Abstract
We present here the results of a systematic bioinformatics analysis of control (C) proteins, a class of DNA-binding regulators that control time-delayed transcription of their own genes as well as restriction endonuclease genes in many type II restriction-modification systems. More than 290 C protein homologs were identified and DNA-binding sites for ∼70% of new and previously known C proteins were predicted by a combination of phylogenetic footprinting and motif searches in DNA upstream of C protein genes. Additional analysis revealed that a large proportion of C protein genes are translated from leaderless RNA, which may contribute to time-delayed nature of genetic switches operated by these proteins. Analysis of genetic contexts of newly identified C protein genes revealed that they are not exclusively associated with restriction-modification genes; numerous instances of associations with genes originating from mobile genetic elements were observed. These instances might be vestiges of ancient horizontal transfers and indicate that during evolution ancestral restriction-modification system genes were the sites of mobile elements insertions.
INTRODUCTION
Type II restriction–modification (R–M) systems are comprised of (i) a restriction endonuclease that recognizes a specific DNA sequence and introduces double-stranded breaks at or around the recognition site and (ii) a methyltransferase (methylase) that recognizes the same DNA sequence and methylates it first on one of DNA strands to produce hemimethylated DNA, and then on the other strand to produce fully methylated DNA. Methylation protects DNA from cleavage by the endonuclease (1,2).
In several cases that have been studied experimentally, bacterial cells carrying R–M genes become resistant to infection by bacteriophages containing unmethylated (unmodified) DNA, which may explain their wide dissemination. R–M systems are often carried on mobile genetic elements capable of horizontal spread between different bacterial species (3). Premature production of endonuclease upon the entry of a genetic element carrying R–M system genes into a naïve host can lead to host DNA degradation and death of the host. To minimize the likelihood of such an outcome, which would also destroy the R–M system and the mobile element that carries it, R–M systems evolved special mechanisms to coordinate expression of their genes, ensuring that endonuclease expression is activated only after the host DNA is fully methylated by the methylase. Several distinct mechanisms of such activation have been described (4).
Eight R–M systems—AhdI (5,6), BamHI (7), BglII (8), Eco72I (9), EcoRV (10), Esp1396I (11), PvuII (12) and SmaI (13)—have been experimentally shown to rely on specialized control (C) proteins (7,12) for coordinated expression of their genes. All C proteins are related through common ancestry and are also distantly related to phage helix-turn-helix DNA-binding transcription factors, including the well-studied phage λ repressor. The structures of three C proteins (C.BclI, C.AhdI and C.Esp1396I) that have been studied by crystallography (14–16) reveal that these proteins form dimers and that each monomer is similar to the DNA-binding domain of the lambda cI repressor, belonging to the Xre family of transcription factors (15).
Genes coding for C proteins are often located upstream of, and partially overlap with, the endonuclease (R) gene (17), forming a single operon. Upstream of and partially overlapping with the CR operon promoter, two C protein-binding sites are located (17). When a C protein dimer binds to the high-affinity promoter-distal site, transcription is activated, leading to increased C protein (and endonuclease) gene expression (6,10). The exact mechanism(s) of activation is not known and may vary in different R–M systems. In the few cases that have been studied, the promoter-distal C protein-binding site is located immediately upstream of the −35 promoter element of the CR operon promoter (6,10,18). Thus, C protein-dependent activation may involve protein-protein contacts between the C protein and the RNA polymerase σ subunit region 4, which specifically recognizes the –35 promoter element (19). C protein binding to the weaker, promoter-proximal site, occludes the –35 element of the CR operon promoter and inhibits transcription (6,15,20), most likely by excluding the RNA polymerase σ subunit region 4 from the –35 element. The dual (activation and repression), concentration-dependent mode of transcription regulation of the CR promoter by C protein ensures a delayed appearance of the endonuclease activity during establishment of C protein-dependent R–M systems in a naïve host and allows to maintain constant steady-state levels of endonuclease during the stable maintenance of R–M system in the host [see, for example, ref. (6) for kinetic modeling of the process]. The high cooperativity of C protein dimer interactions with DNA observed in at least some studied systems (5,6) affords sharp regulatory responses of C protein-dependent autoregulatory loops.
In this work, we used a bioinformatics approach to answer the following questions. First, we wanted to systematically identify genes coding for C protein homologs. Second, we wanted to predict DNA-binding sites of bioinformatically identified C proteins. Lastly, we sought to determine if proteins homologous to C proteins from R–M systems are specific to such systems or are also found in other genetic contexts.
METHODS
The Rebase database contains 48 C protein sequences. One of the proteins (C.MjaVP) is more than twice as long as the rest of the C proteins. Another protein (C.AmaFACHORFAP) resulted from a formal translation of a pseudogene. These two proteins were excluded from the analysis. The remaining 46 C protein sequences were used as queries in the BLAST (21) search against the non-redundant nucleotide database of GenBank (22) (tblastn, threshold 1e–05). This search yielded 245 unique hits, which were considered as genes encoding putative C proteins, or, more exactly, C protein-family regulators (although, naturally, in the absence of experimental data even this general functional assignment is only preliminary). Starts of the genes were manually corrected using the standard bacterial ORF analysis rules and the fact that the average C protein length is about 70 amino acids (aa). Multiple alignment of all 291 proteins (46 proteins from Rebase and 245 hits generated by the BLAST search) was built using the muscle program (23) and the unrooted maximum likelihood tree with molecular clock was constructed using the proml procedure from the PHYLIP package (24). Both programs were run with the default parameters.
The tree was split into several large subfamilies which were analyzed independently. For each group of closely related proteins, short (100 bp) regions upstream of the corresponding genes were aligned using muscle with the default parameters. The following procedure was used to extend this alignment by including upstream regions of more distantly related C protein genes. Genes were added to the alignment one by one in the order dictated by the tree, and at each step the upstream regions were re-aligned. When the alignment started to degrade, such ‘extension’ process was stopped and putative binding motifs were manually predicted by the analysis of the remaining conservation islands. Further, each remaining upstream region was compared with its nearest neighbors on the tree, for which the binding sites had already been predicted. The multiple alignment, which included this remaining region and its tree neighbors, was forced to align the predicted sites and again was analyzed manually. If the conserved island covering the putative site did not deteriorate upon inclusion of the new sequence, the latter was also predicted to be a binding site.
