Abstract
The histone-like nucleoid structuring protein (H-NS) is an important regulator of stress response and virulence genes in gram-negative bacteria. In addition to binding regulatory regions of genes in a structure-specific manner, H-NS also binds in a structure-specific manner to sites in the Tn10 transpososome, allowing it to act as a positive regulator of Tn10 transposition. This is the only example to date of H-NS regulating a transposition system by interacting directly with the transposition machinery. In general, transposition complexes tend to include segments of deformed DNA and given the capacity of H-NS to bind such structures, and the results from the Tn10 system, we asked if H-NS might regulate another transposition system (Tn5) by directly binding the transposition machinery. We show in the current work that H-NS does bind Tn5 transposition complexes and use hydroxyl radical footprinting to characterize the H-NS interaction with the Tn5 transpososome. We also show that H-NS can promote Tn5 transpososome formation in vitro, which correlates with the Tn5 system showing a dependence on H-NS for transposition in vivo. Taken together the results suggest that H-NS might play an important role in the regulation of many different bacterial transposition systems and thereby contribute directly to lateral gene transfer.
INTRODUCTION
The H-NS protein is a global transcriptional repressor that plays an important role in lateral gene transfer (LGT) in gram-negative bacteria by temporarily silencing newly acquired genes (1–5). In addition, there is emerging evidence that H-NS can directly contribute to LGT by acting as a positive regulator of several different DNA transposition systems (6–10). In the case of Tn10, H-NS appears to up-regulate transposition by binding directly to the transposition complex (or transpososome) and altering the conformation of this complex in a manner that promotes intermolecular transposition events while at the same time inhibiting self-destructive intramolecular events (10–12). This represents a new role for H-NS and at the present time it is unclear how many other transposition systems are regulated by H-NS via a direct interaction with the transposition machinery.
H-NS is a highly abundant DNA-binding protein in gram-negative bacteria. It readily forms dimers in solution and can form tetramers and higher order oligomers. H-NS binds regulatory regions of genes and through higher order oligomerization is thought to bring distant segments of DNA together to block RNA polymerase progression (13,14). H-NS preferentially binds bent and deformed DNA sequences (15). The ability of H-NS to recognize structural features of DNA, as opposed to recognition of specific DNA sequences, may make H-NS particularly well suited to regulate transposition reactions because the available evidence is consistent with transpososomes containing distorted DNA structures (16–20). In the current work we have asked if H-NS binds to and influences the activity of the Tn5 transposition machinery.
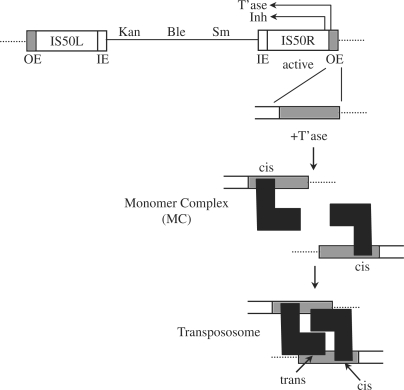
Tn5 is a composite bacterial transposon made up of degenerate copies of IS50 (R and L) that flank genes encoding resistance to the antibiotics kanamycin, streptomycin and bleomycin (Figure 1) (21,22). IS50R encodes a transposase protein that interacts with a pair of outside ends (OE) or an OE and an inside end (IE) to catalyze Tn5 and IS50 transposition, respectively. Like Tn10, Tn5/IS50 transposition occurs by a cut-and-paste mechanism that involves a transposon end hairpin (donor cleavage) intermediate (23,24).
Figure 1.
Structure of Tn5 and pathway for transpososome assembly. Tn5 is a 5.8 kb composite transposon encoding resistance to kanamycin (Kan), bleomycin (Ble) and streptomycin (Sm). These antibiotic resistance genes are flanked by nearly identical copies of the 1.5 kb IS50 insertion sequence. IS50R encodes functional transposase (T’ase) and inhibitor (Inh) proteins. Outside (OE) and inside (IE) sequences contain determinants for transposase binding and are shown as grey and white rectangles, respectively. Dotted line represents flanking donor DNA. A possible pathway for transpososome assembly is shown below the illustration of Tn5. An OE sequence is shown binding a single molecule of transposase (black L-shape) forming a monomer complex (MC). At this stage the transposase-OE contacts are defined as cis contacts. Subsequently, two MCs interact to form a transpososome. This interaction is driven by trans contacts between protein and DNA and protein–protein contacts. In an alternative model (not shown) transposase is transferred to a transposon end sequence in dimer form after initially pairing two non-transposon end DNA segments. The dimer-bound transposon end then captures a second transposon end (54,60).
The overall frequency of Tn5 transposition is just under one event per 105 cells per generation. This relatively low transposition frequency reflects the occurrence of negative regulation primarily at three levels: (1) the expression of the transposase gene; (2) the expression of an inhibitor protein; and (3) the formation of the transpososome (25–29). With regard to the latter, assembly of the transpososome is inefficient. Tn5 transpososome assembly may involve the interaction of two monomer complexes (MCs), each of which consists of a single molecule of transposase bound to a Tn5 end sequence (Figure 1), although there is also evidence for another similar but more complicated assembly pathway (see Discussion section). MC formation is directed by an interaction between the N-terminal DNA-binding domain of transposase and a portion of the transposon end spanning roughly bp 6 to 19 (20,30,31). These contacts are referred to as cis contacts. In contrast, pairing of two MCs is directed by trans contacts involving the C-terminal portion of a monomer bound to one end with bp 1–5 of the partner end. In addition, an α-helix in the C-terminal portion of transposase mediates protein–protein interactions between transposase monomers bound to separate ends (32). Prior to DNA binding, the N- and C-terminal domains of transposase have a tendency to interact with each other and this interaction is inhibitory to both MC formation and transpososome formation (26). In addition to Tn5 transposase being suboptimal for transposition, the OE and IE sequences of Tn5 are also suboptimal for transposition. A mosaic end (ME) sequence made of a combination of OE and IE sequences has been shown to increase the transposition frequency substantially (12- to 45-fold) by increasing transpososome formation (33).
With the above considerations in mind, a highly active in vitro transposition system was developed by employing an optimized end sequence (i.e. the ME) and a hyperactive (HA) transposase (34). This HA transposase contains three mutations: one at position 372 that prevents the inhibitory N- and C-terminal domain interaction (LP372); one at position 56 that prevents synthesis of the inhibitor protein (MA56); and a mutation at position 54 that enhances the transposase-end interaction (EK54) (26,35,36). This system was employed to generate X-ray crystal structures of the Tn5 transpososome, which have provided a wealth of information regarding the protein–DNA and protein–protein interactions that govern transpososome formation (37,38). These structures, as well as footprinting studies (20), reveal that portions of the DNA within the Tn5 transpososome have a distorted DNA structure making this complex a potential substrate for H-NS binding.
We show in the current work that H-NS binds to the Tn5 transpososome in vitro and that under certain conditions H-NS can stimulate transpososome formation. We also show that H-NS is able to bind to the MC, an observation that is consistent with H-NS acting at an early stage to facilitate transpososome formation. Finally, we show that in vivo, the presence of H-NS stimulates the level of Tn5 transposition.
