Abstract
The Wilms’ tumour suppressor protein WT1 plays a central role in the development of the kidney and also other organs. WT1 can act as a transcription factor with highly context-specific activator and repressor functions. We previously identified Brain Acid Soluble Protein 1 (BASP1) as a transcriptional cosuppressor that can block the transcriptional activation function of WT1. WT1 and BASP1 are co-expressed during nephrogenesis and both proteins ultimately become restricted to the podocyte cells of the adult kidney. Here, we have analysed the WT1/BASP1 complex in a podocyte precursor cell line that can be induced to differentiate. Chromatin immunoprecipitation revealed that WT1 and BASP1 occupy the promoters of the Bak, c-myc and podocalyxin genes in podocyte precursor cells. During differentiation-dependent upregulation of podocalyxin expression BASP1 occupancy of the podocalyxin promoter is reduced compared to that of WT1. In contrast, the repressive WT1/BASP1 occupancy of the c-myc and Bak promoters is maintained and these genes are downregulated during the differentiation process. We provide evidence that the regulation of BASP1 promoter occupancy involves the sumoylation of BASP1. Our results reveal a dynamic cooperation between WT1 and BASP1 in the regulation of gene expression during differentiation.
INTRODUCTION
The Wilms’ tumour suppressor protein WT1 was identified on the basis of its frequent mutation in Wilms’ tumours, a paediatric malignancy of the kidneys (1,2). WT1 plays a central role in nephrogenesis and WT1 null mice die in utero, displaying complete agenesis of the kidneys and also several other organs (3,4). WT1 is a nuclear protein containing a zinc finger nucleic acid-binding domain that is alternatively spliced, leading to insertion of an additional three amino acids (KTS) between zinc fingers 3 and 4. This insertion produces a form of WT1 (+KTS) that has a high affinity for RNA, while the –KTS form binds DNA. These and other observations have led to the proposal that the –KTS form of WT1 acts as a transcriptional regulator, while the +KTS form plays a role in RNA processing. Mice that express exclusively either the +KTS or – KTS forms of WT1 show that WT1 –KTS plays a greater role in nephrogenesis and podocyte differentiation (5).
The transcriptional regulatory properties of WT1 are highly context-specific and can manifest as both transcriptional activation and repression (6). Candidate target genes of WT1 include signalling-related factors such as amphiregulin (7,8), sprouty (9), platelet-derived growth factor-A (PDGFA; 10,11), colony-stimulating factor-1 (CSF-1; 12) and Wnt 4 (13,14), regulators of cell growth/survival such as Bcl2 (15), Bak (16,17), c-Myc (18,19) and p21 (20) and also podocyte-specific proteins such as nephrin (21,22) and podocalyxin (23,24). Studies of these putative target genes in physiologically relevant cells are few and thus we have very little understanding of how WT1 regulates its target genes during development.
Several interaction partners have been described for WT1 (6). It is likely that they provide specificity to WT1 function and determine whether WT1 acts as a transcriptional activator or repressor. WT1 contains a suppression domain at its N-terminus that inhibits the function of the transcriptional activation domain (25). We previously identified Brain Acid Soluble Protein 1 (BASP1) as a WT1 transcriptional cosuppressor that mediates this inhibition (26). Very little is known of BASP1 function or structure, but it exists as multiple forms from 30 to 150 kDa that are derived from post-translational modifications and proteolytic processing (26,27). BASP1 is present in the nephrogenic intermediates of the embryonic kidney coincident with WT1 and, like WT1, BASP1 becomes restricted to the podocyte cells of the adult kidney. In the present study we analyse WT1 and BASP1 in a cell line model of podocyte differentiation. Our results demonstrate occupancy by WT1 and BASP1 of the promoters of the endogenous podocalyxin, Bak and c-myc genes. During podocyte differentiation BASP1 dissociates from the podocalyxin promoter by a mechanism involving BASP1 sumoylation and coincident with the transcriptional upregulation of podocalyxin. Conversely, WT1 and BASP1 remain bound to the Bak and c-myc promoters, both of which show differentiation-dependent repression.
MATERIALS AND METHODS
Cell lines, immunofluorescence and transfection
MPC5, HEK 293 and K562 cells were cultured as described previously (26,28,29). Transfection was performed using Lipofectamine 2000 or calcium phosphate as previously (25,30). Luciferase assays were performed as described before (30). Immunofluorescence was performed as described previously (26). Antibodies against WT1 were the C19 antibody (Santa Cruz), PML was H238 (Santa Cruz), tubulin was DM1A (Sigma) and synaptopodin was P19 (Santa Cruz). The affinity-purified rabbit anti-BASP1 antibodies have been described before (26). Sheep anti-BASP1 antibodies were generated against full-length BASP1 by Diagnostics Scotland and affinity purified.
Plasmids and proteins
pcDNA3 BASP1 has been described before (26). BASP1 K78R:K83R was generated using the Quickchange mutagenesis kit (Stratagene). pRSET BASP1, producing 6-Histidine-tagged BASP1 was generated by cloning the full-length human BASP1 cDNA into the plasmid pRSET A (Invitrogen). Bacterial induction and lysate preparation was performed as described (31) using bacteria transformed with SUMO-3 sumoylation machinery (32) and were a kind gift of James Witty and Andy Sharrocks. The podocalyxin luciferase reporter was a kind gift of Daniel Haber (23). Gel filtration was performed using a superose 6 column and Akta apparatus (GE Healthcare). For the gel filtration analysis MPC5 nuclear extracts were prepared as described previously (25). Nuclear extract measuring 0.25 ml was applied to the column at a flow rate of 0.1 ml/min and 25 × 0.5 ml fractions were collected. Proteins were precipitated as described (26) and re-suspended in SDS–PAGE loading dye. Subnuclear extracts were prepared as described previously (33).