To account for a possibility that some sites were missed because of mis-annotation of gene starts or positioning of the site outside of the 100 bp upstream region, all C protein-family genes for which the procedure described above failed to reveal a putative binding site were analyzed further. First, hmmer (http://hmmer.wulst.edu) profiles of candidate sites were built for each constructed alignment (hmmbuild –g, nucleotide mode). Second, the hmmsearch procedure was applied and these profiles were used to scan regions from –100 bp to +50 bp relative to translation starts of putative C protein genes without predicted sites. The best candidates were added to the set of predicted binding sites. However, this procedure resulted in few additional binding sites, showing that the overall approach is a robust one.
The whole set of putative binding sites was split into clusters, which are further referred to as motifs, using the ClusterTree-RS procedure (25). The procedure yielded 10 stable motifs, which contained 181 (90%) of 201 predicted sites. Since ClusterTree-RS generates nested clusters, we were able to subdivide motifs 2 and 6 into motifs 2, 2b, 2c and 6 and 6b, respectively. The resulting motifs 2 and 6 contain the most conserved members of the original motifs, while 2b, 2c and 6b contain sequences more distantly related to motif 2 and motif 6 consensus sequences.
Genome loci containing C protein-family genes with predicted binding sites were studied in more detail. These loci were defined formally as genomic regions from 3000 bp upstream to 3000 bp downstream of each C protein-family gene. Candidate genes were defined as ORFs longer than 150 codons with potential start codons. These genes were translated and compared to the non-redundant protein database (blastp, threshold 1e–06), and the pfam-a seed database (26) (hmmer, global mode, no calibrate-mode, threshold –E 0.01 –Z 1).
All hits were classified into three functional categories: ‘phage-related’, ‘R–M-related’ and ‘the rest’. To do that, a list of relevant pfam families and words in protein annotations was compiled. This list was used to scan the pfam assignments and lists of BLAST hits for each ORF, followed by manual verification of the ORF status. A locus containing at least one phage-related or R–M-related ORF was labeled correspondingly.
To identify hypothetical genes consistently appearing in the vicinity of C protein-family genes, all ORFs were further clustered by similarity using a two-step procedure. At the first step, groups of highly similar ORFs were identified using the standard blastclust procedure (21) (length coverage L = 0.50, identity percentage S = 0.90). Sixty groups which contained three or more sequences were collapsed and only one representative of each group was used for further clustering. At the second step, total pairwise BLAST search (blastp, threshold 1e–10) was performed and clusters were determined by a single linkage procedure. Proteins from the resulting 38 clusters were aligned using muscle (default parameters). However, since the analysis did not reveal any genes significantly associated with the candidate C-protein genes, the clusters were not considered further (data not shown).
At the end, each candidate binding site was assigned the following data: its sequence; site motif (if any); candidate C protein sequence; similar Rebase C proteins that contained this protein in the BLAST similarity search output, see above; map of the genomic locus with all ORFs; list of pfam families which matched the orfs; list of BLAST hits for each ORF. These data were collected into a specially developed database that can be accessed online (http://iitp.bioinf.fbb.msu.ru/vsorokin).
Logos were produced using the weblogo 2.8 package (27). The tree was visualized using the web-based tool iTOL (http://itol.embl.de/).
RESULTS
Identification of new C protein family members
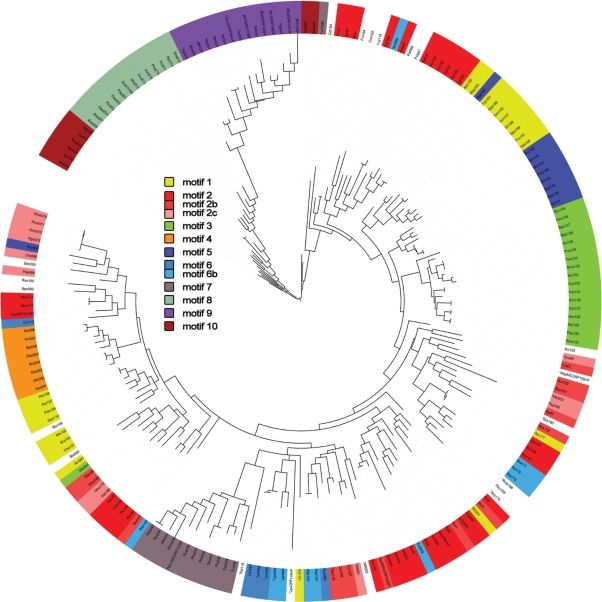
Using 46 annotated C proteins from Rebase as a starting point for database similarity search with BLAST, we obtained 245 additional putative members of the C protein family. The parameters of the search, described in the Materials and methods section, were set such that distant relatives of known C proteins, such as phage repressors, were not retrieved by the search. To identify closely related C protein sequences, an unrooted likelihood tree of all 291 (46 annotated sequences + 245 newly found sequences) members of the family was constructed. A slightly smaller variant of this tree, containing proteins whose binding motifs could be identified (see below), is shown in Figure 1. A version of the tree with bootstrap values resulting from 100 pseudoreplications is available as Supplementary Data. It should be noted that the tree reflects protein, rather than species, evolution as in large parts it does not match accepted taxonomy divisions. This likely indicates that C protein genes are subject to extensive horizontal transfer, an expected result, given the biological function and genetic contexts of known C proteins.
Figure 1.
Maximum likelihood tree built of REBASE and newly discovered putative C proteins. Color indicates C proteins whose predicted binding sites fall into distinct motifs (1 through 10). The subdivided motifs (2, 2b, 2c and 6, 6b) are marked with similar, but not identical colors. The asterisk preceding the protein id indicates a relatively lower level of prediction confidence.
