Abstract
Appropriate control of the chromosome end-replicating enzyme telomerase is crucial for maintaining telomere length and genomic stability. The essential telomeric DNA-binding protein Cdc13p both positively and negatively regulates telomere length in budding yeast. Here we test the effect of purified Cdc13p on telomerase action in vitro. We show that the full-length protein and its DNA-binding domain (DBD) inhibit primer extension by telomerase. This inhibition occurs by competitive blocking of telomerase access to DNA. To further understand the requirements for productive telomerase 3′-end access when Cdc13p or the DBD is bound to a telomerase substrate, we constrained protein binding at various distances from the 3′-end on two sets of increasingly longer oligonucleotides. We find that Cdc13p inhibits the action of telomerase through three distinct biochemical modes, including inhibiting telomerase even when a significant tail is available, representing a novel ‘action at a distance’ inhibitory activity. Thus, while yeast Cdc13p exhibits the same general activity as human POT1, providing an off switch for telomerase when bound near the 3′-end, there are significant mechanistic differences in the ways telomere end-binding proteins inhibit telomerase action.
INTRODUCTION
In 1985, a novel enzyme was identified in ciliates that adds repeated DNA sequence to telomeric oligonucleotides (1). This enzyme, telomerase, is a reverse transcriptase that iteratively adds species-specific, GT-rich, DNA repeats to the 3′ ends of chromosomes. Telomerase has since been identified in a wide range of eukaryotic organisms, including humans, where it plays a prominent role in cancer and a growing number of other diseases (2).
Telomerase is a ribonucleoprotein complex. In the yeast Saccharomyces cerevisiae, the core enzyme consists of the Est2p reverse transcriptase (or TERT) subunit and an 1157-nt RNA, TLC1 (3–7). In addition to the catalytic subunit, the yeast telomerase holoenzyme also includes the Est3 protein, and proteins that probably directly interact with the flexible RNA scaffold, including Est1p, the Ku DNA repair heterodimer, and the Sm7 complex, also involved in snRNA maturation (8–11). In vitro, whether purified from yeast cells or reconstituted in a heterologous system, yeast telomerase synthesizes predominantly a single DNA repeat, after which it remains stably associated with the ssDNA product (5,12). However, in vivo, telomerase can add as little as a few to hundreds of nucleotides to a single telomere in one cell cycle (13), suggesting that additional telomere-associated proteins may modulate activity.
Telomere length homeostasis involves the coordination of telomere lengthening and shortening processes. Although the complete mechanism of telomere length regulation remains to be elucidated, several telomere-associated proteins have been implicated in the process. The observation that targeting the C-terminus of Rap1p to telomeres leads to telomere shortening, and that this is proportional to the number of molecules at a given telomere, led to the ‘protein-counting model’ of telomere length regulation by Rap1p (14). Thus, double-strand DNA-binding factors play a role in length regulation. Additionally, the abundance of limiting telomerase components also appears to provide another level of regulation. There are only ∼29 copies of the TLC1 RNA per haploid yeast cell (∼37 per diploid) and halving the RNA abundance in a TLC1/tlc1Δ heterozygote leads to short telomeres (15). This finding complemented those from mammalian studies (16–19) that had also suggested that telomerase activity can be regulated by the number of active complexes. The function of the yeast ATM kinase homolog, Tel1p, is important for proper telomere length maintenance and is required for repeat addition processivity in vivo (20–22), implicating phosphorylation as critical to telomere length regulation. It has also been proposed that telomeres exist in either an extendible or non-extendible state (13,23), which dictates whether or not telomerase can access a particular telomere end. Recent studies show that the extendible state tends to exist at the chromosome ends with the shortest telomeres, where telomerase is relatively enriched (24,25). Despite the many pathways involved in regulating telomere length, control of telomerase activity by modulating its access to chromosome ends is one of the most fundamentally important.
The essential Cdc13 protein binds to the 3′ telomere overhangs through its structurally and biochemically characterized ssDNA-binding domain (DBD) (26–30). Cdc13p is a chromosome end-capping protein, preventing the CA-rich strand of the telomere from undergoing unregulated nucleolytic degradation (31). In addition, Cdc13p regulates telomere elongation through recruitment of telomerase via its Est1p subunit (32). Recruitment is abolished in the cdc13-2est mutant, which displays shortened telomeres, similar to those of a telomerase-defective strain (7). Cdc13p may also prevent runaway elongation by telomerase, since yeast expressing the cdc13-5 allele, which lacks the C-terminus, exhibit elongated telomeres and long G-strand overhangs (33). Cdc13p is enriched at telomeres during S-phase (24,25), providing additional support for its role in telomere length regulation.
Telomeric ssDNA-binding protein complexes are known to regulate telomerase activity. It was recently shown that the human single-stranded telomere binding protein POT1 (hPOT1) inhibits telomerase activity in vitro on human telomere substrates (34,35). hPOT1 exerts its inhibitory effect when bound within six nucleotides of the 3′-end of a ssDNA oligonucleotide substrate (35). Both full-length protein and the DBD of hPOT1 display identical telomerase inhibitory activity, and DNA-binding activity is required for inhibition (34,35). Telomerase activity is fully restored even in the presence of full-length hPOT1 or its DBD, if there is at least a 6-nt tail beyond the hPOT1-binding site. This suggests that the relative positioning of hPOT1 along the 3′ telomere overhangs can serve as a binary switch for telomerase-accessibility (35). More recent evidence suggests that hPOT1 regulates telomerase in conjunction with a second shelterin component, TPP1. When both hPOT1 and TPP1 are prebound onto a telomeric oligonucleotide substrate, the human telomerase core enzyme displays enhanced processivity (36). While these data suggest a potential telomerase regulatory role for hPOT1, how this is manifested in vivo is not yet understood. Manipulation of hPOT1 by overexpression (37–39) or RNAi suppression of hPOT1 (40–43) leads to a variety of phenotypes, including telomere elongation, 3′ tail structure changes and various DNA damage responses. The complexity of the in vivo data does not provide a clear mechanism for how hPOT1 might regulate telomerase activity, potentially due to indirect effects associated with the concomitant disruption of the large shelterin complex at telomeres upon manipulation of hPOT1 (44,45). In contrast, understanding of the role played by budding yeast Cdc13p in telomere length regulation is somewhat clearer, due to the availability of separation-of-function and temperature-sensitive alleles (32).
As a starting point for evaluating the regulation of telomerase by the host of factors identified through genetic analysis in S. cerevisiae, it is important to know the extent to which Cdc13p binding to telomeric DNA affects its ability to serve as a telomerase substrate. Until recently, such studies were complicated by the inability to reconstitute the yeast telomerase core enzyme in vitro and the limitations associated with using immunopurified telomerase from yeast extracts. The discovery that a miniaturized yeast telomerase RNA component provides reconstituted activity with yeast TERT when coexpressed in rabbit reticulocyte lysates (5) provides immunopurified core enzyme to directly examine the role of telomere end-binding proteins in the regulation of telomere length by telomerase. We show that, unlike hPOT1, Cdc13p inhibits telomerase activity on substrates that have substantial 3′ overhangs. This inhibitory activity is observed for both the DBD of Cdc13p and the full-length protein, although we find differences between how the DBD and Cdc13p inhibit telomerase activity on specific substrates. We propose that three mechanisms for yeast telomerase inhibition by Cdc13p have a fundamental role in defining the non-extendible state of yeast telomeres.
MATERIALS AND METHODS
Cdc13p and Cdc13(DBD) expression and purification
Full-length Cdc13p was expressed using baculovirus in Sf9 cells and purified as previously described (27,32), with the following modifications. Briefly, baculovirus-infected cells were homogenized and Cdc13p was purified to >90% homogeneity using Ni-affinity chromatography (GE Healthcare). Active concentrations of Cdc13p were determined by electrophoretic mobility shift assay (EMSA), and defined as the concentration of Cdc13p required to fully shift 100 nM GTGTGGGTGTG (TEL) probe, which is >100-fold above the measured Kd.
Cdc13(DBD) constructs were overexpressed and purified as described previously, with minor modifications (27). Glycerol and detergents were removed from the lysis buffer, and lysis was carried out by sonication. The protein domain was purified using ion-exchange, Ni-affinity and size-exclusion chromatography (GE Healthcare). Filter binding assays were employed to determine the active concentration of DBD protein for all subsequent assays. Active concentration was defined as the concentration of protein required to fully bind the standard TEL oligonucleotide at a concentration of 200 nM, well above the measured Kd of the DBD•TEL interaction.