RNA and chromatin immunoprecipitation (ChIP) analysis
Total RNA and cDNA were prepared using the Qiagen Rneasy kit and Promega Access reverse transcriptase kit. The primers for RT-PCR were; GAPDH, fwd TGA TGA CAT CAA GAA GGT GGT GAA G, Rev TTC TTG GAG GCC ATG TAG GCC AT; podocalyxin, fwd GAA AGG AGC CCT CTG GAT GA, Rev GGG CTC AGG CAC AAG TAG GT; WT1, fwd GAG ACA YAC AGG TGT GAA ACC ATT, Rev GCC ASS TGG AGT TTG GTC A; PDGFA, fwd GAC GGT CAT TTA CGA GAT ACC TC, Rev CTA CGC CTT CCT GTC TCC TC; CSF1, fwd ATG GAC ACC TGA AGG TCC TG, Rev GTT AGC ATT GGG GGT GTT GT; Bak, fwd CCA CAT CTG GAG CAG AGT C, Rev CCT GCT GGT GGA GGT AAA AA; c-myc, fwd GCG ACT CTG AAG AAG AG, Rev GTT GTG CTG GTG AGT GGA GA. Annealing temperatures for the PCR reactions were; WT1, 53°C; GAPDH, 58°C; podocalyxin, 58°C; PDGFA, 59°C;CSF1, 58°C; Bax, 58°C; c-myc, 58°C.
ChIP was performed using the ChIP kit (Upstate) as before (34). Primers for Podocalyxin ChIP were; promoter, fwd CCT CCC CAA ACG AAT AGG AT, Rev TCC AAC AGC TGC TGA GAC AC, Internal region, fwd CAT CCG TGT CTT CCC GTA GT, Rev TGG ATC AAG TTG GCA GT; PDGFA, fwd TAA GGA TCT GGA GGG TGC AG, Rev CCA AAA AGG GCA TGA GAG TC; CSF, fwd GGT TCC ATG AGG GAC TTG AA, Rev TGT GCA GCC TCC TCA AAG TA; Bak, fwd GAG GGA GCT GCA GAG AGA AC, Rev AAA CCT GGT GGG GAG TAA GG; c-myc, fwd TTC TGA CTC GCT GTA GTA ATT CCA, Rev CCT GGC TCG CAG ATT GTA AG. Annealing temperatures for the PCR reactions were; podocalyxin, 58°C; PDGFA, 59°C;CSF1, 56.5°C; Bax, 56.5°C; c-myc, 59°C.
RESULTS
WT1 and BASP1 expression in podocyte precursor cells
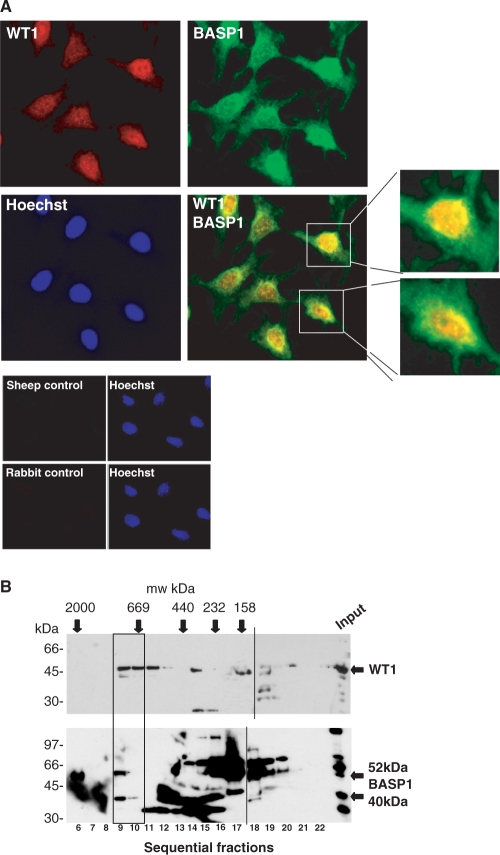
The mouse cell line MPC5 was developed as a model of podocyte differentiation and has previously been shown to express podocyte-specific markers following induction of differentiation (28,35). Figure 1A shows immunofluorescence performed with anti-BASP1 and anti-WT1 antibodies on the exponentially growing MPC5 cells. Both WT1 and BASP1 are primarily nuclear and the merged signal suggests that WT1 and BASP1 show partial co-localization. As we have observed before with a range of cell lines, the MPC5 cells contain several forms of BASP1, which arise from proteolytic processing and post-translational modification, including myristoylation (Figure 1B; lower panel input lane; 26). Our previous work showed that different cells lines express the various forms of BASP1 in different proportions. We demonstrated previously by co-immunoprecipitation that the 40-kDa form of BASP1 (BASP140), but not the 30-kDa form or the large forms (up to 150 kDa), complexes with WT1 in mouse M15 embryonic kidney cells (26). To determine the form of BASP1 that associates with WT1 in MPC5 cells we performed gel filtration with whole nuclear extracts and examined the fractions by immunoblotting with either anti-WT1 or anti-BASP1 antibodies (Figure 1B). WT1 was present in high-molecular-weight complexes of ∼700 kDa. Significantly, and consistent with our previous data, BASP140 was present in the same fractions as WT1 (26). The other forms of BASP1, ranging from 30 kDa to ∼150 kDa, are present in lower molecular weight complexes.
Figure 1.
WT1 and BASP1 localization in MPC5 podocyte precursor cells. (A) MPC5 cells were subject to immunofluorescence with rabbit anti-WT1 (red) and sheep anti-BASP1 (green) antibodies. A merge of the two images is shown in the bottom right panel, along with enlarged images of the two cells indicated. Cells were counterstained with Hoechst. Below, control immunoflourescence with each of the sheep and rabbit secondary antibodies is shown. (B) Gel filtration was performed with nuclear extracts prepared from MPC5 cells and the fractions immunoblotted with either anti-WT1 (top) or rabbit anti-BASP1 antibodies (bottom). Calibration (in kilodalton) is above the autoradiographs, fraction numbers are below and molecular weight markers (in kilodalton) are at left. WT1, BASP140 and BASP152 are each indicated by arrows. The WT1 complexes that co-fractionate with BASP1 are boxed. The dashed line indicates the alignment from two separate gels that were immunoblotted together.
Differentiation of MPC5 cells
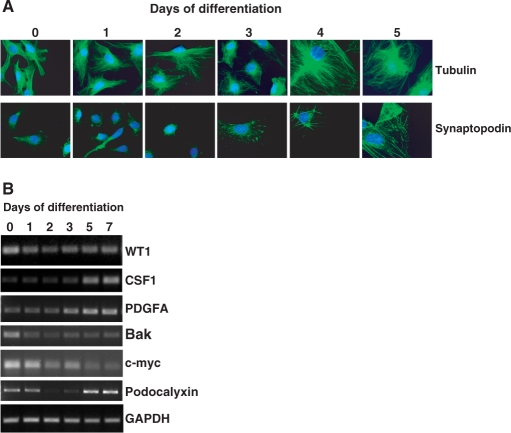
We next transferred the MPC5 cells to 37°C and removed the γ-interferon. This leads to downregulation of the temperature-sensitive T-antigen, the cells cease to proliferate and then undergo differentiation. Figure 2A shows a series of panels of immunofluorescence performed when the MPC5 cells were switched to the non-permissive temperature and allowed to differentiate over 6 days. Analysis of tubulin shows that between Days 3 and 4 the cells enlarge and undergo a significant restructuring within the cytoplasm. Detection of synaptopodin showed an elevation in abundance after Day 3 and assembly into actin fibres. This is a hallmark of podocyte differentiation and is consistent with previous studies with this cell line (28). We also analysed BASP1 and WT1 by immunofluorescence, neither of which showed any observable changes during podocyte differentiation (data not shown).
