Abstract
Zinc-finger proteins (ZFPs) have long been recognized for their potential to manipulate genetic information because they can be engineered to bind novel DNA targets. Individual zinc-finger domains (ZFDs) bind specific DNA triplet sequences; their apparent modularity has led some groups to propose methods that allow virtually any desired DNA motif to be targeted in vitro. In practice, however, ZFPs engineered using this ‘modular assembly’ approach do not always function well in vivo. Here we report a modular assembly scoring strategy that both identifies combinations of modules least likely to function efficiently in vivo and provides accurate estimates of their relative binding affinities in vitro. Predicted binding affinities for 53 ‘three-finger’ ZFPs, computed based on energy contributions of the constituent modules, were highly correlated (r = 0.80) with activity levels measured in bacterial two-hybrid assays. Moreover, Kd values for seven modularly assembled ZFPs and their intended targets, measured using fluorescence anisotropy, were also highly correlated with predictions (r = 0.91). We propose that success rates for ZFP modular assembly can be significantly improved by exploiting the score-based strategy described here.
INTRODUCTION
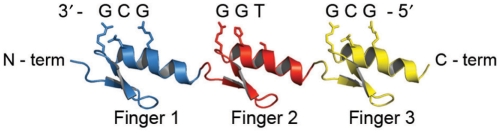
The ability to reliably engineer DNA binding proteins that recognize any desired DNA sequence would provide an unprecedented level of control over genetic information; for example, by allowing the creation of site-specific nucleases that specifically alter genomic DNA (1–5). The C2H2 zinc-finger domain (ZFD) is arguably the best characterized DNA binding motif and offers considerable promise for the rational engineering of site-specific DNA binding proteins (6–11). Zinc-finger proteins (ZFPs) consist of multiple individual ZFDs, each of which typically recognizes adjacent sequence triplets in duplex DNA (Figure 1). An individual ZFD comprises a pair of anti-parallel β-strands and one α-helix, which coordinate a zinc ion through conserved pairs of cysteine and histidine residues. In the canonical three-finger domain of the Zif268 transcription factor, the amino acid side chains at positions −1, +3 and +6 relative to the amino-terminal end of the α-helix typically make base-specific contacts with three adjacent nucleotides within the major groove of double-stranded DNA (12). An aspartic acid residue in the +2 position of the DNA recognition helix can specify a fourth nucleotide, resulting in either target-site overlap with an adjacent module or specification of an additional nucleotide at the 3′-end of the target site (13,14).
Figure 1.
A three-finger ZFP with its DNA target site. A ZFP consisting of three adjacent ZFDs binds its target DNA through contacts between the amino acids of the DNA recognition helices and consecutive nucleotides in the DNA. The protein chain is drawn in the N- to C-terminal direction and the DNA target in the 3′–5′ direction. Note that an ‘unnatural’ extended array is shown to better illustrate the critical amino acid/nucleotide contacts. Structure diagrams were generated using PyMol (http://www.pymol.org).
Several research groups have characterized ZFDs that recognize many of the 64 possible DNA triplets (15–20). Using a ‘modular assembly’ approach, novel ZFPs that recognize variant DNA sites are assembled by simply stringing together individual ZFDs. In practice, however, ZFPs made by modular assembly display a wide range of binding affinities and specificities (15,19,21–23). Although modular assembly has proven useful for some in vivo applications, such as artificial transcription factors, recent work suggests that the success rate of creating artificial zinc-finger nucleases (ZFNs–fusions of engineered zinc fingers to a non-specific nuclease domain) by this method is considerably lower (24,25). These low success rates, together with the inability to predict which ZFPs are likely to function in vivo, have motivated our groups to improve the procedures and design criteria for ZFP engineering (25,26).
The present study was motivated by our observation that among a small set of modularly assembled ZFPs, those that fail to function in vivo are more likely to possess modules previously shown to have relatively low affinity for target DNA. This observation implies that insufficient affinity can contribute to poor function in vivo and also suggested that it might be possible to predict the affinity of a modularly assembled ZFP using existing affinity data for component modules. Here we test these hypotheses and demonstrate that both the in vitro binding affinity and the lack of in vivo activity of a ZFP can be predicted using the energy contributions of its component ZFDs. Our approach for predicting the binding of ZFPs to desired target sequences should improve success rates of modular assembly by guiding investigators away from target sites and ZFP combinations least likely to function in vivo.
MATERIALS AND METHODS
Zinc-finger modules and three-finger arrays (ZFPs)
All ZFDs used in these experiments have been described by the Barbas group (15) and are referred to as ‘Barbas modules’. ZFPs containing desired three-finger (three-module) arrays were assembled by iterative ligation and cloning of restriction fragments encoding ZFDs using reagents and protocols previously described by the Zinc Finger Consortium (http://www.zincfingers.org/) (27). ZFP-encoding fragments were then cloned into vectors for expression as Gal11P-hybrid proteins in the bacterial two-hybrid (B2H) system as previously described (27).
B2H assays
A series of B2H reporter plasmids, each harboring a target binding site for one of 27 different three-finger ZFPs, was constructed by cloning synthetic target oligonucleotides into reporter plasmid pBAC-lacZ as previously described (27). Binding of a Gal11P-ZFP hybrid protein to the target sequence on a B2H reporter plasmid triggers transcriptional activation of a lacZ reporter gene encoding β-galactosidase. In vivo ZFP performance was therefore assayed using a β-galactosidase assay in which ZFP-induced activation of lacZ expression was measured relative to control constructs lacking the ZFP.
Zinc finger–maltose binding protein fusion protein construction, expression and purification
Zinc finger–maltose binding protein (MBP) fusion protein constructs were generated by transferring three-finger arrays, assembled as described above, into pHMTC (28). The MBP fusion plasmids were transformed into BL21 Escherichia coli cells (Invitrogen) using standard chemical transformation procedures (29).