MATERIALS AND METHODS
Chemicals and oligonucleotides were from Sigma. BCA reagent was from Pierce. Growth media was from Becton Dickinson. Enzymes were from New England Biolabs. Radio-nucleotides were from Amersham Biosciences.
Strains and plasmids
Escherichia coli strains DH10B (39) and NK5830 (40) were transduced to hns− by generalized transduction using phage P1 (41) and M182Δhns::KanR (42) as described in (8). MDW320 (27) contains a Tn5-derived transposon with a promoterless lacZ gene inserted into the F’ plasmid pOX38-Gen and this strain was mated to DH10B and DH10BΔhns::KanR to generate strains CRW31 and CRW51, respectively.
Plasmid pDH551 encodes MA56/LP372 transposase and was constructed by ligating an 8078 bp NheI-MfeI backbone fragment from pGRTYB35 (23) to the 815 bp NheI-MfeI fragment from pRZ9905 (43). pDH389 encodes a mini-Tn5-TetR element and was constructed by cloning a BamHI fragment containing Tn10 TetR from pNK861 (44) into BamHI digested pMOD-2 (Epicenter). pDH390 encodes an arabinose-inducible Tn5 transposase gene and was constructed by cloning a ClaI-SalI fragment containing AraC-AraOp-Tn5 HA transposase from pGRAra2 into ClaI-SalI digested pACYC184. pGRAra2 was derived from pBAD18 (45). pDH508 encodes a mini-Tn5-CmR element and was constructed by cloning a BamHI fragment from pNK1210 (46) containing the CmR gene of IS903 into BamHI digested pMOD-2. pDH533 encodes Tn5 transposase (MA56) under the control of its native promoter and a mini-Tn5-CmR element. This plasmid is a derivative of pRZ9905 in which the PshAI fragment encoding the mini-Tn5-CmR gene from pDH508 was ligated into the filled-in BglII site of pRZ9905. pDH548 is a derivative of pRZ9905 in which HA transposase was substituted for the MA56 transposase. This was accomplished by cutting pRZ9905 with MfeI and BsaAI and ligating the 3331 bp backbone to a 1052 bp BsaAI-MfeI fragment encoding HA transposase obtained from pGRTYB35. pDH549 is a derivative of pDH548 in which the mini-Tn5-CmR PshAI fragment of pDH508 was ligated into the filled-in BglII site of pDH548. pDH550 is a derivative of pDH549 encoding MA56/LP372 transposase and was constructed by ligating the large NheI-MfeI backbone fragment from pDH549 to the 815 bp NheI-MfeI fragment from pRZ9905.
Protein purification
Tn5 HA transposase and derivatives were over-expressed as C-terminal chitin binding domain fusions and purified using the IMPACT system (NEB) as described in (47). H-NS (WT and P116S) was over-expressed from pET3a-H-NS in BL21(DE3)hns::KanR and purified as described in (42). StpA was over-expressed from pET3a-StpA in BL21(DE3)hns::KanR and purified as described in (48). Protein concentrations were determined by Bradford assay for Tn5 transposase and by BCA assay for H-NS and StpA.
Formation of transposition complexes
DNA substrates for the assembly of transposition complexes were generated by annealing the following 5′ 32P-labeled, gel-purified oligonucleotides: 20 bp ME—5′ CTGTCTCTTATACACATCTT 3′ (NTS)/5′ AAGATGTGTATAAGAGACAG 3′ (TS); 53 bp ME (WT)—5′ CCCTGCAGGTCGACTGTCTCTTATACACATCTTGAGTGAGTGAGCATGCATGT 3′ (NTS)/5′ ACATGCATGCTCACTCACTCAAGATGTGTATAAGAGACAGTCGACCTGCAGGG 3′ (TS); 53 bp ME (3/6)—5′ CCCTGCAGGTCGACTCTCACTTATACACATCTTGAGTGAGTGAGCATGCATGT 3′ (NTS)/5′ ACATGCATGCTCACTCACTCAAGATGTGTATAAGTGAGAGTCGACCTGCAGGG 3′ (TS). Transposition complexes were assembled by mixing oligonucleotide substrates (0.05 μM) with transposase (0.4 μM) and where indicated, variable amounts of H-NS or StpA in HEPES–KCl buffer for 1 h at 37°C as previously described (47). Where H-NS was added subsequent to transposase, ME DNA and transposase were first incubated for 1 h at 37°C and then H-NS was added and incubation was carried out for an additional 30 min at 25°C. When heparin sulphate was present in assembly reactions it was added before transposase, H-NS or StpA to a final concentration of 2 μM. Samples were mixed with non-denaturing load dye and subjected to electrophoresis on a 5% TAE gel as previously described (49). Gel images were obtained by phosphorimaging using the Storm 680 PhosphorImager (Molecular Dynamics) and species levels were quantified using ImageQuant software (Molecular Dynamics).
Stoichiometric analysis
Large-scale transpososome assembly reactions (275 μl) were set up with H-NS at 0.36 and 1.4 μM, as previously described in the ‘Materials and Methods’ section. Reactions were concentrated using Vivaspin 3K filters (Vivascience) to approximately 65 μl. Samples were loaded in duplicate onto a 5% native polyacrylamide gel. After electrophoresis a portion of the gel with one set of samples was stained with ethidium bromide and this stained gel was used as a guide to identify the position of H-NS-transpososomes in the unstained portion of the gel; we avoided staining the analytical portion of the gel because we found that ethidium bromide can cause proteins to dissociate from DNA. Proteins were eluted out of gel slices containing H-NS-transpososomes by incubating gel slices in 1 ml of elution buffer (0.1% SDS, 250 mM NH4Ac) for 16 h at 42°C. Eluted proteins were then concentrated to approximately 15 μl using Vivaspin 3K filters and the entire sample was subjected to SDS–PAGE using a 12% gel. Proteins were visualized by Coomassie blue staining and levels quantified using the AlphaImager (AlphaInnotech).
Hydroxyl radical footprinting
Transpososomes were assembled as previously described except that the 53 bp ME contained a 5′ 32P-label on either the transferred (TS) or non-transferred (NTS) strand and H-NS was added at a concentration of 0.6 or 1.8 μM (approximately equivalent to reactions in lanes 8 and 10 of Figure 2B). Hydroxyl radical treatment of assembly reactions was performed essentially as described (20), except that the chemical treatment was carried out at 25°C for 10 min and then samples were directly applied to a 5% native polyacrylamide gel. Transpososomes, H-NS-transpososomes and unbound ME DNA were isolated from the wet gel after exposure of the gel to film. DNA was eluted from gel slices in elution buffer (0.1% SDS, 250 mM NH4Ac) for 16 h at 42°C. Eluted DNA was then treated with phenol, and ethanol precipitated. The purified DNA was resuspended in water and denaturing loading dye and the volume adjusted so that 1 μl of each sample contained approximately equal numbers of radioactive counts. Samples were then applied to a 10% high-resolution denaturing gel, along with a Maxim and Gilbert G ladder prepared from the appropriate ME DNA, and a gel image was obtained as described earlier.
