Abstract
RNA 5-methyl and 5-propynyl pyrimidine analogs were substituted into short interfering RNAs (siRNAs) to probe major groove steric effects in the active RNA-induced silencing complex (RISC). Synthetic RNA guide strands containing varied combinations of propynyl and methyl substitution revealed that all C-5 substitutions increased the thermal stability of siRNA duplexes containing them. Cellular gene suppression experiments using luciferase targets in HeLa cells showed that the bulky 5-propynyl modification was detrimental to RNA interference activity, despite its stabilization of the helix. Detrimental effects of this substitution were greatest at the 5′-half of the guide strand, suggesting close steric approach of proteins in the RISC complex with that end of the siRNA/mRNA duplex. However, substitutions with the smaller 5-methyl group resulted in gene silencing activities comparable to or better than that of wild-type siRNA. The major groove modifications also increased the serum stability of siRNAs.
INTRODUCTION
RNA interference (RNAi) represents a powerful molecular strategy for inhibition of gene expression (1–3). This process can be induced by 21–23 nt double-stranded RNAs (dsRNAs), known as short interfering RNAs (siRNAs) (4–6). These oligoribonucleotides (formed by a sense and an antisense strand) are recognized by the RNA-induced silencing complex (RISC) (7,8), a protein complex located in the cytoplasm. After siRNA binding, RISC is activated by ATP and carries out unwinding and strand separation of the siRNA duplex, loading one RNA (the antisense ‘guide’ strand) into the activated RISC complex. The RISC complex uses the guide strand as a template to find the complementary target mRNA (9) and induces endonucleolytic cleavage of the mRNA (10–12), preventing its translation into protein. Despite the significance of RNAi both as a biological tool and as a potential therapeutic strategy, some important challenges remain, including development of strategies for effective siRNA delivery, improving stability against serum nuclease degradation (13) and addressing low sequence specificity that results in undesired ‘off-target’ activities (14,15). To address these limitations and to further understand the mechanism of silencing, several research groups have been actively studying siRNAs with various chemical modifications (16–19). However, most of the focus to date has been on modifying the RNA backbone, and few laboratories have modified the bases of siRNAs, despite their central involvement in target recognition.
In recent studies, we and others (20–22) have begun to investigate the physicochemical factors contributing to mRNA sequence recognition by the RISC complex using siRNAs containing modified nucleobases. In one study, we used nonpolar base replacements to probe steric effects of altering nucleobase shapes on the sequence specificity of mRNA recognition; we found that base shape had a large effect on the preferred target sequence, and that with the unnatural bases, sequence specificity could be increased over that of natural RNA (21). In a different study, we incorporated a nonhydrogen bonding uridine isostere into 11 different positions in place of natural uridine along an RNA guide strand (20). This experiment allowed us to perturb the stability of the duplex in a systematic way. We found that at 9 of 11 positions there was a very close correlation of RNA duplex stability with gene supression activity, such that more stable siRNA duplexes showed better activity in suppressing the target mRNA expression in HeLa cells. In those experiments, the nonpolar analog decreased stability, resulting in lower activity. However, if such a correlation were general, then in principle, increases in RNA duplex stability (as a result of chemical modifications) might increase RNAi activity over that of natural RNA. Such duplex stabilization might add favorable free energy that could compensate for the unwinding of the mRNA target from its native structure. In general, the experiments to date have revealed that both steric effects and thermodynamic stability of siRNAs play substantial roles in their biological activity.
A survey of the literature shows that few nucleobase modifications known to affect stability or sterics have been tested for their effect on siRNA activity. In this context, Rana and co-workers incorporated 5-bromouridine, 5-iodouridine and 2,6-diaminopurine (23) into siRNAs and tested the cellular activity of the modified oligonucleotides (24). The former two substitutions add a small amount of steric size in the RNA major groove, and all three may increase duplex stability to a small extent. It was found that RNAs with these modifications could induce RNAi-directed gene suppression, although to a level significantly lower than the one observed for the wild-type siRNA. On the other hand, several sugar modifications known to stabilize double helices, such as 2′-fluoro and 2′-O-Me substitutions (25), have been studied in siRNAs. In particular, siRNAs having 2′-fluoro substitutions at internal positions along the guide strand have been shown to display near wild-type activity (24).