For protein expression, 5 ml cultures were grown for 16 h at 30°C with agitation in ZFE broth [Luria Broth (LB), 1.11 mM dextrose, 100 µg/ml ampicillin]. Expansion cultures of 10 ml were inoculated from these overnight cultures (1:100 dilution) and grown to an OD600 of 0.5 before a 2 h induction with isopropyl β-D-1-thiogalactopyranoside (IPTG). Cells were harvested by centrifugation for 10 min at 4000g at 4°C and frozen overnight at −20°C. The following day, cells were resuspended in 4 ml WB1 (15 mM HEPES pH 7.8, 200 mM NaCl, 20 µM ZnSO4)/1 mM PMSF/0.1% Nonidet™ P40 (NP-40) and refrozen at −70°C. Cells were then thawed in ice water and centrifuged at 9000g at 4°C for 20 min. To remove remaining nucleic acids, the resulting supernatant was transferred to a new cold tube and polyethyleneimine was added to 0.1%. The supernatant was then incubated for 30 min before a second centrifugation at 16 000 g at 4°C for 30 min.
Amylose beads (NEB) were prepared in 50 µl aliquots in 1.5 ml micro-centrifuge tubes according to manufacturer's instructions. Beads were washed (suspended, spun down and supernatant removed) three times in 1 ml WB1/0.1% NP-40 at 4°C and resuspended in 450 µl WB1. For affinity purification, 1 ml of clarified protein supernatant was added to prepared beads, and incubated at 4°C for 30 min. The slurry was centrifuged and the supernatant was removed. The proteins bound to beads were washed two times with 700 µl WB1/0.1% NP-40 and two times with zinc buffer A (ZBA; 10 mM Tris–HCl, pH 7.5, 90 mM KCl, 1 mM MgCl2, 90 µM ZnCl2)/0.1% NP-40 (15). Purified proteins were then eluted in 200 µl ZBA/0.1% NP-40/40 mM maltose for 30 min at room temperature, with gentle agitation. After elution, beads were centrifuged at 16 000g. The supernatant was transferred to a new cold tube and centrifuged again at 16 000g. The supernatant was transferred to a new cold tube and gently stirred to mix protein. Proteins were stored at −70°C in Axygen MaxymumRecovery™ tubes. Protein concentrations were estimated using a Bradford assay against a bovine serum albumin (BSA) standard in ZBA/0.1% NP-40.
Binding measurements using fluorescence anisotropy
Binding reactions were performed in ZBA/0.1% NP-40/0.1 mg/ml non-acetylated BSA (Sigma) for 30 min on ice with 5 nM target DNA. Target sites (shown in Figure 2a) were formed using hairpin DNA oligonucleotides as described (15). HPLC purified, 3′-6-FAM-labeled oligonucleotides were ordered from Integrated DNA Technologies (Coralville, IA, USA). In each experiment, two serial dilutions of purified ZFP-MBP fusion protein were performed over a range of 1000–0.122 nM. Reported binding affinity values are based on the average of three separate binding experiments, performed on different days, using three separate protein preparations. Fluorescence anisotropy (FA) measurements were made using a Varian Cary Eclipse spectrophotometer in L-format configuration. Each value was based on five measurements averaged over 5 s, using a 490 nm excitation wavelength (5 nm slit width), and 530 nm emission wavelength (20 nm slit width) at 880 V. Background light scattering for each protein sample dilution was measured and subtracted to correct for protein concentration-dependent variation in intensities. Kd values were determined by nonlinear regression (30,31) using Prism (http://www.graphpad.com/prism/Prism.htm).
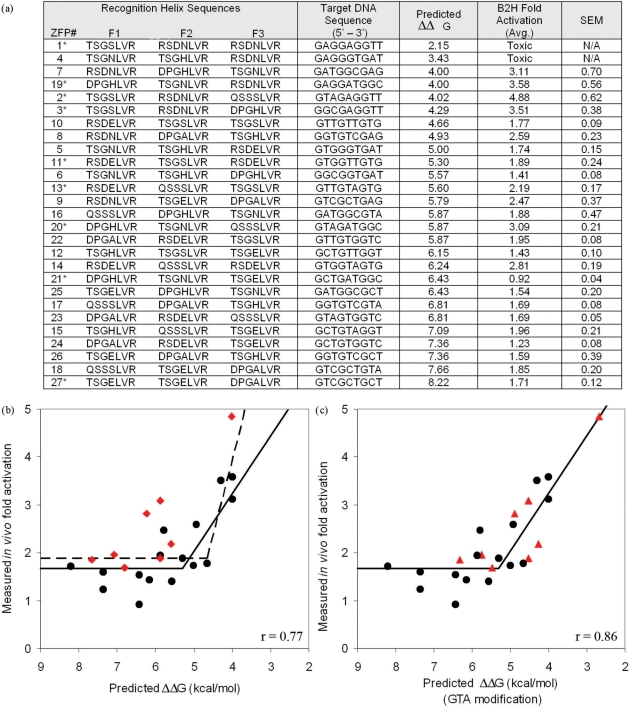
Figure 2.
Predicted binding energies are highly correlated with in vivo activity in a B2H assay. Twenty-seven three-module ZFPs were designed by modular assembly to span a broad range of predicted binding affinities. (a) DNA recognition helix sequences, DNA sequence targets, predicted energies, measured fold-activation in the B2H assay, and standard error of the mean are listed for each construct. Entries are sorted from lowest to highest predicted ΔΔG. Constructs marked with an asterisk were also tested in vitro (Figure 4). (b) ZFP activities in the B2H assay are plotted versus predicted energies. The two constructs with highest predicted affinities were toxic to their host cells and therefore could not be included. Points shown as red diamonds correspond to proteins containing the GTA-specific QSSSLVR module (see text). Best-fit lines from a segmental linear regression model using all points (dashed line, r = 0.77), or excluding red points (solid line, r = 0.86) are shown. (c) Same as (b), except that values indicated by red triangles were adjusted assuming a binding affinity of 2.5 nM, rather than 25 nM, for the GTA-specific QSSSLVR module; this increases the correlation coefficient to 0.86 (see text for details).