Figure 2.
Differentiation of MPC5 cells. (A) Mouse MPC5 cells were differentiated for the times indicated at top. The cells were then fixed and subject to immunofluorescence with either anti-tubulin or anti-synaptopodin antibodies (both in green), counterstaining with Hoechst. (B) MPC5 cells were differentiated for the number of days indicated and total RNA prepared. Semi-quantitative RT-PCR was performed to detect WT1, CSF1, PDGFA, Bak, c-myc, Podocalyxin and GAPDH.
RT-PCR was used to analyze the mRNA levels of WT1, GAPDH (as a control) and a selection of previously proposed WT1 target genes during the differentiation of MPC5 cells (Figure 2B). WT1 mRNA levels showed a small reduction when the cells were transferred to the non-permissive temperature, but were maintained at similar levels during differentiation. The changes in expression of the potential WT1 target genes could be grouped into three categories. Firstly, CSF-1 and PDGFA both showed increases in mRNA level from Day 3 onward of the differentiation process. Secondly, Bak and c-myc both showed differentiation-dependent decreases in mRNA. Finally, podocalyxin mRNA revealed a sharp decrease following induction of differentiation, but gradually recovered over Days 2–5.
Promoter occupancy by WT1 and BASP1
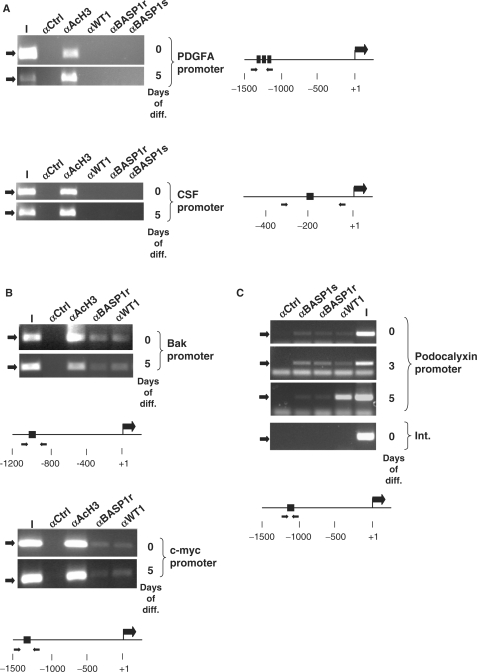
We next determined if the distinct modes of change that we observed in the regulation of the PDGFA and CSF1, Bak and c-myc and podocalyxin genes could be explained by changes in WT1/BASP1 occupancy of their promoter regions. ChIP was performed with anti-WT1 and two different anti-BASP1 antibodies (raised in sheep, BASP1s and rabbit, BASP1r) followed by amplification of the regions of the promoters that contain the proposed WT1-binding sites (schematics for each promoter are shown in Figure 3). First, we tested the PDGFA and CSF1 promoters, both of which were upregulated during MPC5 cell differentiation (Figure 3A). We could not detect binding of WT1 or BASP1 at either the PDGFA or CSF1 promoters, before or after 5 days of differentiation. A control antibody (anti-acetyl histone H3, αAcH3) did however precipitate the CSF1 and PDGFA promoter regions.
Figure 3.
Promoter occupancy by WT1 and BASP1 in differentiating MPC5 cells. (A) ChIP was performed with undifferentiated MPC5 cells (0) or MPC5 cells that had differentiated for 5 days (5). The antibodies used in the ChIP were rabbit anti-GAL4 antibodies (αCtrl), anti-acetylated histone H3 (αAcH3), rabbit anti-WT1 antibodies (αWT1), rabbit anti-BASP1 antibodies (αBASP1r) and sheep anti-BASP1 antibodies (αBASP1s). The resultant samples were subject to amplification by PCR using primers directed to the mouse PDGFA promoter (top) or CSF1 promoter (bottom). I is 1/50 of the input chromatin in each immunoprecipitation. A diagram of each promoter region is shown at right, with the positions of the WT1 DNA-binding sites and forward/reverse primer regions indicated. (B) ChIP was performed as in part A except that the Bak (top) and c-myc (bottom) promoters were analysed. (C) As in (A) except that the analysis was performed with undifferentiated MPC5 cells (0) and cells that had been allowed to differentiate for 3 (3) and 5 (5) days. The bottom panel is amplification of an internal region of the podocalyxin gene (Int.) from ChIP analysis of undifferentiated MPC5 cells.
We next tested the Bak and c-myc promoters, both of which showed downregulation of the mRNA during podocyte differentiation. ChIP revealed that both WT1 and BASP1 were present at the Bak and c-myc promoters in undifferentiated MPC5 cells (Figure 3B). After 5 days of differentiation, WT1 and BASP1 remained bound to the Bak and c-myc promoters. Thus, the ChIP data suggest that the Bak and c-myc genes are bona fide targets of WT1/BASP1 in podocytes, but do not provide an explanation for the downregulation of the activities of these genes during the differentiation process.
Based on the expression profile of podocalyxin during MPC5 cell differentiation we performed ChIP analysis at 0-, 3- and 5-day time points. In the undifferentiated podocyte cells, we detected both WT1 and BASP1 at the podocalyxin promoter (Figure 3C). At Day 3, WT1 and BASP1 were similarly engaged at the podocalyxin promoter. However, we note that, compared to the input chromatin, there was a significant elevation in the efficiency of ChIP, suggesting an increase in promoter occupancy over the cell population. Thus, the reduction in podocalyxin expression upon induction of differentiation is concomitant with an increase in promoter occupancy by WT1 and BASP1. At Day 5, we found that the WT1 ChIP at the podocalyxin promoter remained robust, but the BASP1 ChIP showed a significant reduction (with both BASP1 antibodies) when compared with WT1. Thus the subsequent recovery of podocalyxin expression occurs when the BASP1 ChIP is reduced, but WT1 remains at the promoter.