Telomerase activity assays
Reconstituted yeast telomerase was synthesized in rabbit reticulocyte lysates (Promega), as described (5), using plasmids encoding the Mini-T(500) RNA and ProA-tagged Est2p. At the final step of telomerase immunopurification, IgG Sepharose beads were resuspended 1 : 3 (v/v) with 10 mM Tris (pH 8), 1 mM MgCl2 and 30% (v/v) glycerol. Telomerase from yeast extracts had been isolated previously from cells overexpressing both TLC1 RNA and ProA-tagged Est2p (46). Unless otherwise indicated, all telomerase assays were performed as reported previously (5), except at 27°C for 10 min with final substrate concentration at 0.3 µM. When included, DBD or Cdc13p were preincubated with primer DNA for at least 15 min on ice prior to adding telomerase and radioactive [α32P]-dGTP. All oligonucleotide substrates were commercially PAGE or reverse-phase HPLC-purified. Markers for +1 telomerase products were made essentially as described (5). Telomerase signal (detected by a GE Typhoon Trio phosphorimager) was quantified by using Imagequant TL software (GE Healthcare) to sum the signal from the reaction products (typically +1 through +7 bands) and normalize it to the internal loading and recovery control. To determine the stoichiometry of DBD required for telomerase inhibition, we determined the amount of protein required to inhibit 78% (on average) of the telomerase signal from six completely independent protein preparations. It required 0.81 ± 0.64 (mean ± SD) equivalents of protein to achieve this level of inhibition. The high variability is likely caused by batch-to-batch differences in the quantitation of DBD protein preparations, as the extent of inhibition was highly reproducible for a given preparation.
Binding assays
Filter-binding assays were used to determine equilibrium binding constants for interactions with Cdc13(DBD). The concentration of DNA used for all experiments was 50 pM and the protein concentrations were varied ∼100-fold above and below the measured Kd. Oligonucleotides were 5′-end labeled as described (27). All filter-binding reactions were carried out in high-salt binding buffer (28). The salt-dependence of the Cdc13(DBD) interaction has been previously shown to be log-linear (28), hence the high-salt buffer was used to reduce the affinity of the interactions to the nM range to facilitate more accurate measurements. A Schleiser & Schuell minifold vacuum manifold dot blot apparatus was used, employing a two-filter method (47) consisting of a nitrocellulose filter (GE Infrastructure Water & Process Technologies) to capture the protein and bound DNA, and a HyBond (XL) filter (Amersham Biosciences) to capture free DNA. Membranes were washed twice with 90 µl of binding buffer (without BSA) prior to the addition of 30 µl of each reaction mixture. Membranes were then washed three times with 90 µl of binding buffer (without BSA) then dried and exposed to phosphorimager plates. ImageQuant (GE) software was used to quantify the DNA signal from each membrane filter. The data were normalized to obtain fraction bound and fit to a two-state binding model as described (29). The reported and relative Kd values are the average of triplicate measurements independently repeated three times, reported with standard error.
Equilibrium binding constants for the interactions of telomeric oligonucleotides and Cdc13p were measured using EMSAs. Protein concentrations were varied from 0.3 pM to 3 µM with 5′-end labeled DNA concentration constant at 20 pM. Reactions and gel conditions were as described previously (27,32,48), except that binding was allowed to equilibrate for 30 min. Binding constants were measured by fitting the data to the same two-state binding model as previously described (29). The apparent and relative Kd values reported are averages of triplicate experiments with standard errors.
Snake venom phosphodiesterase exonuclease assays
Experiments were performed as previously described (35), with minor modifications. Binding was performed on ice and 5′-end labeled DNA concentrations were lowered to 0.3 µM due to higher affinity of the Cdc13•DNA interactions, and to match the conditions of telomerase extension assays. For the experiment shown in Figure 4a, the protein•DNA complex was pre-formed at a 10 : 1 molar ratio prior to the addition of Snake venom phosphodiesterase (SVP) (Worthington Biochemical Corporation) to the reactions, whereas for experiments in Figure 4b, the protein concentration was titrated onto each oligonucleotide (see figure legend for ratio of protein : DNA for each lane). The reaction was incubated at 30°C for the indicated time in Figure 4a, and for 12 min in Figure 4b, then stopped with the addition of 50 µM Na2EDTA. Samples were run on 20% denaturing polyacrylamide gels, (1× TBE, 7 M urea) and these were dried and exposed to phosphorimager plates for visualization.
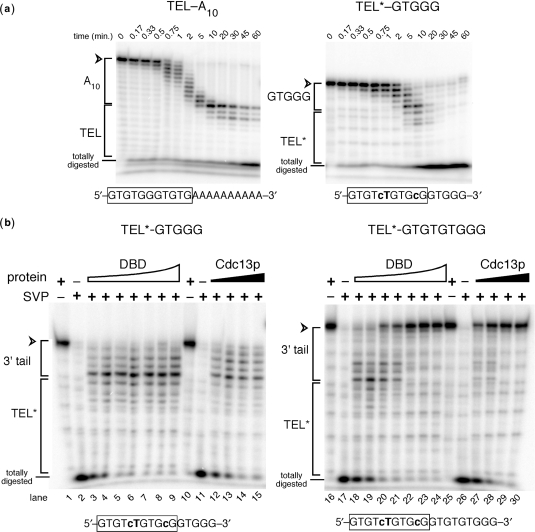
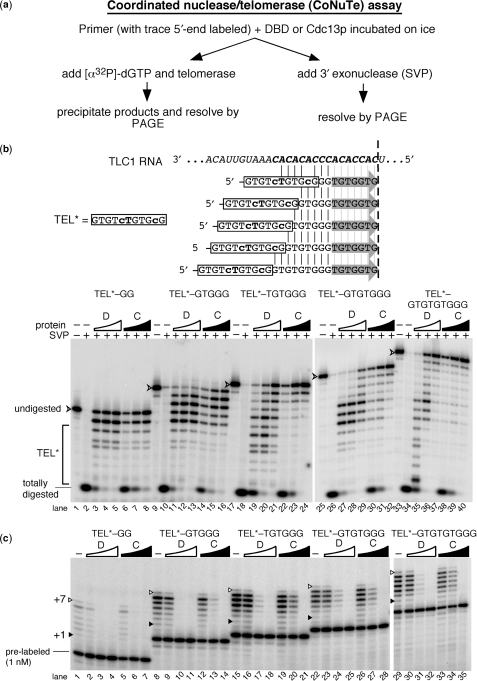
Figure 4.
SVP 3′ exonuclease assays show that Cdc13p and its DBD both bind specifically to the 5′ region of TEL*–GTGGG, but differ in binding site preference on longer oligonucleotides. (a) SVP digestion time course experiments show that Cdc13p DNA-binding protects the TEL region of a TEL–a10 control and 11-nt TEL* sequence in TEL*–GTGGG. Time of incubation with SVP is indicated above. Correspondence of portions of the gel to relative position in the oligonucleotide sequence is indicated to the left: TEL or TEL* with tails of 10 adenosines or –GTGGG, a telomeric sequence complementary to a unique portion of the telomerase template. Totally digested represents the signal resulting from complete SVP digestion of the 5′-end labeled oligonucleotides. (b) Cdc13(DBD) and full length protein bind to TEL* when a GTGGG extension is appended, but Cdc13p selectively binds to a GTGTGTGGG extension. The molar ratios of DBD:DNA (lanes 3–9 and 18–24) are 0.5 : 1, 1 : 1, 1.5 : 1, 2 : 1, 3 : 1, 5 : 1, and 10 : 1. Molar ratios of Cdc13p : DNA (lanes 12–15 and 27–30) are 0.5 : 1, 1 : 1, 1.5 : 1, and 2 : 1.
Coordinated nuclease/telomerase assays
Varying concentrations of Cdc13p or its DBD, diluted with their storage buffers, were incubated with the indicated primer diluted in telomerase reaction buffer (50 mM NaCl, 40 mM Tris (pH 8), 5% (v/v) glycerol, 2.5 mM MgCl2, 0.5 mM spermidine, 0.5 mM DTT). A trace amount of polynucleotide kinase 5′-labeled primer was also included (final concentration of 0.5 nM and 1500 cpm/µl). After binding on ice for 15 min, telomerase reaction components were added, except telomerase and [α32P]-dGTP. The reactions where then split into two tubes and one was used for telomerase assay while mock treatment with telomerase storage buffer was added to the other, which was then incubated with SVP at 27°C for 12 min. SVP digestion products were subsequently run on 12% sequencing-sized denaturing polyacrylamide gels, (1× TBE, 7 M urea) which were then dried and exposed to phosphorimager plates for visualization.
RESULTS
The DNA binding domain of Cdc13p inhibits telomerase
We began by analyzing the effect of the DBD of Cdc13p (Figure 1a) on telomerase activity, since this protein domain is biochemically and structurally well characterized (26–28,30). A standard 15-nt yeast telomeric DNA primer (12,46,49–52), referred to here as TP1, was tested first. This oligonucleotide ends in –GGG, constraining base pairing with yeast telomerase RNA to the unique occurrence of CCC in the template (Figure 1b, bottom). Therefore, yeast telomerase adds up to 7 nt, as has been shown previously for both the wild-type enzyme purified from yeast extracts (6,53) and the reconstituted enzyme with the miniaturized TLC1 RNA, Mini-T(500) (5). Both the reconstituted and the yeast-expressed telomerase were potently inhibited when the DBD of Cdc13p was preincubated with TP1 (Figure 1b).