Identification of putative C protein DNA-binding sites
On the basis of published information about transcription regulation by a few C proteins that have been studied experimentally, we expected that C protein-binding sites would be located in close vicinity to translation start points of C protein genes. We also hypothesized that the binding sites for closely related C proteins will be similar. The following unbiased iterative procedure was used to search for evolutionary conservation of DNA sequences upstream of C protein genes (see also Materials and methods section). The tree of C proteins was split into several large branches, which were analyzed independently. For every branch, upstream (100 bp upstream of manually curated annotated translation start codon) DNA regions for most closely related annotated or putative C protein genes were aligned using muscle. Next, the alignment was extended by inclusion of upstream regions of more distantly related C protein genes from the same branch. At each step, the upstream regions were realigned. When the alignment started to degrade, the ‘extension’ process was terminated. In this way, ∼60% of sequences from each branch of the tree could be aligned. Putative binding motifs were next predicted by visual analysis of conservation islands in the alignments. It is noteworthy that in all cases, only one continuous conserved motif-like element was detected. Some additional sequences, which did not ‘naturally’ fit in the alignments, were subsequently added manually using conserved consensus sequences derived from the iterative procedure. In total, conserved motifs were detected upstream of 201 of the 291 C protein family genes (69.1%). An alignment of the proteins with predicted binding sites is available as Supplementary Data. Of the 46 C protein genes listed in Rebase, 32 (69.5%) contained conserved upstream motifs. These included eight R–M systems where C protein-binding sites have been identified experimentally (C.BamHI, C.BglII, C.PvuII, C.SmaI, C.AhdI, C.MunI, C.EcoRV, C.EcoO109I), as well as 24 additional C protein-dependent R–M systems (C.Pde1222ORF1578P, C.MspMCORF1281P, C.BcnI, C.SptAI, C.SbaI, C.SonORF4P, C.NmeSI, C.VeiORF3519P, C.BstLVI, C.Sse9I, C.Lci22RP, C.SspMR4ORF3202P, C.SgrAI, C.Csp231I, C.BfrYORF1158P, C.BfrLV23P, C.BfrYORF1980P, C.BfaSORF1835P, C.ChuAORF2941P, C.BfaSORF1077P, C.EcoT38I, C.LlaDI, C.SnaBI, C.LgaORF1464); putative C protein-binding sites in some of these systems were also identified earlier by Naderer et al. (28). Most importantly, for eight of the 46 Rebase-listed C protein genes for which C protein-binding sites are known, the conserved upstream motifs coincided with experimentally determined binding sites. Since the binding site information was ignored during our analysis, we conclude that identification of upstream conserved sequences leads to identification of C protein-binding sites with high confidence. The fact that the search procedure did not result in identification of other regulatory elements likely to be present upstream of C protein genes, such as C protein gene promoters, which in cases that have been studied in experiment overlap with C protein-binding sites (6,10,11,18), indicates that the evolutionary conservation of promoter elements is significantly lower than that of the C protein-binding sites (see also below).
The structure of putative C protein-binding sites
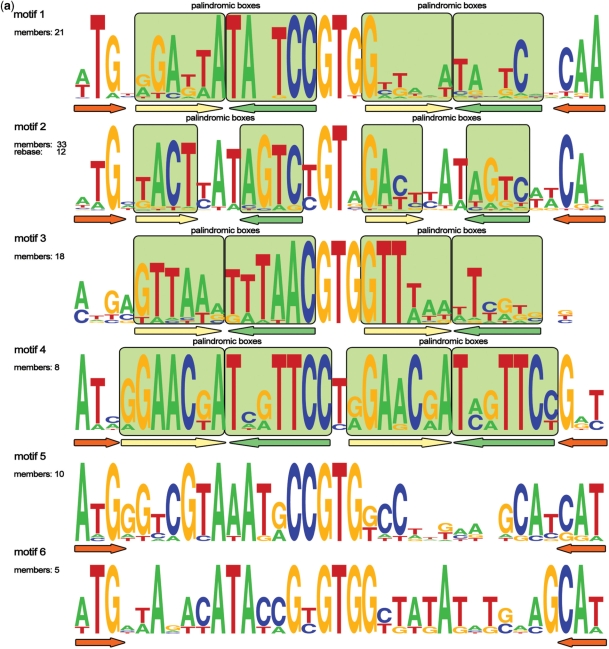
Previous limited analysis of C protein-binding sites by Blumenthal and colleagues identified three types of sites (20). The first type contained six distinct sites including the C.PvuII site and was called non-palindromic; two remaining types were represented by only one site each (of C.EcoRV and C.EcoO109I) and were considered palindromic. A collection of putative C protein-binding sites revealed by our analysis allowed us to extend the previous classification and to identify new motifs (Table 1, Figure 2a–c).
Table 1.
Motifs, the number of candidate C proteins and the gene content of respective loci
| Motifs | Total members | Rebase members | Motif type | Number of loci with RM-related orfs | Number of loci with phage-related orfs |
|---|---|---|---|---|---|
| Motif 1 | 21 | 0 | C.PvuII-like (double-box motif) | 7 | 15 |
| Motif 2 | 33 | 12 | C.PvuII-like (double-box motif) | 21 | 23 |
| Motif 2b | 14 | 6 | C.PvuII-like (double-box motif) | 8 | 10 |
| Motif 2c | 12 | 1 | C.PvuII-like (double-box motif) | 2 | 6 |
| Motif 3 | 18 | 0 | C.PvuII-like (double-box motif) | 0 | 18 |
| Motif 4 | 8 | 0 | C.PvuII-like (double-box motif) | 3 | 7 |
| Motif 5 | 10 | 0 | C.PvuII-like (single-box motif) | 3 | 9 |
| Motif 6 | 5 | 0 | C.PvuII-like (single-box motif) | 0 | 2 |
| Motif 6b | 10 | 0 | C.PvuII-like (single-box motif) | 1 | 7 |
| Motif 7 | 13 | 3 | C.EcoRV-like (palindromic motif) | 8 | 11 |
| Motif 8 | 14 | 2 | C.EcoO109I-like (palindromic motif) | 6 | 7 |
| Motif 9 | 15 | 6 | new (non-palindromic motif) | 7 | 11 |
| Motif 10 | 8 | 0 | new (palindromic motif) | 1 | 4 |
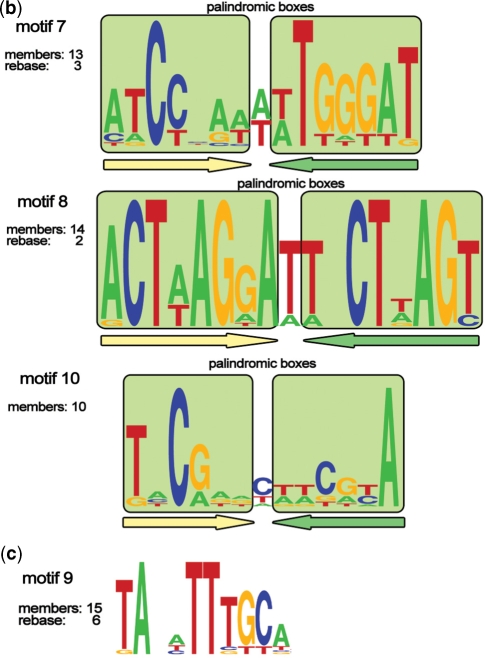
Figure 2.
