Abstract
Toward the expansion of the genetic alphabet, we present an unnatural base pair system for efficient PCR amplification, enabling the site-specific incorporation of extra functional components into DNA. This system can be applied to conventional PCR protocols employing DNA templates containing unnatural bases, natural and unnatural base triphosphates, and a 3′→5′ exonuclease-proficient DNA polymerase. For highly faithful and efficient PCR amplification involving the unnatural base pairing, we identified the natural-base sequences surrounding the unnatural bases in DNA templates by an in vitro selection technique, using a DNA library containing the unnatural base. The system facilitates the site-specific incorporation of a variety of modified unnatural bases, linked with functional groups of interest, into amplified DNA. DNA fragments (0.15 amol) containing the unnatural base pair can be amplified 107-fold by 30 cycles of PCR, with <1% total mutation rate of the unnatural base pair site. Using the system, we demonstrated efficient PCR amplification and functionalization of DNA fragments for the extremely sensitive detection of zeptomol-scale target DNA molecules from mixtures with excess amounts (pmol scale) of foreign DNA species. This unnatural base pair system will be applicable to a wide range of DNA/RNA-based technologies.
INTRODUCTION
The creation of an unnatural base pair, compatible with the natural A–T and G–C base pairs, could expand the genetic alphabet of DNA, thus allowing the present recombinant-based biotechnology to progress to future technologies enabling the site-specific incorporation of extra, functional components into nucleic acids and proteins (1–6). The two additional letters of the third base pair could greatly augment the information capacity and variety of DNA base sequences (7,8). In addition, an increased genetic alphabet could confer new functions to nucleic acids. By joining functional groups to unnatural bases, a great variety of extra components could be site-specifically incorporated into nucleic acids, by replication and transcription mediated by unnatural base pairing (6,7,9–12). To adapt this genetic expansion system to modern biotechnology, faithful complementarity of the unnatural base pairing is critical in polymerase reactions, especially in PCR amplification (13–16).
One precedent unnatural base pair employing nonstandard hydrogen-bonding topology is that between isoguanine (iG) and isocytosine (iC), which exhibited ∼98% fidelity-per-cycle in PCR (17). This system requires 2-thiothymidine triphosphate, instead of the natural thymidine triphosphate, to improve the cognate base pairing selectivity. 2-Thiothymidines are, however, inserted into DNA in place of all of the natural T bases. Another unnatural base pair, between 2-amino-imidazo[1,2-a]-1,3,5-triazin-4(8H)-one (P) and 6-amino-5-nitro-2(1H)-pyridone (Z), is also amplified by PCR, without using 2-thiothymidine triphosphate, with 97.5% fidelity-per-cycle (18).
We have recently developed hydrophobic, unnatural base pairs between 7-(2-thienyl)-imidazo[4,5-b]pyridine (Ds) and pyrrole-2-carbaldehyde (Pa) (12) and between Ds and 2-nitropyrrole (Pn) (19), which have specific shape complementation that differs from those of the A–T and G–C pairs. These unnatural base pairs exhibited high fidelity-per-cycle (>99%) in PCR, with the help of the 3′→5′ exonuclease activity of DNA polymerases (Pol) and in combination with the usual triphosphates and modified triphosphates, γ-amidotriphosphates. The γ-amidotriphosphates of Ds and A increase the selectivity of the cognate pairings in replication, by preventing the noncognate pairings of Ds–Ds and A–Pa, respectively. However, the use of the γ-amidotriphosphates decreases the PCR amplification efficiency and restricts the range of in vivo applications. Thus, the further development of unnatural base pair systems that bypass the need for the γ-amidotriphosphates, but maintain high fidelity (>99.9%), in PCR could pave the way to broader practical applications.
Here, we present an efficient PCR amplification system using an unnatural, hydrophobic base pair, based on that between Ds and 2-nitro-4-propynylpyrrole (Px) (Figure 1), with a set of specific sequences around the Ds–Px pair in DNA templates. The system no longer requires γ-amidotriphosphates, and moreover, it provides the efficient, multiple site-specific incorporation of modified Px bases, which were linked with functional groups at the 4-propynyl position, into the amplified DNA. These efficient surrounding sequences were identified by an in vitro selection method, using a DNA library of random sequences containing the unnatural bases. This system provides highly selective and efficient amplification and functionalization PCR: DNA fragments (0.15 amol) containing the Ds–Px pair were amplified 107-fold by 30 cycles of PCR with >99.9% fidelity-per-cycle.
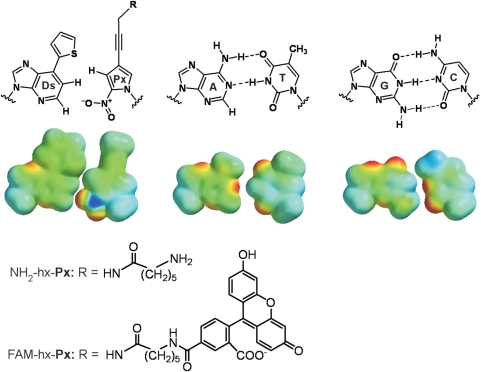
Figure 1.
Structures of the unnatural Ds–Px and natural A–T and G–C pairs. R = aminohexanamide (for NH2-hx-Px) or (fluorescein-5-carboxamido)hexanamide group (for FAM-hx-Px). Space filling models of the base alone (with methyl in place of deoxyribose and R = H in the Px base) are shown, with electrostatic potentials mapped on van der Waals surfaces (PM3 calculations, Spartan ‘06, Wavefunction Inc.).