BASP140 is modified during podocyte differentiation
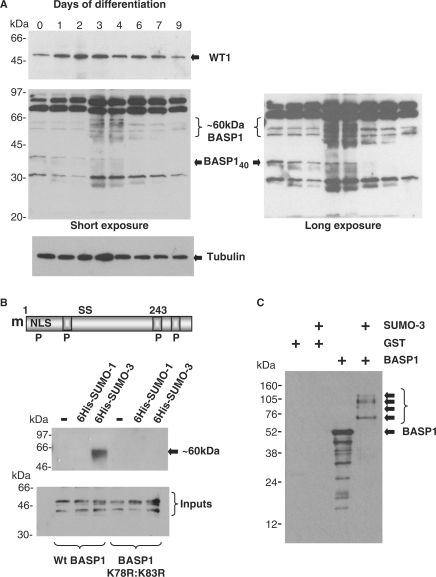
We next sought to determine if the changes in WT1 and BASP1 ChIP that we observed during the course of MPC5 cell differentiation were due to changes in WT1 and/or BASP1 abundance. We first immunoblotted extracts from a differentiation time course with anti-WT1 antibodies (Figure 4A, top panel). The level of WT1 increased over the first 3 days of podocyte differentiation and then gradually returned to the same level as that observed prior to differentiation. We note that our RT-PCR analysis revealed a decrease in WT1 mRNA (Figure 2B) suggesting that the WT1 protein is stabilized upon MPC5 cell differentiation. Indeed, recent studies have shown that WT1 is subject to regulated degradation by the proteasome (36). Analysis of the same differentiation time-course samples with anti-BASP1 antibodies revealed a significant change in the BASP1 profile (Figure 4A, lower panel shown as short and long exposures). Specifically, over Days 3–6, BASP140 showed a dramatic reduction and there is a concomitant appearance of BASP1 immunoreactive bands around the 60-kDa region. Such a size difference is consistent with the potential sumoylation of BASP140. BASP1 contains two highly conserved consensus SUMO-modification sites (K78 and K83; Figure 4B). We generated a double substitution mutant in which these two lysine residues were substituted with arginine (K78R:K83R). Wild-type HA-tagged BASP1 or the BASP1 K78R:K83R derivative were transfected into 293 cells along with either his-tagged SUMO-1 or his-tagged SUMO-3. Lysates were prepared under denaturing conditions and the his-tagged SUMO purified by nickel chelate affinity chromatography. The purified products were resolved by SDS–PAGE and immunoblotted with anti-HA antibodies (Figure 4B). The results show that wild-type BASP1, but not the BASP1 K78R:K83R derivative attaches specifically to SUMO-3, forming a product of ∼60 kDa. To confirm the direct sumoylation of BASP1, we used a bacterial system that expresses the components required to attach SUMO-3. Control bacteria, or bacteria that express SUMO-3, were transformed with a plasmid driving expression of his-tagged BASP1 or, as a control, GST. Three hours after induction, whole-cell lysates were prepared, resolved by electrophoresis and immunoblotted with anti-BASP1 antibodies (Figure 4C). In the presence of SUMO-3, slower migrating forms of BASP1 were observed that are consistent with the single and multiple attachment of SUMO-3 to BASP1. Indeed, SUMO-3 itself contains a consensus sumoylation motif and hence attaches to its substrates as chains of variable length (37).
Figure 4.
BASP1 undergoes modification during podocyte differentiation. (A) MPC5 cells were differentiated for the number of days indicated, whole-cell lysates were then prepared and immunoblotted with anti-WT1 (top), anti-BASP1 (middle) or anti-tubulin (bottom) antibodies. A short and long exposure of the anti-BASP1 blot are shown. Molecular weight markers (in kilodalton) are shown at left. WT1, tubulin, BASP140 and the ∼60-kDa forms of BASP1 are indicated. (B) A schematic of BASP1 indicating the myristoylation motif (m), nuclear localization sequence (NLS), potential phosphorylation sites (P) and conserved potential sumo-modification sites (S). The boxed regions are PEST sequences. Below, plasmids driving expression of wild-type HA-tagged BASP1 or BASP1 K78R:K83R were transfected into 293 cells alone, or with a plasmid driving expression of either his-tagged SUMO-1 or SUMO-3. Forty-eight hours later, extracts were prepared under denaturing conditions and the his-tagged derivatives purified and immunoblotted with anti-HA antibodies. Molecular weight markers (in kilodalton) are shown at left. The autoradiograph below shows an immunoblot with anti-BASP1 antibodies of whole-cell lysates before nickel chelate affinity purification. (C) GST or his-tagged BASP1 were expressed in Escherichia coli that either harbour or lack the apparatus to attach sumo-3. Bacterial lysates were resolved by electrophoresis and immunoblotted with anti-BASP1 antibodies. Molecular weight markers are shown at left. BASP152 and sumo-dependent modified forms of BASP1 are indicated by arrows.
BASP1 sumoylation can affect its subnuclear localization
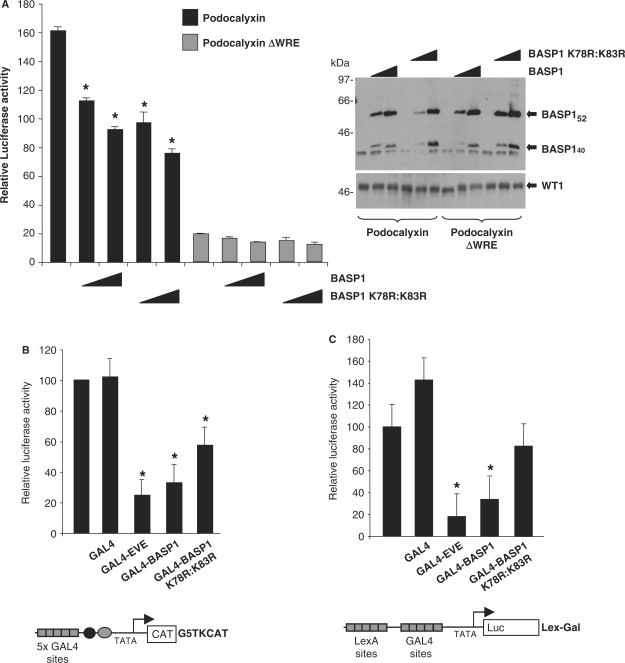
Sumoylation can elicit direct effects on transcriptional regulation (37,38). We therefore tested whether BASP1 (K78R:K83R) exhibited different transcription properties when compared with wild-type BASP1. K562 cells, which have low endogenous BASP1 but contain WT1 (26), were transfected with a luciferase reporter containing the intact podocalyxin promoter or a derivative that lacks the WT1 response element (ΔWRE; 23). The wild-type podocalyxin promoter shows a higher activity than the ΔWRE derivative, consistent with activation of the podocalyxin promoter by WT1 (Figure 5A). As we observed in our previous analyses of the amphiregulin promoter in K562 cells (26), ectopic expression of BASP1 suppresses WT1-mediated transcriptional activation of the podocalyxin promoter. The mutant BASP1 derivative (K78R:K83R) also represses the activity of podocalyxin promoter in a manner dependent upon the WT1 DNA-binding sites. Similar effects were observed when we tested a Bak promoter reporter construct and also an amphiregulin promoter reporter construct (data not shown).