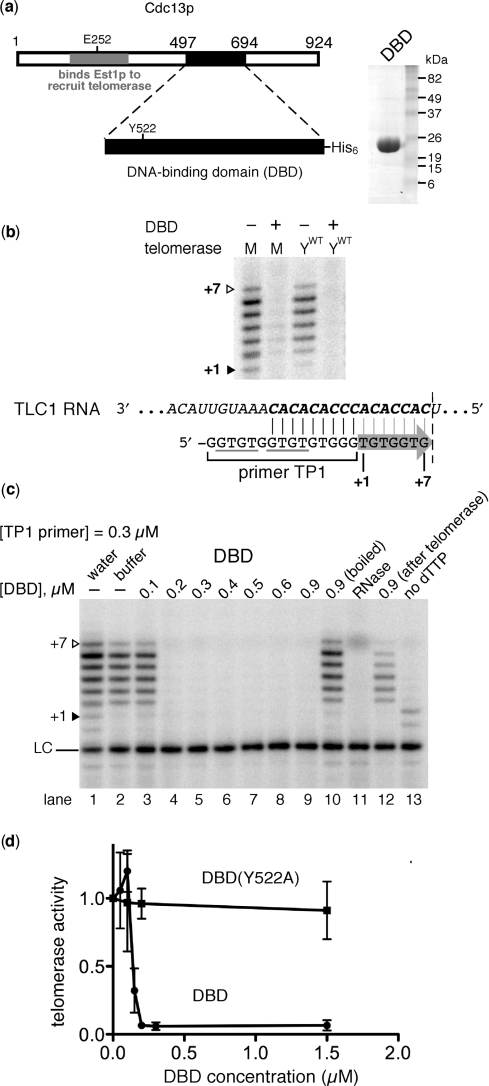
Figure 1.
The DBD of Cdc13p inhibits yeast telomerase. (a) Schematic of Cdc13p. The DBD resides near the C-terminus of the protein (amino acids 497–694). Purified Cdc13(DBD) (24.7 kDa with the His6 tag) is shown on a Coomassie-stained gel (right). E252, residue required for binding Est1p to recruit telomerase (32). Y522, amino acid important for high-affinity DNA-binding (27). (b) DBD inhibits telomerase in vitro. Cdc13(DBD) was incubated with the 15-nt primer TP1 and then telomerase was added along with deoxynucleotides A, C, T and [α-32P]-dGTP. M, reconstituted mini-telomerase. YWT, telomerase from extracts of yeast overexpressing ProA-Est2p and 1157-nt TLC1. The +1 and +7 telomerase extension products are indicated with filled and open triangles, respectively. Primer TP1 aligns with telomerase RNA as shown; arrow denotes template-directed DNA synthesis up to the boundary element (dotted line). Primer concentration was 0.3 µM. Reactions contained 0.6 µM Cdc13(DBD) (+) or its storage buffer (−). Gray lines show instances of GxGT, the essential 5′ binding portion of the Cdc13(DBD) binding consensus. (c) Cdc13(DBD) inhibits telomerase when present at a concentration similar to TP1 primer. Reaction conditions were essentially the same as when using reconstituted mini-telomerase in b. Water and Cdc13(DBD) storage buffer controls are shown in lanes 1 and 2, respectively. Lanes 3–9, increasing amounts of Cdc13(DBD) were incubated with DNA prior to adding telomerase. Lane 10, boiled (denatured) Cdc13(DBD). Lane 11, telomerase incubated for 10 min at 37°C with RNase A to degrade Mini-T RNA. Lane 12, 0.9 µM Cdc13(DBD) added after primer and telomerase were incubated on ice for 10 min. Lane 13, The first nucleotide encoded by the RNA template, dTTP (b), was omitted. LC, loading and recovery control: TP1 primer labeled with 32P at its 5′-end, which increases electrophoretic migration from increased negative charge. (d) DNA-binding-defective point mutant of Cdc13(DBD) does not inhibit telomerase in vitro. Standard deviation from three independent experiments is shown. Experiments were performed as in b and quantitated as described in Materials and Methods section. Primer concentration was 0.3 µM.
Telomerase action was completely inhibited when the concentration of Cdc13(DBD) was approximately equal to that of the DNA primer (Figure 1c). In six telomerase experiments each performed with a different DBD protein preparation, the ratio of DBD : DNA required for inhibition was 1.0 ± 0.8 (mean ± SD). If Cdc13(DBD) was heat-denatured, inhibition was completely abolished (Figure 1c, lane 10). We also tested importance of the order of addition by adding Cdc13(DBD) after telomerase and primer had been pre-incubated for 10 min. In this case, inhibition was intermediate (Figure 1c, lane 12), probably because the oligonucleotide substrate is competed away by the relatively fast and tight binding of the Cdc13(DBD) (27). Lastly, we show that the DNA synthesis observed was clearly due to telomerase activity since it was abolished by RNase and also required dTTP, the first nucleotide predicted to be added by the TLC1 template (Figure 1c, lanes 11 and 13).
If Cdc13(DBD) inhibits telomerase by blocking its access to the DNA substrate, rather than, for example, binding to telomerase, then a mutant DBD with a specific reduction in DNA association should not inhibit effectively. Y522A reduces DNA-binding by the Cdc13(DBD) 650-fold (28). When tested for its effect on telomerase action, DBD (Y522A) did not inhibit (Figure 1d). Lack of inhibition by the Y522A Cdc13(DBD) mutant is not due to inactive protein. At high concentrations, DBD (Y522A) did bind DNA by filter-binding and weak telomerase inhibition was observed when 10-fold more protein (6 µM) was used (data not shown). Thus, DNA binding is required for telomerase inhibition by Cdc13(DBD).
Full-length Cdc13p inhibits telomerase similarly to the DBD
The 198-residue DBD of Cdc13p inhibits telomerase, but the full-length protein is much larger (924 amino acids) and contains several other domains that also participate in telomere maintenance and length regulation (54, 32, 33). Therefore, we purified recombinant full-length Cdc13p to high homogeneity (Figure 2a) (32) and tested its effect on telomerase.
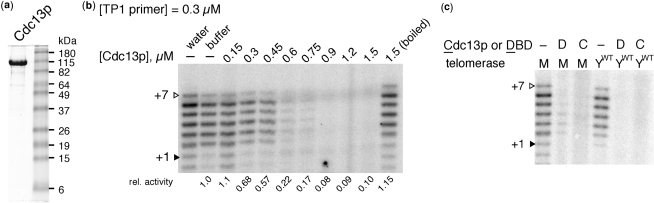
Figure 2.
Full-length Cdc13p inhibits telomerase. (a) Coomassie-stained SDS–PAGE gel of purified Cdc13p (108 kDa with His6 tag). (b) Cdc13p inhibits reconstituted yeast telomerase activity in a concentration dependent manner on TP1 primer. See Figure 1b for alignment of primer with telomerase RNA. Water, Cdc13p storage buffer and boiled Cdc13p negative controls are shown. Relative activity is listed below lanes. (c) Cdc13p (C) and its DBD (D) inhibit wild-type telomerase (YWT) purified from yeast extracts similarly to miniaturized reconstituted telomerase (M). Experimental conditions were the same as in Figure 1b.
When we preincubated Cdc13p with the telomeric primer TP1 (Figure 1b) and then added telomerase, we observed telomerase inhibition very similar to that with the DBD (Figure 2b): the inhibition was, within error, reproducibly stoichiometric with respect to primer and was abolished by heat-denaturing Cdc13p. Cdc13p also inhibited telomerase isolated from yeast overexpressing the core enzyme components Est2p and the full-length TLC1 RNA (Figure 2c). Furthermore, the amount of DBD and Cdc13p required for inhibition scaled with primer concentration (data not shown). Thus, full-length Cdc13p and its DNA binding domain both inhibit telomerase by binding to the DNA substrate, leading us to favor a model where Cdc13p prevents productive telomerase access.