Another class of major groove modification that is known to increase RNA duplex affinity is the substitution of alkyl and alkynyl functionality at the C-5 position of pyrimidine nucleobases (26–30). The methyl group adds ∼23 Å3 of steric bulk in the major groove, while the propynyl group adds ∼53 Å3. Both types of substitution not only add steric size, but also stabilize duplexes. For example, in one context, each methyl in a 12-mer RNA increased the favorable free energy of duplex formation by 0.3 kcal/mol, yielding several kilocalories per moles of total stabilization in short duplexes with multiple substitutions (26). Interestingly, the larger, unsaturated propyne group is expected to exert even greater effects on duplex stabilization (28). This substitution in DNA was shown to increase the melting temperature (Tm) of a DNA:RNA hybrid by 0.9–2.6°C per modification (28,31). Both the methyl and propynyl stabilization effects are likely due to enhanced base stacking propensity (25,31,32,33). The application of propynyl-substituted oligonucleotides as antisense agents in cell cultures has been well documented (34–38). However, to the best of our knowledge, there are no published studies of the effects of the 5-propynyl substitution in siRNAs. As for methyl substitution, we know of no previous reports of methyl effects on ribonucleotides in siRNA. However, Eberle et al. (39) recently described siRNAs containing deoxythymidine substitutions, in which methyl groups and deoxy sugars were incorporated together. Interestingly, they found that four substitutions led to a decrease in off-target effects in plasmacytoid dendritic cells without affecting gene silencing activity. However, they did not test for positional effects. It is well-known that the 5′-half of the siRNA duplex (as defined by the antisense strand) is functionally distinct from the 3′-half, due to the asymmetric nature of siRNA recognition for initiation of unwinding (24,40), and in addition, interactions of the RISC protein(s) with the guide and mRNA are expected to be different at the two ends as well. Thus, a detailed study comparing the positional effects of 5-substitutions of different size on RNAi activity would be valuable in probing major groove steric effects in the active RISC complex.
Here we explore the effects of methyl and propynyl substitution on siRNA duplex stability and on cellular RNAi activity. The goals of these studies are (i) to probe major groove steric effects in the active RISC complex and (ii) to evaluate whether enhanced duplex stability has a positive effect on RNAi activity. Furthermore, we explore the effects of these nucleobase modifications on the sensitivity to siRNA degradation in human serum, since the unnatural C-5 substitution might alter biostability, thus contributing in a different way to the activity.
MATERIALS AND METHODS
Nucleoside phosphoramidite synthesis
The 5-propynylU ribonucleoside was prepared as described (41). For automated RNA synthesis, the 5-dimethoxytrityl, 2′-O-TOM (TOM = [(triisopropylsilyl)oxy]methyl), 3′-O-phosphoramidite derivative was prepared. Details are given in the Supplementary Data.
RNA preparation
Modified and unmodified 21-nt RNAs (passenger strand and guide strands 1–9) were synthesized on the 1 µmol scale on an applied Biosystems 394 synthesizer using 2′-TOM-phosphoramidites (42,43). Acetonitrile (synthesis grade), the 2′-TOM protected phosphoramidite monomers of A, C, G, U and 5-methylC and the 2′-OTBDMS protected phosphoramidite monomer of 5-methylU were from commercial suppliers. 5-Ethylthio-1H-tetrazole was used as activator. The standard coupling time of 6 min was used for the four standard phosphoramidites and an increased coupling time of 15 min was used for the modified 5-propynylU, 5-methylC and 5-methylU phosphoramidites. All oligonucleotides were synthesized in DMT-off mode. After the solid-phase synthesis, the solid support was transferred to a screw-cap vial and incubated at room temperature for 16 h with 1.5 ml methylamine solution (prepared by mixing one volume of 40% aqueous methylamine with one volume of 33% methylamine in ethanol). The vial was then cooled on ice and the supernatant was transferred into a 2 ml eppendorf tube and the support was rinsed with 50% ethanol (2 × 0.25 ml). The combined solutions were evaporated to dryness using an evaporating centrifuge. The residue that was obtained was dissolved in 1 ml of 1 M tetrabutylammonium fluoride (TBAF) in tetrahydrofuran (THF) and rocked at 37°C for 12 h. Then, 1 ml of 1 M Tris–HCl (pH 7.4) was added and the oligonucleotide was desalted on a NAP-25 column using water as the eluant and evaporated to dryness. After purification by 20% denaturing polyacrylamide gel, the oligonucleotides were isolated by the crush and soak method, quantified by absorption at 260 nm and confirmed by MALDI mass spectrometry (Supplementary Data). Sequence-scrambled control siRNAs were synthesized by the protein and nucleic acid (PAN) facility at Stanford University.