Figure 5.
BASP1 exhibits sumo-dependent and sumo-independent functions as a WT1 transcriptional cosuppressor. (A) K562 cells were transfected with 0.5 μg of podocalyxin promoter-luciferase (LUC) reporter with or without (ΔWRE) the WT1-binding sites. Where indicated pcDNA3-HA-BASP1 or pcDNA3-HA-BASP1 K78R:K83R (0.1 μg and 0.5 μg) was also transfected. Forty-eight hours later, cells were harvested and luciferase activity was measured and is presented is presented relative to the luciferase activity of the wild-type podocalyxin reporter in the absence of BASP1. Bars are the mean of three independent experiments with standard deviation. Asterisks denote t-test significance better than 95%. At right an immunoblot of whole-cell lysates with anti-BASP1 antibodies is shown, (B) Human embryonic kidney 293 cells were transfected with the G5TKCAT reporter and either pcDNA3 or pcDNA3- GAL4 (residues 1–94), GAL4-EVE, GAL4-BASP1 or GAL4-BASP1 K78R:K83R. Forty-eight hours later cells were harvested, CAT activity was measured and is presented relative to reporter activity in the absence of a GAL4 fusion protein. Bars are the mean of three independent experiments with standard deviation. (C) As in (B) except that the Lex/Gal luciferase reporter was used and a plasmid driving expression of LexA-VP16 was included.
BASP1 can repress transcription independently of WT1 when linked directly to GAL4 (26). We therefore tested BASP1 K78R:K83R in this context with the ‘high basal’ thymidine kinase reporter (Figure 5B; 25) or a minimal core promoter driven to high levels of expression by LexA-VP16 (Figure 5C; 39), in both cases linked to five GAL4 DNA-binding sites. A GAL4-fusion protein containing the transcriptional repression domain of even skipped (GAL4-EVE) was used as a positive control (40). The results show that, at both promoters, GAL4-BASP1 (K78R:K83R) exhibits a diminished ability to repress transcription when compared to a GAL4-fusion containing wild-type BASP1. Taken together, the data of Figures 5A–C suggest that sumoylation of BASP1 is not required for its ability to block the transcriptional activation domain of WT1 per se, but can potentially affect its intrinsic transcriptional repression activity.
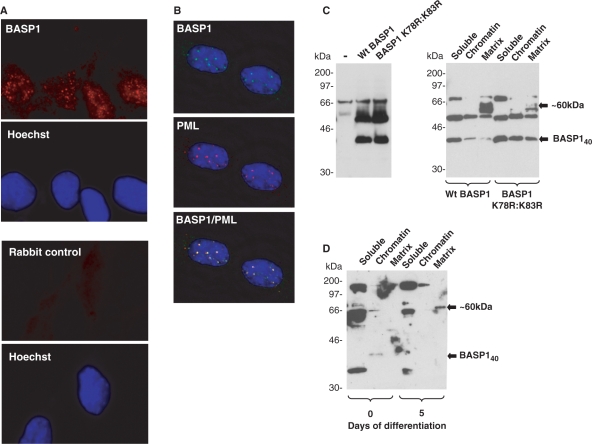
Sumoylation of transcription factors can also result in their re-localization within a cell (e.g. to the PML bodies; 37,38). Interestingly, in our immunofluorescence studies BASP1 showed a distinct speckled pattern within the nucleus (Figure 1A), which we also observed with a different anti-BASP1 antibody (Figure 6A). Co-immunofluorescence with anti-BASP1 and anti-PML antibodies followed by examination using a DeltaVision microscope revealed that BASP1 is indeed present within PML bodies in MPC5 cells (Figure 6B).
Figure 6.
Sumoylation of BASP1 affects its intranuclear localization. (A) Immunofluorescence was performed with MPC5 cells using rabbit anti-BASP1 antibodies (top, red) or secondary antibody alone (bottom). Hoechst staining for each panel is shown below. (B) MPC5 cells were subject to immunofluorescence with sheep anti-BASP1 (green) and rabbit anti-PML (red) antibodies. A merge of the two images is shown in the bottom panel. The images were captured using a Deltavision microscope. (C) Wild-type HA-tagged BASP1 or the mutant derivative (K78R:K83R) were transfected into K562 cells. Forty-eight hours later either whole-cell extracts (left) or nuclei were extracted and subfractionated (right). Samples were resolved by electrophoresis and immunoblotted with anti-BASP1 antibodies. Molecular weight markers (in kilodalton) are shown at left. BASP140 and BASP1 ∼ 60 kDa are indicated by arrows. (D) Undifferentiated MPC5 cells or MPC5 cells that had differentiated for 5 days were analysed as in (C) to detect endogenous BASP1 in the soluble, chromatin and matrix compartments of the nucleus. BASP140 and the ∼60-kDa form of BASP1 are indicated.