(a) Logos of C.PvuII-like (1–6) motifs. The total number of members and the number of REBASE members (if any) are indicated for every motif. Paired palindromic boxes (consensus sequences) are marked with light green squares. Palindromic elements of motifs’ architecture are underlined with colored arrows. Conserved trinucleotides found at the outside of the motifs are underlined with orange arrows. (b) Logos of palindromic (7, 8 and 10) motifs. The total number of members and the number of REBASE members (if any) are indicated for every motif. Paired palindromic boxes (consensus sequences) are marked with light green squares. Palindromic elements of motifs’ architecture are underlined with colored arrows. (c) Logo of new motif 9. The total number of members and the number of REBASE members are shown.
The C.PvuII-like sites were assigned to six related but clearly distinct motifs (motifs 1–6, Figure 2a). All these motifs have the same length (35 bp) and share a common architecture, which consists of three conserved bases at the outside of the motif and some almost invariant positions in the core of the motif. The three bases at the outside boundary of the motif are often complementary. A typical motif consists of two copies of the same palindromic consensus, which we term ‘operator’, with the 5′ copy (with respect to C protein gene translation start point) being much closer to the consensus than the 3′ copy. The two copies are separated by a highly conserved non-symmetrical GTG sequence. Thus, the overall typical motif architecture is Z-X-N-X*-GTG-x-n-x*-Z*, where X denotes internal boxes forming a palindrome, N denotes internal, non-palindromic positions in operators, Z denotes external three-nucleotide boxes and asterisks denote complementary boxes; uppercase denotes highly conserved nucleotides or boxes, while lower-case denotes weakly conserved boxes or nucleotides. It should be noted, however, that some individual conserved putative C protein-binding sites within a group do not match this idealized scheme. Moreover, distinct variations in symmetry patterns of different motifs exist. For instance, in motifs 1, 3 and 5, the overall symmetry is odd (the center of symmetry at the T of the central GTG) while the individual operators have the even symmetry (X-X*) (Figure 2a). On the contrary, the overall symmetry of motif 2 is even (with symmetry center between the G and the T of the central GT), while the operators are odd palindromes (Figure 2a). It is worth mentioning that a sequence identical to motif 2 was previously identified by Vijesurier et al. (29) The outer elements (TG/CA) in a motif 2 sequence from the AhdI system were recently shown to be contacted by the cognate C protein (16). In motif 4, the symmetry of operators extends and includes the internal conserved trinucleotide, in this case, CTG (Figure 2a); resulting in the richest symmetry pattern for putative binding sites identified here, which can be codified as Z3-X6-X6*-T-X6-X6*-Z3*, where numbers denote lengths of respective elements. In motifs 5 and 6, no major symmetry pattern could be observed besides the palindromic ATG-CAT trinucleotides at the outside flanks of the motif. Together, these observations suggest that molecular details of C protein interactions with different motifs are subject to considerable variation.
Our analysis extends the total number of palindromic C.EcoRV-type and C.EcoO109I-type binding sites to 13 (for C.EcoRV-type sites) and 14 (for C.EcoO109I-type sites). The C.EcoRV-type and C.EcoO109I-type binding sites form motifs 7 and 8, respectively. We also identified a new motif of a similar palindromic structural type (motif 10, Figure 2b). The newly identified motif 9 (Figure 2c) also may be tentatively assigned to this type, although its properties are rather unusual. It is short and lacks any discernible symmetry; in particular, it does not have complementary terminal trinucleotides. Thus it might be a case of a false positive. On the other hand, motif 9 is the only highly conserved sequence found upstream of 15 C protein homologs, of which six are annotated in Rebase. The branch of C proteins corresponding to motif 9 is the most distant one in the phylogenetic tree (Figure 1), which may explain the unique properties of the putative binding site. Still, given the uncertainty about this motif, we exclude it from further consideration.
The structure of genomic loci containing C protein family genes
We analyzed 6 kbp genomic loci centered at putative C protein genes. The size of the window was arbitrarily selected; however, type II R–M loci from Rebase are almost half the length of the window selected (BamHI ∼ 2.3 kb; PvuII ∼ 1.8 kb; AhdI ∼ 3.5 kb) and we therefore expected that this analysis should reveal association of putative C proteins with R–M systems genes. Each ORF in the locus was used as a query for the BLAST similarity search against the non-redundant protein database and the search against the curated database of protein families pfam.
The resulting hits were clustered by similarity and classified into R–M-related ORFs (candidate restriction endonucleases and methyltransferases), phage-related ORFs, or neither of the above as described in the Materials and methods section. The results are given in Table 2. Naturally, 100% of C protein genes from Rebase belong to R–M loci. In contrast, only ∼25% of newly defined putative C protein genes belong to such loci. However, the number of putative C protein genes with adjacent phage-related ORFs in Rebase and in newly defined loci is similar (80% and 70%, correspondingly). Twenty-seven of 169 newly defined putative C protein genes have both R–M-related and phage-related genes in their vicinity.
Table 2.
Distribution of candidate C protein genes in loci containing RM and phage-related genes
| Genomic loci | RM-related orfs | Phage-related orfs | Both | Total |
|---|---|---|---|---|
| New C protein- family genes | 39 (23%) | 115 (68%) | 27 (16%) | 169 (100%) |
| C protein genes from Rebase with binding sites predicted | 32 (100.0%) | 26 (78%) | 26 (81%) | 32 (100%) |
Since a conspicuously large number of newly defined putative C protein genes did not contain putative R–M genes, in particular, easily recognizable methyltransferase genes, we addressed a possibility that some branches in our C protein family may in fact contain bona fide phage repressors. Visual analysis of the tree shown in Figure 1 demonstrates that C protein genes from purely phage-related loci are interspersed with those from R–M-related loci, and, with an exception of 18 putative C proteins that are associated with motif 3, all branches contain interspersed phage- and R–M-related loci. This observation argues against a hypothesis that a large proportion of newly identified C protein genes encode phage repressors. High frequency of phage-related genes in the vicinity of putative C protein genes may indicate that a considerable fraction of putative C protein genes identified by our analysis are remnants of previously functional R–M systems destroyed by genome recombination/phage insertion events.