UV-monitored thermal denaturation
Absorbance versus temperature curves of duplexes were measured at 1 µM strand concentration in 15 mM HEPES–KOH (pH 7.4), 1 mM MgCl2 and 50 mM KOAc buffer and in low salt buffer (2 mM sodium phosphate, pH 7.0, 1 mM ethylenediaminetetraacetic acid (EDTA)). Experiments were performed on a Varian Cary UV-vis spectrophotometer equipped with thermoprogrammer. The samples were heated to 95°C, allowed to slowly cool to 25°C, and then warmed during the denaturation experiments at a rate of 1°C/min to 95°C, monitoring absorbance at 260 nm. The data were analyzed by the denaturation curve processing program, MeltWin v. 3.0. Melting temperatures (Tm) were determined by computer-fit of the first derivative of absorbance with respect to 1/T.
RNAi methods
HeLa cells were grown at 37°C in Dulbecco's modified Eagle's medium (DMEM; GIBCO) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 µg/ml streptomycin. Cells were regularly passaged to maintain exponential growth. Twenty-four hours before transfection at 50–80% confluency, mammalian cells were trypsinized and diluted 1:5 with fresh medium without antibiotics (1–3 × 105 cells/ml) and transferred to 24-well plates (500 µl per well). Two luciferase plasmids, Renilla luciferase (pRL-CMV) and firefly luciferase (pGL3) from Promega, were used as reporter and control, respectively. Cotransfection of plasmids and siRNAs was carried out with Lipofectamine 2000 (Life Technologies) as described by the manufacturer for adherent cell lines. Per well, 1.0 µg pGL3-Control, 0.1 µg pRL-CMV and 0.03–26 nM siRNA duplex formulated into liposomes, were applied; the final volume was 600 µl per well. The cells were harvested 22 h after transfection, and lysed using passive lysis buffer, 100 µl per well, according to the instructions of the Dual-Luciferase Reporter Assay System (Promega). The luciferase activities of the samples were measured using a Fluoroskan Ascent FL Luminometer (Thermo Electron Corporation, USA) with a delay time of 2 s and an integrate time of 10 s. The following volumes were used: 20 µl of sample and 30 µl of each reagent (Luciferase Assay Reagent II and Stop and Glo Reagent). The inhibitory effects generated by siRNAs were expressed as normalized ratios between the activities of the reporter (Renilla) luciferase gene and the control (firefly) luciferase gene.
Stability assays
The dsRNAs (14 μM) were incubated in 90% human serum, and the mixture was incubated at 37°C for various lengths of time (0 min, 15 min, 30 min, 120 min and 240 min). At the time points indicated, 1% sodium dodecyl sulfate was added and mixtures were heat-denatured for 5 min at 100°C. siRNAs were isolated by hot phenol extraction in the presence of sodium dodecyl sulfate followed by ethanol precipitation. Re-suspended RNA samples were run on a denaturing 14% polyacrylamide gel containing 20% formamide. RNA bands were visualized with the ‘Stains All’ reagent (Sigma-Aldrich) according to the manufacturer's instructions.
RESULTS AND DISCUSSION
Design and synthesis of modified guide-RNA strands containing 5-methyl and 5-propynyl substitutions
The 5-propynylU (pU), 5-methylU (mU) and/or 5-methylC (mC) monomers (Figure 1) were incorporated into different positions in place of natural rU and rC along a 21-mer RNA guide strand that targets an A-rich site 501–519 of Renilla luciferase mRNA (20,21). The 5-methylU and 5-methylC monomers were obtained from commercial sources. The 5-propynylU monomer was synthesized using known Sonogashira chemistry according to the literature (41,44) (see the Supplementary Data for details). We prepared RNAs containing no substitutions (wt, sequence 1), 5-methyl groups at internal positions [methylC in place of C (2)], propynyl groups at spaced-out positions along the guide strand (3), fully substituted with 11 propynyl groups (4) and substituted with propynyl and methyl groups [methylC and propynylU (5)]. Moreover, we focused attention on predominantly 5′-modified and predominantly 3′-modified siRNA duplexes, in order to localize any possible steric effects. To this end, 5-propynylU or 5-methylU were incorporated within the antisense (guide) strand predominantly on the 5′-half (RNAs 6 and 7) or predominantly on the 3′-half (RNAs 8 and 9).
Figure 1.
The 5-substituted nucleosides employed in this study.