To determine the potential effects of BASP1 sumoylation on its subnuclear distribution we transfected wild-type BASP1 or the BASP1 K78R:K83R mutant derivative into K562 cells (Figure 6C, left panel), prepared subnuclear fractions and analysed them by western blotting with anti-BASP1 antibodies (Figure 6C, right panel). The ∼60-kDa form of BASP1 in the nuclear matrix showed a substantial reduction in cells transfected with the mutant BASP1 K78R:K83R when compared to wild-type BASP1. Significantly, concomitant with the ablation of this nuclear matrix-specific form of BASP1 caused by the K78R:K83R mutation, there was a significant increase in BASP140. Taken together with the data of Figure 4, these results suggest that the ∼60-kDa form of BASP1 arises from sumoylation of BASP140 which then accumulates in the nuclear matrix. We therefore prepared subnuclear fractions from undifferentiated MPC5 cells and also cells that had been allowed to differentiate for 5 days. BASP140 was present in the chromatin fraction of the undifferentiated MPC5 cells (Figure 6D). As expected from the data of Figure 4A, at Day 5 of differentiation, BASP140 is no longer present in any of the fractions. However, a ∼60-kDa form of BASP1 was observed in the matrix fraction of the differentiated cells. These results are consistent with the sumoylation and nuclear redistribution of BASP140 during podocyte differentiation.
DISCUSSION
Here we have analysed WT1 and BASP1 in a cell line model of podocyte differentiation, a process known to be dependent on WT1 function as a transcription factor. WT1 and BASP1 both localize at the Bak, c-myc and podocalyxin gene promoters, providing the first demonstration that BASP1 can localize to gene promoters that are regulated by WT1. We were unable to detect either WT1 or BASP1 at the CSF-1 or PDGFA promoters. It is possible that PDGFA and CSF-1 are not regulated by WT1 in the MPC5 cell line, but we cannot rule out restricted epitope accessibility of WT1 and BASP1 at these specific promoters.
We found that BASP1 can be sumoylated and presented evidence that this is a dynamic process during podocyte differentiation. Sumoylation of BASP140 can result in its redistribution within the nucleus from the chromatin to the nuclear matrix. We also found that, while sumoylation of BASP1 was not required for its function as a WT1 transcriptional cosuppressor per se, a GAL4-BASP1 derivative that was unable to undergo sumo-modification was less effective as a transcriptional repressor. Thus, the major effect of sumoylation of BASP1 is its redistribution within the nucleus. In addition, although not necessary, sumoylation of BASP1 can potentially augment its WT1 transcriptional cosuppressor function.
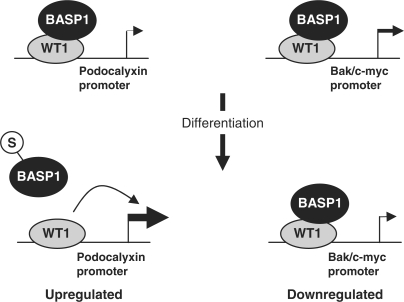
The changes in BASP1 occupancy at the podocalyxin promoter during differentiation that we observed are consistent with the subnuclear re-localization of BASP1 in a sumo-dependent manner. Moreover, there is a direct correlation between the presence of BASP140 in the chromatin, occupancy by BASP1 of the WT1-bound podocalyxin promoter and changes in podocalyxin mRNA that occur between Days 3 and 5 of podocyte differentiation. Our results therefore provide a mechanism by which WT1-mediated transcriptional activation of the podocalyxin gene is modulated by BASP1 during podocyte differentiation (Figure 7, left panel). It will be interesting to determine if the co-activator CBP is recruited to the podocalyxin promoter by WT1, and if this event is regulated by the WT1-BASP1 interaction (41).
Figure 7.
A model of dynamic cooperation between WT1 and BASP1 in transcriptional regulation during podocyte differentiation. WT1 is shown bound to the podocalyxin (left) and Bak or c-myc (right) promoters in undifferentiated MPC5 cells. Upon differentiation, the podocalyxin gene is upregulated while the Bak and c-myc promoters are downregulated. Coincident upon differentiation-dependent sumoylation of BASP1, BASP1 dissociates from the podocalyxin promoter, but remains bound to the Bak and c-myc promoters.
Unlike the podocalyxin gene, the Bak and c-myc genes were downregulated during podocyte differentiation. ChIP analysis revealed that WT1 and BASP1 both remained bound to the Bak and c-myc promoters during the differentiation process (Figure 7, right panel). The maintenance of BASP1 at the Bak and c-myc promoters is consistent with their transcriptional suppression during MPC5 cell differentiation. Although the subnuclear fractionation of the MPC5 cells revealed the depletion of BASP140 from the chromatin and accumulation in the matrix (Figure 6D), the retained ChIP of BASP1 at the Bak and c-myc promoters suggests either that some BASP1 remains within the chromatin, but is below the level of detection in this assay. Whether or not the retained BASP1 is sumoylated remains to be determined, but could potentially contribute to the downregulation of the Bak and c-myc genes during differentiation.
Taken together, our data suggest that WT1/BASP1 cooperation in transcriptional regulation is complex. Although the differentiation state-dependent presence of BASP1 at the podocalyxin and Bak/c-myc promoters is consistent with their upregulation and downregulation respectively, it is not clear why BASP1 should selectively dissociate from specific gene promoters. It is very likely that other factors are involved in the WT1-BASP1 dynamic. Indeed, our gel filtration data (Figure 1A) suggests that the WT1 and BASP1 are contained in complexes of at least 700 kDa.
The MPC5 cells used in this study provide a model for the differentiation of podocytes both morphologically and in their gene expression profile (28). However, we found that at the permissive temperature, when the MPC5 cells are immortalized by expression of T-antigen, the level of podocalyxin expression was elevated and only reduced once the T-antigen was inactivated. This coincides with a sharp increase in WT1 protein. However, other factors could contribute to the elevated level of podocalyxin mRNA in the immortalized MPC5 cells. For example, previous studies have shown that p53 transcriptionally represses the podocalyxin promoter (24). Moreover, the cell-type specific expression of podocalyxin is dependent upon correct methylation of its promoter (42). Both of these processes are sensitive to T-antigen, suggesting that the ground state of podocalyxin expression in MPC5 cells might only be achieved when T-antigen is inactivated.
Previous studies have shown that WT1 can also be sumoylated and that this specifically involves SUMO-1 (43). Our finding that BASP1 is modified by SUMO-3, but not by SUMO-1, provides potential for differential regulation of WT1 and BASP1 by sumoylation. Future studies will focus on how regulatory modifications of WT1 and BASP1 can modulate their cooperation in the control of transcriptional regulation in development and tumourigenesis.
FUNDING
The AICR, Cancer Research UK and the Wellcome Trust; studentships from the BBSRC (to L.M.G. and H.C.); a studentship from the Wellcome Trust (to K.A.); and S.G.E.R. is a Wellcome Trust Senior Fellow. Funding for open access charge: The Wellcome Trust.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Peter Mundel for the MPC5 cell line and Ron Hay for the Sumo plasmids. We are grateful to James Witty and Shen Hsi Yang for advice and reagents regarding the Sumo experiments and to Daniela Hüls for help optimizing the Gal/LexA reporter assays. We thank Neil Perkins, Andy Sharrocks, Paul Shore, Shen Hsi Yang and members of the lab for comments.
