Abstract
The tumor suppressor p53 contributes to the cellular fate after genotoxic insults, mainly through the regulation of target genes, thereby allowing e.g. repair mechanisms resulting in cell survival or inducing apoptosis. Unresolved so far is the issue, which exact mechanisms lead to one or the other cellular outcome. Here, we describe the interferon regulatory factor-2-binding protein-2 (IRF2BP2) as a new direct target gene of p53, influencing the p53-mediated cellular decision. We show that upregulation of IRF2BP2 after treatment with actinomycin D (Act.D) is dependent on functional p53 in different cell lines. This occurs in parallel with the down-regulation of the interacting partner of IRF2BP2, the interferon regulatory factor-2 (IRF2), which is known to positively influence cell growth. Analyzing the molecular functions of IRF2BP2, it appears to be able to impede on the p53-mediated transactivation of the p21- and the Bax-gene. We show here that overexpressed IRF2BP2 has an impact on the cellular stress response after Act.D treatment and that it diminishes the induction of apoptosis after doxorubicin treatment. Furthermore, the knockdown of IRF2BP2 leads to an upregulation of p21 and faster induction of apoptosis after doxorubicin as well as Act.D treatment.
INTRODUCTION
The loss of the tumor suppressor p53 appears to be a crucial event in the development of cancer, since p53 plays an essential role in the cellular stress response program. Germline mutations of p53 lead to a strong cancer predisposition in mice and in humans (1). Various forms of stress such as DNA damage, oncogene activation, hypoxia, viral infection or nutrient deprivation all induce the activation of p53 (2). Besides the regulation of transcription-independent apoptotic pathways (3), p53 mediates many of its key functions by transactivation or transrepression of its target genes. It can recognize and bind to specific DNA sequences, thereby recruiting general and specialized transcriptional coregulators (4). Interestingly, p53 can regulate target genes that promote growth arrest and DNA repair, ultimately leading to cellular survival, as well as target genes that eventually trigger cell death (5). Thus, one of the most important current research questions is how the decision between the different p53-mediated response pathways is being made.
In general, the outcome of the p53-activation appears to depend on the respective cell type, the nature of the stress signal itself or the kind and extent of DNA damage, the presence of survival factors in the cell and, if present, inappropriate activity of oncogenes (6). The activity of p53 itself seems to be influenced by the overall levels of p53, post-translational modifications of p53, the presence or absence of transcriptional cofactors and possible differences in the p53-binding sequences of the potential target genes (4,5).
For the induction of certain subsets of p53 target genes distinct transcriptional factors are required, such as CARM1, PRMT and JMY, that cooperate with the CBP/p300 family of acetyl transferases to activate specific p53 target promoters (7) as well as the long-range chromatin modifier hCAS/CSE1L, which affects different classes of p53 target genes (8).
For the induction of apoptosis, p53 alone appears not to be sufficient, but it seems to require other factors binding to cis-elements in promoters of crucial genes (9) like proteins of the ASPP-family (10) or the NF-κB transcription factor that has been shown to influence the outcome of p53-activity under certain cellular conditions (11). Also, post-translational modifications appear to influence the p53 response. Phosphorylation of serine 46, which was reported to be crucial for p53-induced apoptosis, can be regulated through the antagonizing actions of p53DIP1, a kinase cofactor and WIP1, a phosphatase (12,13). The fact that both proteins are induced by p53 shows how tightly the activity of p53 is regulated. While the binding of p53 to its binding sites requires acetylation of its C-terminus (14,15), the activation of certain apoptotic target genes, like Bax and Puma, is associated with acetylation of lysine 120 within the DNA-binding domain (16).
For the induction of growth arrest other cofactors are responsible, e.g. proteins which mediate inhibition of cell-cycle progression or those that impede on the induction of apoptosis, like Miz (17), Hzf (18) or SLUG (19). Some of these factors are target genes of p53 and although they do not necessarily interact directly with p53, they can form regulatory loops, delaying or inhibiting p53-mediated apoptosis.
Within the interferon regulatory factor (IRF) family of transcription factors that play important roles in antiviral defense, immune response and cell-growth regulation, IRF1 and IRF2 are generally described as a tumor suppressor and an oncoprotein, respectively (20). IRF1 was identified as the first member of the IRF-family being induced upon IFN-γ and activating the transcription of IFN-γ responsive genes. IRF2 is also induced by IFN-γ. While IRF2 can bind to the same target gene sequences as IRF1, it can act as an antagonist of the latter (21,22). In addition to the repression of IRF1 induced target genes, IRF2 has also been shown to promote cell proliferation through the transcriptional activation of the histone H4 gene (23,24). Both IRFs are able to recruit the histone acetyl transferases PCAF and CBP/p300 to target promoters resulting in enhanced transcriptional activity (25). Experiments in nude mice and expression pattern analysis in several different kinds of tumors support the hypothesis that IRF1 can function as a tumor suppressor and IRF2 as an oncoprotein (26–29).
Interestingly, IRF1 can induce the growth arrest mediating p21 gene together with p53 (30). Although a binding site for IRF1 was identified in the p21 promoter (31), Dornan et al. (32) showed that this transactivation depends on the interaction between IRF1 and p300. On the other hand, knockdown of IRF2 or mutation of its repressive C-terminus was shown to upregulate p21 transcription (29,33), suggesting that this part of the protein, at least in part, is responsible for its oncogenic potential. Recently, a new nuclear factor, IRF2BP2, has been identified that interacts with the C-terminal repression domain of IRF2 and that has the potential of an IRF2-dependent transcriptional corepressor (34).
Here, we describe IRF2BP2 as a direct target gene of p53. We show that p53 can bind in vivo and in vitro to a p53 consensus-binding site upstream of the transcriptional start site of the IRF2BP2 gene and that p53 can transactivate its transcription. Functioning as a transcriptional cofactor IRF2BP2 was able to influence the p53-mediated transactivation of target genes as shown by luciferase assays. Upon induction, IRF2BP2 promotes cell-cycle arrest and seems to interfere with p53-mediated apoptosis after chemotherapeutic treatment. When IRF2BP2 is knocked down with small interfering RNA, stress-induced p53-mediated apoptosis increases. We propose a function of the new p53 target gene IRF2BP2 in a feedback-loop, influencing the outcome of p53-activation in the direction of growth arrest instead of apoptosis.
MATERIAL AND METHODS
Cell culture conditions
The human osteosarcoma cell line U2OS expressing wild-type p53, the human osteosarcoma Saos-2 cell line, the human embryonic kidney cell line HEK 293T, the synovial sarcoma Syo-1, the lung adeno-carcinoma NCI-H460 and the breast carcinoma MDA-MB231 cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum at 37°C. The Tet-on inducible expression system (BD Biosciences) was used to generate a cell line that conditionally express TAp73α in Saos-2 cells as described previously (35). To generate p53 knockdown cells the following oligos were cloned into a pSuper-vector, which was transfected into U2OS cells: pSuperp53-s 5′-gatccccgactccagtggtaatctacttcaagagagtagattaccactggagtctttttggaaa-3′ and pSuperp53-as 5′-agcttttccaaaaagactccagtggtaatctactctcttgaagtagattaccactggagtcggg-3′. Transfections were performed using the calcium phosphate precipitation method (BES). Stable clones were selected with 1 µg/ml puromycin (Sigma).
The U2OS cells were treated for chromatin immunoprecipitation (ChIP) experiments and expression analysis, the Syo-1, NCI-H460 and MDA-MB231 cells for expression analysis with 5 nM actinomycin D (Act.D) (Sigma) for 24 h. For FACS analysis U2OS cells were treated either with 2.5 nM Act.D for 12 h, or with 1 nM doxorubicin (Sigma) for 18 or 24 h. IRF2BP2 knockdown cells were treated for 24 or 48 h either with 5 nM Act.D or with 1 nM, doxorubicin, respectively. The inducible Saos-2 cell line was first induced with Doxycyclin (Sigma) for 24 h and then additionally also treated with 5 nM Act.D for another 24 h.