Thermal denaturation studies
First we measured the effects of these substitutions on thermal stability of the siRNA duplex formed by these sequences paired with the passenger strand (Figure 2). Compared to the unmodified RNA sequence, all substituted oligoribonucleotides had greater duplex stability (as evaluated by higher Tm values) when hybridized to their RNA complement (see data in Figure 2 and in Supplementary Table S2). As expected, the incorporation of the propyne group at the position 5 gave rise to a substantially larger stabilization of the RNA duplex compared with the 5-methyl modification. The incorporation of 5-propynylU residues resulted in an increase of 1.3–1.8°C per modification depending on the position of substitution. The contribution of the 5-methylC was smaller (1.0°C per methyl group), and 5-methylU substitution yielded the smallest stabilization, at 0.2–0.7°C per substitution. The rank order of duplex Tm was: eleven 5-propynylU (4) > five 5-propynylU and five methylC (5) > six 5-propynylU on the 3′-half (8) > six 5-propynylU on the 5′-half (6) = five propynylU at spaced-out positions along the guide strand (3) > five 5-methylC at internal positions (2) = six 5-methylU on the 3′-half (9) > six 5-methylU on the 5′-half (7).
Figure 2.
Sequences of modified guide-RNAs (1–9) and of passenger RNA used in the RNAi experiments and Tm data. Sites of propynylU (‘pU’), methylU (‘mU’) and methylC (‘mC’) substitutions are indicated in bold. Tms were measured in 15 mM HEPES–KOH (pH 7.4), 1 mM MgCl2 and 50 mM KOAc. Tm data from lower ionic strength solution are included in the Supplementary Table S2.
Effect of 5-methyl and 5-propynyl substitutions on RNAi activity
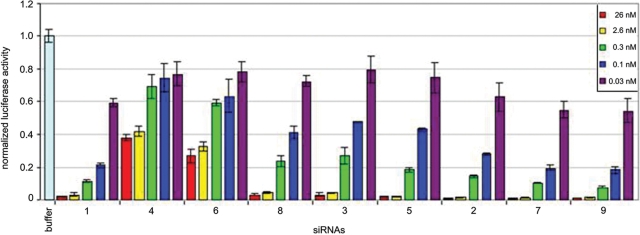
We then carried out separate RNAi studies in HeLa cells with siRNA duplexes containing each of the eight modified guide-RNA strands containing 5-methylU, 5-propynylU and/or 5-methylC substitutions (2–9), as well as with the unmodified (wt) RNA (1). Experiments were carried out in triplicate. The cells were first transfected with dual reporter plasmids that express Renilla luciferase (the target) and nontargeted firefly luciferase as an internal nontargeted control. The effects of the different RNAs on luciferase expression were evaluated after dosing with 0.21–210 ng (0.03–26 nM) of dsRNA in the cell media, and measuring luminescence responses after 22 h. The results, showing Renilla luciferase activity normalized to firefly luciferase, are represented in Figure 3. Surprisingly, 5-propynylU siRNAs disrupted Renilla luciferase-specific RNAi activity despite the strong stabilization that this substitution causes. Incorporation of 11 5-propynylU units within the guide strand (RNA 4) caused loss of most of the interference activity even at the highest siRNA concentration (26 nM). Similar results were observed for the guide RNA containing six 5-propynylU substitutions on the 5′-half (6). Interestingly, and in contrast, incorporation of six 5-propynylU groups on the 3′-half (8) did not disrupt RNAi activity, although gene silencing was not as efficient as for the wild-type RNA. At siRNA concentration of 0.3 nM, Renilla luciferase expression was ∼59% for sequence 6, ∼24% for sequence 8 and ∼12% for the wild-type siRNA (1). These contrasting results support the idea that the major groove of the siRNA/mRNA active substrate is more sterically sensitive in the 5′-half of the antisense strand of the duplex than the 3′-half. This is also consistent with previous observations that mismatches near the 5′-half of a siRNA caused negative effects on RNAi activity, whereas siRNAs mismatched at the 3′ retained a significant level of gene silencing (24). We surmise that close fitting of protein around the guide/mRNA complex may result in more costly disruptions at the 5′-end.
Figure 3.
Plot of gene-specific RNAi activity for 5-propynyl and 5-methyl substituted siRNAs at different sites in the Renilla luciferase mRNA expressed in HeLa cells. Varied amounts of RNA were added as shown.
In contrast, the 5-methyl substitution did not disrupt RNAi activity. When 5-methylC was incorporated into positions 6, 8, 9, 12 and 15 in place of natural C along the guide RNA strand containing five 5-propynylU substitutions (3), gene silencing was more efficient than that observed for 3, which contained the propynyl groups alone (81% knockdown for 5 versus 73% for 3). Moreover, guide RNAs having five 5-methylC substitutions at internal positions (2), six 5-methylU residues on the 5′-half or six 5-methylU's on the 3′-half (7 and 9, respectively) showed activity comparable to or better than that of the wild-type siRNA (Figure 4). For example, at a siRNA concentration of 0.3 nM, Renilla luciferase gene silencing was 86.0 ± 0.8% for sequence 2, 89.0 ± 0.3% for 7, 92.1 ± 0.3% for 9 and 88 ± 0.4% for the wild-type siRNA (1). At all siRNA concentrations from 0.1 nM to 26 nM, the methylated RNAs 7 and 9 showed small but statistically significant increases in gene supression activity.