RESULTS
To test the hypothesis that binding energy contributions of individual ZFDs can be used to predict the in vitro binding affinities and in vivo performance of extended ZFP arrays, 27 three-module ZFPs were constructed by assembling various GNN-specific modules previously characterized by the Barbas group (15). ZFP compositions were chosen to systematically explore a wide range of predicted binding affinities and to test the influence of context on module performance. As shown in Table 1, ZFDs were divided into three affinity classes based on their reported affinity constants measured in a fixed context, namely as fingers in the middle position of a three-finger Zif268 variant (15). Modules comprising Zif268 variants with Kd values <10 nM were categorized as ‘strong’, Kd = 10–30 nM as ‘moderate’ and Kd > 30 nM as ‘weak’ (Table 1). Using three different modules to represent each binding class, all possible combinations of strong, moderate and weak affinity modules for a three-module ZFP were assembled. To allow direct comparisons among proteins that differ by a single module, ZFPs were designed in subgroups in which only one finger position was varied (Table 2).
Table 1.
ZFP variants that differ in the middle (F2) position bind targets with variable affinities
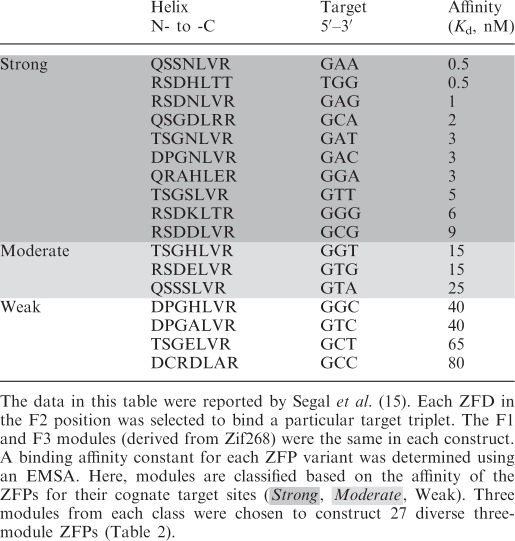 |
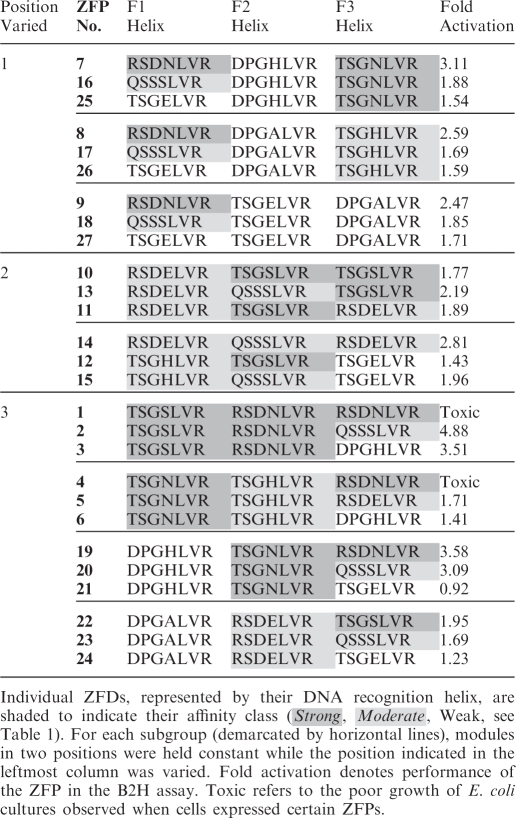
Table 2.
Single module substitutions in ZFPs alter target affinity
 |
Predicting relative binding energies for modularly designed ZFPs
If one assumes that the binding energy of a three-finger ZFP (ΔG°ZFP) is equal to the sum of the binding energies of its three component ZFDs (ΔG°ZFD) [Equation (1)], it follows that the difference in binding energy between any two ZFPs is the sum over the positions of the difference in binding energy between the modules at each position [Equation (2)].
| 1 |
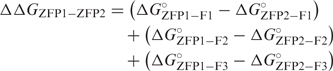 |
2 |
Because the ZFDs used in this study were evaluated in the middle (F2) position of a three-finger ZFP, and because the other fingers (F1 and F3) were constant in all these ZFPs, the differences in measured binding constants among these constructs should be attributable to the differences in binding energy between the F2 ZFDs. Thus, ΔΔG can be calculated between any two ZFDs by using the identity relating Gibbs free energy to Kd [Equation (3), RT = 0.58].
| 3 |
To compare binding affinity measurements with predicted values, the predicted ΔΔG was calculated as the difference between each ZFP and a standard (STD) ZFP composed entirely of the F2 domain of parental C7 (15).
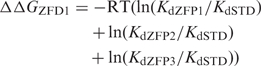 |
4 |
Thus, using Equation (4) and binding constants for ZFP variants published by the Barbas group (15), we predicted ΔΔG values for 27 novel modularly assembled ZFPs constructed using Barbas GNN modules (Figure 2). Predicted ΔΔG values ranged from 2.1 kcal/mol for ZFP #1, containing three strong modules to 8.2 kcal/mol for ZFP #27 containing three weak modules.
In vivo activities of ZFPs are highly correlated with predicted binding energies
To evaluate in vivo binding of the 27 modularly assembled ZFPs to their cognate DNA targets, we used a quantitative B2H assay (32). In this assay, binding of a ZFP to its target site activates transcription of a lacZ reporter positioned downstream of an adjacent promoter. Thus, ZFP DNA-binding activity can be assessed by quantifying β-galactosidase activity in ZFP-expressing cells relative to control cells that do not express the ZFP. We chose to use the B2H system as an assay because recently published studies have shown that absence of ZFP activity in this system is an excellent predictor for failure of these proteins to function as ZFNs in human cells (24–26). For 25 of the 27 ZFPs tested, the level of lacZ activation observed was in excellent agreement with predicted energies (Figure 2a). Expression of the two ZFPs with the strongest predicted binding energy was toxic to cells, preventing analysis of these constructs. Several models describing the relationship between predicted and measured activity were evaluated, with segmental linear regression providing the best fit (r = 0.77; Figure 2b, dashed line). Inspection of the data revealed that the GTA-specific module (QSSSLVR) was present in most ZFPs that exhibited significantly greater activation than predicted (Figure 2b, red diamonds). Excluding ZFPs containing this module from the analysis increased the correlation coefficient to 0.86 (Figure 2b, solid line).