MATERIALS AND METHODS
Chemical synthesis of nucleotides, single-nucleotide insertion experiments and DNA sequences for experiments
Materials, experimental procedures and data for chemical synthesis of the unnatural base triphosphates and DNA fragments, single-nucleotide insertion experiments and DNA sequences are available in the Supplementary Material.
In vitro selection of DNA sequences for efficient PCR amplification
In vitro selection was performed using a 55-mer single-stranded DNA library containing randomized NNNDsNNN sequences. In the initial round of PCR, 2 pmol of the library were used as a template for PCR (200 μl scale), and thus, all possible sequences (46 = 4096) could be present in the initial library. PCR was carried out in a reaction buffer (20 mM Tris–HCl pH 8.8, 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4 and 0.1% Triton X-100) with 2.5 μM FAM-hx-dPxTP, 50 μM dDsTP, 0.3 mM each natural dNTP (N = A, G, C and T), 1 μM each of a 5′-primer (20-mer, 5′-GATAATACGACTCACTATAG-3′) and a 3′-primer (24-mer, 5′-TTTCACACAGGAAACAGCTATGAC-3′) and 0.04 U/μl DeepVent DNA polymerase (NEB). The PCR cycle was as follows: 94°C, 0.5 min; 45°C, 0.5 min; 65°C, 4 min. After 10 cycles of PCR, the full-length amplified products were purified by gel electrophoresis (10% polyacrylamide—7 M urea gel) and then were quantified from their absorbance at 260 nm.
For the isolation of DNA fragments containing FAM-hx-Px, ∼20 pmol of the amplified DNA fragments were mixed with an anti-fluorescein IgG solution (20 μl) (Invitrogen, A-889) and incubated for 1 h on ice in a phosphate-buffered saline (100 μl). Then, the IgG-bound DNA fragments were recovered by filtration, using a Microcon YM-100 filter (Millipore). After phenol extraction and ethanol precipitation, ∼1 pmol of the recovered DNA was used as a template for the next round of PCR (200 μl).
After five rounds of in vitro selection, about 1 pmol of the recovered DNA was amplified by eight cycles of PCR (25 μl), in a reaction buffer with 50 μM NH2-hx-dPxTP, 50 μM dDsTP, 0.3 mM each natural dNTP, 1 μM each of 5′-primer (40-mer, 5′-CGTTGTAAAACGACGGCCAGGATAATACGACTCACTATAG-3′) and 3′-primer (24-mer, 5′-TTTCACACAGGAAACAGCTATGAC-3′) and 0.02 U/μl DeepVent DNA polymerase, and then the PCR products were purified by electrophoresis on 8% polyacrylamide—7 M urea gel for direct sequencing. For comparison, the original library (55-mer, 1 pmol) was also similarly amplified and purified. In the conventional cloning method, the recovered DNA fragments were amplified by eight cycles of PCR using Premix Taq (Ex Taq version; TaKaRa), without the unnatural base substrates, and were cloned using the TA cloning vector of the TOPO TA Cloning Kit Dual Promoter (Invitrogen). Plasmid DNAs were isolated and were sequenced using a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) on a DNA sequencer (model 3100, Applied Biosystems). Selected sequences of each clone are shown in Supplementary Figure 1.
Analysis of PCR amplification products
PCR was performed in a reaction buffer (20 mM Tris–HCl pH 8.8, 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4 and 0.1% Triton X-100) with 2.5 μM FAM-hx-dPxTP or 50 μM NH2-hx-dPxTP, 50 μM dDsTP, 0.3 mM each natural dNTP, 1 μM of 32P-labeled (or nonlabeled) 5′-primer (40-mer), 1 μM of nonlabeled 3′-primer (24-mer), 0.6 nM–0.6 fM single-stranded DNA template and DeepVent DNA Pol (0.02 U/μl). The PCR cycle was as follows: 94°C, 0.5 min; 45°C, 0.5 min; 65°C, 4 min. The amplified products were analyzed by denaturing 15% PAGE, and the radioactivity and fluorescence intensity of the DNA fragments on the gel were quantified by a bio-imaging analyzer, FLA-7000 (FUJIFILM). For the detection of nonlabeled PCR products, the DNA fragments on the gel were stained with SYBR Green II. For the sequencing analysis, the PCR products were purified by denaturing 8–15% PAGE, unless otherwise noted.
DNA sequencing
The cycle sequencing reaction (20 μl) was performed with the Cycle Sequencing Mix (8 μl) from the BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems), containing ∼0.3 pmol of the template and 4 pmol of the sequencing primer (20-mer), in the presence of 40 pmol or 1 nmol of 4-propynyl-1-(β-D-ribofuranosyl)pyrrole-2-carbaldehyde 5′-triphosphate (dPa′TP) (40 pmol for DNA fragments containing one Ds base and 1 nmol for DNAs with two Ds bases) or 1 nmol of ddPa′TP. After 25 cycles of PCR (96°C, 10 s; 50°C, 5 s; 60°C, 4 min), the residual dye terminators were removed from the reaction with CENTRI-SEP columns (Princeton Separations), and the solutions were dried. The residues were resuspended in a formamide solution (4 μl) and were fractionated on the ABI 377 DNA sequencer, using a 7% polyacrylamide-6 M urea gel. The sequence data were analyzed with the Applied Biosystems PRISM sequencing analysis v3.2 software.