Figure 4.
Plot comparing RNAi activity of methylated siRNAs (2, 7, 9) to unmodified siRNA (1). Data are from Figure 3, plotted on a scale that allows for ready comparison of highest activity RNAs. Note the consistently greater gene supression by 7 and 9 relative to 1.
To ensure the specificity of the observed effects, sequence-scrambled siRNAs (as negative controls) were used. Cells cotransfected with scrambled versions of the unmodified siRNA 1 and scrambled versions of two of the more active chemically modified siRNAs (2 and 9) (see sequences in Supplementary Figure 1) showed levels of Renilla luciferase activity similar to those of cells transfected with plasmids alone (see data in Supplementary Figure 2). The results indicate that the siRNAs used in this study do indeed specifically induce inhibition of expression of the target gene.
The above results showed that the significant increase in thermal stability caused by the 5-propynyl substitution did not have positive effects on activity. As a result, there is no general correlation of Tm with gene suppression ability (Figure 5). However, in contrast to this, the less stabilizing 5-methyl substitution resulted in siRNAs displaying activity at least comparable to that of the unmodified siRNA (Figure 4). Thus the methyl substitution, possibly as a result of its positive influence on stability, favorably affects biological activity, while the larger propynyl group does not.
Figure 5.
Dual plot comparing helix stability (Tm) and RNA gene knockdown activity (percent suppression) with 5-propynyl and 5-methyl substitutions along the guide strand of siRNA duplexes at different positions. Suppression data are for siRNA concentrations of 0.3 nM.
We hypothesize that this difference between methyl and propynyl substitution may result from steric effects in the RNA duplex. Rana and co-workers reported that RNAi absolutely requires A-form helix formation between target mRNA and its guiding antisense strand (40). Structurally, methyl and propynyl functionalities located at the position 5 of pyrimidine nucleosides would lie in the major groove of the dsRNA. Our results show that both of these groups stabilize the RNA duplex, and thus likely do not affect major groove structure strongly. However, the RNAi activities suggest that the propynyl substituent, as a bulky group, may sterically hinder protein contacts in the major groove around the 5′-half of the active RISC complex. In contrast, the methyl group is substantially smaller (by ∼2.6 Å in length) (45,46), and appears to be well tolerated at all regions of the active siRNA. A recent crystal structure of the PIWI domain of the Ago 2 protein (a key component of RISC) in complex with a 16-nt siRNA-like duplex (47) reveals potential steric clashes between bulky substituents at the C-5 position of U1 and U2 residues and the protein. This structure contains a highly conserved metal-binding site that recognizes the 5′-end of the guide strand. Methyl or propynyl groups introduced at the C-5 position of U1 and U2 residues would point toward the Ile142 and the Tyr124 residues of the PIWI domain, respectively (Figure 6). The distances between the C-5 atom of U1 and the Cγ atom of Ile142 (3.99 Å) and between the C-5 atom of U2 and the hydroxyl group of Tyr124 (4.07 Å) suggest that a 5-propynyl substitution might cause a serious steric conflict, whereas a 5-methyl group might be well tolerated. Based on these observations we speculate that such steric clashes might lead to loss of important interactions between the 5′-end of the guide strand and the protein residues involved in its recognition (47,48). On the other hand, 3′-nucleobases of the guide strand are not likely involved in direct contacts with the protein. A crystal structure of the PAZ domain of Ago 2 in complex with a 9-mer siRNA-like duplex, which mimics the 3′-overhanging end of a guide strand bound to a target mRNA (pdb code 1SI2) (49), reveals interactions predominantly between the phosphodiester backbone of the 3′-end of the guide strand and the protein. Thus, based on the above described hypotheses one could expect negative effects on RNAi activity for siRNAs having five bulky substituents at positions 1 and 2 of the guide strand. This hypothesis needs further exploration but may provide a satisfactory explanation to the observation that RNAi activity was significantly low for guide-siRNAs having 5-propynyl substitutions at positions 1 and 2 (4 and 6), markedly higher for siRNA 3, which has a 5-propynyl substitution at position 2 and a natural U nucleobase at position 1, and much higher activity for predominantly 3′-propynyl-modified siRNA 8.
Figure 6.
Models of interactions between the PIWI domain and the 5′-nucleobases of a siRNA-like guide strand. Adapted from the crystal structure of a Piwi protein from Archaeoglobus fulgidus (AfPiwi) in complex with a siRNA-like duplex (pdb code 2bgg). The figures were prepared with PyMOL (http://pymol.sourceforge.net/).