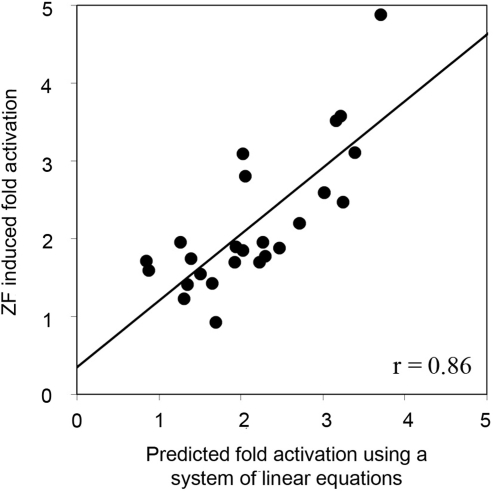
The predictions described above relied on published in vitro binding affinities for ZFPs in which modules were evaluated in a fixed context (15) to estimate binding contributions of individual modules. In an alternate approach, we predicted ZFP performance by solving individual module contributions as component variables of a system of linear equations. Briefly, in constructing the 27 different three-finger proteins, nine ZFDs were used approximately 8–10 times (approximately three times at each of the possible three positions, Table 1). Assuming that the energy contributions of individual ZFDs in a ZFP are additive, the B2H activity of each ZFP was considered to result from its particular combination of modules (Supplementary Figure 1). Individual module contributions were calculated for each ZFP using a leave-one-out linear system solution. Expected lacZ activation in the B2H assay for each of the ZFPs was then predicted by summing individual module contributions. As shown in Figure 3, expected levels of activation computed in this manner were highly correlated with actual B2H activity measurements (r = 0.86).
Figure 3.
ZFDs contribute additively to B2H activity, independent of context and position. For each ZF protein, the expected contribution to B2H activity from each of its component modules was estimated by solving a system of linear equations representing the other 24 proteins (see text). Comparison of actual versus predicted B2H activity (expressed as relative fold-activation in the B2H assay) reveals a high correlation (r = 0.86).
The energy contributions computed using a system of linear equations to analyze in vivo activity data from the B2H assay indicate that the GTA-specific QSSSLVR module binds with higher affinity than previous Kd estimates. This is consistent with our conclusion based on inspection of energies computed from in vitro binding constants (Figure 2b). We estimated a new value for this module by calculating the Kd that optimizes the correlation of predicted energies with the B2H data. This approach resulted in an estimated Kd of 2.5 nM for this module, 10-fold lower than the previously reported value of 25 nM (15). Incorporating this new estimate improved correlation between the in vitro energy model and in vivo fold activation data (r = 0.86, Figure 2c).
To directly evaluate the effects of individual module affinities on in vivo performance, sets of related ZFPs designed to vary at a single module position were analyzed for differences in B2H activity (Table 2). For all three sets of ZFPs in which the F1 position was varied (while F2 and F3 were fixed), the greatest in vivo activity was observed when the F1 position contained a high affinity module; the least activity was observed with a low affinity module in this position. The same trend was observed for all four groups in which the F3 position was varied while the F1 and F2 fingers were fixed. For sets in which the F2 position was varied, only one strong module (TSGSLVR) and one moderate affinity module (QSSSLVR) were tested. In these cases, the moderate affinity module outperformed the high affinity module. These results suggest that the effect of single module substitutions on relative binding affinity can be predicted reliably in most cases.
In summary, three lines of analysis: (i) predictions based on in vitro binding constants for modules in a fixed context, (ii) predictions derived from a system of linear equations based on in vivo performance and (iii) analysis of the effects of various single finger substitutions in vivo, demonstrate that in vivo performance for ZFPs can be predicted based on DNA-binding affinities of individual ZFDs.
In vitro DNA binding affinities of ZFPs are highly correlated with predicted binding energies
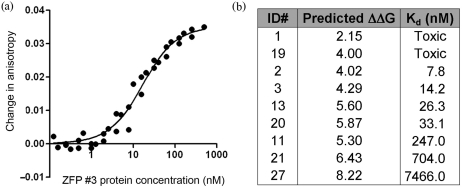
Our success in estimating the activities of ZFPs in the B2H assay suggested that our scoring scheme could be applied more generally to predict in vitro ZFP affinities. To test whether activation measured in the B2H assay directly reflects DNA binding affinity for the desired target site, 9 of the 27 engineered proteins, along with a control Zif268 protein, were chosen for in vitro binding affinity measurements (Figure 4). Kd values were determined using fluorescence anisotropy (FA), a rapid and reproducible solution-based DNA binding assay that allows computation of the bound fraction of a fluorescently labeled ligand, based on the decrease in its rotational velocity due to binding (33,34).
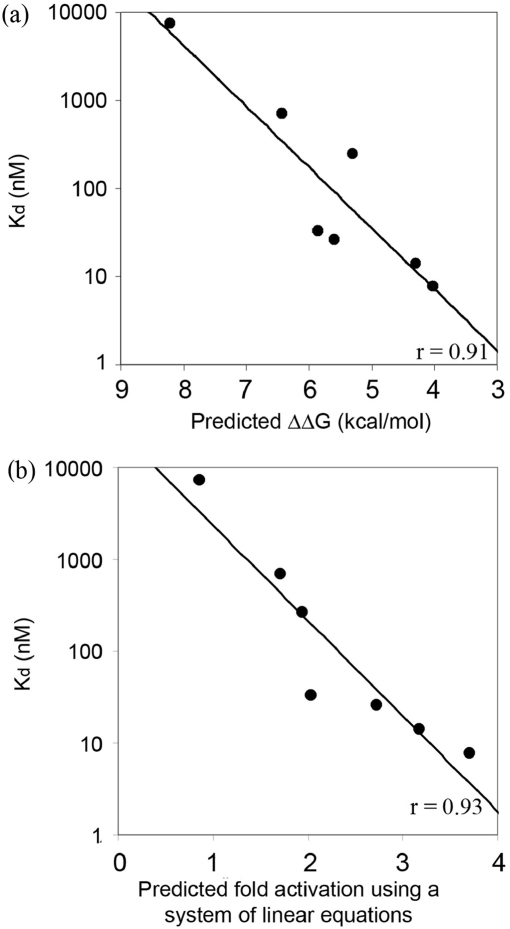
Figure 4.