ChIP
The ChIP was basically done as described by Denissov et al. (36). Briefly, cells were crosslinked for 30 min in 1% formaldehyde at room temperature. Crosslinking was stopped by adding 125 mM glycine. The cells were washed three times, resuspended in lysis buffer and sonicated using a Bioruptor sonicator (Diagenode) for 15 min at high power, 30 s ON, 30 s OFF. Antibody incubation of chromatin was performed overnight at 4°C in incubation buffer supplemented with 0.1% BSA with protein A/G-Sepharose beads (Santa Cruz) and 1 μg of antibody. For U2OS cells, DO1 antibody (BD PharMingen) was used to immunoprecipitate p53. For ChIP experiments in the Saos-2 inducible cell line, BL906 (Abcam) was used for p73α. Beads were washed sequentially with four different washbuffers at 4°C. Precipitated chromatin was eluted from the beads in 1% SDS and 0.1 M NaHCO3 at room temperature for 20 min. Protein–DNA crosslinks were reversed at 65°C for 4 h in the presence of 0.2 M NaCl, after which DNA was isolated by phenol–chloroform extraction and ethanol precipitated with 10 µg of glycogen. Real-time (RT) PCR was performed using the SYBR Green mix (Biorad) with the MyIQ machine (Biorad). Primers used for RT PCR are available upon request. Enrichment of the ChIP material was calculated as recovery over an unspecific control (myoglobin exon 2).
Electrophoretic mobility shift assay (EMSA)
TnT T7 Quick Coupled Transcription/Translation System (Promega) was used for in vitro expression of p53 starting with 500 ng of pcDNA3-p53. 3 ng of radioactively labeled oligonucleotides derived from the motifs 1–4 or the p53-binding motif of the p21-promoter were incubated with IVT p53 and 1 μl of p53 antibody (pAb421; CalBiochem). 150 ng of unlabeled wild-type or mutant p21-oligonucleotide was used as competitor where indicated.
RNA isolation and RT-PCR
Total RNA was extracted using the RNeasy Mini Kit according to protocol (Qiagen). For cDNA synthesis, retrotranscription was performed using 1 μg of RNA with oligo dT anchor primers, dNTPS, DTT, buffer and Superscript Retrotranscriptase (Invitrogen). The cDNA was analyzed by RT-PCR using a MyIQ machine (Biorad). Primers used for RT-PCR are available upon request. After quantitative PCR the obtained values were normalized to GAPDH and induction of target genes was calculated against the untreated control cells.
Construction and transfections of plasmids
Full length cDNA of IRF2BP2A was subcloned from pEF.IRF-2BP2A, a kind gift of S. Goodbourn, into pcDNA3.1-FLAG using BamHI and XhoI.
The IRF2BP2–luciferase construct was made by PCR amplification of a 500 bp region out of genomic DNA from U2OS cells using the primers 5′-ggagtcaccgtatactttacttttca-3′ and 5′-ttttgaagcctctgacttcg-3′ linked to attachment sites of the Gateway Clonase system (Promega). The binding site was subcloned into a promoter-containing pGL3-vector by using Gateway clonase. The mutation of the central p53-binding motif was introduced into the Gateway entry clone using the Quickchange Kit (Stratagene).
U2OS and Saos cells were transiently transfected using the calcium phosphate precipitation method (BES).
Transactivation assays
Saos and U2OS cells were transiently cotransfected with 500 ng p53-responsive luciferase reporter constructs containing either a sequence derived from the identified p53-binding site upstream of the IRF2BP2 gene, a p21- (a gift of Y. Tu) or a Bax-promoter (a gift of M. Oren), 50 ng pRL-TK Renilla reporter (Promega), 2 μg pcDNA3.1FLAG-IRF2BP2, 200 ng pcDNA3.1FLAG-p53 (a gift of K. Vousden) by calcium phosphate transfection. Empty pcDNA6 was used to equalize the amount of transfected cDNA. Cells were lyzed and luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) according to protocol.
Cell-cycle analysis and quantification of apoptosis
Transfected and non-transfected cells treated as described above were prepared for analysis as follows: cells were grown in 10 μM BrdU for 1 h, before fixation in ethanol. Cells were then stained with anti-BrdU-antibody (Dako) and propidium iodide (Sigma) for 30 min at room temperature. DNA content was analyzed by flow cytometry (Becton Dickinson FACScan). The data was analyzed using CellQuest Pro software.
RNAi-mediated knockdown of IRF2BP2
Stable knockdown of IRF2BP2 was mediated by lentiviral infection of the following oligos targeting mRNA sequences of IRF2BP2, which were cloned into the pLKO-vector, containing a puromycin resistance gene flanked by LTRs used for genomic integration: IRF2BP2 si1 5′-caacggcttctccaagctaga-3′; IRF2BP2 si2 5′-gcagttgcaagaacagcaagg-3′; IRF2BP2 si3 5′-aactgcttgaattgtatatat-3′. pLKO-Luciferase si 5′-cgtacgcggaatacttcga-3′ served as control. 1.8 μg of the single pLKO–si constructs, 1.8 μg packaging vector R8.91 and 300 ng VSV-G were transfected into HEK 293T cells to produce viral particles using Lipofectamin2000 (Invitrogen). After 24 h the supernatant was filtrated and then used to transduce target U2OS cells. The transduction was repeated after 24 h. On Day 3 the selection was started with 1 µg/ml puromycin (Sigma). Transient knockdown of IRF2BP2 was achieved by using Dharmacon ON-TARGETplus siRNAs, according to manufacturer's manual.
RESULTS
Identification of IRF2BP2 as a direct target gene of p53
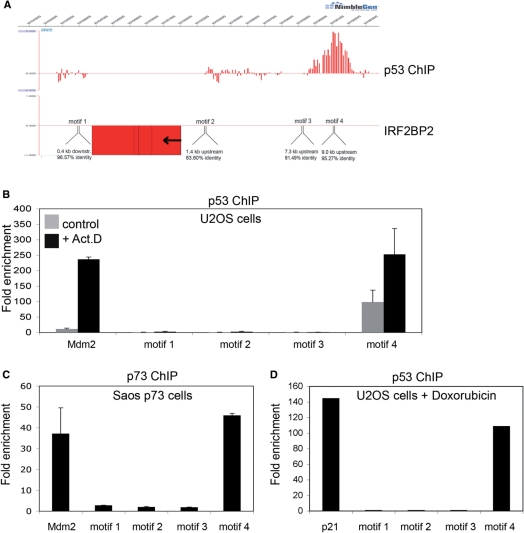
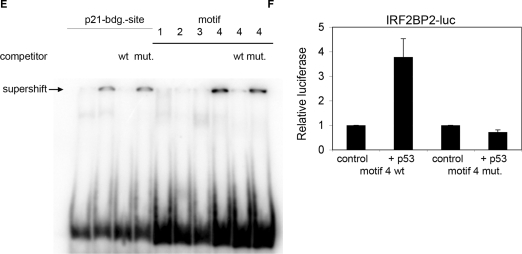
To obtain more insight into the molecular mechanisms how the activation of p53 upon genotoxic stress can lead to growth arrest and DNA repair or apoptosis, we have previously performed a systematic analysis to identify new binding sites for p53 in the human genome applying the ChIP-on-chip technique on Act.D treated U2OS cells (35). One of the interesting newly identified target genes was IRF2BP2, an interacting partner of IRF2 (34). As shown in Figure 1A, a p53-binding site was found 9 kb upstream of the promoter of the IRF2BP2 gene. Within this binding site we could identify the p53 consensus binding motif, consisting of two halfsites with the sequence RRRCWWGYYY (37). Scanning the surrounding sequences of the IRF2BP2 gene for more potential p53 consensus binding motifs with the p53MH algorithm (38), we found four different motifs, each of them with a score of more than 80% of the score of the ideal p53-binding motif (Figure 1A and Table 1). These included a motif (motif 4) upstream of the transcriptional start site, corresponding to the site which was found by ChIP-on-chip. Since we had found only this last motif bound by p53 in the ChIP-on-chip analysis we wanted to know if the other predicted sites were really not bound in vivo by endogenous p53. To this end, we performed targeted ChIPs using an anti-p53 antibody to precipitate p53–chromatin complexes from U2OS cells treated for 24 h with 5 nM Act.D and PCR-primers for all potential binding sites. We verified that only the sequence for motif 4 is bound by p53 with a significant enrichment (Figure 1B). The binding strength of p53 towards the motif 4 is comparable to its binding strength to the known target gene Mdm2 (Figure 1B). In addition, we found that p53 is also bound to motif 4 in untreated U2OS cells, although to a much lesser extent (Figure 1B). To obtain a complete analysis of the 5-kb region upstream of the IRF2BP2 transcriptional start site we designed primers every 500 bp and tested for further p53 binding in this region. In the whole region no p53 binding was observed (Supplementary Figure 1). To analyze if IRF2BP2 is a common target gene of the p53 family, we compared the binding properties of p53 with those of the closely related protein p73α. For this purpose, we used a Saos osteosarcoma cell line stably expressing p73α under the control of an inducible promoter (35). After the induced expression of p73α, the cells were treated with 5 nM Act.D for 24 h prior to chromatin isolation. DNA bound by p73α was precipitated using anti-p73 antibody and RT-PCR was performed with the indicated primers. We verified that the p53-binding site upstream of the IRF2BP2 gene is bound by p73α in this Saos cell line upon Act.D treatment to the same extent as the known binding site of Mdm2 (Figure 1C). To test whether binding of p53 to the binding site upstream of IRF2BP2 is restricted to Act.D treatment or a more general response, we also performed targeted ChIP from U2OS cells treated for 24 h with 1 nM doxorubicin (Figure 1D). We found that the IRF2BP2-binding site was also strongly bound by p53 upon doxorubicin treatment. Next we tested if one or more of the identified motifs were directly bound by p53. Therefore we performed EMSA on radioactive labeled nucleotides spanning each of the predicted p53-binding sites, motifs 1–4, (Figure 1E). The p21-binding motif was used as a positive control. After incubation of the different nucleotides together with p53 and an anti-p53 antibody, a supershift could be observed after incubation of p53 with motif 4, but not with the motifs 1–3. To show the specificity of the binding an excess of unlabeled p21-binding motif was added, either the wild-type or a mutant sequence (as indicated). To analyze whether the identified p53-binding site upstream of the IRF2BP2 gene also has a potential transactivating property, we cloned a 500-bp fragment containing the sequence around motif 4 into a luciferase vector. This vector contains a basic promoter and therefore can be used for testing potential enhancer activity. Upon cotransfection of this pGL3-IRF2BP2-luc vector together with p53 into Saos cells, luciferase activity increased 3–4-fold, compared to the luciferase vector transfected alone (Figure 1F). A mutation in the central binding motif for p53 from CATG to TATA abolished the transactivation, thus proving a specific and functional p53-binding site.