Stability of modified siRNAs in human serum
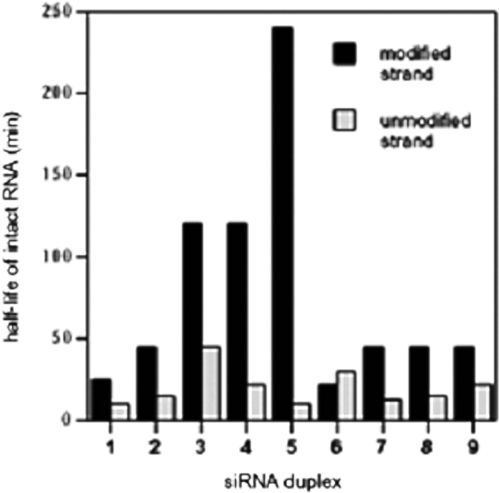
Since the above experiments showed that siRNAs modified with 5-substituted nucleobases could effectively mediate RNAi activity to levels comparable to wild-type RNA, it was of interest to study the serum stability of these oligonucleotides. To carry out such studies, unmodified or modified dsRNAs were incubated in 90% human serum. At various time points, siRNAs were extracted, analyzed on a 20% polyacrylamide gel under denaturating conditions and visualized by staining (Supplementary Data). Unmodified double-stranded siRNA (wt, 1) showed low stability with ∼50% and 0% of the original siRNA remaining intact through 30 min and 2 h in serum, respectively. Double-stranded siRNAs with six 5-methylU's on the 5′-half and six 5-methylU on the 3′-half (7 and 9, respectively) demonstrated an increase in stability over the course of the experiment, with half-lives twice that of unmodified guide strand (Figure 7). Similar results were observed for strand 2, which contained five 2-methylC residues at internal positions. Interestingly, double-stranded siRNA having five 5-methylC and 5-propynylU residues along the antisense strand (5) displayed strongly enhanced stability, with the original siRNA population remaining intact for the duration of the experiment (4 h). In order to determine the effect of the 5-methyl substitutions on serum stability, the 5-methylC residues of strand 5 were replaced by natural cytidine. Although the resulting modified strand (3) was significantly more stable than the wild-type siRNA (1), it showed strongly decreased stability as compared with the siRNA containing five 5-methylC and five 5-propynylU residues (5), showing the positive effect of propynyl groups. Finally, the results for strand 11, which was fully substituted with 11 propynyl groups, were similar to those observed for 3. Thus, we can conclude that, in general, incorporation of 5-methyl and 5-propynyl substitutions along the guide strand cause an increase in serum stability. The above results further suggest that the half-life of the resulting modified siRNAs likely depends also on the position of the substitution along the guide strand.
Figure 7.
Oligoribonucleotide [modified (guide) and unmodified (passenger) strands] half lives in 90% human serum, from stained gel data. Half-life is defined as the time at which full-length RNA staining density is half that of the density at time zero. Error is estimated at ± 30%.
We and others (39) have noted that incorporation of 5-methyl substitutions into guide-siRNAs did not disrupt RNAi activity, while in contrast, previous studies with 5-bromo- and 5-iodo-substitutions revealed lowered activity of the siRNAs containing them (24). The origin of this difference is unclear at present, and may be due either to chemical differences associated with methyl versus halogen substitution, or to differences in the RNA targets in the two studies. Although a number of base substitutions have been studied previously in RNAi, only the methyl substitution has shown to yield gene suppression activity comparable to that of substituted RNA.
CONCLUSIONS
In summary, we have evaluated steric and stability effects on RNAi activity by use of siRNAs modified with propynyl and methyl functionalities at the C-5 position of pyrimidine nucleobases. Our results suggest that at the 5′-half of the guide RNA, large increases in the size of the functionality at the position 5 of pyrimidine nucleobases can be detrimental to RNAi activity. Despite the strong stabilizing effects of the 5-propynyl modification, this bulky substitution caused negative effects in RNAi activity, probably due to disruption of interactions in the major groove of the active RISC complex. However, the smaller 5-methyl substitution did not adversely affect gene silencing activity. Furthermore, this modification contributed positively to stability of siRNAs in human serum. Thus, our work offers an avenue for the design of future RNA agents for biomedical applications of siRNAs, and adds support to the notion that modified nucleobases can play an important role in biological activity and sequence selectivity of siRNAs (16–22,24).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (GM072705). Funding for open access charge: National Institutes of Health (GM072705).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the National Institutes of Health (GM072705) for support. M.T. acknowledges support from Beatriu de Pinós Fellowship (AGAUR, Generalitat de Catalunya, Spain).