Determining binding affinity constants using fluorescence anisotropy. (a) A representative in vitro binding isotherm obtained using FA. Data points for each ZFP were collected using three separate purified protein preparations, each assayed for binding activity on a different day. Curve fitting was performed using Prism. (b) Kd values for seven modularly assembled ZFPs, determined in FA experiments. Note that two ZFPs were toxic to host cells, preventing purification of proteins in quantities required for in vitro analysis.
As shown in Figure 5, binding affinity constants determined by FA were highly correlated with predicted energies. The two ZF proteins with highest predicted affinities were toxic to bacterial cells, prohibiting purification of sufficient quantities of protein for in vitro analysis. Energies computed based on previously published in vitro affinity measurements for modules in a fixed context (15) were proportional to the log of Kd's measured in our experiments (r = 0.91) (Figure 5a). As before, assuming a Kd of 2.5 nM for the QSSSLVR module significantly improved the correlation (r = 0.97). Predicted in vivo activation levels generated by the leave-one-out linear system method were also highly correlated with experimentally determined binding constants (r = 0.93; Figure 5b). Thus, results obtained using a rapid and reliable spectroscopic method suggest that ZFP binding affinities measured in vitro generally correspond to results obtained in vivo using the B2H system. This demonstrates that our rule-based strategy can be used to predict ZFP DNA binding affinity.
Figure 5.
In vitro affinity constants for ZFPs are highly correlated with predictions. Using affinity constants for seven ZFPs determined by FA (Figure 4b), log(Kd) is plotted against: (a) predicted energy, expressed as ΔΔG in kcal/mol (r = 0.91) and (b) predicted B2H activity based on a leave-one-out system of linear equations analysis (r = 0.93).
A binding energy threshold for ZFP function in vivo
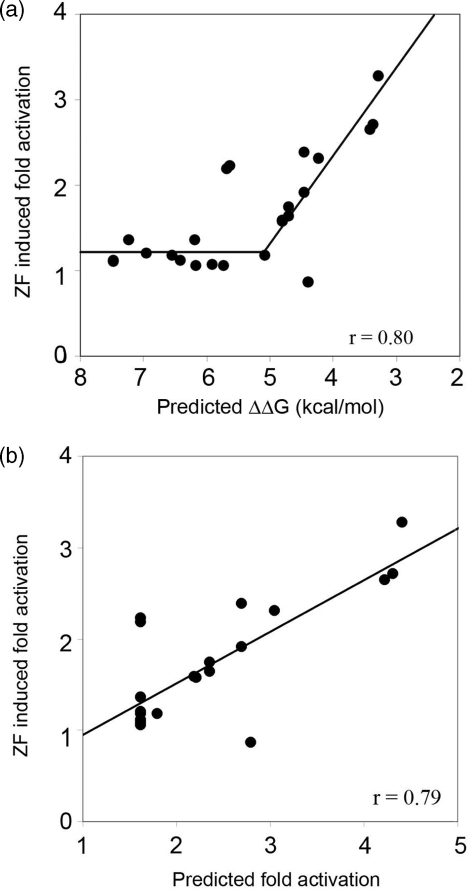
To evaluate the generality of this rule-based approach, we calculated predicted energies for another set of modularly assembled ZFPs that had been previously evaluated using the B2H system (25). From 168 modularly assembled ZFPs, we selected all ZFPs comprising GNN or TGG modules for which published in vitro DNA binding affinity constants are available [measured in the F2 position of the standard Zif268 variant backbone (15)]. As shown in Figure 6a, based on a segmental linear regression model, binding energies for 24 of these 26 ZFPs are highly correlated (r = 0.80) with reported B2H activity measurements. These results are also in excellent agreement with the results described above and shown in Figure 2b, although slightly higher activation levels were uniformly observed in the latter experiments. Notably, both sets of experiments identify a ΔΔG of ∼5 kcal/mol (corresponding to a Kd of ∼100 nM) as the threshold for zinc-finger function in vivo (in bacterial cells). We also used the scoring function generated from the B2H experiments performed by us (and shown in Figure 2c) to predict B2H activity for the 24 ZFPs evaluated by Ramirez et al. (25). Again, the predicted and measured fold-activation scores were in close agreement, with a correlation coefficient of 0.79 (Figure 6b). Taken together, these results suggest that the scoring function developed and evaluated may be generally applicable to ZFPs assembled using the Barbas lab GNN modules.
Figure 6.
Predicted ZFP performance agrees with in vivo activity for an independently generated set of ZFPs. Data shown are for 24 of 26 ZFPs containing characterized GNN or TGG modules, constructed and evaluated by Ramirez et al. (25) (a) A segmental linear regression model provides an excellent fit of in vivo ZF-induced fold activation measured in the B2H assay with predicted binding energies (r = 0.80). (b) ZF-induced fold activation values measured in the B2H assay for 24 ZFPs from Ramirez et al. (25) are also highly correlated (r = 0.79) with predicted fold-activation levels calculated based on a scoring function derived from the segmental linear regression model fit for the 25 ZFPs shown in Figure 2a (see text for details). Note: predictions for 2 of 26 ZFPs containing characterized GNN or TGG modules from Ramirez et al. (25) were considered outliers (values were outside the range included in these graphs); they were not included in the regression analysis.
DISCUSSION
Using a rule-based strategy that combines experimentally determined binding energies of individual ZFDs, we were able to compute binding energies for ZFPs made from a particular set of well-characterized GNN modules (15). We also showed that these predicted binding energies are in excellent agreement (r = 0.91; Figure 5a) with binding affinity constants measured directly in vitro. Furthermore, we showed a strong correlation between these computed binding energies and ZFP activities in a B2H system for two different sets of modularly assembled three-finger ZFPs. This is an important advance because a ZFP that lacks activity in the B2H system will also have a high probability of failing to function as a ZFN in human cells (24–26). Thus, using only our scoring method, researchers can now identify target sites that will have a high probability of failing to yield functional zinc-finger arrays by the method of modular assembly. Our rule-based strategy will thus allow researchers to focus their modular assembly efforts on a smaller number of target sites with a higher probability of success.