PCR amplification with foreign DNA fragments and isolation of FAM-labeled target DNA fragments
PCR amplification, as described above, was performed using two types of DNA fragments as templates: one is a 55-mer DNA mixture (0.15 nM each, DNA S2, Cont2, Cont3 and Cont4) and the other is the mixture of DNA S2 (6 fM) and random 100-nt sequences consisting of only the natural bases (60 nM). A 20 μl portion of the PCR solution was filtered with a Microcon YM-30 filter (Millipore), using wash/binding buffer (20 mM Tris–HCl pH 7.6, 0.5 M NaCl, 10 mM MgCl2), to remove the excess unincorporated dNTPs. The filtrate was mixed with 40–80 μg of Streptavidin magnetic beads (NEB) bound with anti-fluorescein IgG (5–10 μg), and the mixture was incubated for 0.5–1 h on ice (∼30 μl volume). After washing the beads twice with 100 μl of the wash/binding buffer, the bound DNA fragments were eluted by adding 20 μl of 0.1 mM EDTA (pH 8.0) and heating at 75°C for 30 s. For the sequencing analysis, a 5–10 μl aliquot of the eluted solution was used as the template.
RESULTS
Development of the unnatural base pair system
The Px base was designed as a pairing partner of Ds (Figure 1), by adopting the concepts of hydrophobicity and specific shape and electrostatic complementarity. The nitro group of Px excludes the noncognate A–Px pairing by electrostatic repulsion between the 1-nitrogen of A and the nitro group of Px (19). In addition, the propynyl group (20) of Px increases its π-electronic hydrophobicity to improve its incorporation efficiency into DNA opposite Ds in templates. The hydrophobicity of the base moiety of the incoming triphosphates enhances their interactions with the hydrophobic protein side chains of DNA Pol and with the 3′-end base of primer strands (21–23).
We chemically synthesized two modified Px triphosphates, which were functionalized with an aminohexanamide group (NH2-hx-dPxTP) or a (fluorescein-5-carboxamido)hexanamide group (FAM-hx-dPxTP) linked to the 4-propynyl position of Px (Figure 1 and Supplementary Methods). Fluorescence excitation and emission maxima of FAM-hx-dPxTP are 496 nm and 522 nm (quantum yield: 32%), respectively. The high efficiency and selectivity of the Ds–Px pairing were supported by kinetic studies (24,25) of the cognate and noncognate pairings, using NH2-hx-dPxTP and templates containing Ds with the exonuclease-deficient Klenow fragment of Escherichia coli DNA Pol I (Supplementary Table 1). We expected to bypass the use of the γ-amidotriphosphate of Ds for preventing the Ds–Ds pairing, by the help of the 3′→5′ exonuclease activity of the polymerases (12).
In vitro selection of DNAs that are efficiently amplified in the unnatural base pair system
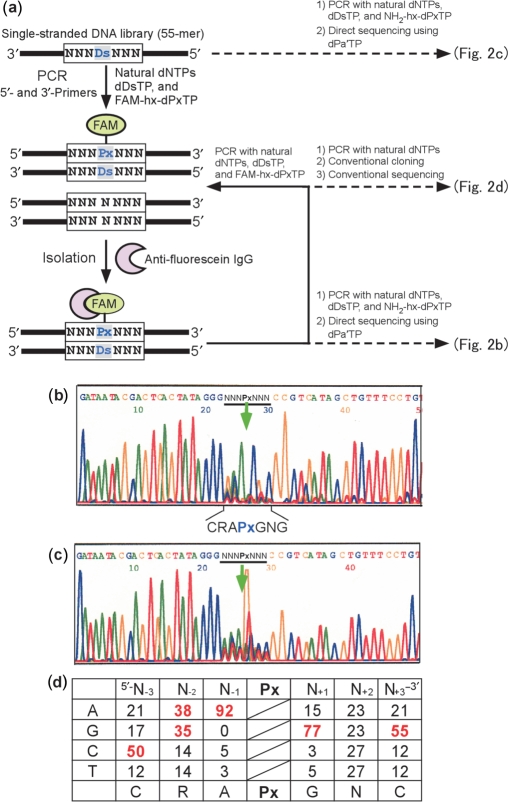
To attain further efficiency and selectivity of the Ds–Px pairing in PCR, we scrutinized the dependency on the natural base sequences surrounding the Ds–Px pair in DNA templates. Through our studies, we noticed that the natural-base sequences around unnatural bases in templates affect the replication efficiency. Thus, we performed an in vitro selection (26,27) using a DNA library (55-mer) containing a random natural-base sequence with Ds, NNNDsNNN (where N is A, G, C or T) (Supplementary Table 2), to comprehensively determine which sequences facilitate efficient PCR involving the Ds–Px pair (Figure 2a). The library was amplified by the 3′→5′ exonuclease-proficient Deep Vent DNA Pol with the unnatural base substrates, dDsTP and FAM-hx-dPxTP, besides the natural dNTPs. After 10 cycles of PCR, the amplified products containing FAM-hx-Px were isolated by using anti-fluorescein IgG. This PCR-isolation procedure was repeated, and after five rounds of selection, the Ds-containing strands of the library were directly sequenced by a previously established method, involving dideoxynucleotide chain-termination supplemented with an unnatural base triphosphate (dPa′TP), which is another pairing partner of Ds for DNA sequencing (12,19). The sequence bias of the random region (Figure 2b) was observed in the direct sequencing pattern of the library after the five-round selection, as compared with that of the original library (Figure 2c). In the sequencing, the unnatural base position is indicated as a gap; the peak that corresponds to the unnatural base position disappeared, because of the absence of the unnatural base dye terminator (12,19).
Figure 2.