Figure 1.
Identification of a p53-binding site upstream of the IRF2BP2 gene. (A) ChIP-on-chip profile from U2OS cells expressing p53. A region on chromosome 1 including the IRF2BP2 gene is visualized using Signalmap (Nimblegen Systems Inc.). Cells were treated with 5 nM Act.D for 24 h prior to chromatin isolation and ChIP with anti-p53 antibody. In the upper track the log 2 ratio of p53 ChIP material over input signal is shown. Every bar represents one probe on the array. The lower track displays the location of the IRF2BP2 gene including a schematic representation of p53 motifs identified by the p53MH algorithm in the vicinity of the IRF2BP2 gene. Also shown are their locations relative to the IRF2BP2 gene. Motif 4 corresponds to the p53-binding site previously identified by ChIP-on-chip. (B) Binding of endogenous p53 in U2OS cells to the putative p53-binding motifs. Cells were treated with 5 nM Act.D for 24 h or left untreated prior to chromatin isolation. Targeted ChIP was performed with primers for Mdm2 and for sequences spanning the predicted motifs. Shown is the enrichment in fold over the negative control (myoglobin). Error bars represent the standard deviation of three independent experiments. (C) Saos cells expressing p73α were induced with Doxycyclin for 24 h prior to treatment with 5 nM Act.D for another 24 h before chromatin was isolated. Targeted ChIP was performed using an anti-p73 antibody. Extent of binding of p73α was determined by RT-PCR with primers used in (B). Error bars represent the SD of three independent experiments. (D) U2OS cells were treated with 1 nM doxorubicin for 24 h, before chromatin was isolated. Targeted ChIP was done as in (B). (E) EMSA shows specific binding of p53 to the DNA-sequence containing motif 4. Labeled oligonucleotides spanning either the p53-binding site in the p21-promoter or DNA sequences of the IRF2BP2 surrounding regions containing the motifs 1–4 were incubated with in vitro translated p53. In all lanes except lane 1 p53-antibody was added to induce a supershift of the protein–DNA complexes. An unlabeled oligo containing the wild-type or a mutant p53-binding site of the p21-promoter was used as a competitor to show specificity of binding. (F) The motif 4 containing p53-binding site can function as an enhancer. Saos cells were transfected with a luciferase-construct containing either the wild-type or a mutant DNA sequence derived from motif 4, alone or together with p53. Shown is the fold activation of the IRF2BP2 luciferase construct over the vector alone. Error bars represent the standard deviation of three independent experiments.
Table 1.
Putative p53-binding motifs close to the IRF2BP2 gene
| Sequence | Location relative to IRF2BP2-gene | Identity score (%) | |
|---|---|---|---|
| Motif 1 | TGGCATGCCC <6 bp> CAACATGCCC | 0.4 kb downstream | 96.57 |
| Motif 2 | GGACTAGCCT <8 bp> CACCAAGTGC | 1.4 kb upstream | 83.60 |
| Motif 3 | AAACATGCCT <9 bp> ACACATGTGA | 7.3 kb upstream | 81.49 |
| Motif 4 | AAACATGTCA <0 bp> GGACATGCCT | 9.0 kb upstream | 95.27 |
Thus, we verified an in vivo binding site for p53 upstream of the IRF2BP2 gene, which had originally been identified by our ChIP-on-chip approach. Although more binding motifs were predicted by the p53MH algorithm, we found that they were not bound by p53 in U2OS cells after Act.D as well as doxorubicin treatment. Furthermore, this newly identified p53-binding site could also be bound by p73α. In addition, we found that this sequence is also bound in vitro by p53 and it can potentially function as a p53-dependent enhancer sequence as shown by luciferase assays.
Activated p53 increases the expression of IRF2BP2
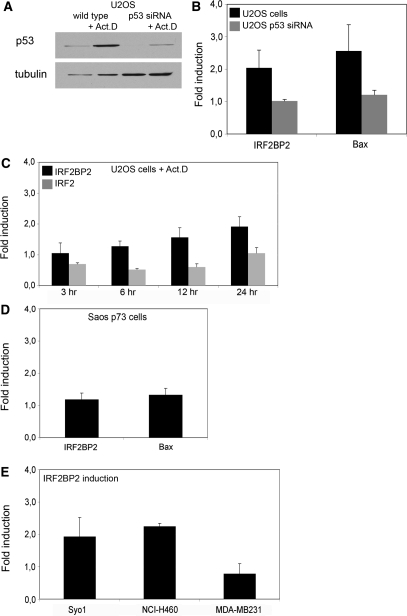
Next we examined whether p53 binding also leads to a change in IRF2BP2 expression. To test to which extent the regulation of IRF2BP2 expression is dependent on p53, we compared its expression change in wild-type U2OS cells with the expression change in U2OS p53 knockdown cells upon stress treatment. Although the knockdown of p53 is not complete in these cells, the p53 protein levels are markedly decreased (Figure 2A). The respective cells were either treated with 5 nM Act.D for 24 h or left untreated. In treated cells the expression of IRF2BP2 increases 2-fold compared to untreated cells, which is comparable to the p53-induced activation of the known p53 target gene Bax under the same physiological conditions (Figure 2B). However, the cells containing the p53 knockdown do not show increased mRNA levels of IRF2BP2 nor Bax upon treatment, thus suggesting a p53-dependent induction of IRF2BP2. A similar induction of IRF2BP2 mRNA could be observed after treatment with Etoposide (data not shown). To further validate that the observed induction of IRF2BP2 indeed is mediated directly through p53, we studied the kinetics of its expressional change (Figure 2C). The level of IRF2BP2 mRNA increases 6 h after stabilization of p53 and reaches its highest level after 24 h of treatment, showing kinetics which could reflect a direct response to p53 and which are comparable to the kinetics of Bax mRNA induction upon the same treatment (data not shown). In our previous study (35) we also found a binding site for p53 close to the IRF2 gene. Because IRF2 is a protein directly interacting with IRF2BP2 (34), we were interested to analyze whether the expression of IRF2 also changed over time upon treatment with Act.D. We observed a decrease of IRF2 expression to 50% during the first 12 h of treatment. However, after 24 h the levels of IRF2 mRNA were restored to the same relative mRNA levels as of unstressed cells (Figure 2C). Since p73α could bind to the upstream sequence of IRF2BP2 as well, we wanted to know whether p73α was also able to induce the expression of the IRF2BP2 gene. Therefore we analyzed RNA isolated from the p73α-expressing Saos cells, which had been treated with Act.D. Although p73α can bind the same site as p53, induction of p73α does not lead to an elevated expression of IRF2BP2 mRNA (Figure 2D), while known target genes like GADD45 or Mdm2 are upregulated after p73α-induction (data not shown).