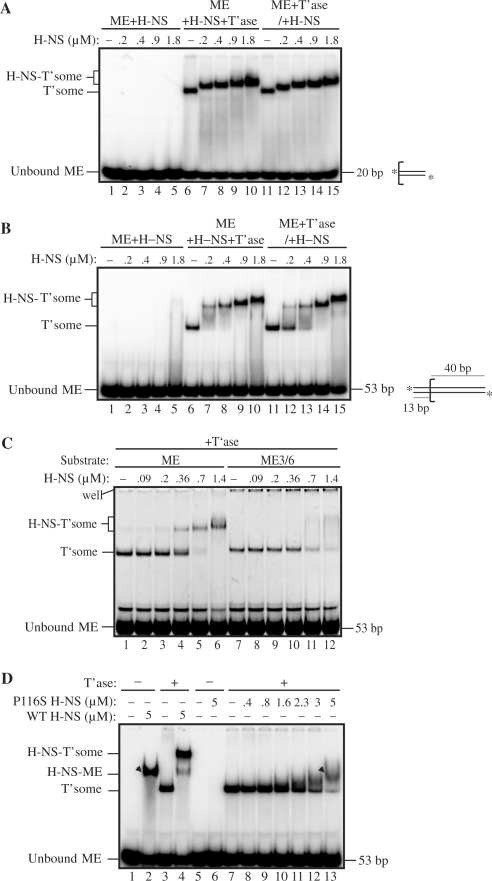
Figure 2.
Electrophoretic mobility-shift assays with Tn5 transpososomes and H-NS. (A) and (B) H-NS mobility-shifts of transpososomes formed with 20 and 53 bp ME substrates, respectively. Where indicated, H-NS was added to transpososome assembly reactions either at the same time as transposase (ME+T’ase+H-NS) or after transposase (i.e. subsequent to transpososome formation) (ME+T’ase/+H-NS). An illustration of the respective substrates is shown beside the gel image; the bracket defines the beginning of the ME sequence and asterisks indicate the position of 32P-labels. (H-NS-T'some) H-NS bound transpososome; (T'some) transpososome. (C) H-NS mobility-shift of a transpososome formed with a ME substrate containing a G to C and a T to A mutation at positions 3 and 6, respectively (ME3/6). (D) H-NS mobility-shift with P116S H-NS. The arrowheads show the mobility of a non-specific H-NS-ME complex (lane 2) and a P116S H-NS-transpososome complex (lane13). H-NS binds the ME in the absence of transposase only at relatively high (⩾5 μM) H-NS concentrations. Similarly, P116S H-NS binds to the transpososome only at relatively high concentrations (⩾3 μM). Note that in (A)–(D) HA transposase was used.
In vitro excision assay
Transposition time-courses following donor cleavage were carried out by initially forming transpososomes with or without HNS (2 μM) and/or heparin (2 μM) as described in the ‘Materials and Methods’ section. MgCl2 (10 mM) was added to the each transpososome sample and the reactions were incubated at 37°C for up to 16 h. At the indicated time points (0, 1, 3, 16 h) equal volume aliquots were removed, treated with phenol and the DNA was recovered by ethanol precipitation. DNA was resuspended in denaturing loading dye and analyzed on an 8% high-resolution denaturing gel. The amount of transpososome formed per reaction condition was quantified using ImageQuant software (Molecular Dynamics) based on the percentage of transpososome signal compared to the total amount of signal in the entire lane. Similarly, ImageQuant was used to quantify the amount of cleavage products generated in each reaction at each time point as a percentage of the total signal in the entire lane.
In vivo transposition assays
The papillation assay was carried out by transforming CRW31 (hns+) and CRW51 (hns−) with pGRPET2 (ApR) and selecting for transformants on MacConkey-lactose plates containing 100 μg/ml ampicillin. pGRPET2 (34) is a pET-21d derivative encoding HA transposase under the control of a T7 promoter and was used to achieve a relatively low level of transposase expression in vivo in strains lacking the T7 RNA polymerase gene. Plates were incubated for up to 4 days at 37°C. The mating out assay where transposase was provided in cis was performed as described in (50) except that donor transformants (pDH533 or pDH549 in NK5830 and NK5830Δhns::KanR) were selected on M9 plates supplemented with glucose, arginine, ampicillin (50 μg/ml) and chloramphenicol (20 μg/ml). In addition, HB101 was used as the recipient and mating mixtures were pelleted and resuspended in 0.85% saline before plating on M9 plates supplemented with glucose, leucine and streptomycin (150 μg/m) for total exconjugants, or glucose, leucine, streptomycin (150 μg/ml) and chloramphenicol (20 μg/ml) for transposition events. For the mating out assay where transposase was provided in trans pDH389 and pDH390 were co-transformed into NK5830 and NK5830Δhns::KanR and transformants were selected as above. Transformants were grown overnight in SOC media to inhibit transposase synthesis. Cells were then pelleted, washed with saline and subcultured (1 : 20 dilution) into 1 ml of M9 media supplemented with arginine and 0.2% arabinose; growth in arabinose induces transposase synthesis. Donors were then grown for 4 h on fast roll followed by 4 h on slow roll. After mixing donor cultures (1 ml) with 2.5 ml of HB101 recipient cells (OD600 = 0.6), cultures were grown for an additional hour on slow roll. Mating mixtures were then processed as above and plated on M9 plates Supplemented with glucose, leucine and streptomycin (150 μg/ml) for total exconjugants, or glucose, leucine, streptomycin (150 μg/ml) and tetracycline (10 μg/ml) for transposition events.
RESULTS
H-NS binds with high specificity to the Tn5 transpososome
To determine if H-NS binds the Tn5 transpososome, we performed electrophoretic mobility shift assays (EMSA) with purified H-NS and mixtures containing short linear 32P-labeled ME DNA and the HA transposase. In one arrangement we incubated H-NS, HA transposase and ME DNA (ME+T’ase+H-NS) and in another we pre-mixed HA transposase and ME DNA before adding H-NS (ME+T’ase/+H-NS). In the former arrangement there is the potential for H-NS to bind either the transpososome or a pre-transpososome MC. We also performed incubations with H-NS and only the ME (ME+H-NS). Also, the size of the ME DNA was varied in different experiments in order to determine the minimal end sequence requirements for H-NS binding.