Overview of in vitro selection using a Ds-containing DNA library. (a) Scheme for in vitro selection using the Ds-containing DNA library, by FAM-hx-Px incorporation into PCR products and isolation with an anti-fluorescein antibody. A chemically synthesized, single-stranded DNA library containing an NNNDsNNN sequence (55-mer) was amplified by 10 cycles of PCR, in the presence of natural dNTPs, dDsTP and FAM-hx-dPxTP, with DeepVent DNA Pol. After selection of the PCR products containing FAM-hx-Px by binding with an anti-fluorescein antibody, the isolated DNA fragments were used as a template for the next round of PCR amplification and selection. For direct sequencing of the library after five rounds of selection, the isolated DNA fragments were amplified in the presence of NH2-hx-dPxTP, instead of FAM-hx-dPxTP (Figure 3a), by eight cycles of PCR. Direct sequencing was performed in the presence of 2 μM dPa′TP, using a BigDye terminator v1.1 Cycle Sequencing kit. For the sequencing of clones after five rounds of selection, the library was amplified in the presence of only the natural dNTPs with Taq DNA polymerase, and the PCR products were used for TOPO TA cloning. (b) Sequencing of the DNA library after five rounds of selection. (c) Sequencing of the initial library. The arrow indicates the unnatural base position. (d) Probability (%) of occurrence at each position of the selected 66-clone sequences (Supplementary Figure 1). Bases with an occurrence rate of ≥35% among the clones are colored red.
Each sequence in the selected library was determined by a conventional cloning method. The library after the five-round selection was further amplified by Taq Pol without the unnatural base substrates, to forcibly replace the unnatural bases in the library with the natural bases for the following cloning procedures. We then isolated 66 clones and obtained their sequences (Figure 2d and Supplementary Figure 1). The sequence pattern tendency was quite similar to that obtained by the direct sequencing (Figure 2b).
PCR amplification involving the Ds–Px pair
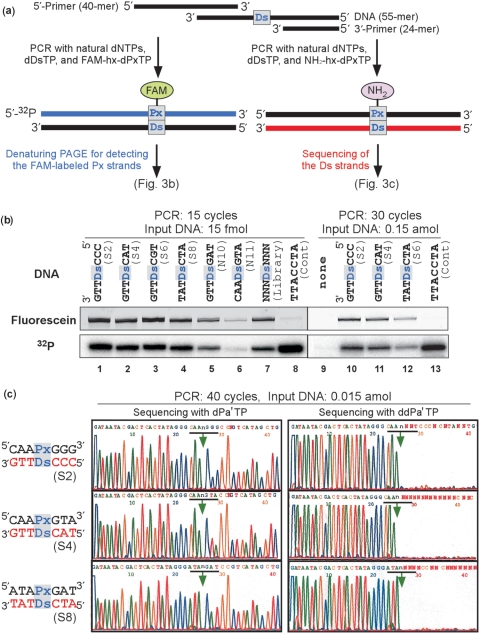
We chemically synthesized Ds-containing fragments of eight isolated sequences (DNA S1–S8) among the clones, as well as other designed sequences (DNA N9–N12) (Supplementary Tables 2 and 3) that were not obtained by the selection, and examined their amplification efficiency using FAM-hx-dPxTP or NH2-hx-dPxTP with a 32P-labeled 5′-primer (Figure 3 and Supplementary Table 3). The amplified DNA fragments were analyzed by detecting their FAM fluorescence and 32P radioactivity on a gel. Small amounts (15 fmol for 15-cycle PCR and 0.15 amol for 30-cycle PCR) of the input DNA fragments containing Ds (DNA S2, S4, S6 and S8) were efficiently amplified and detected fluorescently and radioactively (Figure 3b, lines 1–4 and 10–12). A DNA fragment containing a 3′-TTACCTA-5′ sequence without Ds (Cont), amplified in the presence of FAM-hx-dPxTP, was detected radioactively, but was very scarcely detected fluorescently (Figure 3b, lines 8 and 13). This indicates that FAM-hx-dPxTP was selectively incorporated opposite Ds in templates. The amplification efficiency of DNA N11, with the complementary sequence of DNA S4, was significantly decreased (Figure 3b, line 6), and DNA N10, which has a point mutation at the 5′-neighbor of Ds in DNA S4, was amplified with much lower efficiency (Figure 3b, line 5), relative to that of DNA S4. These results suggested that the sequences obtained by in vitro selection exhibited high amplification efficiency and selectivity for the Ds–Px pairing.
Figure 3.
PCR amplification of DNA fragments (55-mer) containing one Ds base. (a) Scheme for PCR amplification in the presence of FAM-hx-dPxTP or NH2-hx-dPxTP with DeepVent DNA Pol. To detect DNA fragments containing FAM-hx-Px, the PCR products were analyzed by denaturing PAGE. DNA fragments amplified with NH2-hx-dPxTP and dDsTP were used for sequencing of the Ds strands. (b) Polyacrylamide-gel analysis of the DNA fragments amplified by 15 or 30 cycles of PCR with the 32P-labeled 5′-primer, in the presence of FAM-hx-dPxTP and dDsTP. The fluorescence of the FAM-labeled full-length products on a gel was detected with a bio-imaging analyzer, FLA-7000, and the radioactivity of the full-length products on the same gel was analyzed by autoradiography. The fold amplification of each DNA fragment is summarized in Table S3. (c) Sequencing of the 40-cycle amplified DNA fragments, in the presence of dPa′TP (2 μM) or ddPa′TP (50 μM). The arrows indicate the unnatural base position.