Figure 2.
Upregulation of IRF2BP2 after p53 induction. (A) Induction of p53 in U2OS wild-type or p53 knockdown cells. Cells were treated with 5 nM Act.D for 24 h, before whole cell extracts were prepared and protein levels of p53 and tubulin as loading control were determined. (B) Expression changes in U2OS wild-type and p53 knockdown cells after treatment with 5 nM Act.D for 24 h, compared to untreated cells. Whole RNA was extracted and quantitative RT-PCR was performed, with primers for IRF2BP2 and Bax. Upregulation is shown after normalization against GAPDH and calculated over the untreated control. Error bars represent the SD of three independent experiments. (C) Kinetics of expression changes of IRF2BP2 and IRF2 upon Act.D treatment. U2OS cells were treated for the indicated time points with 5 nM Act.D before whole RNA was isolated, followed by quantitative RT-PCR for IRF2BP2 and IRF2 expression. Error bars result from two independent experiments. (D) Induction of target genes by p73α in Saos t.o. cells induced for 24 h and treated with 5 nM Act.D for another 24 h prior to RNA isolation. Error bars represent the SD of three independent experiments. (E) Human cell lines containing wild-type p53 (Syo1, NCI-H460) or no functional p53 (MDA-MB231) were treated with 5 nM Act.D for 24 h. Changes in the levels of IRF2BP2 mRNA are shown after normalization to GAPDH and calculated against untreated controls. Error bars result from two independent experiments.
As expression levels of a certain mRNA species may vary between several cell types we analyzed more cell lines for their expression of IRF2BP2 upon stress treatment: synovial sarcoma cell line Syo-1 and NCI-H460 cells derived from a lung adeno-carcinoma, both containing wild-type p53 and a breast carcinoma cell line, MDA-MB231 without functional p53. The expression of IRF2BP2 was analyzed under the same conditions as used before (Figure 2E). The induction of IRF2BP2 mRNA in the Syo-1 and NCI-H460 cell lines was comparable to the one observed in U2OS cells. Strikingly, in MDA-MB231 cells the level of IRF2BP2 mRNA did not increase upon Act.D treatment and those cells do not contain functional p53. Thus, it appears that the expression change of IRF2BP2 mRNA upon Act.D treatment seems to depend on the activation of functional p53 in several different tumor cell lines.
IRF2BP2 can influence the transactivation function of p53
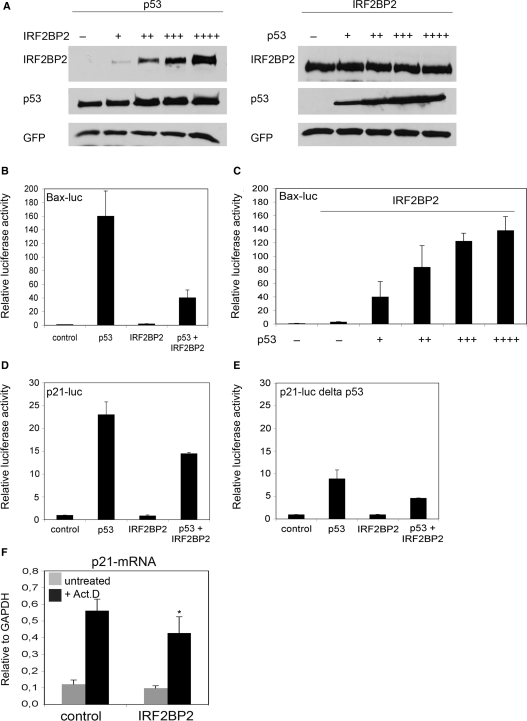
Recently the IRF2BP2 related protein IRF2BP1 was identified as an ubiquitin ligase (39). Since IRF2BP2 shares the C3H4 RING finger with IRF2BP1, which mediates the transfer of ubiquitin molecules and since p53 is regulated through ubiquitination (40), we decided to test whether IRF2BP2 could influence the protein stability of p53 by transfecting U2OS cells with p53 and increasing amounts of IRF2BP2 or vice versa (Figure 3A). We could not detect that IRF2BP2 changes the levels of cotransfected p53 nor that p53 could change the protein levels of transfected IRF2BP2. To unravel the functional consequence of the IRF2BP2 upregulation, we investigated whether IRF2BP2 could have an influence on the transactivation of p53-dependent promoters, which would be comparable to its described function as a transcriptional corepressor of IRF2 (34). Therefore we transfected U2OS or Saos cells with p53 and IRF2BP2 expression constructs together with different luciferase reporter constructs containing known target gene sequences. Neither p53 nor IRF2BP2 have an effect on the basic pGL3–luciferase construct (pGL3–luc), which does not contain a p53-response element (data not shown). The cotransfection of p53 together with a Bax–luciferase construct (Bax–luc) leads to a 150 times higher luciferase activity of the Bax–luc construct than the transfection of the reporter construct alone (Figure 3B). Very interestingly, the IRF2BP2 overexpression alone has no effect on the transactivation of the Bax–luc construct, but upon cotransfection of p53 and IRF2BP2, we could observe an inhibition of the p53-mediated activity of Bax–luc up to 75% (Figure 3B). To test whether this effect was dose-dependent we increased the amounts of transfected p53 from 200 ng to 2000 ng, while holding the IRF2BP2 levels equal. At the highest amount of cotransfected p53, the luciferase level is restored to the level reached with p53 alone (Figure 3C). To test whether the inhibition mediated by IRF2BP2 was specific for the Bax–luc construct, we also tested a p21–luciferase construct (p21–luc) in cotransfections with p53 and IRF2BP2. Also with the p21–luc construct we observed that the p53-mediated transactivation was impeded by the cotransfection of IRF2BP2 (Figure 3D). We also tested a p21–luc construct in which the distal, high-affinity binding site of p53 in the p21 promoter was deleted, p21–luc delta p53 (41). As expected, the p53-transactivation of this deletion construct was reduced to less than 50% of the activity of the wild-type construct, although a certain luciferase activity was still seen probably due to the low-affinity binding site of p53 present in the p21-promoter (Figure 3E). Interestingly, IRF2BP2 coexpression was able to reduce also the p53-mediated transactivation of this deletion luciferase construct. Thus, the effect of IRF2BP2 on the transactivation activity of p53 does not seem to be dependent exclusively on the high-affinity binding site of the p21 promoter. To analyze the effect of IRF2BP2 on the expression of endogenous genes regulated by p53, we transfected cells with IRF2BP2 and subsequently treated them with Act.D before changes in the mRNA-levels were analyzed. We found a slight, but significant downregulation of p21–mRNA in IRF2BP2 transfected cells compared to the control cells (Figure 3F). Thus, IRF2BP2 seems to have a repressing effect on the transactivation function of p53 towards its target genes and this repression can be overcome by increasing the protein levels of p53, without changing the protein stability of p53.
Figure 3.