In eight cases, two putative C proteins genes were found within 3 kb from each other (Table 3). Such ‘paired’ C protein genes may result from large-scale genomic rearrangements, such as duplications of C protein genes-containing loci or multiple insertions of C protein genes-containing elements in the same genomic location. The fact that two out of eight paired C protein genes loci encode C proteins associated with different motifs, while the other six loci contain genes encoding putative C proteins whose predicted binding sites belong to same motifs, indicates that both scenarios are realized. While known C proteins control simple autoregulatory loops individually, it is attractive to speculate that some clustered C protein genes may jointly control more complex regulatory circuits.
Table 3.
Loci containing two C protein genes
| Putative C protein gene 1 | Start | End | Binding motif class of C protein 1 | Putative C protein gene 2 | Start | End | Binding motif class of C protein 2 | Genbank ID | Distance |
|---|---|---|---|---|---|---|---|---|---|
| Mlo243 | 54253 | 54495 | Motif 7 | Mlo093 | 54714 | 54923 | Motif 4 | AL672113 | 219 |
| Bxe227 | 453842 | 454198 | Motif 8 | Bxe226 | 453455 | 453781 | Motif 8 | CP000272 | 61 |
| Ccr197 | 2919529 | 2919741 | Motif 2b | Ccr196 | 2919837 | 2920043 | Motif 2b | AE005673 | 96 |
| Mlo092 | 4943838 | 4944044 | Motif 4 | Mlo105 | 4944350 | 4944562 | Motif 1 | BA000012 | 306 |
| Plu192 | 157322 | 157555 | Motif 2 | Plu191 | 154190 | 154423 | Motif 2 | BX571873 | 2899 |
| Brl149 | 2459664 | 2459906 | Motif 5 | Brl147 | 2458276 | 2458518 | Motif 5 | CP000085 | 1146 |
| Bps148 | 521079 | 521321 | Motif 5 | Bps146 | 522475 | 522717 | Motif 5 | BX571966 | 1154 |
We also wondered whether any of the 39 putative R–M genes associated with predicted C protein genes correspond to known R–M systems from Rebase. To address this possibility, the coordinates and Genbank IDs of R–M-related genes associated with a particular putative C protein gene were compared with Genbank IDs of bona fide R–M systems genes present in REBASE. The match of Genbank IDs and coordinates would indicate that R–M genes associated with a putative C protein gene are identical to genes of an R–M system from REBASE that, however, lacks an annotated C protein gene. The results of this analysis are shown in Table 4. As can be seen, 14 of the 39 putative R–M loci correspond to already known REBASE R–M systems. In 2 of 14 cases (Hne199, Gur068), the newly discovered putative C proteins correspond to a short uncharacterized ORF of a Rebase-annotated R–M system. Our independently identified putative C protein gene Ent115 corresponds to the recently annotated C.Esp1396I. In all remaining cases, the discovered putative C protein genes are adjacent to Rebase-annotated R–M systems (which, however, are not annotated as having any genes other than the restriction endonuclease or methyltransferase genes). Our analysis predicts that these R–M systems are in fact C protein-dependent.
Table 4.
Genomic co-occurrence of C protein genes and known Rebase R–M systems
| No. | Putative system ID | Motif | Organism description | Genbank ID | Putative C protein start-end | REBASE system ID | Gene annotation | Gene start-end | Distance |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Pst154 | Motif 1 | Pseudomonas stutzeri A1501, complete genome | CP000304.1 | 749928–750170 | PstA1501ORF647P | Methylase | 743080–745056 | 4872 |
| 2 | Lho088 | Motif 2c | Laribacter hongkongensis plasmid pHLHK8, complete sequence | AY858987.1 | 1735–1983 | LhopHLHKP | Restrictase | 3832–4806 | 1849 |
| 3 | Nha178 | Motif 2b | Nitrobacter hamburgensis X14, complete genome | CP000319.1 | 2773100–2773321 | NhaXORF2515P | Methylase | 2770295–2771002 | 2098 |
| 4 | Hch122 | Motif 2b | Hahella chejuensis KCTC 2396, complete genome | CP000155.1 | 2547264–2547500 | HchORF2488P | Methylase | 2547622–2548812 | 122 |
| 5 | Hne199 | Motif 2 | Hyphomonas neptunium ATCC 15444, complete genome | CP000158.1 | 2696435–2696641 | HneORF2545P | Unannotated short protein | 2696432–2696641 | 0 |
| 6 | Swi193 | Motif 2 | Sphingomonas wittichii RW1, complete genome | CP000699.1 | 1752458–1752658 | SwiRWORF1578P | Methylase | 1754713–1756164 | 2055 |
| 7 | Nwi091 | Motif 4 | Nitrobacter winogradskyi Nb-255, complete genome | CP000115.1 | 922287–922499 | NwiORF847P | Methylase | 924490–925248 | 1991 |
| 8 | Ent115 (C.Esp1396I) | Motif 2 | Enterobacter sp. RFL1396 plasmid pEsp1396, complete sequence | AF527822.1 | 1481–1717 | Esp1396I | Recently annotated C protein | 1481–1717 | 0 |
| 9 | Lpn060 | Motif 6b | Legionella pneumophila str. Corby, complete genome | CP000675.1 | 226951–227193 | LpnCMrrP | Restrictase | 233982–234959 | 6789 |
| 10 | Cef187 | Motif 2 | Corynebacterium efficiens plasmid pCE3 DNA, complete sequence | AP005226.1 | 16363–16605 | CefpCE3MrrP | Restrictase | 10596–11558 | 5047 |