Validation of a system to control the DNA-binding register of Cdc13(DBD)
We set out to determine if addition of a free 3′ DNA tail would restore telomerase activity to a Cdc13(DBD)- or Cdc13p-bound oligonucleotide. Binding of the Cdc13(DBD) to telomeric sequence occurs through a minimal consensus binding site containing a GxGT motif at the 5′ end of an otherwise GT-rich 11-mer sequence (29). Thus, longer deoxyoligonucleotides representing degenerate sequence of yeast telomeres typically contain multiple consensus Cdc13(DBD) binding sites. This degeneracy of yeast telomeres and the minimal GxGT DBD-binding motif complicate determining the ssDNA tail beyond bound Cdc13(DBD) needed for telomerase action, since simply adding telomeric sequence to a minimal DBD-binding oligonucleotide creates multiple overlapping protein-binding registers (see next section). Therefore, to test how close to the DNA 3′-end Cdc13(DBD) must be in order to inhibit telomerase action, we designed four potential telomerase substrates predicted to restrict DBD binding to the single 11-nt register at the 5′-end, allowing for a defined protruding 3′ tail. Based on prior knowledge of Cdc13(DBD) binding to DNA, our designed sequences are expected to have only one high-affinity consensus site (Figure 3a) even if lengthened at the 3′-end by ∼9 nt. As expected from the lack of perfect sequence-complementarity to telomerase RNA, these oligonucleotides were suboptimal telomerase substrates, but their ability to serve as telomerase primers was improved by adding –GG or –TGG to their 3′-ends, which increases pairing to TLC1 (Supplementary Figure 1 and data not shown). Because TEL* permitted the most telomerase activity, we used this sequence as the basis for design of primer substrates with 3′ tails of increasing length.
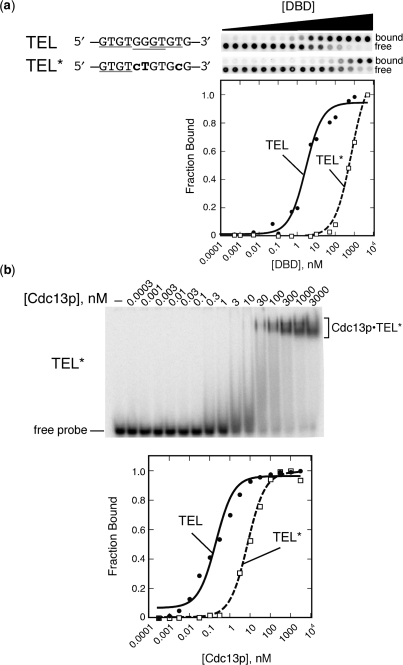
Figure 3.
Design of a single GxGT-containing mutant sequence, TEL*, that still permits binding by Cdc13(DBD) and full-length Cdc13p. (a) Like TEL, mutant TEL* binds to Cdc13(DBD), although with reduced affinity (see also Table 1). Left, schematic of mutations (bold) engineered into the canonical 11-nt Cdc13(DBD) binding site, TEL, to generate TEL*, which has only one remaining GxGT consensus site (indicated by gray bars). Right, representative membranes and quantified data from dot blot filter-binding experiments to determine the affinity of Cdc13(DBD) for the TEL and TEL* sequences. Note that filter binding was performed under high-salt (750 mM KCl) conditions (28) to reduce binding of Cdc13(DBD) to TEL, which is otherwise so tight (Kd = 3 pM at 75 mM monovalent salt) that it is technically difficult to measure accurately. (b) Like DBD, full-length Cdc13p also binds TEL* but with reduced affinity relative to TEL. A representative gel-shift using 5′-end labeled TEL* is shown (top). Cdc13p•TEL*, the protein–DNA complex in the nondenaturing gel; free probe is the unbound 5′-end labeled oligonucleotide. A graphical representation of the quantified gel shift signal is shown (bottom). All curves were fitted to a two-state binding model, as described in the Materials and Methods section.
As anticipated based on its binding consensus, Cdc13(DBD) bound to TEL*, although 220-fold more weakly than to TEL (Figure 3a and Table 1). Some recovery of binding affinity was observed when additional Gs were appended to the 3′-end of TEL*; the mechanism of this recovery is currently under investigation. Given the 3 pM Kd of Cdc13(DBD) for TEL in low salt (27), binding to the TEL-derivative, TEL*, is predicted to be sub-nanomolar under the conditions of our telomerase experiments, and thus Cdc13(DBD) would bind all of the primer (300 nM) when present at equimolar concentration (see Table 1; n.b. higher Kd values shown in Table 1 are from experiments done at 750 mM KCl to facilitate quantitative analysis (28) given the very tight interaction).
Table 1.
Binding of TEL and TEL*-based sequences to Cdc13(DBD)
| Oligonucleotide | Sequence | Kd,appa (nM) | Relative Kdb |
|---|---|---|---|
| TEL | GTGTGGGTGTG | 2.8 ± 0.6 | − |
| TEL* | GTGTcTGTcTG | 620 ± 120 | 220 |
| TEL*-a10 | GTGTcTGTcTGaaaaaaaaaa | 380 ± 57 | 140 |
| TEL*-GG | GTGTcTGTcTGGG | 120 ± 19 | 43 |
| TEL*-GGG | GTGTcTGTcTGGGG | 33 ± 5.7 | 12 |
| TEL*-GTGGG | GTGTcTGTcTGGTGGG | 11 ± 1.3 | 4 |
| TEL*-GTGTGGG | GTGTcTGTcTGGTGTGGG | 24 ± 4.3 | 8.6 |
| TEL*-GTGTGTGGG | GTGTcTGTcTGGTGTGTGGG | 13 ± 1.8 | 4.6 |
aBinding experiments were performed in 750 mM KCl to attenuate the picomolar affinity of Cdc13(DBD) for TEL sequence to the nanomolar range. Kd values shown are averages of at least three independent apparent Kd measurements ± standard error.
bKd values presented are relative to the Kd of the TEL oligonucleotide.
Full-length Cdc13p also bound well to the TEL* oligonucleotide (Figure 3b and Table 2). As reported previously, Cdc13p binds telomeric DNA with ∼100-fold weaker affinity relative to Cdc13(DBD) (27), a trend observed throughout the series of oligonucleotides used in this study (Tables 1 and 2). Cdc13p has the same consensus binding sequence as its DBD; however, it exhibits higher tolerance for changes at specific positions within the canonical oligonucleotide GTGTGGGTGTG (Tables 1, 2 and J.N.R. and D.S.W. unpublished results). The fact that Cdc13p has a similar binding consensus to DBD provides additional evidence that the major DNA-binding activity originates from the DBD (residues 497–694; Figure 1a) (27,48). Binding of Cdc13p to TEL* is consistent with these biochemical features identified on fully telomeric substrates. Cdc13p bound more weakly to TEL* than to TEL, although it was less impacted by the sequence changes (26-fold versus 220-fold). This suggests that there is a role for other portions of the protein in modulating Cdc13p interactions with DNA.
Table 2.
Cdc13p binding to non-cognate telomeric sequences shows similar binding trends to the Cdc13(DBD) with increased tolerance to non-telomeric nucleotides
| Oligonucleotide | Sequence | Kd,appa (nM) | Relative Kdb |
|---|---|---|---|
| TEL | GTGTGGGTGTG | 0.43 ± 0.17 | − |
| TEL* | GTGTcTGTcTG | 11 ± 3.8 | 26 |
| TEL*-a10 | GTGTcTGTcTGaaaaaaaaaa | 7.4 ± 2.4 | 17 |
| TEL*-GG | GTGTcTGTcTGGG | 0.81 ± 0.03 | 1.9 |
| TEL*-GGG | GTGTcTGTcTGGGG | 2.2 ± 0.7 | 5.1 |
| TEL*-GTGGG | GTGTcTGTcTGGTGGG | 3.1 ± 0.8 | 7.2 |
| TEL*-GTGTGGG | GTGTcTGTcTGGTGTGGG | 0.81 ± 0.15 | 1.9 |
| TEL*-GTGTGTGGG | GTGTcTGTcTGGTGTGTGGG | 0.3 ± 0.3 | 0.83 |
aKd values are reported as apparent Kd values and are averages of at least three independent measurements ± standard error.
bRelative Kd values presented are relative to the Kd of the TEL oligonucleotide.
We next addressed whether the TEL* sequence indeed constrains protein binding to the 5′ region of the primer DNA, even when multiple telomeric nucleotides are added at its 3′-end. We used a 3′-specific exonuclease, snake venom phosphodiesterase (SVP) (35,55), to determine the extent of protection by Cdc13p or DBD. First, TEL*–GTGGG and a control oligonucleotide containing a tract of adenosines as a 3′ tail, TEL–a10, were incubated with Cdc13p and then digested by SVP in a 60-min time course—the 5′ nucleotides were protected in a Cdc13p-dependent manner (Figure 4a). Thus, TEL was protected as expected by Cdc13p after the −a10 3′ tail was trimmed off by SVP. This assay shows that Cdc13p was specifically targeted to the engineered TEL* sequence when five telomeric 3′ nucleotides were appended, because after 10 min of exonuclease action the 5′ 12 nucleotides remained protected (Figure 4a). The transience of the TEL* protection probably originates from a relatively quick off-rate of the full-length protein (27) for the mutant TEL* consensus. These results show that ∼10-min incubations with SVP are optimal for studying Cdc13p•TEL* interactions.