IRF2BP2 is able to influence the transactivation of p53-responsive promoters. (A) Neither p53 nor IRF2BP2 have an influence on each other's protein stability. U2OS cells were transfected either with p53 and increasing amounts IRF2BP2 or vice versa and 24 h later the indicated proteins were detected by western Blot. GFP staining was used to ensure equal transfection efficiency. (B) IRF2BP2 can impede on the p53-transactivation of Bax-luc. A luciferase reporter construct containing the Bax-promoter was transfected into U2OS cells together with p53, IRF2BP2 or an empty vector, as indicated. Luciferase activity was normalized to Renilla. Error bars represent the SD of three independent experiments. (C) Higher p53 levels can reverse the effect of IRF2BP2 on Bax–luc. Increasing amounts of p53-plasmid were transfected, ranging from 200 ng to 2000 ng, together with Bax-luc and 2000 ng of IRF2BP2 where indicated. (D) IRF2BP2 can impede on the p53-transactivation of p21–luc. Saos cells were transfected with a luciferase-construct containing the p21-promoter together with p53, IRF2BP2 or empty vector where indicated. (E) The high-affinity site of p53 is not required for IRF2BP2 function. Saos cells were transfected with p53, IRF2BP2 or empty vector where indicated together with a luciferase construct containing the p21-promoter lacking the high-affinity binding site for p53. (F) Overexpressed IRF2BP2 can influence the expression of p21. U2OS cells were either tranfected with empty vector or IRF2BP2. Cells were treated with 5 nM Act.D for 24 h before RNA was isolated and mRNA of p21 was analyzed by qPCR. Error bars represent the SD of three independent experiments. The asterisk indicates statistical significance shown by Student's t-test (P < 0.05) compared to control transfection.
Overexpression of IRF2BP2 influences the induction of apoptosis after genotoxic stress
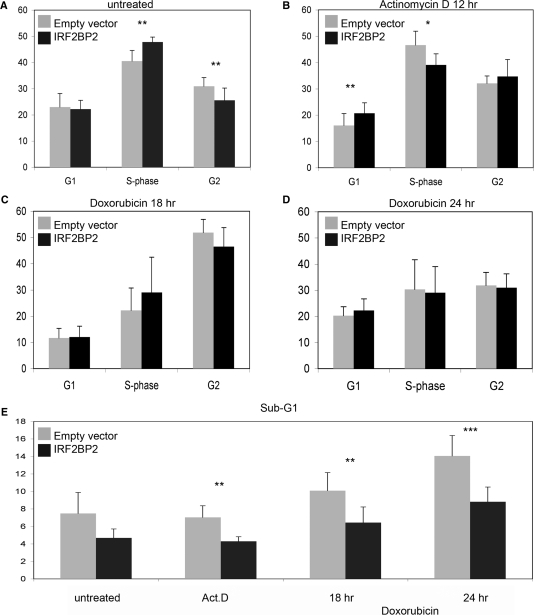
Since IRF2BP2 was described as a binding partner of IRF2, which is involved in the positive regulation of the cell cycle, we wanted to examine a possible role of IRF2BP2 during cell cycle regulation, especially upon chemotherapeutic treatment, since we had found IRF2BP2 to be upregulated during the p53 response to Act.D. Two major response pathways mediated by p53 after genotoxic insults are growth arrest and apoptosis. To stimulate these responses we treated U2OS cells transfected with IRF2BP2 either with 2.5 nM Act.D or 1 nM doxorubicin, for 12–24 h to induce growth arrest or apoptosis, respectively. The efficiency of tranfection was monitored by GFP, to ensure that at least 60% of the cells were transfected. To discriminate between the different phases of the cell-cycle BrdU staining was performed. In untreated IRF2BP2 transfected cells, a slight but significant increase in the S-phase population (48% in IRF2BP2 transfected cells versus 41% in the control cells) and a decrease of cells in the population in G2-phase (26% in IRF2BP2 transfected cells versus 31% in the control cells) can be observed compared to the control cells transfected with empty vector DNA only (Figure 4A). Upon Act.D treatment we observed a slight increase of S-phase in the control cells (from 41% to 47%), whereas IRF2BP2 transfected cells show a statistical significant decrease in S-phase (from 47% to 39%) (Figure 4B). Furthermore, the G1-population of the control cells was reduced from 23% to 16%, which was not seen in the IRF2BP2 transfected cells. On the other hand we observed an increase in the G2-phase of Act.D-treated, IRF2BP2 transfected cells (from 26% to 35%). The treatment of cells with doxorubicin for 18 h or 24 h induces several changes in the cell-cycle distribution, without significant differences between the IRF2BP2- and the control transfected cells (Figure 4C and D). However, doxorubicin leads after 18 h to a strong increase in the G2-phase in both transfected cell populations, up to 50%, whereas the populations of G1- and S-phase cells decrease. This is reversed after 24 h of treatment with doxorubicin, the amounts of cells in each phase of the cell cycle vary now between 20–30%.
Figure 4.
Cell-cycle changes after overexpression of IRF2BP2. Cell-cycle analysis of U2OS cells transfected either with an empty vector or with an IRF2BP2-expression construct. Transfection efficiency was monitored by GFP and BrdU-staining was performed for analysis. (A) The transfected cells were left untreated prior to harvest. (B) After transfection the cells were treated with 2.5 nM Act.D for 12 h prior to cell-cycle analysis. (C) Treatment of the transfected cells with 1 nM of doxorubicin for 18 h. (D) Treatment of the transfected cells with 1 nM of doxorubicin for 24 h. (E) Determination of levels of apoptosis after transfection of cells with IRF2BP2 or empty vector. Sub-G1 population was measured after treatment of cells by propidium iodide staining. All SD result from three biological replicas, each performed in duplicate. Statistical significance was determined by Student's t-test (*P < 0.05; **P < 0.01; ***P < 0.001).
Finally, we examined the induction of apoptosis after treatment in cells transfected with IRF2BP2 compared to cells transfected with empty vector only. After transfection and treatment, PI-staining was performed and the Sub-G1 population was determined (Figure 4E). Upon treatment with Act.D the levels of apoptosis do not increase compared to untreated cells, although the Sub-G1 population in the IRF2BP2 transfected cells is significantly smaller compared to the control transfection (4% in IRF2BP2 transfection; 7% in the control cells). The treatment with doxorubicin induces apoptosis already after 18 h, which increases even more after 24 h. This occurs mainly in the cells transfected with empty vector to a total of 14% after 24 h of treatment. In IRF2BP2 transfected cells the population of apoptotic cells is significantly smaller, even after 24 h less than 9% of the cells undergo programmed cell death.
From this we concluded, that IRF2BP2 can induce some changes in the cell cycle of living cells, mainly it changes the S-phase population, which increases in untreated and decreases in Act.D treated cells, compared to control transfections. Interestingly, IRF2BP2 appears to impede or diminish the induction of cell death after apoptotic stimulation.
More rapid apoptosis after chemotherapeutic treatment in IRF2BP2 knockdown cells
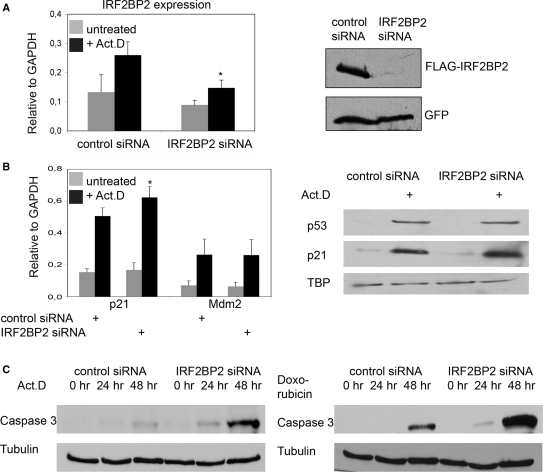
To investigate the function of IRF2BP2 during cell cycle under more physiological conditions, we downregulated the endogenous levels of IRF2BP2 by introducing siRNAs against three different regions of the IRF2BP2 mRNA. We monitored the efficiency of the knockdown by analyzing the transcript levels of IRF2BP2 in U2OS cells 24 h after treatment with 5 nM Act.D and observed a significant reduction of IRF2BP2 expression, especially after Act.D treatment compared to the cells transfected with non-targeting siRNAs (Figure 5A). Furthermore, we analyzed the knockdown efficiency by western blotting. Due to the lack of a specific IRF2BP2 antibody we transfected FLAG-IRF2BP2 into the knockdown and the control cells. Figure 5A shows that the knockdown of the transfected protein is under these circumstances almost complete. Equal levels of transfection were monitored by GFP cotransfection.
Figure 5.