The high selectivity of the Ds–Px pairing in PCR was also verified by sequencing of the amplified DNA. Even with only 15 zeptomol of the input DNA, the amplified DNAs were detected by 40 cycles of PCR using NH2-hx-dPxTP and dDsTP, in which the Ds–Px pair was conserved (Figure 3c). We performed two type dideoxy sequencing reactions, in the presence of either dPa′TP or dideoxy-Pa′TP (ddPa′TP). In the sequencing with dPa′TP, the Ds position in the templates was detected as a gap on the sequencing peak patterns. In the sequencing with ddPa′TP but without dPa′TP, the sequencing reaction terminated at the Ds position in the templates by the ddPa′TP incorporation opposite Ds, and the subsequent peaks disappeared on the sequencing peak patterns (Figure 3c). If the Ds–Px pair was replaced with the natural base pairs to some degree, then the subsequent peak heights following the Ds position would be increased significantly above background, depending on the degree of the mutation in PCR, in the sequencing with ddPa′TP. In fact, the sequencing patterns of the amplified DNAs after 40-cycles of PCR were nearly identical to those of the original DNA, indicating the faithful Ds–Px pairing in the PCR amplification. The replacement rates from the unnatural to the natural base pairs at the Ds–Px sites were <1% (>99.9% fidelity-per-cycle), as determined by comparison to the authentic sequencing patterns of control DNAs (12,19).
The amplification efficiency of each sequence was assessed quantitatively by the radioactivity of each amplified product (Figure 3b and Supplementary Table 3). The Ds–Px pair system exhibited high amplification efficiency, even with FAM-hx-dPxTP: 15 fmol of DNA S1–S8 were 300-fold amplified by 15 cycles of PCR, and 0.15 amol of DNA (DNA S2, S4 and S7) were 107-fold amplified by 30 cycles of PCR. These PCR amplification efficiencies involving the Ds–Px pair were only 2-fold lower than that involving only the natural base pairs (DNA Cont). Besides these sequences, we found that a 5′-CACPxGTA-3′/3′-GTGDsCAT-5′ sequence (DNA N12) also exhibited high efficiency and selectivity. Thus, the efficient sequences for PCR can be summarized as 5′-CRMPxGNR-3′/3′-GYKDsCNY-5′ (where R = A or G, M = A or C, N = A, G, C or T, Y = C or T and K = G or T).
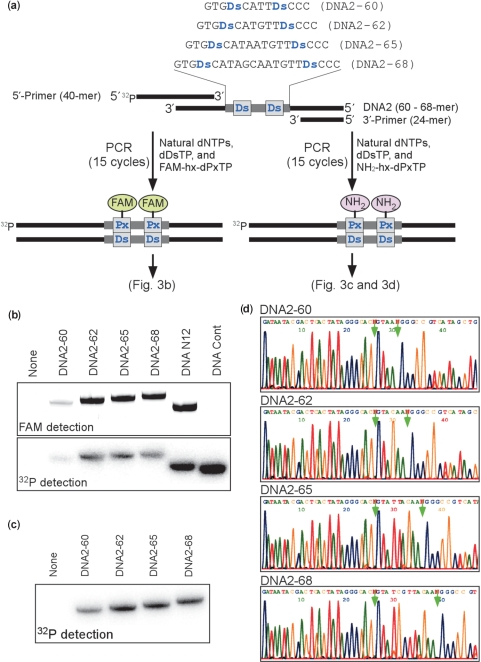
Using the efficient sequences, we next performed PCR with DNA templates containing two Ds bases, which were separated by 4, 6, 9 or 12 natural bases (Figure 4a and Supplementary Table 2). Each DNA template was amplified by 15 cycles of PCR with dDsTP and FAM-hx-dPxTP or with dDsTP and NH2-hx-dPxTP. The DNA templates with at least six natural bases inserted between the two Ds bases exhibited highly efficient amplification, and were 200- and 500-fold amplified by the incorporation of FAM-hx-Px (Figure 4b) and NH2-hx-Px (Figure 4c), respectively. DNA Cont, which lacks Ds bases, was 670-fold amplified in the presence of FAM-hx-dPxTP and dDsTP, but the FAM-hx-Px incorporation was hardly observed in the amplified DNA (Figure 4b, right-side line). The sequences of the amplified DNAs were confirmed by sequencing in the presence of dPa′TP (Figure 4d).
Figure 4.
PCR amplification of DNA fragments containing two Ds bases (60-, 62-, 65- or 68-mer). (a) Scheme for the PCR amplification in the presence of FAM-hx-dPxTP or NH2-hx-dPxTP with DeepVent DNA Pol. The amplified DNA fragments containing FAM-hx-Px were detected by denaturing PAGE. (b) Polyacrylamide-gel analysis of the PCR products amplified by 15 cycles of PCR with the 32P-labeled 5′-primer, in the presence of FAM-hx-dPxTP and dDsTP. The fluorescence of the FAM-labeled full-length products on the gel was detected with an FLA-7000 bio-imager, and the radioactivity of the full-length products on the same gel was analyzed by autoradiography. (c) Polyacrylamide-gel analysis of the PCR products amplified by 15 cycles of PCR with the 32P-labeled 5′-primer, in the presence of NH2-hx-dPxTP and dDsTP. (d) Sequencing of the 15-cycle amplified DNA fragments, in the presence of dPa′TP (50 μM). The PCR products amplified in the presence of NH2-hx-dPxTP and dDsTP were used for sequencing of the Ds-strands. The arrows indicate the unnatural base position.