To compare the binding position of Cdc13(DBD) with that of full-length Cdc13p on TEL*-based oligonucleotides, we performed titrations from substoichiometric protein levels to >2 : 1 protein-to-DNA ratios. For TEL*–GTGGG, the DBD and Cdc13p showed indistinguishable patterns of SVP 3′ nuclease protection of the TEL* sequence (Figure 4b, left). However, when we titrated these proteins on TEL*-based primers with longer 3′ tails, such as TEL*–GTGTGTGGG, we observed protection at the 3′-end. At low concentrations, Cdc13(DBD) bound the TEL* sequence at the 5′-end and, at higher concentrations, protected a 3′ site. Cdc13p, on the other hand, protected the 3′-end of TEL*–GTGTGTGGG from SVP digestion even at substoichiometric levels, suggesting that the 9-mer tail binds Cdc13p more tightly than TEL* (Figure 4b, right). Thus, Cdc13p and its DBD can be directed to the 5′-end of TEL* in the context of a telomeric substrate with short tails. However, longer telomeric tails create additional alternate binding sites for Cdc13p and the DBD, thus restricting the tail length that can be examined using this approach.
Coordinated nuclease protection and telomerase experiments unveil modes of Cdc13p inhibition
We recognized that the most direct assessment of end structure and its correlation with telomerase activity could be achieved if the telomerase and exonuclease assays were performed on the same Cdc13p•DNA complex. Therefore, we developed a coordinated assay that essentially eliminates variability that exists when exonuclease and telomerase assays are performed separately on independently formed protein•DNA complexes. This was achieved by doing the two assays in parallel: each particular Cdc13p•DNA binding reaction is split and one portion is incubated with telomerase and the other with SVP 3′ nuclease (Figure 5a). We refer to this technique as the CoNuTe (coordinated nuclease/telomerase) assay. A trace amount of 5′ 32P-labeled primer is included in the initial binding reaction to allow subsequent monitoring of protection by DBD/Cdc13p from SVP. It also serves as a loading control for telomerase assays. Tracer oligonucleotide is not detectably extended since it is at least 600-fold less abundant than the cold primer.
Figure 5.
A coordinated assay reveals that Cdc13p and its DBD inhibit telomerase similarly via distinct DNA interactions. (a) CoNuTe assay scheme used to generate data in b and c. (b) SVP assay shows that Cdc13(DBD) (D) preferentially protects TEL* even in the context of long 3′ telomeric appendages, while Cdc13p (C) protects the 3′ region of substrates with ⩾6-nt tails. Oligonucleotides tested are shown: TEL* region is boxed and vertical lines indicate complementarity to TLC1 RNA shown above, while gray arrows indicate the 7 TLC1-endoded nucleotides predicted to be added to each by telomerase (results for which are shown in c). SVP results are shown below. V-shaped arrowhead indicates completely undigested 5′-end labeled primer. TEL*, region of gel with SVP products 11 nucleotides or shorter. Arrowhead indicates full-length 5′-end labeled oligonucleotide. Totally digested, DNA not protected by binding to DBD or Cdc13p; serves as an indicator for fraction complex formation. Concentrations of Cdc13(DBD) were 0.15 µM, 0.3 µM and 0.6 µM and for Cdc13p were 0.3 µM, 0.6 µM and 1.2 µM. (c) Telomerase is consistently inhibited by the DBD and Cdc13p bound to TEL*-based oligonucleotides containing 2–9-nt appendages. Pre-labeled 5′-end-labeled primers, included in the reaction for the SVP experiment shown in (b), are indicated for the shortest primer and serves as a recovery and loading control for telomerase activity assays. The +1 and +7 telomerase products are indicated as in Figures 1 and 2 (gray arrows in b). See legend b for concentrations of DBD and Cdc13p.
We applied the CoNuTe assay to a series of TEL*-based telomeric DNA substrates with differing lengths (Figure 5b). As observed in the SVP digestion assay alone (Figure 4), SVP analysis of the protein•DNA complex used in the telomerase assay showed that, at approximately stoichiometric protein : DNA concentrations, Cdc13(DBD) was indeed bound to the 5′ TEL* region, as designed. With a telomeric tail of –TGTGGG or longer, protection of the 3′-end of the oligonucleotide occurred when Cdc13(DBD) was present at high concentration (Figure 5b, lanes 29 and 37), suggesting that Cdc13(DBD) binds these long DNA tails secondarily to the TEL* sequence. This is consistent with binding experiments that suggest short oligonucleotides are bound by Cdc13(DBD) with a high nanomolar Kd (J.N.R. and D.S.W., unpublished results). Gel shift analyses support this being a second binding event and not simply repositioning of Cdc13(DBD) on the DNA (J.N.R. and D.S.W., unpublished results).
Full-length Cdc13p showed binding similar to Cdc13(DBD) on TEL*–GG and TEL*–GTGGG but, even at concentrations below the level required for binding all of the DNA (as judged by intensity of the ‘totally digested’ signal in Figure 5b), Cdc13p protected the 3′ terminus of TEL*–TGTGGG and longer oligonucleotides (Figure 5b). The 3′ protection by Cdc13p is not due to a bona fide ‘end-binding’ preference by the full-length protein, but rather is more likely caused by a slight alteration in the sequence and length preferences for binding by full-length Cdc13p compared to its DBD alone (J.N.R. and D.S.W. unpublished results).
With well-characterized tail structures in hand, we tested telomerase activity on these primers in the presence and absence of bound Cdc13p and Cdc13(DBD). In their absence, telomerase was active on all TEL*-based primers, though lower activity was seen with TEL*–GG given its limited ability to anneal to the telomerase RNA template (Figure 5b and Supplementary Figure 1). We found that both Cdc13p and Cdc13(DBD) inhibited telomerase action on every primer tested. The DBD and full-length Cdc13p inhibited telomerase action similarly, despite the differences in binding to these telomeric substrates (Figure 5c). Residual telomerase activity correlated well with the fraction of unbound primer present (as measured by the fraction of primer fully digested in our SVP assay; Supplementary Figure 2 online), suggesting the bound state is fully inhibitory. In the presence of a short, accessible 5-nt tail, no telomerase activity was observed. Cdc13p and DBD remain 5′-localized in this condition, suggesting that an available 5-nt protruding end is too short to support telomerase activity. With the 7-nt tail –GTGTGGG, Cdc13(DBD) bound the 5′-end preferentially, whereas Cdc13p bound the 3′-end of this DNA, yet telomerase was similarly inhibited in both cases. While 3′-end-binding by Cdc13p is likely to physically occlude telomerase access, the inhibition observed in the presence of Cdc13(DBD) suggests that even a tail of seven accessible nucleotides is insufficient to support telomerase action. Telomerase was also fully inhibited when a 9-nt tail was appended to TEL*, although both Cdc13p and DBD are bound relatively closer to the 3′-end (cf. Figure 5b lanes 27–29 with lanes 35–37), inhibiting telomerase in a similar manner (cf. Figure 5c lanes 23–25 with 30–32).
Thus, our CoNuTe experiments with TEL*-based primers show that Cdc13p and its DBD alone each potently inhibit telomerase even when bound rather distant from the 3′ terminus. Furthermore, these results show that a 7-nt 3′ tail beyond a DBD binding site is still too short for productive telomerase access to DNA. However, establishing the tail length that would allow for recovery of telomerase activity to protein-bound primers is not possible with the TEL* design as longer telomeric tails necessarily create a second protein binding site, as observed with a 9-nt tail. To further explore the minimal 3′ tail length required for telomerase access we employed a different approach to constraining the protein binding register, as follows.
Action at a distance
Since Cdc13(DBD) and Cdc13p bind the 3′-end of TEL*-based oligonucleotides with long extensions, we designed an alternate set of primers for determining the minimal length of DNA required to recover telomerase activity on Cdc13p/DBD-bound primers. In this design, a canonical 11-nt Cdc13p/DBD binding TEL site at the 5′-end is separated from a robust telomerase-annealing substrate sequence –TGTGGG at the 3′-end using a non-telomeric spacing sequence (tracts of deoxyadenosines). Control experiments showed that a TGTGGG hexamer was readily extended by telomerase but was not bound tightly by Cdc13p (data not shown). This approach differs from the TEL*-based oligonucleotide series, shown in Figure 5, in three key ways: (i) the region complementary to TLC1 (–TGTGGG) is invariant at the 3′ end of all primers of the series and should hybridize similarly to TLC1 RNA within the telomerase active site (Figure 6a); (ii) there is no theoretical limit to the length of spacer nucleotides that can be inserted between the 3′-end and the Cdc13p/DBD binding site since deoxyadenosines do not create a new DBD or Cdc13p site, and (iii) as a result, the high-affinity TEL sequence, as opposed to TEL*, can be used.