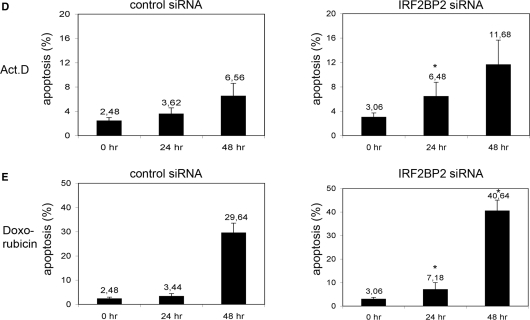
Knockdown of IRF2BP2 renders cells sensitive to chemotherapeutic stress. (A) Knockdown of IRF2BP2 in U2OS cells using lentiviral vectors or ON-TARGETplus oligo siRNAs. Efficiency of the knockdown was monitored by analyzing the transcript-levels of IRF2BP2 relative to GAPDH and by transient retransfection of 500 ng FLAG-IRF2BP2 followed by western blotting with anti-FLAG antibody. GFP-staining was used to monitor transfection efficiency. SD was derived from three biological replicas. Asterisk indicates statistical significance shown by Student's t-test (P < 0.05) compared to non-targeting siRNA. (B) Expression analysis showing the induction of p21 and Mdm2 mRNA in the control and the IRF2BP2 knockdown cells after treatment with 5 nM Act.D for 24 h. SD was derived from three independent experiments. Asterisk indicates statistical significance shown by Student's t-test (P < 0.05) compared to non-targeting siRNA. Western blot showing the levels of p53 and p21 in the IRF2BP2 knockdown and the control siRNA cells. TBP was used as a loading control. Whole cell extracts of the IRF2BP2 knockdown and the control cells were prepared after treatment with either Act.D or doxorubicin and 15 μg of protein from each sample were used for western blot and stained against the indicated proteins. (C) Activation of Caspase 3 after chemotherapeutic treatment. IRF2BP2 knockdown or control cells were treated for the indicated times with 5 nM Act.D (left panel) or with 1 nM doxorubicin (right panel) before Caspase 3 levels were analyzed. Tubulin was used as loading control. (D) Quantification of apoptosis determined by FACS analysis of the control (left) and the IRF2BP2 knockdown (right) cells. Cells were treated for the indicated times with 5 nM Act.D prior to harvest and propidium iodide staining for FACS. Apoptotic Sub-G1 population is shown as percentage of all cells. SD was derived from three independent experiments. Asterisks indicate statistical significance shown by Student's t-test (P < 0.05) compared to the non-targeting siRNA. (E) Quantification of apoptosis after treatment with 1 nM doxorubicin. After the indicated time points IRF2BP2 knockdown and control cells were harvested and propidium iodide staining was performed. Apoptotic Sub-G1 population is shown as percentage of all cells. SD was derived from three independent experiments. Asterisks indicate statistical significance shown by Student's t-test (P < 0.05) compared to the non-targeting siRNA.
First, we analyzed whether the p21 and Mdm2 expression levels were influenced by the knockdown of IRF2BP2. We observed a small, but reproducible and significant increase of p21 expression levels upon Act.D treatment in the IRF2BP2 knockdown cells in comparison to the control cells. The Mdm2 expression level, on the other hand, stays the same in IRF2BP2 knockdown as well as the control cells (Figure 5B). To test the possibility that IRF2BP2 influences protein stability of p53 itself in a feedback-loop comparable to the one of Mdm2, we analyzed whether the protein levels of p53 and p21 were affected in the IRF2BP2 knockdown cells (Figure 5B). In untreated cells very low levels of p53 and p21 are present, without any difference between the IRF2BP2 knockdown and the control cells. After treatment with Act.D, p53 and also p21 accumulates. While p53 levels seem to be very similar in the knockdown and the control cells, we observed slightly higher p21 protein levels in the IRF2BP2 knockdown cells upon Act.D treatment. Thus, IRF2BP2 knockdown most likely does not change the stability of p53 or Mdm2, but slightly increases the levels of p21 mRNA and protein.
Since overexpressed IRF2BP2 had an influence on the cell cycle, we also examined the effect of IRF2BP2 knockdown followed by stress treatment. Using BrdU staining we wanted to detect differences in the cell cycle between control and IRFBP2 knockdown cells, but we could not observe any significant changes in the distribution of cells in G1-/S- or G2-phase (data not shown). Very interestingly, we found again significant differences in the induction of apoptosis (Figure 5C). Control and IRF2BP2 knockdown cells were treated either with 5 nM Act.D or with 1 nM doxorubicin for the indicated time points, before the activation of Caspase 3 was analyzed. After both treatments the cells containing the IRF2BP2 knockdown show higher levels of active Caspase 3 already after 24 h. To quantify this effect we measured the Sub-G1 population by FACS after propidium iodide staining. Under unstressed conditions the IRF2BP2 knockdown and the control siRNA transfection show similar levels of spontaneous apoptosis (Figure 5D). After the induction of a stress response, mediated through 5 nM Act.D, the knockdown of IRF2BP2 leads to a slight accumulation of apoptotic cells after 24 h, this population increases after 48 h in total almost 4-fold compared to the untreated cells. The respective increase of the apoptotic population in the control cells is significantly smaller upon Act.D treatment. When treated with 1 nM doxorubicin for 24 h IRF2BP2 knockdown cells already display twice the amount of apoptotic cells compared to the control (Figure 5E). After 48 h of treatment with doxorubicin the control and the knockdown cells accumulate high levels of apoptotic cells, with the amount of apoptotic cells being 10% higher in the IRF2BP2 knockdown cells. Our interesting observation that the IRF2BP2 knockdown cells are more sensitive to chemotherapeutic treatment and undergo apoptosis more rapidly hints at a role of endogenous IRF2BP2 in the inhibition of cell death.
DISCUSSION
In this study we identified the transcriptional coregulator IRF2BP2 as a new, direct target gene of p53. The binding site we found was bound in vivo and in vitro by p53 and it had transactivating potential. We showed that IRF2BP2 participates in the genotoxic response mediated by p53, influencing the stress response pathways of the cells. We observed an upregulation of IRF2BP2 mRNA after treatment with Act.D in different cell systems containing functional p53, but not in cells without functional p53.
Analyzing the function of IRF2BP2 after its overexpression as well as its knockdown, we found that it influences the cellular response after genotoxic insults. Its overexpression without stress treatment seems to slightly increase the population of cells in S-phase. This might be due to an IRF2BP2-mediated stabilization of IRF2 containing complexes at promoters like the one for the histone H4-gene (23). This gene is regulated during cell cycle with a peak in transcription during early S-phase (23,24). When U2OS cells are exposed to doxorubicin, the overexpression of IRF2BP2 seems to delay the induction of apoptosis. On the other hand cells overexpressing IRF2BP2 display an reduced S-phase population after low doses of Act.D. In line with this, the knockdown of IRF2BP2 leads to increasing amounts of apoptotic cells after doxorubicin treatment and also after treatment with Act.D, which normally rather causes growth arrest. This points towards an important function of IRF2BP2 in the decision between cellular survival and programmed cell death. From this data the question rises for which purpose p53 induces IRF2BP2, a factor that appears to delay or diminish apoptosis. Here we suggest a role for IRF2BP2 in maintaining a cell growth arrest state, which might allow the cells to repair damaged DNA.
After the identification of IRF2BP2 as an interaction partner of IRF2, it had been speculated that they act together to inhibit the tumor suppressor function of IRF1 by impeding on the IRF1 mediated induction of p21 (31). Our IRF2PB2 knockdown data seem to support this model: We observe a slight upregulation of p21 expression and more apoptosis after chemotherapeutic treatments in IRF2BP2 knockdown cells. These observations are very similar to the effect of the IRF2 knockdown cells in which the p21 expression is also upregulated and more spontaneous apoptosis is found (29). Thus, while physiological levels of IRF2 can upregulate the anti-apoptotic Bcl-2 (29), the knockdowns of IRF2 as well as the knockdown of IRF2BP2 seem to have a pro-apoptotic effect.