PCR amplification in the presence of DNA fragments containing only natural nucleotides
The site-specific incorporation of modified Px bases into DNA by PCR would be a powerful tool for DNA-based biotechnology. We developed a highly sensitive DNA amplification detection system, by reducing the background caused by contamination with foreign DNAs. In this system, a target DNA containing Ds was amplified with FAM-hx-dPxTP and dDsTP in the presence of other foreign DNAs consisting of only the natural bases, and then the amplified target DNA was isolated and detected by using anti-fluorescein IgG. To evaluate this system, we examined two types of reactions. The first was DNA S2 mixed with an equivalent amount of three DNA fragments with the same primer sequences as DNA S2, but different 11-base sequences without Ds (Figure 5), and the second was DNA S2 mixed with a 107-fold excess amount of a DNA mixture composed of random 100-nt sequences consisting of only the natural bases (Figure 6).
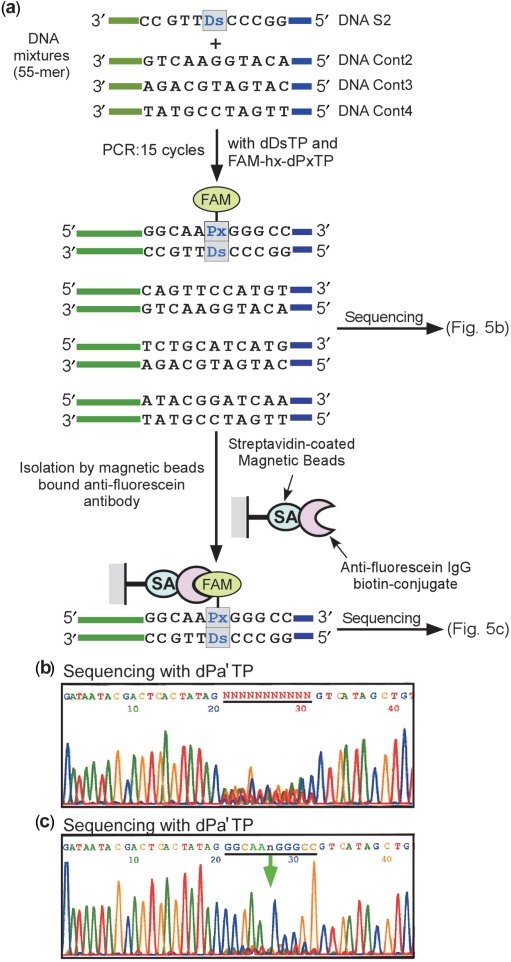
Figure 5.
Detection of a target DNA molecule within foreign DNA fragments with the same primer sequences. (a) Overview of the purification of the DNA fragments containing Ds. After 15 cycles of PCR amplification of four different DNA fragments (55-mer, 0.15 nM each; one fragment contains one Ds base and the others contain no Ds bases) in the presence of FAM-hx-dPxTP and dDsTP, the amplified products were sequenced both directly and after purification using an anti-fluorescein antibody bound to magnetic beads. (b) Direct sequencing of the 15-cycle amplified DNA mixture, after removal of primers and dNTPs by gel purification, in the presence of dPa′TP (2 μM). (c) Sequencing of the DNA mixtures after purification of the FAM-labeled PCR products, in the presence of dPa′TP (2 μM).
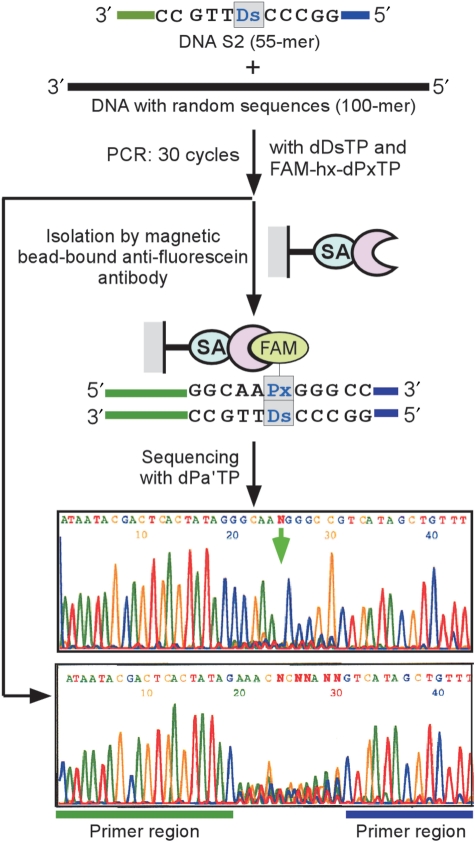
Figure 6.
Detection of a target DNA molecule within foreign DNA fragments with random sequences. Overview of the detection of the target DNA (DNA S2, 55-mer) containing Ds. A Ds-containing DNA fragment (DNA S2, 55-mer, 6 fM) and foreign DNA fragments with random sequences (100-mer, 60 nM) were amplified by 30 cycles of PCR in the presence of FAM-hx-dPxTP and dDsTP. After the amplification, the FAM-labeled PCR products were isolated by the magnetic bead-bound anti-fluorescein antibody, and then were sequenced in the presence of dPa′TP (2 μM). Prior to the antibody isolation, the amplified DNA fragments corresponding to the 55-mer were purified by gel electrophoresis and were also sequenced.