| 11 | Gur068 | Motif 2 | Geobacter uraniumreducens Rf4, complete genome | CP000698.1 | 1337269–1337499 | GurRORF1135P | HTH-domain protein | 1337269–1337499 | 0 |
| 12 | Pgi032 | Motif 9 | Porphyromonas gingivalis W83, complete genome | AE015924.1 | 595797–595997 | PgiTORF544P | Methylase | 596192–598138 | 195 |
| 13 | Bth033 | Motif 9 | Bacteroides thetaiotaomicron VPI-5482, complete genome | AE015928.1 | 5932179–5932379 | BthVORF4518P | Restrictase | 5934256–5936961 | 1877 |
| 14 | Nha244 | Motif 7 | Nitrobacter hamburgensis X14, complete genome | CP000319.1 | 883322–883612 | NhaXORF803P | Methylase | 885888–887198 | 2276 |
Regulatory mechanisms
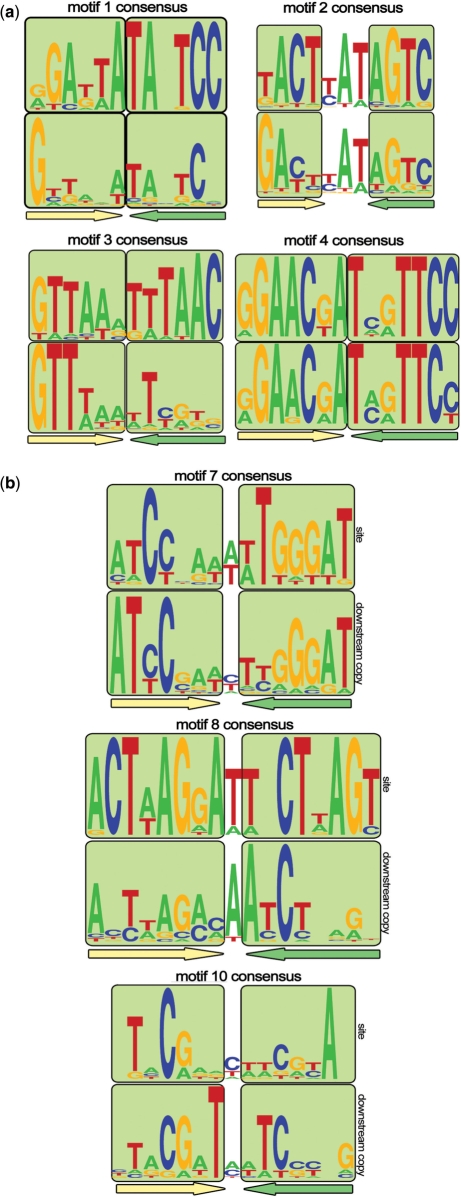
C proteins bind DNA as dimers (5,6). Thus, their binding sites should be palindromic. In addition, in all cases that have been investigated experimentally, two adjacent C protein dimer binding sites are present in the regulatory regions of the C protein genes. The upstream site has a higher affinity for the C protein dimer and the interaction with this site activates transcription of the C protein gene (6,10). The downstream site has lower affinity for C protein dimer and the interaction with this site decreases transcription of the C protein gene. These general considerations lead to certain constraints that putative C protein-binding sites should conform to (assuming that the regulatory mechanisms in the newly identified cases are similar to those already described). A complete C protein-binding site should contain a direct repeat of a palindromic sequence. Cooperative interaction of a C protein with its two binding sites necessitates that a distance between the binding sites is conserved. In C.PvuII-like motifs 1–6, a highly conserved GTG trinucleotide is observed between the putative palindromic operators. The special geometry and/or bendability of this trinucleotide may promote cooperative interactions between bound C protein dimers. Indeed, mutational and structural analysis of C protein interactions with two motifs of this group (16,20), confirmed the importance of this trinucleotide in cooperative C protein binding and the existence of a bend in this sequence when bound by two C protein dimers. The analysis presented above demonstrates that four out of six C.PvuII-like motifs (1, 2, 3 and 4) contain two copies of the operator (Figure 3a).
Figure 3.
(a) Logos of 1–4 motifs’ consensus sequences. Palindromic boxes are marked with light green squares and underlined with colored arrows. The upper logo represents the 5′ (distal) copy, while the lower logo represents the 3′ (proximal) copy. (b) Logos of palindromic (7, 8 and 10) motifs’ consensus sequences. Palindromic boxes are marked with light green squares and underlined with colored arrows. The upper logo corresponds to annotated binding sites, while the lower logo corresponds to their weak downstream copies.
Motifs 7, 8 and 10, as defined by our procedure, are single palindromes. One of the reasons why the second operator was not identified could be that the initial procedure implied a fixed distance between the operators. Thus, we used scanning of the 100 bp regions upstream of putative C protein genes with profiles constructed from already identified sites, and also performed additional manual searches for conservation islands. This resulted in identification of additional, downstream conservation islands containing weaker copies of the same motif (Figure 3b). As expected, no preferred length of the spacer between the sites could be seen (data now shown).
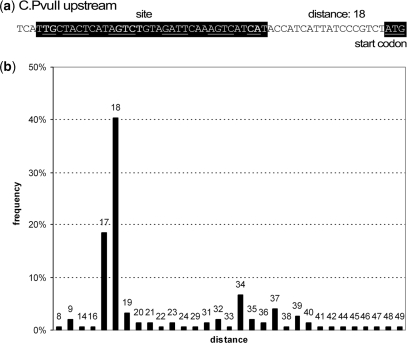
In several cases that have been studied in experiment, the C protein RNA is leaderless (10,18), i.e. the transcription start site coincides with or is located 1–3 nucleotides upstream of adenosine (or guanosine) of C protein ORF initiating ATG (or GTG) codon. Presumably, the less efficient translation of a leaderless message (30) causes a delay in C protein-dependent activation of transcription of the toxic restriction endonuclease gene located downstream of the C protein gene (31). In the cases where the existence of leaderless C protein mRNA has been experimentally demonstrated, the distance between the C protein-binding site and the C protein ORF initiating codon is necessarily short, the typical spacer length being 18 nt (Figure 4a). We considered all instances of sites forming motifs 1–6 and calculated the distance between the motif and the C protein start codon. The results are presented in Figure 4b, where, indeed, a very strong maximum at a distance of 17–18 nt between the C protein gene start codon and the putative C protein-binding site is evident. We take this result as a strong indication that leaderless translation is a common feature of C protein regulation.
Figure 4.