The results in Figure 2A show that with a 20 bp ME, H-NS binds to the Tn5 transpososome irrespective of whether the transpososome was formed before or after H-NS addition (compare lane 6 with lanes 7–10 and lane 11 with lanes 12–15). Based on our titration data (Figure 2 and data not shown) we estimate that a molar excess of H-NS to transpososome of 20-fold is optimal for the formation of the H-NS-transpososome. As the concentration of H-NS present in the reaction increased the mobility of the H-NS-transpososome further decreased, implying that at the lower H-NS concentrations H-NS binding sites within the transpososome were not saturated; note that additional binding sites might be located within the DNA or the protein components of the transpososome—the latter might include H-NS oligomerization. Operationally, we refer to the reduced mobility forms of the H-NS-transpososome as ‘supershifted’ forms. Notably, at the same range of H-NS concentrations used to detect H-NS binding to the transpososome, we did not detect H-NS binding to free ME DNA (lanes 1–5). These results indicate that H-NS binds the transpososome with considerably higher affinity than it binds unbound ME DNA. These results also show that a transpososome formed with the least amount of ME sequence required for transposase binding (20 bp) contains sufficient determinants for H-NS binding. ME DNA was also substituted in these reactions with an authentic OE DNA substrate and similar H-NS binding was observed (data not shown).
Similar results were obtained with a 53 bp substrate containing 40 bp of ME DNA and 13 bp of flanking donor DNA (Figure 2B), although for this substrate there is some indication that the order of H-NS addition did have an impact on the amount of H-NS-transpososome formed. When ME DNA, transposase and H-NS were added at the same time there was as much as a 2-fold increase in the amount of H-NS-transpososome formed versus when H-NS was added after ME and transposase (compare lanes 7–8 and 12–13 in Figure 2B). This raises the possibility that H-NS is capable of binding the MC and facilitating transpososome assembly (see also Figures 4 and 5).
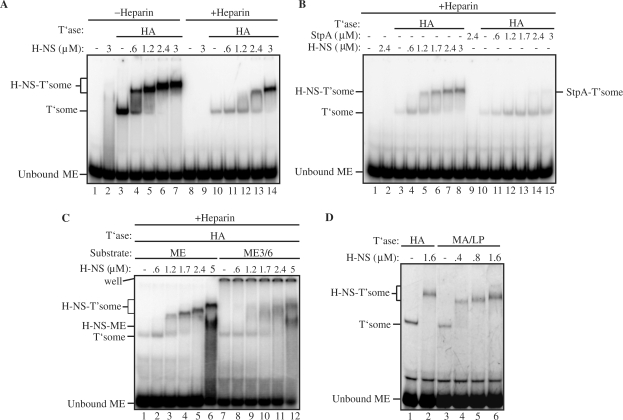
Figure 4.
Effect of H-NS on transpososome formation under heparin challenge conditions or in the presence of MA/LP transposase. In (A) and (B) transpososome assembly reactions were carried out with 32P-labeled 53 bp ME substrate following the ‘ME+T’ase+H-NS’ regimen, in the presence and absence of heparin. Note that in (B) either H-NS or StpA were added to the reactions. The same reaction set-up was used in (C) except that in addition, the ME3/6 substrate described in Figure 2C was used. (D) H-NS was added to transpososome assembly reactions carried out with HA transposase and MA/LP transposase (MA/LP). Species are labeled as in Figure 2. (StpA-T'some) StpA transpososome.
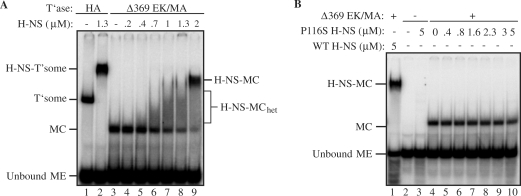
Figure 5.
Electrophoretic mobility-shift assays of transpososomes formed with Δ369 EK/MA transposase and H-NS. (A) H-NS mobility-shifts with transpososomes formed with either HA transposase or Δ369 EK/MA transposase. (B) Mobility shift with Δ369 EK/MA and either wild-type (WT) or P116S H-NS. H-NS-transpososomes in (A) and (B) were assembled under the ‘ME+T’ase+H-NS’ regimen, and the substrate is the same as in Figure 2B. Species are labeled as in Figure 2. (MC) monomer complex; (H-NS-MC) H-NS monomer complex; (H-NS-MC het) a heterogeneous mixture of H-NS monomer complexes.
Strikingly, there was a significant reduction in the ability of H-NS to bind a transpososome formed with the 53 bp ME DNA containing mutations at positions 3 and 6 (compare lanes 4–6 to 10–12 in Figure 2C). Positions 3 and 6 have been shown to contribute to trans contacts with transposase (37) and accordingly we found that these mutations reduce the amount of transpososome formed by about 50%. That these mutations reduce the binding of H-NS to the transpososome is suggestive of the region immediately adjacent to the transposon terminus (i.e. where trans contacts occur with the transposase) providing important determinants for H-NS binding.
A mutant form of H-NS (P116S H-NS) has been identified that has lost the capacity to bind DNA in a structure-specific manner but retains non-specific DNA binding (51,52). We used this mutant to ask if H-NS binding to the Tn5 transpososome involves a DNA structure-specific interaction between H-NS and the transpososome. We show in Figure 2D that P116S H-NS has a greatly reduced binding affinity for the Tn5 transpososome, as even at the highest concentration of P116S H-NS used (500-fold molar excess relative to the transpososome), only a small mobility shift was observed (compare lanes 2 and 13); this species has a greater gel mobility relative to the H-NS-transpososome generated with wild-type H-NS (lane 4) and is only observed at high H-NS concentrations (⩾5 μM) in our standard reaction conditions. This result supports the idea that H-NS recognizes specific structural determinants in the Tn5 transpososome that are not present in the unbound ME DNA. An analogous experiment in the Tn10 system led to the same conclusion (11).
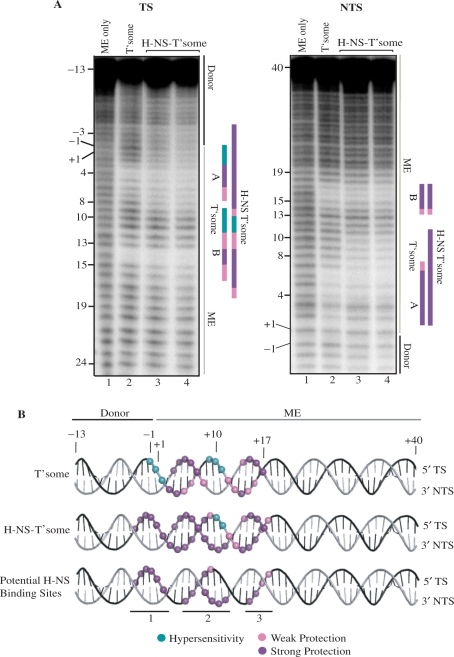
Hydroxyl radical footprinting of H-NS-transpososomes
We characterized the protein–DNA contacts within the H-NS-transpososome by performing hydroxyl radical DNA footprinting as described in the ‘Materials and Methods’ section. Incorporation of H-NS into the transpososome significantly altered the hydroxyl radical footprint as regions of hydroxyl radical protection typically observed in the transpososome were expanded, and regions of hypersensitivity were protected from cleavage (Figure 3A). In our transpososome footprint there are two zones of protection attributable to transposase: zone A includes nucleotides 3–7 on the TS and nucleotides 2–7 on the NTS; zone B includes nucleotides 12–16 on the TS and nucleotides 14–17 on the NTS. As well, residues 2-(-1) and 9–11 on the TS show hypersensitivity to hydroxyl radicals. These results match well with the results of previously reported hydroxyl radical footprinting experiments performed on the Tn5 transpososome and contact data inferred from the Tn5 transpososome crystal structure (20,37). In the H-NS-transpososome footprints, protection in zone A was extended in both directions to include residues 3-(-4) and 8–9 on the TS and 8–11 on the NTS. This includes loss of hydroxyl radical hypersensitivity at residues 2-(-1) and residue 9 of the TS. In addition, protection in zone B was extended from residues 17–18 on the TS. Overall, the footprinting data suggests that H-NS may have as many as three distinct binding sites within the transpososome (Figure 3B). Notably, estimates from stoichiometric analysis of the H-NS-transpososome formed at 1.4 μM H-NS (similar to the H-NS-transpososome formed for each footprint shown in lanes 4 of Figure 3A) suggest that at least four dimers of H-NS may be present per transpososome (Supplementary Figure 1).