In the first case, the DNA mixtures were amplified by 15 cycles of PCR with dDsTP and FAM-hx-dPxTP (Figure 5). The direct sequencing of the amplified products yielded a random base pattern at the 11-base region (Figure 5b) because of the foreign DNAs. Then, by using anti-fluorescein IgG bound to streptavidin-coated magnetic beads, the target DNA containing the Ds–Px pair was isolated. The sequencing of the amplified products after the isolation step revealed a clear pattern corresponding to that of DNA S2 (Figure 5c). In the second case, the mixtures were amplified by 30 cycles of PCR with dDsTP and FAM-hx-dPxTP (Figure 6). The amplified DNAs containing the Ds–Px pair were isolated with the IgG-streptavidin beads, and the DNA S2 sequence was clearly identified by sequencing. Without the isolation step, the amplified DNAs generated a randomized sequence pattern, even after their gel purification. Thus, the functionalization PCR by FAM-hx-Px incorporation into DNA is useful for detecting amplified target DNA with high signal-to-noise ratios, by reducing the background and the false positives.
DISCUSSION
Here, we have described an efficient, unnatural base pair system, suitable for practical use towards the expansion of the genetic alphabet. The hydrophobic Ds–Px pair can function in PCR with a 3′→5′ exonuclease-proficient DNA Pol. Increasing the hydrophobicity and adjusting the shape and electrostatic complementarity of the unnatural bases achieved high fidelity of the cognate pairings, thus bypassing the need for any modified triphosphates in PCR. In addition to the intrinsic Ds–Px pairing selectivity, the sequences around the unnatural bases could be optimized for high efficiency and selectivity of PCR with this system. We found that the adjacent natural base sequences of the unnatural base pair affect both the efficiency and selectivity of PCR amplification, and the efficient sequences were identified. Interestingly, not only the base positions adjacent to the unnatural base but also the second and third base positions influenced the amplification efficiency. This might be caused by the fitting of each sequence with the polymerases during PCR elongation, including the proof-reading by the exonuclease activity of the polymerases (28,29). By using the sequence information, researchers can now construct efficient sequences involving the unnatural base pair for functionalization PCR of interest. Our results showed that the sequence dependency is influenced by the structures of the unnatural base moieties of Ds and Px, rather than by those of the functional groups, such as FAM and amino, attached to Px. Thus, a wide variety of functional groups that are linked to Px could be utilized with this system.
The Ds–Px system will provide useful tools for a wide range of DNA/RNA-based technologies, such as sensitization for immuno-PCR assays (30,31), DNA steganography (32), DNA arrays and multiplexes (7), DNA-based nanomaterials and in vitro selection and evolution methods (26,27). For immuno-PCR assays and DNA steganography, the site-specific incorporation of FAM-hx-Px into amplified DNA can significantly reduce the background signals caused by contamination with foreign DNAs, by isolating the target FAM-containing DNA with anti-fluorescein IgG beads, as we demonstrated here. In addition, the unnatural bases increase the information capacity of the target DNA. Although there are some biases in the DNA sequences around the Ds–Px pair for pursuing high amplification efficiency, other sequences are also permissible for PCR involving the Ds–Px pair. Furthermore, the Ds–Px pair can be applied to RNA-based technologies by combining it with other unnatural base pairs, such as the Ds–Pa pair, which exhibit high efficiency and selectivity for the site-specific incorporation of functional groups into RNA by transcription (12).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by a Grant-in-Aid for Scientific Research (KAKENHI 19201046 to I.H., 18710197 to M.K.) from the Ministry of Education, Culture, Sports, Science and Technology and by the Targeted Proteins Research Program and RIKEN Structural Genomics/Proteomics Initiative, the National Project on Protein Structural and Functional Analyses, Ministry of Education, Culture, Sports, Science and Technology of Japan. Funding for open access charge: KAKENHI 19201046.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Akira Sato for synthesizing nucleoside derivatives.
REFERENCES
- 1.Bain JD, Switzer C, Chamberlin AR, Benner SA. Ribosome-mediated incorporation of a nonstandard amino acid into a peptide through expansion of the genetic code. Nature. 1992;356:537–539. doi: 10.1038/356537a0. [DOI] [PubMed] [Google Scholar]
- 2.Benner SA, Burgstaller P, Battersby TR, Jurczyk S. Did the RNA world exploit an expanded genetic alphabet? In: Gesteland RF, Cech T, Atkins JF, editors. The RNA World. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 163–181. [Google Scholar]
- 3.Ogawa AK, Wu T, McMinn DL, Liu J, Schultz PG, Romesberg FE. Efforts toward the expansion of the genetic alphabet: information storage and replication with unnatural hydrophobic base pairs. J. Am. Chem. Soc. 2000;122:3274–3287. [Google Scholar]
- 4.Hirao I, Ohtsuki T, Fujiwara T, Mitsui T, Yokogawa T, Okuni T, Nakayama H, Takio K, Yabuki T, Kigawa T, et al. An unnatural base pair for incorporating amino acid analogs into proteins. Nat. Biotechnol. 2002;20:177–182. doi: 10.1038/nbt0202-177. [DOI] [PubMed] [Google Scholar]