REFERENCES
- 1.Rivera MN, Haber DA. Wilms’ tumour: connecting tumourigenesis and organ development in the kidney. Nat. Rev. Cancer. 2005;5:699–712. doi: 10.1038/nrc1696. [DOI] [PubMed] [Google Scholar]
- 2.Yang L, Han Y, Saurez Saiz F, Minden MD. A tumor suppressor and oncogene: the WT1 story. Leukemia. 2007;21:868–876. doi: 10.1038/sj.leu.2404624. [DOI] [PubMed] [Google Scholar]
- 3.Hohenstein P, Hastie ND. The many facets of the Wilms' tumour gene, WT1. Hum. Mol. Genet. 2006;15:196–201. doi: 10.1093/hmg/ddl196. [DOI] [PubMed] [Google Scholar]
- 4.Scholz H, Kirschner KM. A role for the Wilms' tumor protein WT1 in organ development. Physiology. 2005;20:54–59. doi: 10.1152/physiol.00048.2004. [DOI] [PubMed] [Google Scholar]
- 5.Hammes A, Guo J, Lutsch G, Leheste J, Landrock D, Ziegler U, Gubler M, Schedl A. Two splice variants of the Wilms’ tumour 1 gene have distinct functions during sex determination and nephron formation. Cell. 2001;106:319–329. doi: 10.1016/s0092-8674(01)00453-6. [DOI] [PubMed] [Google Scholar]
- 6.Roberts SGE. Transcriptional regulation by WT1 in development. Curr. Opin. Genet. Dev. 2005;15:542–547. doi: 10.1016/j.gde.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 7.Lee SB, Huang K, Palmer R, Truong VB, Herzlinger D, Kolquist KA, Wong J, Paulding C, Yoon SK, Gerald W, et al. The Wilms tumor suppressor WT1 encodes a transcriptional activator of amphiregulin. Cell. 1999;98:663–673. doi: 10.1016/s0092-8674(00)80053-7. [DOI] [PubMed] [Google Scholar]
- 8.Kim HS, Kim MS, Hancock AL, Harper JC, Park JY, Poy G, Perantoni AO, Cam M, Malik K, Lee SB. Identification of novel wilms' tumor suppressor gene target genes implicated in kidney development. J. Biol. Chem. 2007;282:16278–16287. doi: 10.1074/jbc.M700215200. [DOI] [PubMed] [Google Scholar]
- 9.Gross I, Morrison DJ, Hyink, D.P. Georgas K, English MA, Mericskay M, Hosono S, Sassoon D, Wilson PD, Little M, et al. The receptor tyrosine kinase regulator Sprouty1 is a target of the tumor suppressor WT1 and important for kidney development. J. Biol. Chem. 2003;278:41420–41430. doi: 10.1074/jbc.M306425200. [DOI] [PubMed] [Google Scholar]
- 10.Gashler AL, Bonthron DT, Madden SL, Rauscher FJ, Collins T, Sukhatme VP. Human platelet-derived growth factor A chain is transcriptionally repressed by the Wilms tumor suppressor WT1. Proc. Natl Acad. Sci. USA. 1992;89:10984–10988. doi: 10.1073/pnas.89.22.10984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang ZY, Madden SL, Deuel TF, Rauscher FJ. The Wilms' tumor gene product, WT1, represses transcription of the platelet-derived growth factor A-chain gene. J. Biol. Chem. 1992;267:21999–22002. [PubMed] [Google Scholar]
- 12.Harrington MA, Konice B, Song A, Xia XL, Fredericks WJ, Rauscher FJ. Inhibition of colony-stimulating factor-1 promoter activity by the product of the Wilms' tumor locus. J. Biol. Chem. 1993;268:21271–21275. [PubMed] [Google Scholar]
- 13.Davies JA, Ladomery M, Hohenstein P, Michael L, Shafe A, Spraggon L, Hastie N. Development of an siRNA-based method for repressing specific genes in renal organ culture and its use to show that the Wt1 tumour suppressor is required for nephron differentiation. Hum. Mol. Genet. 2004;13:235–246. doi: 10.1093/hmg/ddh015. [DOI] [PubMed] [Google Scholar]
- 14.Sim EU, Smith A, Szilagi E, Rae F, Ioannou P, Lindsay MH, Little MH. Wnt-4 regulation by the Wilms' tumour suppressor gene, WT1. Oncogene. 2002;21:2948–2960. doi: 10.1038/sj.onc.1205373. [DOI] [PubMed] [Google Scholar]
- 15.Mayo MW, Wang CY, Drouin SS, Madrid LV, Marshall AF, Reed JC, Weissman BE, Baldwin AS. WT1 modulates apoptosis by transcriptionally upregulating the bcl-2 proto-oncogene. EMBO J. 1999;18:3990–4003. doi: 10.1093/emboj/18.14.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrison DJ, English MA, Licht JD. WT1 induces apoptosis through transcriptional regulation of the proapoptotic Bcl-2 family member Bak. Cancer Res. 2005;65:8174–8182. doi: 10.1158/0008-5472.CAN-04-3657. [DOI] [PubMed] [Google Scholar]
- 17.Kim MK, Mason JM, Li C-M, Berkofsky-Fessler W, Jiang L, Choubey D, Grundy PE, Tycko B, Licht JD. A pathologic link between Wilms tumor suppressor gene, WT1, and IFI16. Neoplasia. 2008;10:69–78. doi: 10.1593/neo.07869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han Y, San-Marina S, Liu J, Minden MD. Transcriptional activation of c-myc proto-oncogene by WT1 protein. Oncogene. 2004;23:6933–6941. doi: 10.1038/sj.onc.1207609. [DOI] [PubMed] [Google Scholar]
- 19.Udtha M, Lee SJ, Alam R, Coombes K, Huff V. Upregulation of c-MYC in WT1-mutant tumors: assessment of WT1 putative transcriptional targets using cDNA microarray expression profiling of genetically defined Wilms' tumors. Oncogene. 2003;22:3821–3826. doi: 10.1038/sj.onc.1206597. [DOI] [PubMed] [Google Scholar]
- 20.Englert C, Maheswaran S, Garvin AJ, Kreidberg J, Haber DA. Induction of p21 by the Wilms' tumor suppressor gene WT1. Cancer Res. 1997;57:1429–1434. [PubMed] [Google Scholar]
- 21.Guo G, Morrison DJ, Licht JD, Quaggin SE. WT1 activates a glomerular-specific enhancer identified from the human nephrin gene. J. Am. Soc. Nephrol. 2004;15:2851–2856. doi: 10.1097/01.ASN.0000143474.91362.C4. [DOI] [PubMed] [Google Scholar]