Figure 3.
Hydroxyl radical footprinting of H-NS-transpososomes. (A) Transferred (TS) and non-transferred (NTS) strand footprints are shown. Transpososome assembly reactions were set up as described in the ‘Materials and Methods’ section with the 53 bp ME substrate (Figure 2B) labeled with 32P at the 5′ terminus of either the TS or the NTS. Samples in lanes 3 and 4 were from H-NS-transpososome assembly reactions performed with 0.6 and 1.8 μM H-NS, respectively (these reflect the H-NS mobility shifts of lanes 8 and 10, respectively in Figure 2B). The +1 position is the first base pair of the ME and the –1 position is the first base pair of the flanking donor DNA. Regions of strong and weak hydroxyl radical protection are indicated by purple and pink vertical bars, respectively. Regions of hydroxyl radical hypersensitivity are represented by vertical green bars. Binding sites of transposase, referred to in the text as zones A and B, are labeled. (B) Positions of hydroxyl radical protection and hypersensitivity are summarized on helical representations of DNA for the transpososome and H-NS-transpososome. The three potential binding sites for H-NS (sites 1–3) are shown on the bottom helical representation. These sites are defined by differences in hydroxyl radical protection and hypersensitivity in the transpososome versus the H-NS transpososome.
H-NS facilitates transpososome assembly
Over the course of studying the interactions between H-NS and the Tn5 transpososome we frequently observed an increase in the yield of transpososome when H-NS was included in transpososome assembly reactions as opposed to being added to a mixture of pre-formed transpososome (compare lanes 6 and 10 in Figure 2B). This led us to speculate that H-NS might increase the efficiency of transpososome formation. We also reasoned that it might be difficult to observe a strong effect of H-NS on transpososome assembly under our standard reaction conditions because the in vitro Tn5 system has been optimized for transpososome assembly (see Introduction section). We therefore asked if H-NS might have a more substantial impact on transpososome assembly under conditions where it is more difficult for the transpososome assembly to take place.
One way we reduced the efficiency of transpososome formation was by performing assembly reactions in the presence of heparin, a low molecular weight polyanion. Heparin is expected to compete with the ME DNA for binding of transposase and therefore its addition should reduce transpososome formation. Consistent with this expectation, we show in Figure 4A that addition of heparin reduced transpososome formation by 22-fold (compare lanes 3 and 10). Importantly, when the identical reaction was carried out in the presence of H-NS, the efficiency of transpososome formation was greatly increased. At the highest concentration of H-NS used, transpososome formation was increased 11-fold relative to the heparin-treated sample that did not receive H-NS (compare lanes 10 and 14). In contrast, in the absence of heparin the increase in transpososome formation was less than 2-fold (compare lanes 3 and 7). Moreover, the vast majority of the transpososome detected in reactions with heparin were in the H-NS-bound form. This is suggestive of H-NS increasing transpososome assembly by binding directly to the transpososome (as opposed to titrating out heparin and thus acting in an indirect manner).
Further evidence that H-NS is acting directly to increase transpososome formation comes from two additional experiments. In one experiment, we used StpA instead of H-NS to look for stimulation of transpososome assembly in the presence of heparin. StpA is a paralogue of H-NS, sharing 58% amino acid identity in E. coli (42). However, StpA is more basic than H-NS with a predicted pI of 9.08 compared to 5.25 for H-NS, and thus is expected to bind significantly more heparin relative to H-NS (42,53). If an increase in transpososome formation was simply the result of H-NS titrating heparin out of the reaction, then StpA should be more effective than H-NS at promoting transpososome formation. However, when StpA was added instead of H-NS, only a moderate increase in transpososome formation was observed (3-fold as opposed to 17-fold when H-NS was used in this experiment), suggesting that the H-NS effect is direct (Figure 4B). It is likely that the relatively small StpA enhancement in transpososome formation observed in this experiment is indirect because only a small percentage (approximately 5%) of the transpososome formed in the StpA reaction was in an StpA-bound form.
In a second experiment we asked if H-NS could rescue transpososome formation in a reaction where ME3/6 (described in Figure 2C) was used as substrate. Recall that the ME3/6 transpososome was largely defective for binding H-NS. We failed to see a significant increase in transpososome formation (<2-fold) in this reaction (Figure 4C), suggesting that H-NS binding to the transpososome is required for the stimulation in transpososome formation seen in the presence of H-NS under heparin competition conditions.
An alternative approach to reducing the efficiency of transpososome formation was to utilize a form of transposase that contains the wild-type residue at position 54, but still contains the MA56 and LP372 mutations, hereafter referred to as MA/LP transposase. The glutamate at position 54 hinders transposase-end interactions and impedes transpososome formation. The efficiency of transpososome assembly was reduced 3-fold when this form of transposase was used (compare lanes 1 and 3 in Figure 4D). In the presence of H-NS the efficiency of transpososome formation in the MA/LP reaction was increased by up to 2.5-fold (compare lanes 3 and 6), which coincided with the formation of H-NS-transpososome.
Taken together, the results in this section show that H-NS can stimulate transpososome formation and that this stimulation correlates with H-NS binding to the transpososome. It is also apparent that the region where transposase makes contacts in trans with the ME is not only critical for H-NS binding but also important in H-NS-directed stimulation of transpososome formation.
The above stimulation of transpososome formation by H-NS is only functionally significant if the H-NS bound form of the transpososome is capable of undergoing the chemical steps in transposition. We tested this by adding MgCl2 to a reaction equivalent to that shown in lane 14 of Figure 4A (i.e. a transpososome assembly reaction carried out in the presence of heparin and H-NS). Upon incubating this reaction for varying amounts of time and then analyzing the DNA on a denaturing gel, we detected the expected distribution of transposon excision products (Supplementary Figure 2). Furthermore, the efficiency of excision product formation, per amount of transpososome formed, was comparable with that observed in a control reaction performed without heparin and H-NS. Thus, we conclude that H-NS does not have an inhibitory effect on transposon excision in the Tn5 system.