- 5.Delaney JC, Henderson PT, Helquist SA, Morales JC, Essigmann JM, Kool ET. High-fidelity in vivo replication of DNA base shape mimics without Watson-Crick hydrogen bonds. Proc. Natl Acad. Sci. USA. 2003;100:4469–4473. doi: 10.1073/pnas.0837277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirao I. Placing extra components into RNA by specific transcription using unnatural base pair systems. BioTechniques. 2006;40:711–717. doi: 10.2144/000112187. [DOI] [PubMed] [Google Scholar]
- 7.Johnson SC, Marshall DJ, Harms G, Miller CM, Sherrill CB, Beaty EL, Lederer SA, Roesch EB, Madsen G, Hoffman GL, et al. Multiplexed genetic analysis using an expanded genetic alphabet. Clin. Chem. 2004;50:2019–2027. doi: 10.1373/clinchem.2004.034330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Z, Hutter D, Sheng P, Sismour M, Benner SA. Artificially expanded genetic information system: a new base pair with an alternative hydrogen bonding pattern. Nucleic Acids Res. 2006;34:6095–6101. doi: 10.1093/nar/gkl633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tor Y, Dervan PB. Site-specific enzymatic incorporation of an unnatural base, N6-(6-aminohexyl)isoguanosine, into RNA. J. Am. Chem. Soc. 1993;115:4461–4467. [Google Scholar]
- 10.Moriyama K, Kimoto M, Mitsui T, Yokoyama S, Hirao I. Site-specific biotinylation of RNA molecules by transcription using unnatural base pairs. Nucleic Acids Res. 2005;33:e129. doi: 10.1093/nar/gni128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawai R, Kimoto M, Ikeda S, Mitsui T, Endo M, Yokoyama S, Hirao I. Site-specific fluorescent labeling of RNA molecules by specific transcription using unnatural base pairs. J. Am. Chem. Soc. 2005;127:17286–17295. doi: 10.1021/ja0542946. [DOI] [PubMed] [Google Scholar]
- 12.Hirao I, Kimoto M, Mitsui T, Fujiwara T, Kawai R, Sato A, Harada Y, Yokoyama S. An unnatural hydrophobic base pair system: site-specific incorporation of nucleotide analogs into DNA and RNA. Nat. Methods. 2006;3:729–735. doi: 10.1038/nmeth915. [DOI] [PubMed] [Google Scholar]
- 13.Piccirilli JA, Krauch T, Moroney SE, Benner SA. Enzymatic incorporation of a new base pair into DNA and RNA extends the genetic alphabet. Nature. 1990;343:33–37. doi: 10.1038/343033a0. [DOI] [PubMed] [Google Scholar]
- 14.Kool ET. Synthetically modified DNAs as substrates for polymerases. Curr. Opin. Chem. Biol. 2000;4:602–608. doi: 10.1016/s1367-5931(00)00141-1. [DOI] [PubMed] [Google Scholar]
- 15.Henry AA, Romesberg FE. Beyond A, C, G and T: augmenting nature's alphabet. Curr. Opin. Chem. Biol. 2003;7:727–733. doi: 10.1016/j.cbpa.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 16.Hirao I. Unnatural base pair systems for DNA/RNA-based biotechnology. Curr. Opin. Chem. Biol. 2006;10:622–627. doi: 10.1016/j.cbpa.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 17.Sismour AM, Benner SA. The use of thymidine analogs to improve the replication of an extra DNA base pair: a synthetic biological system. Nucleic Acids Res. 2005;33:5640–5646. doi: 10.1093/nar/gki873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Z, Sismour AM, Sheng P, Puskar NL, Benner SA. Enzymatic incorporation of a third nucleobase pair. Nucleic Acids Res. 2007;35:4238–4249. doi: 10.1093/nar/gkm395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirao I, Mitsui T, Kimoto M, Yokoyama S. An efficient unnatural base pair for PCR amplification. J. Am. Chem. Soc. 2007;129:15549–15555. doi: 10.1021/ja073830m. [DOI] [PubMed] [Google Scholar]
- 20.McMinn DL, Ogawa AK, Wu Y, Liu J, Schultz PG, Romesberg FE. Efforts toward expansion of the genetic alphabet: DNA polymerase recognition of a highly stable, self-pairing hydrophobic base. J. Am. Chem. Soc. 1999;121:11585–11586. [Google Scholar]
- 21.Ollis DL, Brick P, Hamlin R, Xuong NG, Steitz TA. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature. 1985;313:762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- 22.Doublié S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 23.Kiefer JR, Mao C, Braman JC, Beese LS. Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal. Nature. 1998;391:304–307. doi: 10.1038/34693. [DOI] [PubMed] [Google Scholar]
- 24.Goodman MF, Creighton S, Bloom LB, Petruska J. Biochemical basis of DNA replication fidelity. Crit. Rev. Biochem. Mol. Biol. 1993;28:83–126. doi: 10.3109/10409239309086792. [DOI] [PubMed] [Google Scholar]
- 25.Kimoto M, Yokoyama S, Hirao I. A quantitative, non-radioactive single-nucleotide insertion assay for analysis of DNA replication fidelity by using an automated DNA sequencer. Biotechnol. Lett. 2004;26:999–1005. doi: 10.1023/b:bile.0000030047.32578.82. [DOI] [PubMed] [Google Scholar]
- 26.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 27.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 28.Kunkel TA, Wilson SH. DNA polymerases on the move. Nat. Struct. Biol. 1998;5:95–99. doi: 10.1038/nsb0298-95. [DOI] [PubMed] [Google Scholar]
- 29.Morales JC, Kool ET. Functional hydrogen-bonding map of the minor groove binding tracks of six DNA polymerases. Biochemistry. 2000;39:12979–12988. doi: 10.1021/bi001578o. [DOI] [PubMed] [Google Scholar]
- 30.Hendrickson ER, Truby TMH, Joerger RD, Majarian WR, Ebersole RC. High sensitivity multianalyte immunoassay using covalent DNA-labeled antibodies and polymerase chain reaction. Nucleic Acids Res. 1995;23:522–529. doi: 10.1093/nar/23.3.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niemeyer CM, Adler M, Wacker R. Immuno-PCR: high sensitivity detection of proteins by nucleic acid amplification. Trends Biotechnol. 2005;23:208–216. doi: 10.1016/j.tibtech.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 32.Clelland CT, Risca V, Bancroft C. Hiding messages in DNA microdots. Nature. 1999;399:533–534. doi: 10.1038/21092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.