According to a current model of the p53 response, low doses of cellular stress that occur more frequently do not lead to apoptosis although they activate p53. Higher doses of stress can damage the cell irreversibly, leading to apoptotic cell death (6). Both pathways seem to be influenced by groups of cofactors, driving the cellular outcome towards one or the other decision depending on the kind and extent of genotoxic stress. The induction of growth arrest requires other cofactors than the apoptotic pathway, and some of them are transcriptional targets of p53, thereby forming feedback loops and regulating the outcome of p53-activation. An important factor is the zinc-finger containing protein Hzf, which is found in a transcriptional complex with p53 and can target this complex preferentially to promoters of genes involved in growth arrest, but not in apoptosis (18). Since the induction of growth arrest should allow DNA-repair and eventually cell survival, it also requires a temporary inhibition of apoptotic induction. Thus, notably, other target genes of p53 are involved in the inhibition of apoptosis, like the transcriptional repressor SLUG that can repress the expression of puma, a BH3-only protein, which is a key mediator of apoptosis in hematopoietic progenitor cells (19).
We found here that IRF2BP2 might function in a comparable way, since it was able to inhibit p53-mediated p21- and Bax–luc transactivation, but this inhibition of Bax–luc transactivation could be overcome if higher levels of p53 were present. A similar mechanism has also been described for Hzf, where after prolonged p53-activation, Hzf is degraded and p53-activity is rendered towards pro-apoptotic genes (18). This could happen for example upon higher doses of stress leading to an accumulation of p53, thereby passing a certain apoptotic threshold followed by the induction of programmed cell death (5).
Since we could not detect a direct interaction between p53 and IRF2BP2 (data not shown) it is possible that the regulation of p53 target genes by IRF2BP2 is mediated through IRF2 to which the transcriptional factor IRF2BP2 binds. We observed a downregulation of IRF2 around 2-fold after 6 h of Act.D treatment. IRF2 is an antagonist of the growth suppressor IRF1, which is involved in the induction of p21 (30) and the ratio of IRF2:IRF1 is believed to be an important determinant in the regulation of cell growth (42). Besides the activation of H4-transcription (23), IRF2 is able to stimulate the transcription of different regulatory subunits of checkpoint kinases, like cyclin D1 or cyclin B1 and it was also shown to upregulate the anti-apoptotic Bcl-2 protein, displaying features of a proto-oncogene (29,43). Furthermore, high levels of IRF2 keep IRF1 from entering the nucleus (29). The concerted regulation of IRF2 and IRF2BP2 might display a mode of dual action of p53 through the upregulation of IRF2BP2 on the one hand and the downregulation of IRF2 on the other hand. Thereby the balance between growth stimulating and inhibiting activities of the IRF1:IRF2 ratio could be changed (26) and growth arrest pathways could be positively influenced.
It has been reported, that both IRF1 and IRF2 directly activate or repress target genes. The binding element both proteins compete for is known as PRDI/CCE (23,44) or ISRE (34) and this element has been implicated to have activating as well as repressing functions. We found several copies of it in the here used luciferase constructs by Match-analysis using the Transfac-database (Transfac professional 11.1). Furthermore, Coccia et al. verified one out of five putative ISRE-elements 1.2 kb upstream of the p21 transcriptional start site as a true binding site for IRF1/2 (31). Surprisingly the motif in the p21 promoter found by Coccia et al. (31) seems to be dispensable for the IRF1 mediated enhancement of p21 activation by p53, since this enhancement was shown to be the result of an interaction of IRF1 with p300 leading to increased p53 acetylation and higher transcriptional activity towards p21 (32). Interestingly, the domain of p300 interacting with IRF1 maps in the same region as the interacting domains of JMY and p300, maybe leading to an mutual exclusive activation of either pro-apoptotic or pro-growth arrest target genes (32,44). A further possible mechanism of repression of p53 target genes might involve the PU.1 protein. This transcriptional cofactor reduces the transcriptional activity of p53 and it directly interacts with IRF2 (45,46). Besides a high degree of homology in their DNA-binding domain, the C-terminus of IRF2 is unique compared to IRF1 and it can recruit IRF2BP2 to DNA, thereby mediating transcriptional repression towards artificial promoter constructs (34). Nonetheless, the exact circumstances leading to activation or repression through IRF1 or IRF2 are still unclear. Thus, the IRF2BP2 induction by p53 could be the direct link between the p53 pathways upon cellular stress and the for the cell cycle progression so important balance between the IRF1 and IRF2 activities.
Thus, the here observed induction of IRF2BP2 expression after p53-activation appears to play an interesting role to influence the cellular stress response. As a possible mechanism we propose that IRF2BP2 modulates p53 transcriptional activity, by decreasing the p53-mediated p21- and Bax-transactivation until the p53 levels increase and the IRF2BP2-mediated repression is reversed. Our findings suggests that IRF2BP2 might be part of a new feedback-loop for the cells after p53 activation, increasing the threshold for induction of apoptosis, but also inhibiting the progression of cell cycle, possibly to allow repair of damaged DNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Dutch Cancer Foundation (KWF) supported the work of M.K. (KUN 2003-2926) and of L.S. (KUN 2003-2926 and KUN 2005-3347). S.J.v.H. and M.L. were supported from NWO (Vidi 846.05.002). Funding for open access charge: KWF and NWO.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Stephen Goodbourn for IRF2BP2-constructs, Yugang Tu and Moshe Oren for the p53-responsive luciferase constructs and Karen Vousden for providing us with p53-expression plasmids. In addition, we thank Henk Stunnenberg and Robert Akkers for suggestions and Waseem Akhtar for experimental help.
REFERENCES
- 1.Lozano G, Zambetti GP. What have animal models taught us about the p53 pathway? J. Pathol. 2005;205:206–220. doi: 10.1002/path.1704. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 3.Moll UM, Wolff S, Speidel D, Deppert W. Transcription-independent pro-apoptotic functions of p53. Curr. Opin. Cell Biol. 2005;17:631–636. doi: 10.1016/j.ceb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 4.Laptenko O, Prives C. Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ. 2006;13:951–961. doi: 10.1038/sj.cdd.4401916. [DOI] [PubMed] [Google Scholar]
- 5.Vousden KH. Outcomes of p53 activation – spoilt for choice. J. Cell Sci. 2006;119:5015–5020. doi: 10.1242/jcs.03293. [DOI] [PubMed] [Google Scholar]
- 6.Vousden KH, Lu X. Live or let die: the cell's response to p53. Nature Rev. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 7.Coutts AS, La Thangue NB. The p53 response: emerging levels of co-factor complexity. Biochem. Biophys. Res. Commun. 2005;331:778–785. doi: 10.1016/j.bbrc.2005.03.150. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka T, Ohkubo S, Tatsuno I, Prives C. hCAS/CSE1L associates with chromatin and regulates expression of select p53 target genes. Cell. 2007;130:638–650. doi: 10.1016/j.cell.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 9.Qian H, Wang T, Naumovski L, Lopez CD, Brachmann RK. Groups of p53 target genes involved in specific p53 downstream effects cluster into different classes of DNA binding sites. Oncogene. 2002;21:7901–7911. doi: 10.1038/sj.onc.1205974. [DOI] [PubMed] [Google Scholar]
- 10.Samuels-Lev Y, O'Connor DJ, Bergamaschi D, Trigiante G, Hsieh JK, Zhong S, Campargue I, Naumovski L, Crook T, Lu X. ASPP proteins specifically stimulate the apoptotic function of p53. Mol. Cell. 2001;8:781–794. doi: 10.1016/s1097-2765(01)00367-7. [DOI] [PubMed] [Google Scholar]