We believe that our results also provide one potential explanation for the discrepancy between the overwhelming success rates for a previous in vitro report (35) and the low in vivo success rates observed for ZFPs in the recent study of Ramirez et al. (25): many of the modules used to perform modular assembly likely possess low affinities. Our data suggest, in fact, that 30–50% of potential three-finger ZFPs made wholly from the Barbas GNN modules will fail to function in the B2H system, a result in agreement with the recently published results of Ramirez et al. (25).
Although our results demonstrate that the energy contributions of individual ZFDs in a ZFP array are additive, we also believe they lend additional support to the notion that context is an important parameter that should be accounted for when engineering multi-finger ZFPs (i.e. that one single ZFD module will not always be optimal or adequate for recognition of its cognate 3-bp subsite in different multi-finger ZFP contexts). For example, our data show that although a weak finger will sometimes be found in a nonfunctional ZFP array (if it is joined together with other weak affinity ZFDs), it will also sometimes be found in functional arrays when paired with stronger affinity ZFDs. In addition, our data show that although strong fingers will sometimes be found in functional ZFPs, they can be found in nonfunctional ZFPs. Furthermore, the use of three strong fingers in a ZFP can lead to toxicity in E. coli cells. Although the precise mechanism of this toxicity is unclear, a reasonable hypothesis is that excessively high affinity leads to binding to related but off-target sequences with sufficient affinity to cause biological consequences (essentially, excessive affinity leading to problems of specificity). Thus, our data further re-enforce the ideas that individual ZFDs do not function completely independently and that the specific attributes of neighboring fingers do matter in the context of engineering a multi-finger ZFP.
The importance of context-dependent effects also suggests that identification of additional ZFDs with variable affinities for GNN triplets may be needed if the efficiency of modular assembly is to be improved. If such ZFDs were available, it might be possible to achieve higher success rates for modular assembly by creating several ZFPs for a given target site so as to identify a combination that balances affinities (and presumably, specificities) of its component ZFDs. A related point is that our findings also suggest one possible reason why more complex selection-based methods that account for context-dependent effects [e.g. the OPEN method recently described by Joung and colleagues (26,32)] may be more successful than modular assembly: these methods are able to balance the overall affinity and specificity of the final ZFP array by identifying optimal combinations from various ZFDs with a range of affinities and specificities for their target 3-bp subsites.
The strong correlations among predicted binding energies, in vivo activities, and in vitro binding affinity constants for the ZFPs analyzed in this work suggests that our rule-based approach might be extended to evaluate arrays assembled using GNN modules from other sources (17,19) and non-GNN modules. We have not yet evaluated such modules, but our work demonstrates two ways this could be achieved: (i) by directly measuring in vitro binding constants for modules in the F2 position of a standardized ZFP framework and (ii) by computing individual module contributions to ZFP binding as component variables of a system of linear equations that describe their activities (measured in vivo in this work, but in vitro binding constants could also be used). The energy scoring scheme proposed here will allow researchers to determine whether a modular assembly strategy is likely to be feasible for specific targets of interest, based on currently available well-characterized modules, or whether an alternative selection-based engineering strategy should be considered.
A recent study on the use of ZFNs for homologous recombination cited lack of specificity as a primary determinant of ZFN-mediated toxicity in human cells (24). A likely mechanism for ZFN-induced toxicity is through binding to genomic sequences similar to the desired target sequence. As noted above, we observed toxicity in bacterial cells for several ZFPs, even in the absence of a fused nuclease domain, suggesting that ZFP binding to certain sites in genomic DNA can be toxic, particularly for high affinity ZFPs. Although this is the first published report of such toxicity in bacterial cells that we are aware of, it has been observed previously for several other sites (Joung,J.K. unpublished data). However, bacterial expression of ZFPs with affinities in the pM range, with no toxic effects, has also been reported (19,36,37). High-throughput chip or microfluidics-based DNA binding experiments (38–41) could be used to obtain affinity and specificity data for virtually every possible target site for a given ZFP, providing additional insight into ZFP-induced toxicity and into the fundamental rules that govern the affinity and specificity of DNA recognition by zinc-finger DNA binding proteins.
A correlation between ZFP binding constants measured in vitro and functional activity measured in vivo has also been observed by others using different reporter systems (37). A similar degree of correlation was observed using the B2H system in our study (Supplementary Figure 2). Our results further demonstrate that measurable ZFP activity in an in vitro binding assay does not necessarily translate into adequate function in vivo, in agreement with Beerli et al. (42). However, the energy threshold we determined for ZFP activity in vivo, using B2H assays, corresponds to a Kd of ∼100 nM, and thus differs from the estimated threshold Kd of ∼10 nM reported as the minimum affinity necessary for ZFP function in mammalian cells (42). The significance of this difference between thresholds determined in bacterial and mammalian cells is difficult to evaluate, given that functional assays and Kd measurements were performed in different laboratories using different assays and with ZFPs containing different numbers of fingers.
Stormo and colleagues (43–47) have shown that the DNA-binding specificity of ZFPs can be effectively predicted from additive energy contributions of individual residues that make base-specific contacts with target site nucleotides. Our results complement this idea by demonstrating that the affinity of ZFPs also can be predicted, using affinity data for component modules. Our application of the binding energy additivity concept differs somewhat from that used by Stormo to predict specificity in that it assumes additivity of energy contributions at the individual finger rather than individual residue level. Also, our approach implicitly includes energetic contributions of residues that are not directly involved in base contacts (e.g. phosphate contacts), as well as energetic contributions resulting from context-dependent effects that presumably occur among recognition helix residues within each finger.