REFERENCES
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 3.Bumcrot D, Manoharan M, Koteliansky V, Sah DWY. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat. Chem. Biol. 2006;2:711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang G, Reinhart BJ, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Genes Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 8.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 9.Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 10.Meister G, Landthaler M, Patkaniwska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 12.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 13.Hoerter JAH, Walter NG. Chemical modification resolves the asymmetry of siRNA strand degradation in human blood serum. RNA. 2007;13:1887–1893. doi: 10.1261/rna.602307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchad J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 15.Alemán LM, Doench J, Sharp PA. Comparison of siRNA-induced off-target RNA and protein effect. RNA. 2007;13:385–395. doi: 10.1261/rna.352507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amarzguioui M, Holen T, Babaie E, Prydz H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003;31:589–595. doi: 10.1093/nar/gkg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manoharan M. RNA interference and chemically modified small interfering RNAs. Curr. Opin. Chem. Biol. 2004;8:570–579. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fedorov Y, Anderson EM, Birmingham A, Reynolds A, Karpilow J, Robinson K, Leake D, Marshall WS, Khvorova A. Off-target effects by siRNA can induce toxic phenotype. RNA. 2006;12:1188–1196. doi: 10.1261/rna.28106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Somoza A, Kool ET. The roles of hydrogen bonding and sterics in RNA interference. Angew. Chem. Int. Ed. 2006;45:4994–4997. doi: 10.1002/anie.200601311. [DOI] [PubMed] [Google Scholar]
- 21.Somoza A, Silverman AP, Miller RM, Chelliserrykattil J, Kool ET. Steric effects in RNA interference: probing the influence of nucleobase size and shape. Chem. Eur. J. 2008;14:7978–7987. doi: 10.1002/chem.200800837. [DOI] [PubMed] [Google Scholar]
- 22.Xia J, Noronha A, Toudjarska I, Li F, Akinc A, Braich R, Frank-Kamenetsky M, Rajeev KG, Egli M, Manoharan M. Gene silencing activity of siRNAs with a ribo-difluorotoluyl nucleotide. ACS Chem. Biol. 2006;1:176–183. doi: 10.1021/cb600063p. [DOI] [PubMed] [Google Scholar]
- 23.Saenger W. In: Principles of Nucleic Acid Structure. Cantor CR, editor. New York: Springer; 1984. pp. 116–158. [Google Scholar]
- 24.Chiu Y-L, Rana T. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cummins LL, Owens SR, Risen LM, Lesnik EA, Freier SM, McGee D, Guinosso CJ, Cook PD. Characterization of fully 2'-modified oligoribonucleotide hetero- and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res. 1995;23:2019–2024. doi: 10.1093/nar/23.11.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang S, Kool ET. Origins of the large differences in stability of DNA and RNA helixes: C-5 methyl and 2'-hydroxyl effects. Biochemistry. 1995;34:4125–4132. doi: 10.1021/bi00012a031. [DOI] [PubMed] [Google Scholar]
- 27.Han H, Dervan PB. Sequence-specific recognition of double helical RNA and RNA.DNA by triple helix formation. Proc. Natl Acad. Sci. USA. 1993;90:3806–3810. doi: 10.1073/pnas.90.9.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Froehler BC, Wadwami S, Terhorst TJ, Gerrard SR. Oligodeoxynucleotides containing C-5 propyne analogs of 2′-deoxyuridine and 2′-deoxycytidine. Tetrahedron Lett. 1992;33:5307–5310. [Google Scholar]
- 29.Seela F, Sirivolu VR. DNA containing side chains with terminal triple bonds: base-pair stability and functionalization of alkynylated pyrimidines and 7-deazapurines. Chem. Biodivers. 2006;6:509–514. doi: 10.1002/cbdv.200690054. [DOI] [PubMed] [Google Scholar]
- 30.Seela F, Ramzaeva N, Leonard P, Chen Y, Debelak H, Feiling E, Kröschel R, Zulauf M, Wenzel T, Fröhlich T, et al. Phosphoramidites and oligonucleotides containing 7-deazapurines and pyrimidines carrying aminopropargyl side chains. Nucleosides, Nucleotides, Nucleic Acids. 2001;20:1421–1424. doi: 10.1081/NCN-100002568. [DOI] [PubMed] [Google Scholar]