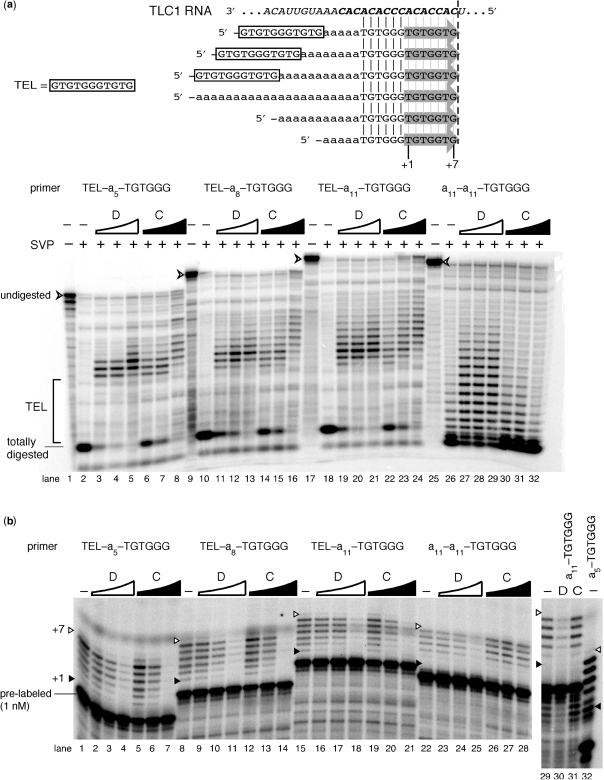
Figure 6.
‘Spacer’ tracts of deoxyadenosines separate the DBD- and Cdc13p-binding site (TEL) and a telomerase-extendible 3′-end, providing further insight into Cdc13p inhibitory mechanisms. (a) Cdc13p, like DBD, preferentially binds TEL, but it also loads towards the 3′-end at higher protein : DNA stoichiometry. SVP data are shown from CoNuTe assay [telomerase results from this CoNuTe experiment are shown in (b)]. Protein concentrations used here and in CoNuTe telomerase assay below are the same as in Figure 5. (b) Both Cdc13p and its DBD inhibit telomerase on TEL-An–TGTGGG oligonucleotides with 5, 8 or 11-nt spacers. See data in (a) for SVP results from this CoNuTe experiment, legend of Figure 5a for concentrations of DBD and Cdc13p. Primers a11-a11-TGTGGG, a11-TGTGGG and a5-TGTGGG serve as controls.
Using this series of primers in the CoNuTe assay, we examined effects of DBD and Cdc13p. As anticipated, both proteins bound preferentially to the 5′ TEL region based on SVP protection (Figure 6a). Cdc13(DBD) inhibited telomerase action substantially, though not completely (13 ± 3% residual activity), on all of the TEL–an–TGTGGG (n = 5, 8, 11) primers (Figure 6b), even when the telomerase-annealing site was as far as 11-nt away from the DBD consensus binding site. SVP analysis showed that Cdc13(DBD) did not extensively protect the 3′-end at concentrations that were largely inhibitory. Thus, by binding to TEL at the 5′-end, Cdc13(DBD) is effectively inhibiting telomerase activity from 5–11 single-stranded nucleotides away, producing an inhibitory effect when bound at a significant distance from the telomerase annealing site, which we refer to as action at a distance.
Cdc13p was an even more potent inhibitor of telomerase than the DBD for TEL–an–TGTGGG (n = 5, 8, 11) substrates, leading to complete loss of telomerase action on all primers tested (Figure 6b, lanes 7, 14, and 21). We propose that this inhibition originates from a combination of action at a distance as well as more direct inhibition due to a concentration-dependent mode of 3′ end binding. At higher protein concentrations, Cdc13p shifts 3′ in these telomerase-refractory complexes (Figure 6a, lanes 8, 16 and 24). Protection, however, is not observed on a control primer a11–a11–TGTGGG (Figure 6a, lanes 30–32) lacking the TEL binding sequence, suggesting the hexamer telomeric portion does not support stable Cdc13p association. The concentration-dependence of this 3′ shift in SVP protection suggests that once Cdc13p is bound to the 5′ TEL sequence of these oligonucleotides, it can facilitate the addition of a second protein. We looked for evidence of a second binding event by gel shift and, although we readily observe the initial binding of Cdc13p onto these primers, a higher mobility band was not observed at high Cdc13p protein concentrations (data not shown). This may be because this secondary binding interaction is rather weak (in the micromolar range) and is not stable during electrophoresis.
Close comparison of data from the control experiments provides some insights into the mechanism of the inhibition at a distance observed for both the DBD and Cdc13p. We observe that the activity of telomerase on TEL–a11–TGTGGG is 47 ± 16% higher than the same oligonucleotide with no TEL sequence at the 5′-end (a11–a11–TGTGGG; Figure 6b, lane 15 versus 22). This suggests that the TEL sequence, even 11 nt away from –TGTGGG 3′-end, has the ability to enhance telomerase activity. When Cdc13(DBD) binds to TEL, it likely restricts the contribution of TEL to telomerase activity, providing a potent alternative mechanism of inhibition.
For all of the control primers with no TEL sequence at the 5′-end (a11–a11–TGTGGG, a11–TGTGGG and a5–TGTGGG), DBD could partly inhibit telomerase at the highest protein concentration tested in the CoNuTe experiment. SVP results from the CoNuTe assays also show some binding to the tract of deoxyadenosines by DBD (Figure 6a, lanes 27–29), which is not observed for Cdc13p at these concentrations, probably due to the intrinsic affinity differences between full-length and the DBD which carry over to nonspecific interactions. The Kd for DBD interaction with hexamer TGTGGG is very close to the working concentration of primer in the CoNuTe reactions (300 nM) (data not shown). Thus, we conclude that the DBD is inhibiting telomerase through a combination of low-affinity interactions for these primers.
DISCUSSION
Studies in various organisms have shown that the primary chromosome end-binding proteins play two direct roles in regulating genome stability: (i) protecting the chromosome end, which otherwise would elicit a DNA-damage response, and (ii) regulating telomerase access to the 3′ terminus, thereby controlling chromosome end synthesis (31,32,35,56,57). Here we have shown that Cdc13p, as well as its DNA-binding domain alone, block telomerase access, thus inhibiting its action. This result is reminiscent of studies with human POT1 and ciliate telomere end binding protein (TEBPα/β), which have also been shown to inhibit telomerase in vitro (34,35,56). However, Cdc13p inhibition differs from these cases in multiple ways, as discussed below.
Cdc13p can inhibit telomerase by three different modes, whereas DBD exhibits two
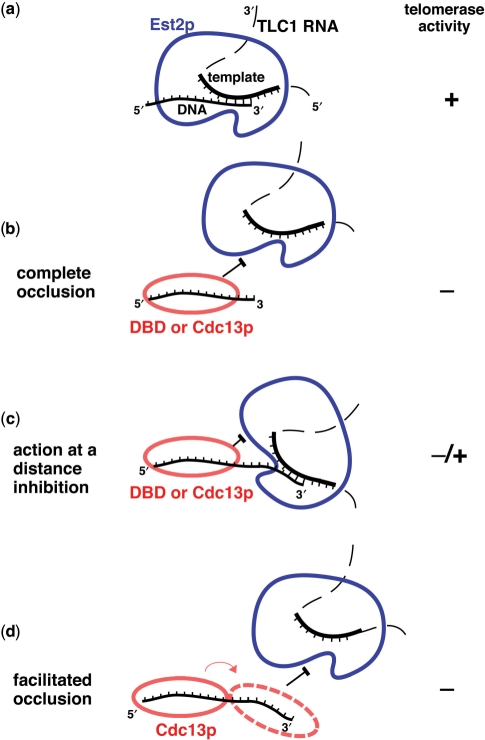
Our data reveal three related mechanisms of Cdc13p inhibition of telomerase (Figure 7). The first of the inhibitory modes occurs when the telomerase active site is simply blocked from productively accessing the 3′-end (‘complete occlusion’) (Figure 7b). This activity is readily explained in context of the structure of Cdc13(DBD) bound to TEL sequence (26): the bases are all buried in a highly aromatic protein•DNA interface, and DNA hybridization with the telomerase RNP could only occur upon gross structural rearrangement. This scenario applies to most of the inhibitory situations we have described herein, such as with primer TP1 in Figures 1 and 2, using the DBD and Cdc13p, respectively. This occlusion mechanism is supported by the observation that the DNA-binding defective Y522A point-mutant of the Cdc13(DBD) does not inhibit telomerase activity (Figure 1d). We also observed that residual telomerase activity is proportional to amount of unbound primer when it is available in excess over protein (Supplementary Figure 2). Furthermore, the fact that more Cdc13(DBD) and Cdc13p are required to inhibit telomerase action as primer concentration is increased supports a straightforward model of competitive binding of protein to DNA that regulates telomerase action.
Figure 7.