The apparent simplicity of modular assembly has contributed to the current focus on C2H2 ZFDs as the domains of choice for designing custom DNA binding proteins. Our results make it possible, for the first time, to reliably identify prospective binding sites that are unlikely to yield functional ZFDs by modular assembly using a set of GNN-specific finger modules. The rule-based strategy presented here can provide accurate guidance for both in vitro binding affinities and in vivo functionality for engineered ZFPs by computing energy contributions of individual ZFDs. We have updated the Zinc Finger Targeter (ZiFiT) web server (http://bindr.gdcb.iastate.edu/ZiFiT) (48) so that it now provides users with a list of potential ZFP-target site pairs for a desired genomic sequence, scored according to the procedures developed and validated in this work.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (GM066387 to D.D.); National Science Foundation (DBI0501678 to D.F.V.); National Institutes of Health (GM069906 and GM078369 to J.K.J.); and graduate research assistantships provided by United States Department of Agriculture (MGET 2001-52100-11506, NSF IGERT0504304 and ISU's Center for Integrated Animal Genomics (CIAG). Funding for open access charge: National Science Foundation (DBI0501678).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of our groups and colleagues, especially Fengli Fu, Deepak Reyon, David Wright, Ronnie Winfrey, Ben Lewis, Bob Farnham, Abd Elhamid Azzaz, Les Miller, Gaya Amarasinghe and Vasant Honavar and the referees for their helpful suggestions and valuable feedback. We also thank Guru Rao for the use of his spectrophotometer.
REFERENCES
- 1.Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandrasegaran S. Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005;33:5978–5990. doi: 10.1093/nar/gki912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klug A. Towards therapeutic applications of engineered zinc finger proteins. FEBS Lett. 2005;579:892–894. doi: 10.1016/j.febslet.2004.10.104. [DOI] [PubMed] [Google Scholar]
- 3.Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat. Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 4.Wu J, Kandavelou K, Chandrasegaran S. Custom-designed zinc finger nucleases: what is next? Cell. Mol. Life Sci. 2007;64:2933–2944. doi: 10.1007/s00018-007-7206-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cathomen T, Joung JK. Zinc-finger nucleases: the next generation emerges. Mol. Ther. 2008;16:1200–1207. doi: 10.1038/mt.2008.114. [DOI] [PubMed] [Google Scholar]
- 6.Desjarlais JR, Berg JM. Use of a zinc-finger consensus sequence framework and specificity rules to design specific DNA binding proteins. Proc. Natl Acad. Sci. USA. 1993;90:2256–2260. doi: 10.1073/pnas.90.6.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jamieson AC, Wang H, Kim SH. A zinc finger directory for high-affinity DNA recognition. Proc. Natl Acad. Sci. USA. 1996;93:12834–12839. doi: 10.1073/pnas.93.23.12834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beerli RR, Segal DJ, Dreier B, Barbas C.F., 3rd Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl Acad. Sci. USA. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 10.Pabo CO, Peisach E, Grant RA. Design and selection of novel Cys2His2 zinc finger proteins. Annu. Rev. Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- 11.Segal DJ. The use of zinc finger peptides to study the role of specific factor binding sites in the chromatin environment. Methods. 2002;26:76–83. doi: 10.1016/S1046-2023(02)00009-9. [DOI] [PubMed] [Google Scholar]
- 12.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 13.Elrod-Erickson M, Rould MA, Nekludova L, Pabo CO. Zif268 protein-DNA complex refined at 1.6 A: a model system for understanding zinc finger-DNA interactions. Structure. 1996;4:1171–1180. doi: 10.1016/s0969-2126(96)00125-6. [DOI] [PubMed] [Google Scholar]
- 14.Miller JC, Pabo CO. Rearrangement of side-chains in a Zif268 mutant highlights the complexities of zinc finger-DNA recognition. J. Mol. Biol. 2001;313:309–315. doi: 10.1006/jmbi.2001.4975. [DOI] [PubMed] [Google Scholar]
- 15.Segal DJ, Dreier B, Beerli RR, Barbas C.F., 3rd Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5'-GNN-3' DNA target sequences. Proc. Natl Acad. Sci. USA. 1999;96:2758–2763. doi: 10.1073/pnas.96.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolfe SA, Greisman HA, Ramm EI, Pabo CO. Analysis of zinc fingers optimized via phage display: evaluating the utility of a recognition code. J. Mol. Biol. 1999;285:1917–1934. doi: 10.1006/jmbi.1998.2421. [DOI] [PubMed] [Google Scholar]
- 17.Liu Q, Xia Z, Zhong X, Case CC. Validated zinc finger protein designs for all 16 GNN DNA triplet targets. J. Biol. Chem. 2002;277:3850–3856. doi: 10.1074/jbc.M110669200. [DOI] [PubMed] [Google Scholar]
- 18.Dreier B, Beerli RR, Segal DJ, Flippin JD, Barbas C.F., 3rd. Development of zinc finger domains for recognition of the 5'-ANN-3' family of DNA sequences and their use in the construction of artificial transcription factors. J. Biol. Chem. 2001;276:29466–29478. doi: 10.1074/jbc.M102604200. [DOI] [PubMed] [Google Scholar]
- 19.Bae KH, Kwon YD, Shin HC, Hwang MS, Ryu EH, Park KS, Yang HY, Lee DK, Lee Y, Park J, et al. Human zinc fingers as building blocks in the construction of artificial transcription factors. Nat. Biotechnol. 2003;21:275–280. doi: 10.1038/nbt796. [DOI] [PubMed] [Google Scholar]
- 20.Dreier B, Fuller RP, Segal DJ, Lund CV, Blancafort P, Huber A, Koksch B, Barbas C.F., 3rd Development of zinc finger domains for recognition of the 5′-CNN-3′ family DNA sequences and their use in the construction of artificial transcription factors. J. Biol. Chem. 2005;280:35588–35597. doi: 10.1074/jbc.M506654200. [DOI] [PubMed] [Google Scholar]
- 21.Alwin S, Gere MB, Guhl E, Effertz K, Barbas C.F., 3rd., Segal DJ, Weitzman MD, Cathomen T. Custom zinc-finger nucleases for use in human cells. Mol. Ther. 2005;12:610–617. doi: 10.1016/j.ymthe.2005.06.094. [DOI] [PubMed] [Google Scholar]