(a) The structure of a region upstream of a typical C.PvuII-like C protein gene. The binding site and the ATG start codon are marked with black color. The palindromic elements of the site are underlined. (b) The histogram of distances between the candidate binding sites and start codons of C protein genes. Only C.PvuII-like motifs (1, 2, 2b, 2c, 3, 4, 5, 6, 6b) are considered. Horizontal axis and numbers above the bars: distances, vertical axis: frequency of such distance.
The presence of a pool of C protein genes whose mRNA is translated through a leaderless mechanism prompted us to bioinformatically search for conserved promoter elements upstream of these genes, since the distance between the promoter element and transcription start (defined by the position of the initiating codon) should be fixed. However, we failed to identify any reliable –10 promoter consensus elements in sequences preceding initiation codons of apparently leaderless putative C protein genes, nor did not observe any over-representation of TATAAT-like hexamers in the region preceding the putative transcription start point areas of these genes (data not shown). This agrees with the absence of easily identifiable –10 promoter elements in known CR operon promoters (6,10,20) and supports a model, which posits that these promoters are weak and require C protein binding for activity.
DISCUSSION
Using a combination of phylogenetic footprinting and bioinformatic motif searches, we have identified 201 putative C protein-binding sites, 181 (90.0%) of which fall into ten distinct motifs. The remaining binding sites do not belong to motifs, however, most of them resemble C.PvuII-like motifs (1–6): the sites are also 35 bp long, some sites contain the central GTGG tetranucleotide and some of them contain self-complementary trinucleotides at the outside flanks.
The genes of Rebase-annotated C proteins are preceded only by sites belonging to motifs 2, 7, 8 and 9. Among REBASE R–M systems with previously unannotated C proteins, we observed three C protein-homologous genes preceded by sites from other motifs: motif 1 (Pst154), motif 4 (Nwi091) and motif 6b (Lpn060). Thus, the apparent limitation in the kinds of C protein-binding sites present in bona fide R–M systems is likely because of a bias in experimental analysis centered on a number of close homologs rather than some biological reason. Still, there remains a possibility that a group of identified candidate transcription factors from the C protein family are not involved in the regulation of R–M systems.
One hundred and sixty nine sites belong to new motifs that have not been described previously. Despite the fact that we have not used experimental data during our searches, all eight experimentally verified C protein-binding sites were identified correctly, indicating that our search procedure is robust. C proteins with putative binding motifs from the same class typically cluster in the phylogenetic tree (Figure 1). The few exceptions could be caused by both low reliability of deep branches of the tree or by bona fide convergent evolution of the motif. A more detailed analysis of co-evolution of C proteins and their binding motifs is required to characterize the molecular events and their structural consequences in detail. This work is currently ongoing in our laboratories.
The motifs identified in this work can be subdivided into two types, and the first type consists of two subtypes. The C.PvuII-like motifs 1, 2, 3 and 4 are characterized by rich symmetries. This group of motifs comprises two palindromic operators separated by highly conserved spacers and framed by highly conserved complementary trinucleotides. The related motifs 5 and 6 retain some features of the former subtype (conserved nucleotides in the middle, short inverted repeats at the termini), but do not contain operators. The symmetry and conservation beyond the palindromic operators suggest that there exist additional functional and/or structural forces shaping the motif. Indeed, the observed pattern of conservation is in good agreement with the recently published structure of a complex of two dimers of the C.Esp1396I protein bound to the binding site (motif 2) (16). In the structure, the outside complementary trinucleotides Z-Z* (A)TG-CA(T) form extensive contacts with the protein. Similarly, the highly conserved, non-symmetrical central dinucleotide (G)TG also contacts the protein. In contrast, no direct interactions with palindromic operators are evident, though the recognition helix of the helix-turn-helix motif of each C.Esp1396I monomer is positioned in the major grove of operator half-sites. The occurrence of a large number of highly conserved positions that do not appear to form contacts with the protein makes it likely that factors such as structural constraints on the DNA or additional modes of protein binding (i.e. as single dimers or even monomers) may be involved in shaping the motif.
The second major type of sites is formed by palindromic motifs 7, 8 and 10, each consisting of a single operator with a downstream weaker copy located at variable distance from the initially identified one. On the basis of the only biochemically studied example, of C.EcoRV (10), motif 7, interactions of C proteins with such binding sites are not characterized by highly cooperative interactions between C protein dimers observed with C.PvuII-like sites with fixed distances between the operators.
Although genomic contexts of some identified candidate C proteins contain genes encoding putative restriction endonucleases and methylases, in the majority of cases no such genes could be identified. On the other hand, the immediate vicinity of many candidate C proteins genes contains phage- or transposon-related genes such as resolvases, integrases, transposases, recombinases and other genes annotated as phage-, plasmid- and conjugation-related. This leaves open a possibility that some of identified C protein-family factors regulate functions other than restriction-modification. Although C proteins share distant homology and structural similarity with phage repressors (15), our database search was sufficiently restrictive, and the pool of putative C proteins was not contaminated with bona fide phage repressors.
Despite the lack of a universal association of putative C protein genes with R–M systems, essential features of autoregulatory loops controlled by characterized C proteins from known R–M systems such as (i) the presence of two binding sites, a high affinity one leading to activation of transcription of C protein gene as well as any gene that is coupled to it, and another, low affinity site whose occupancy leads to transcriptional repression and (ii) translation from leaderless transcripts appears to be a common feature of at least C protein genes associated with motifs 1–6. In R–M systems, these features allow highly regulated, time-delayed expression of the highly toxic restriction endonuclease. It remains to be determined whether newly identified C protein genes that are not associated with any R–M systems also control expression of genes toxic to the cell.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This study was partially supported by grants from the Howard Hughes Medical Institute (55005610 to M.G.), the Russian Foundation of Basic Research (09-04-01098-a to V.S.) and National Intitutes of Health (RO1 GM59295 to K.S.). M.G. and K.S. are partially supported by grants from the program ‘Molecular and Cellular Biology’ of the Russian Academy of Sciences.
Conflict of interest statement: None declared.
ACKNOWLEDGEMENTS
We are grateful to Anna Karyagina, Andrei Alexeevsky and Sergei Spirin for useful discussions.