- 22.Wagner N, Wagner KD, Xing Y, Scholz H, Schedl A. The major podocyte protein nephrin is transcriptionally activated by the Wilms' tumor suppressor WT1. J. Am. Soc. Nephrol. 2004;15:3044–3051. doi: 10.1097/01.ASN.0000146687.99058.25. [DOI] [PubMed] [Google Scholar]
- 23.Palmer RE, Kotsianti A, Cadman B, Boyd T, Gerald W, Haber DA. WT1 regulates the expression of the major glomerular podocyte membrane protein Podocalyxin. Curr. Biol. 2001;11:1805–1809. doi: 10.1016/s0960-9822(01)00560-7. [DOI] [PubMed] [Google Scholar]
- 24.Stanhope-Baker P, Kessler PM, Li W, Agarwal ML, Williams BR. The Wilms tumor suppressor-1 target gene podocalyxin is transcriptionally repressed by p53. J. Biol. Chem. 2004;279:33575–33585. doi: 10.1074/jbc.M404787200. [DOI] [PubMed] [Google Scholar]
- 25.McKay LM, Carpenter B, Roberts SGE. Regulation of the Wilms' tumour suppressor protein transcriptional activation domain. Oncogene. 1999;18:6546–6554. doi: 10.1038/sj.onc.1203046. [DOI] [PubMed] [Google Scholar]
- 26.Carpenter B, Hill KJ, Charalambous M, Wagner KJ, Lahiri D, James DI, Anderson JS, Schumacher V, Royer-Pokora B, Mann M, et al. BASP1 is a transcriptional cosuppressor for the Wilms’ tumour suppressor protein WT1. Mol. Cell. Biol. 2004;24:537–549. doi: 10.1128/MCB.24.2.537-549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mosevitsky MI. Nerve ending “signal” proteins GAP-43, MARCKS, and BASP1. Int. Rev. Cytol. 2005;245:245–325. doi: 10.1016/S0074-7696(05)45007-X. [DOI] [PubMed] [Google Scholar]
- 28.Mundel P, Reiser J, Zuniga Mejia Borja A, Pavenstadt H, Davidson GR, Kriz W, Zeller R. Rearrangements of the cytoskeleton and cell contacts induce process formation during differentiation of conditionally immortalized mouse podocyte cell lines. Exp. Cell Res. 1997;236:248–258. doi: 10.1006/excr.1997.3739. [DOI] [PubMed] [Google Scholar]
- 29.Richard DJ, Schumacher V, Royer-Pokora B, Roberts SGE. Par 4 is a coactivator for a splice-isoform specific transcriptional activation domain in WT1. Gene Dev. 2001;15:328–339. doi: 10.1101/gad.185901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng W, Roberts SGE. A core promoter element downstream of the TATA box that is recognized by TFIIB. Gene Dev. 2005;19:2418–2423. doi: 10.1101/gad.342405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glossop JA, Dafforn TR, Roberts SGE. A conformational change in TFIIB is required for activator-mediated assembly of the preinitiation complex. Nucleic Acids Res. 2004;32:1829–1835. doi: 10.1093/nar/gkh504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mencia M, de Lorenzo V. Functional transplantation of the sumoylation machinery into. Protein Expr. Purif. 2004;37:409–418. doi: 10.1016/j.pep.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 33.Lee JH, Skalnik DG. CpG-binding protein is a nuclear matrix- and euchromatin-associated protein localized to nuclear speckles containing human trithorax. Identification of nuclear matrix targeting signals. J. Biol. Chem. 2002;277:42259–42267. doi: 10.1074/jbc.M205054200. [DOI] [PubMed] [Google Scholar]
- 34.Elsby LM, O’Donnell AJM, Green LM, Sharrocks AD, Roberts SGE. Assembly of TFIIB at a promoter in vivo requires contact with RNA polymerase II. EMBO R. 2006;7:898–903. doi: 10.1038/sj.embor.7400767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shankland SJ, Pippin JW, REiser J, Mundel P. Podocytes in culture: past, present, and future. Kidney Int. 2007;72:26–36. doi: 10.1038/sj.ki.5002291. [DOI] [PubMed] [Google Scholar]
- 36.Makki MS, Heinzel T, Englert C. TSA downregulates Wilms tumor gene 1 (Wt1) expression at multiple levels. Nucleic Acids Res. 2008;36:4067–4078. doi: 10.1093/nar/gkn356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hay RT. SUMO: a history of modification. Mol. Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 38.Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 39.Yang SH, Bumpass, D.C. Perkins ND, Sharrocks AD. The Ets domain transcription factor Elk-1 contains a novel class of repression domain. Mol. Cell. Biol. 2002;22:5036–5046. doi: 10.1128/MCB.22.14.5036-5046.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKay LM, Carpenter B, Roberts SGE. Evolutionary conserved mechanism of transcriptional repression by even skipped. Nucleic Acids Res. 1999;27:3064–3070. doi: 10.1093/nar/27.15.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang W, Lee SB, Palmer R, Ellisen LW, Haber DA. A functional interaction with CBP contributes to transcriptional activation by the Wilms tumor suppressor WT1. J. Biol. Chem. 2001;276:16810–16816. doi: 10.1074/jbc.M009687200. [DOI] [PubMed] [Google Scholar]
- 42.Butta N, Larrucea S, Alonso S, Rodriguez RB, Arias-Salgado EG, Ayuso MS, Gonzalez-Manchon C, Parrilla R. Role of transcription factor Sp1 and CpG methylation on the regulation of the human podocalyxin gene promoter. BMC Mol. Biol. 2006;7:17–25. doi: 10.1186/1471-2199-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smolen GA, Vassileva MT, Wells J, Matunis MJ, Haber DA. SUMO-1 modification of the Wilms' tumor suppressor WT1. Cancer Res. 2004;64:7846–7851. doi: 10.1158/0008-5472.CAN-04-1502. [DOI] [PubMed] [Google Scholar]