Models for the relationship between telomeric DNA binding and telomerase inhibition by Cdc13p. (a) In the absence of any telomere-binding protein, telomerase binds to its substrate and readily extends it via reverse transcription of its RNA template. (b) Complete occlusion model for inhibition of telomerase. The DNA-binding domain as well as full-length Cdc13p are capable of occluding telomerase access to the 3′-end of its substrate, even when additional 3′ nucleotides protrude beyond the Cdc13•DNA interaction interface. (c) Action at a distance inhibition. Although part of telomerase interaction with DNA is blocked by DBD or Cdc13p, some telomerase activity is achieved, presumably via interaction of the catalytic site and the very 3′-end. (d) Facilitated occlusion model. Cdc13p binds secondarily towards the 3′-end of the DNA primer in a concentration-dependent manner, leading to more potent inhibition. This is stepwise, effectively inhibiting first as indicated in c (action at a distance) and then, additionally, as shown in b (complete occlusion), when Cdc13p loads also on the 3′-end, facilitated by first loading at a more 5′ position.
The second mode of observed inhibition unveils an apparent sensitivity of yeast telomerase to Cdc13p or DBD positioned far upstream from the 3′-end on long DNA substrates (Figure 7c). This ‘action at a distance’ inhibition is less efficient than the direct occlusion inhibition, but nevertheless it represents a novel activity that provides insights into the mechanism of telomerase action and its regulation. Telomerase activity, which is enhanced by presence of 5′ telomeric sequence, is reciprocally reduced by DBD and Cdc13p when they are bound to this region of these primers (Figure 6). With only six telomeric nucleotides at the 3′-end of these oligonucleotides, this 5′ TEL•telomerase interface is particularly important for productive substrate-enzyme interaction. Binding of TEL by Cdc13(DBD) then blocks this activity-stimulating interaction, reducing telomerase activity (Figure 7c). Since this region of the primer almost certainly does not directly anneal to telomerase RNA, the inhibition by distally 5′-bound Cdc13(DBD) must be mediated through alternative interactions. One possibility is that binding to the 5′-end of the primers inhibits the ability of the primer to interact with the telomerase ‘anchor site’ (58), which has been reported to positively and negatively affect yeast telomerase activity (59). The anchor site of yeast telomerase may be similar to that of other telomerases, possessing multiple sites that bind DNA based on its specific length and sequence (60,61).
The third mode of telomerase inhibition is exhibited by Cdc13p but not its DBD alone, implicating other domains of the protein in mediating this activity. The SVP protection results for Cdc13p from the CoNuTe assay shown in Figure 6a demonstrate a pattern of increased 3′ protection at the highest Cdc13p concentrations. Furthermore, complete telomerase inhibition does not appear until there is this 3′ shift in SVP protection pattern (Figure 6). Thus, it seems that after Cdc13p initially binds to the 5′ region of these oligonucleotides, it facilitates additional 3′ DNA protection from nuclease and also telomerase action in a Cdc13p concentration-dependent manner. These results suggest an additional mode of inhibition, which we refer to as facilitated occlusion (Figure 7d). How Cdc13p protects additional 3′ sequences once bound to TEL and whether this effect is specific to these particular substrates is not yet entirely clear. It may involve binding of a second protein, but we have not directly demonstrated this.
Telomerase regulation by the major telomere end-binding proteins: yeast Cdc13p in perspective
Cdc13p and human POT1 both inhibit telomerase activity in vitro by binding DNA and occluding access of telomerase to its substrate [(35) and this work]. Human telomerase can access hPOT1•DNA substrates productively when 6 nt or more are protruding past the protein binding site at the 3′-end (35). This activity is exhibited by both the full-length POT1 protein and its DNA-binding domain. However, in budding yeast the telomeric ssDNA-binding protein serves additional functions. A 7-nt protruding tail beyond the bound Cdc13(DBD) is a relatively nonproductive substrate for yeast telomerase. In fact, even when Cdc13p or its DBD alone were bound as far away as 17 nt from the 3′-end of a telomerase substrate, telomerase activity was blocked. These data suggest that the role of Cdc13p in regulating telomerase activity is more multi-faceted than that exhibited by hPOT1.
In vivo, Cdc13p is both a positive and negative regulator of telomere length (23,62,63). A truncated version of Cdc13p lacking the C-terminal domain, which follows the DBD, leads to elongated telomeres (33). This cdc13-5 allele may behave like the DBD alone in vitro, because the N-terminus of Cdc13p has not been implicated in DNA binding, but rather in protein–protein interactions to recruit the telomerase holoenzyme to telomeres (63). With the development of quantitative assays and a reconstituted minimal system, we have a better understanding of the role of Cdc13p on telomerase function. Our inability to find a primer that, upon binding of Cdc13p, is a robust telomerase substrate suggests that the positive regulation role of Cdc13p is mediated in conjunction with other telomere factors. Approximately 273 genes have been shown to have a direct or indirect role in mediating telomere length (64,65). Many of these elicit their effects indirectly, but some are direct regulators. Est1p, a component of telomerase that interacts with Cdc13p to recruit telomerase to telomeres, is a prime candidate to overcome the basal repressive activity of Cdc13p. Like Est1p, Est3p is telomerase-associated and essential for telomerase function in vivo (53,66). Stn1p and Ten1p also interact with Cdc13p and display telomere length phenotypes (62). The in vitro system presented here will be a powerful tool to dissect the role of telomere-associated factors in mediating telomere length homeostasis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Institutes of Health [GM59414 to D.S.W., GM28039 to T.R.C., K99 GM80400 to D.C.Z., and T32 GM65103 (NIH/CU Molecular Biophysics Traineeship awarded) to J.N.R.]. Funding for open access charge: The National Institutes of Health [GM59414 and R00 GM80400].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to V. Lundblad (Salk Institute, CA) for the Cdc13p expression plasmid and to J. A. Goodrich for critical reading of the manuscript. We thank L. Sherman and the tissue culture core facility at the University of Colorado Health Sciences Cancer Center for baculovirus amplification and protein expression, A. G. Seto for telomerase purified from yeast and E. R. Podell for help with SVP experiments.
REFERENCES
- 1.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 2.Garcia CK, Wright WE, Shay JW. Human diseases of telomerase dysfunction: insights into tissue aging. Nucleic Acids Res. 2007;35:7406–7416. doi: 10.1093/nar/gkm644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singer MS, Gottschling DE. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 4.Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 5.Zappulla DC, Goodrich K, Cech TR. A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nat. Struct. Mol. Biol. 2005;12:1072–1077. doi: 10.1038/nsmb1019. [DOI] [PubMed] [Google Scholar]
- 6.Cohn M, Blackburn EH. Telomerase in yeast. Science. 1995;269:396–400. doi: 10.1126/science.7618104. [DOI] [PubMed] [Google Scholar]
- 7.Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zappulla DC, Cech TR. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc. Natl Acad. Sci. USA. 2004;101:10024–10029. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seto AG, Livengood AJ, Tzfati Y, Blackburn EH, Cech TR. A bulged stem tethers Est1p to telomerase RNA in budding yeasts. Genes Dev. 2002;16:2800–2810. doi: 10.1101/gad.1029302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- 11.Peterson SE, Stellwagen AE, Diede SJ, Singer MS, Haimberger ZW, Johnson CO, Tzoneva M, Gottschling DE. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat. Genet. 2001;27:64–67. doi: 10.1038/83778. [DOI] [PubMed] [Google Scholar]
- 12.Prescott J, Blackburn EH. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev. 1997;11:2790–2800. doi: 10.1101/gad.11.21.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teixeira MT, Arneric M, Sperisen P, Lingner J. Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- 14.Marcand S, Gilson E, Shore D. A protein-counting mechanism for telomere length regulation in yeast. Science. 1997;275:986–990. doi: 10.1126/science.275.5302.986. [DOI] [PubMed] [Google Scholar]
- 15.Mozdy AD, Cech TR. Low abundance of telomerase in yeast: implications for telomerase haploinsufficiency. RNA. 2006;12:1721–1737. doi: 10.1261/rna.134706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erdmann N, Liu Y, Harrington L. Distinct dosage requirements for the maintenance of long and short telomeres in mTert heterozygous mice. Proc. Natl Acad. Sci. USA. 2004;101:6080–6085. doi: 10.1073/pnas.0401580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Armanios MY, Chen JJ, Cogan JD, Alder JK, Ingersoll RG, Markin C, Lawson WE, Xie M, Vulto I, Phillips J.A., 3rd, et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N. Engl. J. Med. 2007;356:1317–1326. doi: 10.1056/NEJMoa066157. [DOI] [PubMed] [Google Scholar]