H-NS binds to a Tn5 MC
We can think of two general ways in which H-NS could promote transpososome formation in the Tn5 system. H-NS might increase the stability of the transpososome or it might play a role in transpososome assembly. The latter possibility is much more likely because the Tn5 transpososome is very stable and over the time course of our experiments is not expected to dissociate to any significant degree (54,55). We have tested the idea that H-NS facilitates transpososome assembly by asking if H-NS binds to the MC, a species that may either be a bona fide assembly intermediate or closely resemble such an intermediate (Figure 1 and see ‘Discussion’ section). Under standard in vitro reaction conditions the MC is unstable and thus difficult to work with. In fact, analysis of this species requires the utilization of a mutant form of transposase (Δ369 EK/MA) that lacks 108 amino acids from the C-terminus, which includes the dimerization subdomain, and cannot go on to form a transpososome (30,32). Importantly, formation of the MC with Δ369 EK/MA displays many of the DNA sequence requirements for transpososome formation and thus provides a reasonable model for the study of a transpososome precursor (20).
In assembly reactions with Δ369 EK/MA a MC is readily detected (lane 3, Figure 5A). Addition of H-NS to the Δ369 EK/MA MC resulted in a mobility shift, clearly showing that H-NS is able to bind this species. At the lower H-NS concentrations the H-NS bound form of the complex constitutes a heterogeneous mixture of species as indicated by the presence of a smear above the MC (lanes 5–8). At the highest H-NS concentration (lane 9) there is a dramatic shift in the distribution of H-NS-bound MC towards a more homogeneous species. Given the extent of the mobility shift, which is almost as large as that observed for the transpososome versus free ME DNA (compare lanes 9 and 2), and the concentration-dependence of this shift, it is likely that H-NS is forming higher order oligomers in this complex. Notably, H-NS binding to the MC occurs at concentrations of H-NS that appear to be insufficient to form a complex with the unbound ME. This implies that the MC has binding determinants for H-NS that are not present in unbound ME DNA. Consistent with this, we show in Figure 5B that P116S H-NS failed to bind to the MC.
Disruption of the hns gene reduces the frequency of Tn5 transposition in vivo
The observations presented thus far raise the possibility that H-NS may act as a positive regulator of Tn5 transposition in vivo. We have tested this idea by measuring the relative transposition frequency of Tn5 in isogenic hns+/hns− strains of E. coli. This was done using two well-established transposition assays, a papillation assay and a mating out assay.
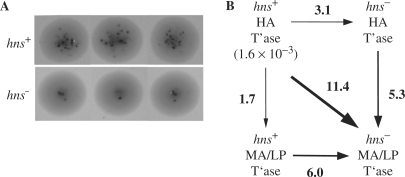
In the papillation assay used here, the experimental read-out for transposition is a ‘LacZ turn-on’ event that results from mobilization of a Tn5 derivative encoding a truncated lacZ gene into an expressed gene. When plated on MacConkey lactose indicator plates, cells that have had a Tn5 transposition event that results in LacZ turn-on form red papillae (or outgrowths) on a background of LacZ− cells (white). The number of LacZ+ papillae formed per colony is roughly proportional to the frequency of Tn5 transposition within the colony (56). When we compared the average number of papillae formed in the hns+ and hns− strains expressing HA transposase encoded by a multicopy plasmid, we observed roughly 5-fold fewer LacZ+ papillae per colony in the hns− strain. Representative colonies are shown in Figure 6A. Note that we have compared the number of papillae formed in the two strains when colonies were of a similar size; the hns− strain grew at approximately half the rate of the hns+ strain.
Figure 6.
In vivo transposition assays in isogenic hns+/hns− E. coli strains. (A) Relative transposition frequency of mini-Tn5-lacZ in isogenic hns+ and hns− strains as measured using a papillation assay. Three colonies of isogenic hns+ and hns− strains transformed with a plasmid encoding HA transposase are shown. The dark spots are LacZ+ papillae; the number of papillae formed per colony provides a measure of the relative frequency of transposition of a mini-Tn5-lacZ transposon within the colony. Note that the photographs were taken when colonies were roughly the same size, which corresponded to 50 and 90 h of growth, respectively, for the hns+ versus the hns− strain. (B) Box diagram showing the fold-changes in transposition frequency in cis as measured by mating out for hns+ and hns− strains expressing HA transposase or MA/LP transposase. Transposition frequencies were calculated by dividing the number of SmRCmR colonies (transposition events) by the number of SmR colonies (total exconjugants) obtained per 0.1 ml of mating mix. Transposition frequencies represent an average from at least two independent experiments. The transposition frequency for the hns+/HA transposase is shown. Arrows point in the direction of the fold-decrease (e.g. hns+/HA transposase supported transposition at a 3.1-fold higher level relative to hns−/HA transposase).
In the mating out assay the frequency of transposon insertions into an F’ plasmid is measured by mating a transposon-containing donor strain (F+) with a suitable recipient strain (F−) and then selecting for recipient cells that have acquired the transposon. A relative transposition frequency is then obtained by dividing the number of transposon-containing exconjugants by the number of total exconjugants. In one experiment, we transformed into isogenic hns+ and hns− strains a plasmid encoding a mini-Tn5-TetR element and a compatible plasmid encoding HA transposase under the control of an arabinose inducible promoter. In this set-up transposase synthesis is induced by growth in arabinose. This system measures Tn5 transposition in trans since the mini-Tn5 element and the transposase source are on separate plasmids. The results presented in Table 1 show an increase in transposition of about 3-fold in the hns+ versus the hns− strain under conditions of arabinose induction.
Table 1.
In vivo transposition of mini-Tn5-TetR catalyzed in trans by HA transposase in hns+/hns− strains
| Transposase source | Strain | Transposition frequencya | Normalized frequencyb |
|---|---|---|---|
| pDH390 | NK5830 | 8.1 (±0.0200) × 10−2 | 1.00 |
| pDH390 | NK5830hns::KanR | 2.6 (±0.0097) × 10−2 | 0.32 |
Relative transposition frequencies were calculated by dividing the number of SmRTetR colonies (transposition events) by the number of SmR colonies (total exconjugants) obtained per 0.1 ml of mating mix. Transposition frequencies represent an average value obtained from three independent experiments wherein matings with at least eight different donor transformants were carried out in each experiment.
Transposition frequencies were normalized to the level of transposition catalyzed by the HA transposase in the hns+ strain.
We also tested the idea that utilization of HA transposase might make Tn5 transposition less sensitive to the H-NS status of the cell. We constructed plasmids containing either HA transposase (pDH549) or MA/LP transposase (pDH550), and a mini-Tn5-CmR element, and used these plasmids in a mating out experiment. In this case, transposase expression was under the control of the native Tn5 transposase promoter and transposition occurs in cis because the transposase and the mini-Tn5 element are on the same plasmid. The EK54 mutation was chosen for reversion to wild-type because we previously showed that H-NS can increase the level of transpososome formed in vitro with the MA/LP transposase (Figure 4D). The results show that the defect in transposition is twice as large (6-fold versus 3-fold) in the hns− versus the hns+ strain in the presence of MA/LP compared to the HA transposase (Figure 6B). This result is consistent with the possibility that H-NS partially compensates for the suboptimal transposase.