- 31.Freier SM, Altmann KH. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 1997;25:4429–4443. doi: 10.1093/nar/25.22.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gyi JI, Gao D, Conn GL, Trent JO, Brown T, Lane AN. The solution structure of a DNA·RNA duplex containing 5-propynylU and C; comparison with 5-Me modifications. Nucleic Acids Res. 2003;31:2683–2693. doi: 10.1093/nar/gkg356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonham MA, Brown S, Boyd AL, Brown PH, Bruckenstein DA, Hanvey JC, Thomson SA, Pipe A, Hassman F, Bisi JE, et al. An assessment of the antisense properties of RNase H-competent and steric-blocking oligomers. Nucleic Acids Res. 1995;23:1197–1203. doi: 10.1093/nar/23.7.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wagner RW, Matteucci MD, Lewis JG, Gutierrez AJ, Moulds C, Froehler BC. Antisense gene inhibition by oligonucleotides containing C-5 propyne pyrimidines. Science. 1993;260:1510–1513. doi: 10.1126/science.7684856. [DOI] [PubMed] [Google Scholar]
- 35.Moulds C, Lewis JG, Froehler BC, Grant D, Huang T, Milligan JF, Matteucci MD, Wagner RW. Site and mechanism of antisense inhibition by C-5 propyne oligonucleotides. Biochemistry. 1995;34:5044–5053. doi: 10.1021/bi00015a015. [DOI] [PubMed] [Google Scholar]
- 36.Flanagan WM, Kothavale A, Wagner RW. Effects of oligonucleotide lenght, mismatches and mRNA levels on C-5 propyne-modified antisense potency. Nucleic Acids Res. 1996;24:2936–2941. doi: 10.1093/nar/24.15.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner RW, Matteucci MD, Grant D, Huang T, Froehler BC. Potent and selective inhibition of gene expression by an antisense heptanucleotide. Nat. Biotechnol. 1996;14:840–844. doi: 10.1038/nbt0796-840. [DOI] [PubMed] [Google Scholar]
- 38.Flanagan WM, Wolf JJ, Olson P, Grant D, Link KY, Wagner RW, Matteucci MD. A cytosine analog that confers enhanced potency to antisense oligonucleotides. Proc. Natl Acad. Sci. USA. 1999;96:3513–3518. doi: 10.1073/pnas.96.7.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eberle F, Gieβler K, Deck C, Heeg K, Peter M, Richert C, Dalpke AH. Modifications in small interfering RNA that separate immunostimulation from RNA interference. J. Immunol. 2008;180:3229–3237. doi: 10.4049/jimmunol.180.5.3229. [DOI] [PubMed] [Google Scholar]
- 40.Chiu Y-L, Rana TM. RNAi in human cells: basic structural and functional features of small interfering RNA. Mol. Cell. 2002;10:549–561. doi: 10.1016/s1097-2765(02)00652-4. [DOI] [PubMed] [Google Scholar]
- 41.Ghilagabaer S, Hunter WN, Marquez R. Efficient coupling of low boiling point alkynes and 5-iodonucleosides. Tetrahedron Lett. 2007;48:483–486. [Google Scholar]
- 42.Pitsch S, Weiss PA, Jenny L, Stutz A, Wu X. Reliable chemical synthesis of oligoribonucleotides (RNA) with 2′-O-[(triisopropylsilyl)oxy]methyl (2′-O-tom)-protected phosphoramidites. Helv. Chim. Acta. 2001;84:3773–3795. [Google Scholar]
- 43.Jin S, Miduturu CV, McKinney DC, Silverman SK. Synthesis of amine- and thiol- modified nucleoside phosphoramidites for site-specific introduction of biophysical probes into RNA. J. Org. Chem. 2005;70:4284–4299. doi: 10.1021/jo050061l. [DOI] [PubMed] [Google Scholar]
- 44.Berry DA, Jung K.-Y, Wise DS, Sercel AD, Pearson WH, Mackie H, Randolph JB, Somers RL. Pyrrolo-dC and pyrrolo-C: fluorescent analogs of cytidine and 2′-deoxycytidine for the study of oligonucleotides. Tetrahedron Lett. 2004;45:2457–2461. [Google Scholar]
- 45.Cygler M, Anderson WF, Giziewicz J, Robins MJ. The crystal and molecular structure of 5-(propyn-1-yl)-1-(β-D-arabinofuranosyl)uracil. A very short C≡C triple bond. Can. J. Chem. 1984;62:147–152. [Google Scholar]
- 46.Young DW, Tollin P, Wilson HR. The crystal and molecular structure of thymidine. Acta Cryst. 1969;B25:1423–1432. [Google Scholar]
- 47.Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song J-J, Joshua-Tor L. Argonaute and mRNA–getting into the groove. Curr. Opin. Struct. Biol. 2006;16:5–11. doi: 10.1016/j.sbi.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 49.Ma J-B, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.