- 11.Ryan KM, Ernst MK, Rice NR, Vousden KH. Role of NF-kappaB in p53-mediated programmed cell death. Nature. 2000;404:892–897. doi: 10.1038/35009130. [DOI] [PubMed] [Google Scholar]
- 12.Okamura S, Arakawa H, Tanaka T, Nakanishi H, Ng CC, Taya Y, Monden M, Nakamura Y. p53DINP1, a p53-inducible gene, regulates p53-dependent apoptosis. Mol. Cell. 2001;8:85–94. doi: 10.1016/s1097-2765(01)00284-2. [DOI] [PubMed] [Google Scholar]
- 13.Takekawa M, Adachi M, Nakahata A, Nakayama I, Itoh F, Tsukuda H, Taya Y, Imai K. p53-inducible wip1 phosphatase mediates a negative feedback regulation of p38 MAPK-p53 signaling in response to UV radiation. EMBO J. 2000;19:6517–6526. doi: 10.1093/emboj/19.23.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 15.Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, Berger SL. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herold S, Wanzel M, Beuger V, Frohme C, Beul D, Hillukkala T, Syvaoja J, Saluz HP, Haenel F, Eilers M. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol. Cell. 2002;10:509–521. doi: 10.1016/s1097-2765(02)00633-0. [DOI] [PubMed] [Google Scholar]
- 18.Das S, Raj L, Zhao B, Kimura Y, Bernstein A, Aaronson SA, Lee SW. Hzf Determines cell survival upon genotoxic stress by modulating p53 transactivation. Cell. 2007;130:624–637. doi: 10.1016/j.cell.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu WS, Heinrichs S, Xu D, Garrison SP, Zambetti GP, Adams JM, Look AT. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell. 2005;123:641–653. doi: 10.1016/j.cell.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen H, Hiscott J, Pitha PM. The growing family of interferon regulatory factors. Cytokine Growth Factor Rev. 1997;8:293–312. doi: 10.1016/s1359-6101(97)00019-1. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka N. [Functional role of IRF–family transcription factors] Tanpakushitsu Kakusan Koso. 2000;45:1551–1559. [PubMed] [Google Scholar]
- 22.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 23.Vaughan PS, Aziz F, van Wijnen AJ, Wu S, Harada H, Taniguchi T, Soprano KJ, Stein JL, Stein GS. Activation of a cell-cycle-regulated histone gene by the oncogenic transcription factor IRF-2. Nature. 1995;377:362–365. doi: 10.1038/377362a0. [DOI] [PubMed] [Google Scholar]
- 24.Vaughan PS, van der Meijden CM, Aziz F, Harada H, Taniguchi T, van Wijnen AJ, Stein JL, Stein GS. Cell cycle regulation of histone H4 gene transcription requires the oncogenic factor IRF-2. J. Biol. Chem. 1998;273:194–199. doi: 10.1074/jbc.273.1.194. [DOI] [PubMed] [Google Scholar]
- 25.Masumi A, Wang IM, Lefebvre B, Yang XJ, Nakatani Y, Ozato K. The histone acetylase PCAF is a phorbol-ester-inducible coactivator of the IRF family that confers enhanced interferon responsiveness. Mol. Cell. Biol. 1999;19:1810–1820. doi: 10.1128/mcb.19.3.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harada H, Kitagawa M, Tanaka N, Yamamoto H, Harada K, Ishihara M, Taniguchi T. Anti-oncogenic and oncogenic potentials of interferon regulatory factors-1 and -2. Science. 1993;259:971–974. doi: 10.1126/science.8438157. [DOI] [PubMed] [Google Scholar]
- 27.Lowney JK, Boucher LD, Swanson PE, Doherty GM. Interferon regulatory factor-1 and -2 expression in human melanoma specimens. Ann. Surg. Oncol. 1999;6:604–608. doi: 10.1007/s10434-999-0604-4. [DOI] [PubMed] [Google Scholar]
- 28.Connett JM, Badri L, Giordano TJ, Connett WC, Doherty GM. Interferon regulatory factor 1 (IRF-1) and IRF-2 expression in breast cancer tissue microarrays. J. Interferon Cytokine Res. 2005;25:587–594. doi: 10.1089/jir.2005.25.587. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Liu DP, Chen PP, Koeffler HP, Tong XJ, Xie D. Involvement of IFN regulatory factor (IRF)-1 and IRF-2 in the formation and progression of human esophageal cancers. Cancer Res. 2007;67:2535–2543. doi: 10.1158/0008-5472.CAN-06-3530. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka N, Ishihara M, Lamphier MS, Nozawa H, Matsuyama T, Mak TW, Aizawa S, Tokino T, Oren M, Taniguchi T. Cooperation of the tumour suppressors IRF-1 and p53 in response to DNA damage. Nature. 1996;382:816–818. doi: 10.1038/382816a0. [DOI] [PubMed] [Google Scholar]
- 31.Coccia EM, Del Russo N, Stellacci E, Orsatti R, Benedetti E, Marziali G, Hiscott J, Battistini A. Activation and repression of the 2-5A synthetase and p21 gene promoters by IRF-1 and IRF-2. Oncogene. 1999;18:2129–2137. doi: 10.1038/sj.onc.1202536. [DOI] [PubMed] [Google Scholar]
- 32.Dornan D, Eckert M, Wallace M, Shimizu H, Ramsay E, Hupp TR, Ball KL. Interferon regulatory factor 1 binding to p300 stimulates DNA-dependent acetylation of p53. Mol. Cell. Biol. 2004;24:10083–10098. doi: 10.1128/MCB.24.22.10083-10098.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rubinstein YR, Driggers PH, Ogryzko VV, Thornton AM, Ozato K, Pontzer CH. An IFN regulatory factor-2 DNA-binding domain dominant negative mutant exhibits altered cell growth and gene expression. Oncogene. 2000;19:1411–1418. doi: 10.1038/sj.onc.1203435. [DOI] [PubMed] [Google Scholar]
- 34.Childs KS, Goodbourn S. Identification of novel co-repressor molecules for interferon regulatory factor-2. Nucleic Acids Res. 2003;31:3016–3026. doi: 10.1093/nar/gkg431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smeenk L, van Heeringen SJ, Koeppel M, van Driel MA, Bartels SJ, Akkers RC, Denissov S, Stunnenberg HG, Lohrum M. Characterization of genome-wide p53-binding sites upon stress response. Nucleic Acids Res. 2008;36:3639–3654. doi: 10.1093/nar/gkn232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denissov S, van Driel M, Voit R, Hekkelman M, Hulsen T, Hernandez N, Grummt I, Wehrens R, Stunnenberg H. Identification of novel functional TBP-binding sites and general factor repertoires. EMBO J. 2007;26:944–954. doi: 10.1038/sj.emboj.7601550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat. Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 38.Hoh J, Jin S, Parrado T, Edington J, Levine AJ, Ott J. The p53MH algorithm and its application in detecting p53-responsive genes. Proc. Natl Acad. Sci. USA. 2002;99:8467–8472. doi: 10.1073/pnas.132268899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kimura M. IRF2-binding protein-1 is a JDP2 ubiquitin ligase and an inhibitor of ATF2-dependent transcription. FEBS Lett. 2008;582:2833–2837. doi: 10.1016/j.febslet.2008.07.033. [DOI] [PubMed] [Google Scholar]
- 40.Horn HF, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26:1306–1316. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]
- 41.Tu Y, Wu W, Wu T, Cao Z, Wilkins R, Toh BH, Cooper ME, Chai Z. Antiproliferative autoantigen CDA1 transcriptionally up-regulates p21(Waf1/Cip1) by activating p53 and MEK/ERK1/2 MAPK pathways. J. Biol. Chem. 2007;282:11722–11731. doi: 10.1074/jbc.M609623200. [DOI] [PubMed] [Google Scholar]
- 42.Harada H, Taniguchi T, Tanaka N. The role of interferon regulatory factors in the interferon system and cell growth control. Biochimie. 1998;80:641–650. doi: 10.1016/s0300-9084(99)80017-0. [DOI] [PubMed] [Google Scholar]
- 43.Xie RL, van Wijnen AJ, van der Meijden CM, Stein JL, Stein GS. Forced expression of the interferon regulatory factor 2 oncoprotein causes polyploidy and cell death in FDC-P1 myeloid hematopoietic progenitor cells. Cancer Res. 2002;62:2510–2515. [PubMed] [Google Scholar]
- 44.Shikama N, Lee CW, France S, Delavaine L, Lyon J, Krstic-Demonacos M, La Thangue NB. A novel cofactor for p300 that regulates the p53 response. Mol. Cell. 1999;4:365–376. doi: 10.1016/s1097-2765(00)80338-x. [DOI] [PubMed] [Google Scholar]
- 45.Huang W, Horvath E, Eklund EA. PU.1, interferon regulatory factor (IRF) 2, and the interferon consensus sequence-binding protein (ICSBP/IRF8) cooperate to activate NF1 transcription in differentiating myeloid cells. J. Biol. Chem. 2007;282:6629–6643. doi: 10.1074/jbc.M607760200. [DOI] [PubMed] [Google Scholar]
- 46.Tschan MP, Reddy VA, Ress A, Arvidsson G, Fey MF, Torbett BE. PU.1 binding to the p53 family of tumor suppressors impairs their transcriptional activity. Oncogene. 2008;27:3489–3493. doi: 10.1038/sj.onc.1211004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.