DISCUSSION
We have shown that H-NS binds with high specificity to the Tn5 transpososome. Characterization of the H-NS interaction with the transpososome by DNA footprinting analysis provided evidence that H-NS binds in very close proximity to transposase and stoichiometric analysis indicated that up to four H-NS dimers can bind the transpososome. H-NS binding to the Tn5 transpososome was shown to affect the in vitro transposition reaction in a positive way. Under conditions where transpososome formation was rendered suboptimal, the presence of H-NS increased the yield of transpososome formed and this transpososome was shown to be catalytically competent. H-NS was also shown to bind a MC, raising the possibility that this host factor can act early in the transpososome assembly process. Finally, we have shown genetically that H-NS has the potential to act as a positive regulator of Tn5 transposition in vivo as the frequency of Tn5 transposition was reduced in an hns disruption strain.
H-NS interactions with the transpososome
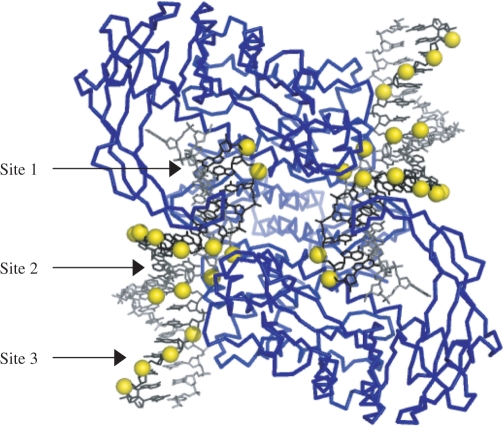
We used EMSA to show that H-NS binds preferentially to the Tn5 transpososome. At H-NS concentrations that did not significantly affect the mobility of ME DNA, we saw a full shift in the transpososome to a form with a lower electrophoretic mobility. We examined the determinants for H-NS binding to the transpososome by increasing the ME DNA size and adding flanking donor DNA. We observed that H-NS did not require the presence of flanking donor DNA in the transpososome in order to bind, but did require the presence of at least 20 bp of ME DNA (i.e. the minimum amount of transposon required for transpososome formation). Given that transposase makes contacts over most of the ME sequence (20,37), this observation suggested that H-NS must be in close association with transposase in the context of the transpososome, an inference that was confirmed by DNA footprinting studies. We observed an expansion of the ‘transposase footprint’ when H-NS was incorporated into the transpososome, as summarized in Figure 3B. We infer from this data that there may be as many as three distinct H-NS binding sites within the transpososome (listed as sites 1–3 in Figure 3B). Site 1 includes residues (-4) to 2 of the TS (i.e. the proximal region of zone A), site 2 includes residues 5–9 of the TS and 8–11 of the NTS (i.e. the distal region of zone A), and site 3 includes residues 15 to 18 of the TS (i.e. the distal region of zone B). Interestingly, there is evidence that all three of these potential H-NS binding sites include DNA with a distorted structure (Figure 7). Site 1 includes a DNA kink between residues 1 and 2 and work from solution studies revealed hydroxyl radical hypersensitivity extending into the flanking donor DNA (20,37,57). Site 2 is immediately adjacent to a 41° bend in the helical axis of the DNA between residues 11–12 (37). Site 3 is coincident with a second region of hydroxyl radical hypersensitivity that extends from residues 16–20 (20). This correspondence of potential H-NS binding sites inferred from footprinting studies with regions of deformed DNA structure fits well with the known preference of H-NS binding to deformed DNA structures.
Figure 7.
Three-dimensional representation of the Tn5 transpososome with potential H-NS binding sites indicated. A model of the Tn5 transpososome formed with the HA transposase and 20 bp OEs is shown (37,61) along with potential H-NS binding sites 1 to 3. Note that all three sites include portions of the DNA that are not encompassed by transposase on at least one surface. Transposase is shown as blue. The OE's DNA strands are depicted in black and grey for the transferred strand and non-transferred strands, respectively. Yellow spheres indicate the position of the phosphate backbone where H-NS binding may occur.
Based on the structure of the Tn5 transpososome we can infer that the major groove of the DNA is available for H-NS binding in sites 1 and 3 and the minor groove is available in site 2. Interestingly, the hydroxyl radical protection pattern (Figures 3 and 7) is consistent with H-NS binding in both the major and minor groove in different segments of transpososome. Notably, it remains controversial as to whether H-NS binds to the major or the minor groove (58,59), although we have previously detected H-NS binding to the Tn10 transpososome using a minor groove-specific DNA footprinting reagent (10,11). A possible complicating factor in the current analysis is that by binding immediately adjacent to transposase, or possibly interacting with transposase, as has been shown in the Tn10 system (10), H-NS may change the contacts transposase makes with the Tn5 end DNA. Thus, at this point we do not know if specific hydroxyl radical protections shown in Figure 7 are a consequence of only H-NS contacts. Additional studies will be required to sort out the mode of H-NS binding in the Tn5 transpososome.
H-NS also bound with high specificity to a Tn5 MC formed with ME DNA and the Δ369 EK/MA transposase. The fact that H-NS selectively bound the MC suggests that H-NS is not restricted to binding the fully assembled transpososome in the Tn5 system. We infer that transposase binding to the ME is likely sufficient to generate determinants for H-NS binding that are not present in free ME DNA. However, at this point it is unclear if the potential H-NS binding determinants described above for the transpososome are also present in the MC. Nevertheless, our finding that P116S H-NS failed to bind the MC and the transpososome with high specificity reinforces our conclusion that deformed DNA structures constitute important H-NS binding determinants in both complexes.
We found that one of the potential H-NS binding sites (Site 1) was critical for H-NS binding to the transpososome. When positions 3 and 6 of the ME DNA were mutated, we noted a small decrease in transpososome levels formed, however these mutations significantly reduced H-NS binding to the transpososome. While we have not yet determined if both mutations are necessary to prevent H-NS binding, it is intriguing that at least one mutation (bp 3) is at a position expected to be part of a distorted DNA structure. It is also interesting that mutations within only one of the three potential H-NS binding sites would significantly abrogate H-NS binding to the transpososome. One possible explanation for this is that H-NS binding to the proximal portion of zone A affects the conformation of the transpososome in a manner that permits additional H-NS binding events. This would be analogous to the situation in the Tn10 system where it appears that H-NS binding to a distorted DNA structure in the flanking donor DNA portion of the transpososome is critical for additional H-NS binding events within the terminal inverted repeat (11).
Functional consequences of H-NS binding to the Tn5 transpososome
Our binding assays showed that addition of H-NS had a small positive effect on the in vitro Tn5 transpososome assembly when the reaction was carried out under conditions that have been optimized for transpososome formation (i.e. with the HA transposase and the ME). In principle, this optimization might render the transpososome assembly reaction insensitive to host factors. With this in mind we set out to identify a means of making transpososome formation less efficient. We found that the addition of the polyanion heparin greatly reduced transpososome formation. Heparin is expected to compete with the ME DNA for transposase binding, thereby making it more difficult for the transpososome to form. At a heparin concentration sufficient to reduce transpososome formation by roughly 22-fold, we found that addition of H-NS resulted in an 11-fold increase in transpososome formation. Thus, under these reaction conditions, transpososome formation became strongly dependent on the presence of H-NS. Importantly, control reactions were included that demonstrated that H-NS binding to the transpososome was a prerequisite for stimulation of transpososome formation in the presence of heparin.