- 22.Beumer K, Bhattacharyya G, Bibikova M, Trautman JK, Carroll D. Efficient gene targeting in Drosophila with zinc-finger nucleases. Genetics. 2006;172:2391–2403. doi: 10.1534/genetics.105.052829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segal DJ, Crotty JW, Bhakta MS, Barbas C.F., 3rd., Horton NC. Structure of Aart, a designed six-finger zinc finger peptide, bound to DNA. J. Mol. Biol. 2006;363:405–421. doi: 10.1016/j.jmb.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 24.Cornu TI, Thibodeau-Beganny S, Guhl E, Alwin S, Eichtinger M, Joung JK, Cathomen T. DNA-binding specificity is a major determinant of the activity and toxicity of zinc-finger nucleases. Mol. Ther. 2008;16:352–358. doi: 10.1038/sj.mt.6300357. [DOI] [PubMed] [Google Scholar]
- 25.Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, Winfrey RJ, Sander JD, Fu F, Townsend JA, et al. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat. Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wright DA, Thibodeau-Beganny S, Sander JD, Winfrey RJ, Hirsh AS, Eichtinger M, Fu F, Porteus MH, Dobbs D, Voytas DF, et al. Standardized reagents and protocols for engineering zinc finger nucleases by modular assembly. Nat. Protoc. 2006;1:1637–1652. doi: 10.1038/nprot.2006.259. [DOI] [PubMed] [Google Scholar]
- 28.Ryder SP, Frater LA, Abramovitz DL, Goodwin EB, Williamson JR. RNA target specificity of the STAR/GSG domain post-transcriptional regulatory protein GLD-1. Nat. Struct. Mol. Biol. 2004;11:20–28. doi: 10.1038/nsmb706. [DOI] [PubMed] [Google Scholar]
- 29.Seidman CE. Transformation using calcium chloride. In: Ausubel FM, editor. Current Protocols in Molecular Biology. 1997. Vol. I Unit 1.8. John Wiley & Son, Inc., New York. [Google Scholar]
- 30.Lundblad JR, Laurance M, Goodman RH. Fluorescence polarization analysis of protein-DNA and protein-protein interactions. Mol. Endocrinol. 1996;10:607–612. doi: 10.1210/mend.10.6.8776720. [DOI] [PubMed] [Google Scholar]
- 31.LiCata VJ, Wowor AJ. Applications of fluorescence anisotropy to the study of protein-DNA interactions. Methods Cell Biol. 2008;84:243–262. doi: 10.1016/S0091-679X(07)84009-X. [DOI] [PubMed] [Google Scholar]
- 32.Hurt JA, Thibodeau SA, Hirsh AS, Pabo CO, Joung JK. Highly specific zinc finger proteins obtained by directed domain shuffling and cell-based selection. Proc. Natl Acad. Sci. USA. 2003;100:12271–12276. doi: 10.1073/pnas.2135381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Veprintsev DB, Fersht AR. Algorithm for prediction of tumour suppressor p53 affinity for binding sites in DNA. Nucleic Acids Res. 2008;36:1589–1598. doi: 10.1093/nar/gkm1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayouka Z, Rosenbluh J, Levin A, Maes M, Loyter A, Friedler A. Peptides derived from HIV-1 Rev inhibit HIV-1 integrase in a shiftide mechanism. Biopolymers. 2008;90:481–487. doi: 10.1002/bip.20930. [DOI] [PubMed] [Google Scholar]
- 35.Segal DJ, Beerli RR, Blancafort P, Dreier B, Effertz K, Huber A, Koksch B, Lund CV, Magnenat L, Valente D, et al. Evaluation of a modular strategy for the construction of novel polydactyl zinc finger DNA-binding proteins. Biochemistry. 2003;42:2137–2148. doi: 10.1021/bi026806o. [DOI] [PubMed] [Google Scholar]
- 36.Yang WP, Wu H, Barbas C.F., 3rd Surface plasmon resonance based kinetic studies of zinc finger-DNA interactions. J. Immunol. Methods. 1995;183:175–182. doi: 10.1016/0022-1759(95)00048-f. [DOI] [PubMed] [Google Scholar]
- 37.Kang JS. Correlation between functional and binding activities of designer zinc-finger proteins. Biochem. J. 2007;403:177–182. doi: 10.1042/BJ20061644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bulyk ML. DNA microarray technologies for measuring protein-DNA interactions. Curr. Opin. Biotechnol. 2006;17:422–430. doi: 10.1016/j.copbio.2006.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berger MF, Philippakis AA, Qureshi AM, He FS, Estep P.W., 3rd, Bulyk ML. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maerkl SJ, Quake SR. A systems approach to measuring the binding energy landscapes of transcription factors. Science. 2007;315:233–237. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- 41.Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beerli RR, Dreier B, Barbas C.F., 3rd Positive and negative regulation of endogenous genes by designed transcription factors. Proc. Natl Acad. Sci. USA. 2000;97:1495–1500. doi: 10.1073/pnas.040552697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stormo GD, Fields DS. Specificity, free energy and information content in protein-DNA interactions. Trends Biochem. Sci. 1998;23:109–113. doi: 10.1016/s0968-0004(98)01187-6. [DOI] [PubMed] [Google Scholar]
- 44.Benos PV, Bulyk ML, Stormo GD. Additivity in protein-DNA interactions: how good an approximation is it? Nucleic Acids Res. 2002;30:4442–4451. doi: 10.1093/nar/gkf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benos PV, Lapedes AS, Stormo GD. Probabilistic code for DNA recognition by proteins of the EGR family. J. Mol. Biol. 2002;323:701–727. doi: 10.1016/s0022-2836(02)00917-8. [DOI] [PubMed] [Google Scholar]
- 46.Liu J, Stormo GD. Quantitative analysis of EGR proteins binding to DNA: assessing additivity in both the binding site and the protein. BMC Bioinformatics. 2005;6:176. doi: 10.1186/1471-2105-6-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu J, Stormo GD. Context-dependent DNA recognition code for C2H2 zinc-finger transcription factors. Bioinformatics. 2008;24:1850–1857. doi: 10.1093/bioinformatics/btn331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sander JD, Zaback P, Joung JK, Voytas DF, Dobbs D. Zinc Finger Targeter (ZiFiT): an engineered zinc finger/target site design tool. Nucleic Acids Res. 2007;35:W599–W605. doi: 10.1093/nar/gkm349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.