC4(XPF) excision repair proteins. Proc. Natl Acad. Sci. USA. 1994;91:5017–5021. doi: 10.1073/pnas.91.11.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuraoka I, Morita EH, Saijo M, Matsuda T, Morikawa K, Shirakawa M, Tanaka K. Identification of a damaged-DNA binding domain of the XPA protein. Mutat. Res. 1996;362:87–95. doi: 10.1016/0921-8777(95)00038-0. [DOI] [PubMed] [Google Scholar]
- 58.Buchko GW, Ni S, Thrall BD, Kennedy MA. Structural features of the minimal DNA binding domain (M98-F219) of human nucleotide excision repair protein XPA. Nucleic Acids Res. 1998;26:2779–2788. doi: 10.1093/nar/26.11.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saijo M, Kuraoka I, Masutani C, Hanaoka F, Tanaka K. Sequential binding of DNA repair proteins RPA and ERCC1 to XPA in vitro. Nucleic Acids Res. 1996;24:4719–4724. doi: 10.1093/nar/24.23.4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.