REFERENCES
- 1.Bickle TA, Krüger DH. Biology of DNA restriction. Microbiol. Rev. 1993;57:434–450. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King G, Murray NE. Restriction enzymes in cells, not eppendorfs. Trends Microbiol. 1994;2:465–469. doi: 10.1016/0966-842x(94)90649-1. [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001;29:3742–3756. doi: 10.1093/nar/29.18.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagornykh M, Bogdanova E, Protsenko A, Severinov K. Regulation of expression of type II restriction-modification systems genes. Russ. J. Genet. 2008;44:606–615. doi: 10.1134/s1022795408050037. [DOI] [PubMed] [Google Scholar]
- 5.Streeter SD, Papapanagiotou I, McGeehan JE, Kneale GG. DNA footprinting and biophysical characterization of the controller protein C.AhdI suggests the basis of a genetic switch. Nucleic Acids Res. 2004;32:6445–6453. doi: 10.1093/nar/gkh975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogdanova E, Djordjevic M, Papapanagiotou I, Heyduk T, Kneale G, Severinov K. Transcription regulation of type II restriction-modification system AhdI. Nucleic Acids Res. 2008;36:1429–1442. doi: 10.1093/nar/gkm1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sohail A, Ives CL, Brooks JE. Purification and characterization of C.BamHI, a regulator of the BamHI restriction-modification system. Gene. 1995;157:227–228. doi: 10.1016/0378-1119(94)00698-r. [DOI] [PubMed] [Google Scholar]
- 8.Anton BP, Heiter DF, Benner JS, Hess EJ, Greenough L, Moran LS, Slatko BE, Brooks JE. Cloning and characterization of the BglII restriction-modification system reveals a possible evolutionary footprint. Gene. 1997;187:19–27. doi: 10.1016/s0378-1119(96)00638-5. [DOI] [PubMed] [Google Scholar]
- 9.Rimseliene R, Vaisvila R, Janulaitis A. The eco72IC gene specifies a trans-acting factor which influences expression of both DNA methyltransferase and endonuclease from the Eco72I restriction-modification system. Gene. 1995;157:217–219. doi: 10.1016/0378-1119(94)00794-s. [DOI] [PubMed] [Google Scholar]
- 10.Semenova E, Minakhin L, Bogdanova E, Nagornykh M, Vasilov A, Heyduk T, Solonin A, Zakharova M, Severinov K. Transcription regulation of the EcoRV restriction modification system. Nucleic Acids Res. 2005;33:6942–6951. doi: 10.1093/nar/gki998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cesnaviciene E, Mitkaite G, Stankevicius K, Janulaitis A, Lubys A. Esp1396I restriction-modification system: structural organization and mode of regulation. Nucleic Acids Res. 2003;31:743–749. doi: 10.1093/nar/gkg135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tao T, Bourne JC, Blumenthal RM. A family of regulatory genes associated with type II restriction-modification systems. J. Bacteriol. 1991;173:1367–1375. doi: 10.1128/jb.173.4.1367-1375.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ives CL, Sohail A, Brooks JE. The regulatory C proteins from different restriction-modification systems can crosscomplement. J. Bacteriol. 1995;177:6313–6315. doi: 10.1128/jb.177.21.6313-6315.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawaya MR, Zhu Z, Mersha F, Chan SH, Dabur R, Xu SY, Balendiran GK. Crystal structure of the restriction-modification system control element C.Bcll and mapping of its binding site. Structure. 2005;13:1837–1847. doi: 10.1016/j.str.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 15.McGeehan JE, Streeter SD, Papapanagiotou I, Fox GC, Kneale GG. High-resolution crystal structure of the restriction-modification controller protein C.AhdI from Aeromonas hydrophila. J. Mol. Biol. 2005;346:689–701. doi: 10.1016/j.jmb.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 16.McGeehan JE, Streeter SD, Thresh SJ, Ball N, Ravelli RB, Kneale GG. Structural analysis of the genetic switch that regulates the expression of restriction-modification genes. Nucleic Acids Res. 2008;36:4778–4787. doi: 10.1093/nar/gkn448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bart A, Dankert J, van der Ende A. Operator sequences for the regulatory proteins of restriction modification systems. Mol. Microbiol. 1999;31:1277–1278. doi: 10.1046/j.1365-2958.1999.01253.x. [DOI] [PubMed] [Google Scholar]
- 18.Knowle D, Lintner RE, Touma YM, Blumenthal RM. Nature of the promoter activated by C.PvuII, an unusual regulatory protein conserved among restriction-modification systems. J. Bacteriol. 2005;187:488–497. doi: 10.1128/JB.187.2.488-497.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson CA, Weinman O, Trester-Zedlitz ML, Darst SA. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol. Cell. 2002;9:527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- 20.Mruk I, Rajesh P, Blumenthal RM. Regulatory circuit based on autogenous activation-repression: roles of C-boxes and spacer sequences in control of the PvuII restriction-modification system. Nucleic Acids Res. 2007;35:6935–6952. doi: 10.1093/nar/gkm837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs, Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Rapp BA, Wheeler DL. Genbank. Nucleic Acids Res. 2000;28:15–18. doi: 10.1093/nar/28.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Felsenstein J. PHYLIP – Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 25.Stavrovskaia ED, Makeev VI, Mironov AA. ClusterTree-RS: the binary tree algorithm for identification of co-regulated genes by clustering regulatory signals. Mol. Biol. 2006;40:524–532. [PubMed] [Google Scholar]
- 26.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naderer M, Brust JR, Knowle D, Blumenthal RM. Mobility of a restriction-modification system revealed by its genetic contexts in three hosts. Bacteriol. 2002;184:2411–2419. doi: 10.1128/JB.184.9.2411-2419.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vijesurier RM, Carlock L, Blumenthal RM, Dunbar JC. Role and mechanism of action of C. PvuII, a regulatory protein conserved among restriction-modification systems. J. Bacteriol. 2000;182:477–487. doi: 10.1128/jb.182.2.477-487.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moll I, Grill S, Gualerzi CO, Blasi U. Leaderless mRNAs in bacteria: surprises in ribosomal recruitment and translational control. Mol. Microbiol. 2002;43:239–246. doi: 10.1046/j.1365-2958.2002.02739.x. [DOI] [PubMed] [Google Scholar]
- 31.Mruk I, Blumenthal RM. Real-time kinetics of restriction-modification gene expression after entry into a new host cell. Nucleic Acids Res. 2008;36:2581–2593. doi: 10.1093/nar/gkn097. [DOI] [PMC free article] [PubMed] [Google Scholar]