- 18.Hathcock KS, Hemann MT, Opperman KK, Strong MA, Greider CW, Hodes RJ. Haploinsufficiency of mTR results in defects in telomere elongation. Proc. Natl Acad. Sci. USA. 2002;99:3591–3596. doi: 10.1073/pnas.012549799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cristofari G, Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006;25:565–574. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lustig AJ, Petes TD. Identification of yeast mutants with altered telomere structure. Proc. Natl Acad. Sci. USA. 1986;83:1398–1402. doi: 10.1073/pnas.83.5.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hector RE, Shtofman RL, Ray A, Chen BR, Nyun T, Berkner KL, Runge KW. Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol. Cell. 2007;27:851–858. doi: 10.1016/j.molcel.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 22.Chang M, Arneric M, Lingner J. Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev. 2007;21:2485–2494. doi: 10.1101/gad.1588807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans SK, Lundblad V. Positive and negative regulation of telomerase access to the telomere. J. Cell Sci. 2000;113 (Pt 19):3357–3364. doi: 10.1242/jcs.113.19.3357. [DOI] [PubMed] [Google Scholar]
- 24.Bianchi A, Shore D. Increased association of telomerase with short telomeres in yeast. Genes Dev. 2007;21:1726–1730. doi: 10.1101/gad.438907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sabourin M, Tuzon CT, Zakian VA. Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol. Cell. 2007;27:550–561. doi: 10.1016/j.molcel.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke DS. Conserved structure for single-stranded telomeric DNA recognition. Science. 2002;296:145–147. doi: 10.1126/science.1068799. [DOI] [PubMed] [Google Scholar]
- 27.Anderson EM, Halsey WA, Wuttke DS. Delineation of the high-affinity single-stranded telomeric DNA-binding domain of Saccharomyces cerevisiae Cdc13. Nucleic Acids Res. 2002;30:4305–4313. doi: 10.1093/nar/gkf554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson EM, Halsey WA, Wuttke DS. Site-directed mutagenesis reveals the thermodynamic requirements for single-stranded DNA recognition by the telomere-binding protein Cdc13. Biochemistry. 2003;42:3751–3758. doi: 10.1021/bi027047c. [DOI] [PubMed] [Google Scholar]
- 29.Eldridge AM, Halsey WA, Wuttke DS. Identification of the determinants for the specific recognition of single-strand telomeric DNA by Cdc13. Biochemistry. 2006;45:871–879. doi: 10.1021/bi0512703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eldridge AM, Wuttke DS. Probing the mechanism of recognition of ssDNA by the Cdc13-DBD. Nucleic Acids Res. 2008;36:1624–1633. doi: 10.1093/nar/gkn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 33.Chandra A, Hughes TR, Nugent CI, Lundblad V. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 2001;15:404–414. doi: 10.1101/gad.861001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelleher C, Kurth I, Lingner J. Human protection of telomeres 1 (POT1) is a negative regulator of telomerase activity in vitro. Mol. Cell. Biol. 2005;25:808–818. doi: 10.1128/MCB.25.2.808-818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lei M, Zaug AJ, Podell ER, Cech TR. Switching human telomerase on and off with hPOT1 protein in vitro. J. Biol. Chem. 2005;280:20449–20456. doi: 10.1074/jbc.M502212200. [DOI] [PubMed] [Google Scholar]
- 36.Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 37.Armbruster BN, Linardic CM, Veldman T, Bansal NP, Downie DL, Counter CM. Rescue of an hTERT mutant defective in telomere elongation by fusion with hPot1. Mol. Cell. Biol. 2004;24:3552–3561. doi: 10.1128/MCB.24.8.3552-3561.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loayza D, De Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;423:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 39.Colgin LM, Baran K, Baumann P, Cech TR, Reddel RR. Human POT1 facilitates telomere elongation by telomerase. Curr. Biol. 2003;13:942–946. doi: 10.1016/s0960-9822(03)00339-7. [DOI] [PubMed] [Google Scholar]
- 40.Yang Q, Zheng YL, Harris CC. POT1 and TRF2 cooperate to maintain telomeric integrity. Mol. Cell. Biol. 2005;25:1070–1080. doi: 10.1128/MCB.25.3.1070-1080.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hockemeyer D, Sfeir AJ, Shay JW, Wright WE, de Lange T. POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J. 2005;24:2667–2678. doi: 10.1038/sj.emboj.7600733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004;18:1649–1654. doi: 10.1101/gad.1215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veldman T, Etheridge KT, Counter CM. Loss of hPot1 function leads to telomere instability and a cut-like phenotype. Curr. Biol. 2004;14:2264–2270. doi: 10.1016/j.cub.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 44.Liu D, O'Connor MS, Qin J, Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J. Biol. Chem. 2004;279:51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- 45.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 46.Seto AG, Umansky K, Tzfati Y, Zaug AJ, Blackburn EH, Cech TR. A template-proximal RNA paired element contributes to Saccharomyces cerevisiae telomerase activity. RNA. 2003;9:1323–1332. doi: 10.1261/rna.5570803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong I, Lohman TM. A double-filter method for nitrocellulose-filter binding: application to protein-nucleic acid interactions. Proc. Natl Acad. Sci. USA. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hughes TR, Weilbaecher RG, Walterscheid M, Lundblad V. Identification of the single-strand telomeric DNA binding domain of the Saccharomyces cerevisiae Cdc13 protein. Proc. Natl Acad. Sci. USA. 2000;97:6457–6462. doi: 10.1073/pnas.97.12.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prescott JC, Blackburn EH. Telomerase RNA template mutations reveal sequence-specific requirements for the activation and repression of telomerase action at telomeres. Mol. Cell. Biol. 2000;20:2941–2948. doi: 10.1128/mcb.20.8.2941-2948.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ji H, Platts MH, Dharamsi LM, Friedman KL. Regulation of telomere length by an N-terminal region of the yeast telomerase reverse transcriptase. Mol. Cell. Biol. 2005;25:9103–9114. doi: 10.1128/MCB.25.20.9103-9114.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peng Y, Mian IS, Lue NF. Analysis of telomerase processivity: mechanistic similarity to HIV-1 reverse transcriptase and role in telomere maintenance. Mol. Cell. 2001;7:1201–1211. doi: 10.1016/s1097-2765(01)00268-4. [DOI] [PubMed] [Google Scholar]
- 52.Boule JB, Vega LR, Zakian VA. The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature. 2005;438:57–61. doi: 10.1038/nature04091. [DOI] [PubMed] [Google Scholar]
- 53.Lingner J, Cech TR, Hughes TR, Lundblad V. Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc. Natl Acad. Sci. USA. 1997;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Theobald DL, Cervantes RB, Lundblad V, Wuttke DS. Homology among telomeric end-protection proteins. Structure. 2003;11:1049–1050. doi: 10.1016/s0969-2126(03)00183-7. [DOI] [PubMed] [Google Scholar]
- 55.Razzell WE, Khorana HG. Studies on polynucleotides. III. Enzymic degradation; substrate specificity and properties of snake venom phosphodiesterase. J. Biol. Chem. 1959;234:2105–2113. [PubMed] [Google Scholar]
- 56.Froelich-Ammon SJ, Dickinson BA, Bevilacqua JM, Schultz SC, Cech TR. Modulation of telomerase activity by telomere DNA-binding proteins in Oxytricha. Genes Dev. 1998;12:1504–1514. doi: 10.1101/gad.12.10.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 58.Hammond PW, Lively TN, Cech TR. The anchor site of telomerase from Euplotes aediculatus revealed by photo-cross-linking to single- and double-stranded DNA primers. Mol. Cell. Biol. 1997;17:296–308. doi: 10.1128/mcb.17.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lue NF, Peng Y. Negative regulation of yeast telomerase activity through an interaction with an upstream region of the DNA primer. Nucleic Acids Res. 1998;26:1487–1494. doi: 10.1093/nar/26.6.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Finger SN, Bryan TM. Multiple DNA-binding sites in Tetrahymena telomerase. Nucleic Acids Res. 2008;36:1260–1272. doi: 10.1093/nar/gkm866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wyatt HD, Lobb DA, Beattie TL. Characterization of physical and functional anchor site interactions in human telomerase. Mol. Cell. Biol. 2007;27:3226–3240. doi: 10.1128/MCB.02368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grandin N, Reed SI, Charbonneau M. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 1997;11:512–527. doi: 10.1101/gad.11.4.512. [DOI] [PubMed] [Google Scholar]
- 63.Qi H, Zakian VA. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated Est1 protein. Genes Dev. 2000;14:1777–1788. [PMC free article] [PubMed] [Google Scholar]
- 64.Gatbonton T, Imbesi M, Nelson M, Akey JM, Ruderfer DM, Kruglyak L, Simon JA, Bedalov A. Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet. 2006;2:e35. doi: 10.1371/journal.pgen.0020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Askree SH, Yehuda T, Smolikov S, Gurevich R, Hawk J, Coker C, Krauskopf A, Kupiec M, McEachern MJ. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc. Natl Acad. Sci. USA. 2004;101:8658–8663. doi: 10.1073/pnas.0401263101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hughes TR, Evans SK, Weilbaecher RG, Lundblad V. The Est3 protein is a subunit of yeast telomerase. Curr. Biol. 2000;10:809–812. doi: 10.1016/s0960-9822(00)00562-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.