We also reduced the efficiency of transpososome formation by using a form of transposase that did not include one of the mutations (EK54) that leads to hyperactivity. In this situation there was a 2.5-fold increase in transpososome formation when H-NS was added to the reaction. Substitution of lysine for glutamate at position 54 creates a favorable base-specific cis contact with thymine 10 and likely removes an unfavorable contact between E54 and the phosphate backbone at positions 10 to 12 (33). It is intriguing that putative H-NS binding sites 2 and 3 are located within a few residues on either side of positions 10–12. H-NS binding to sites 2 and 3 could alter the DNA structure in a manner that prevents the unfavorable contact between E54 and positions 10–12 from forming. Alternatively, H-NS binding to these sites may cause a conformational change in transposase that partially shields the phosphate backbone from E54. In either case, H-NS would make the transposase-ME interaction less sensitive to the nature of the amino acid at position 54.
It is intriguing that H-NS can both stimulate transpososome formation and bind the MC. These observations lead us to propose that the association between H-NS and the MC might assist in transpososome assembly. It is well established that H-NS is capable of forming bridges through self-oligomerization between separate or distantly spaced DNA molecules (13,14). H-NS-H-NS interactions could therefore help drive the association between two MCs. While it is clear that this type of interaction has a minimal effect on the Tn5 system with the HA transposase, it may be important in assembly reactions carried out with wild-type transposase (see discussion later). Notably, there is evidence from studies on longer DNA molecules (relative to that used in the current study) that transpososome assembly in the Tn5 system may be more complicated than outlined above and in Figure 1. It has been shown that transposase is capable of synapsing non-transposon end segments and that a transposase dimer formed during this interaction can be transferred to a transposon end sequence. Subsequently, the transposase dimer would capture the second transposon end, thereby forming a stable transpososome (54,60). As described earlier, H-NS could facilitate the initial pairing of separate, non-transposon end DNA segments. Alternatively, or in addition, H-NS might promote the capture of the second transposon end by helping to stabilize a bend in the intervening DNA between the two transposon ends.
H-NS acts as a positive regulator of Tn5 transposition in vivo
We used two different transposition assays to determine if the absence of H-NS has an impact on the Tn5 transposition frequency in vivo. We found that in the absence of H-NS Tn5 transposition was reduced approximately 5-fold in the papillation assay and up to 11-fold in the mating out assay. These results are consistent with H-NS being a positive regulator of Tn5 transposition in vivo. Furthermore, we have provided evidence that is consistent with this positive role working at the level of transpososome assembly. We found that addition of H-NS to an in vitro assembly reaction with MA/LP transposase partially compensated for the absence of the EK54 mutation, a mutation that is known to increase transposase binding to Tn5 end sequences. In addition, we found that the fold-decrease in transposition frequency in vivo in an hns− strain encoding a transposase lacking the EK54 mutation was larger than in the same strain encoding a transposase with the EK54 mutation. If H-NS acts indirectly to promote Tn5 transposition, it is not obvious why the absence of the EK54 mutation would produce this larger effect.
A broader role for H-NS in lateral gene transfer
The work presented here provides the second documented case of H-NS promoting a transposition reaction through a direct interaction with a transposition complex. In the Tn10 system the available evidence is consistent with H-NS promoting transposition by binding to the initial transpososome and ultimately promoting a conformational change in this complex that inhibits self-destructive intramolecular transposition events and promotes intermolecular transposition events. While H-NS appears to stimulate Tn5 transposition by a different mechanism, the common thread in the two systems is that H-NS binds selectively to transpososomes and this raises the possibility that the activity of many other transposition systems may be modulated by H-NS. If this turns out to be the case, then H-NS would play an even broader role in lateral gene transfer than has previously been predicted through the xenogeneic silencing model because transposons are major drivers of lateral gene transfer.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institutes of Health Research (FRN 11281 to D.B.H.); NSERC CGS M studentship (to C.R.W.). Funding for open access charge: Canadian Institutes of Health Research (FRN 11281).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank D. Edgell for providing comments on the manuscript. We also thank W.S. Reznikoff for providing numerous ‘Tn5 system’ reagents, N. Kleckner for providing plasmids pNK1210 and pNK861, K. Derbyshire for providing M182Δhns::KanR and M. Belfort for providing BL21(DE3)hns::KanR and pET3a-StpA.
REFERENCES
- 1.Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JC. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006;2:e81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 3.Navarre WW, McClelland M, Libby SJ, Fang FC. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 2007;21:1456–1471. doi: 10.1101/gad.1543107. [DOI] [PubMed] [Google Scholar]
- 4.Doyle M, Fookes M, Ivens A, Mangan MW, Wain J, Dorman CJ. An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science. 2007;315:251–252. doi: 10.1126/science.1137550. [DOI] [PubMed] [Google Scholar]
- 5.Oshima T, Ishikawa S, Kurokawa K, Aiba H, Ogasawara N. Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res. 2006;13:141–153. doi: 10.1093/dnares/dsl009. [DOI] [PubMed] [Google Scholar]
- 6.Shiga Y, Sekine Y, Kano Y, Ohtsubo E. Involvement of H-NS in transpositional recombination mediated by IS1. J. Bacteriol. 2001;183:2476–2484. doi: 10.1128/JB.183.8.2476-2484.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rouquette C, Serre MC, Lane D. Protective role for H-NS protein in IS1 transposition. J. Bacteriol. 2004;186:2091–2098. doi: 10.1128/JB.186.7.2091-2098.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swingle B, O'Carroll M, Haniford D, Derbyshire KM. The effect of host-encoded nucleoid proteins on transposition: H-NS influences targeting of both IS903 and Tn10. Mol. Microbiol. 2004;52:1055–1067. doi: 10.1111/j.1365-2958.2004.04051.x. [DOI] [PubMed] [Google Scholar]
- 9.Twiss E, Coros AM, Tavakoli NP, Derbyshire KM. Transposition is modulated by a diverse set of host factors in Escherichia coli and is stimulated by nutritional stress. Mol. Microbiol. 2005;57:1593–1607. doi: 10.1111/j.1365-2958.2005.04794.x. [DOI] [PubMed] [Google Scholar]
- 10.Wardle SJ, O'Carroll M, Derbyshire KM, Haniford DB. The global regulator H-NS acts directly on the transpososome to promote Tn10 transposition. Genes Dev. 2005;19:2224–2235. doi: 10.1101/gad.1338905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ward CM, Wardle SJ, Singh RK, Haniford DB. The global regulator H-NS binds to two distinct classes of sites within the Tn10 transpososome to promote transposition. Mol. Microbiol. 2007;64:1000–1013. doi: 10.1111/j.1365-2958.2007.05708.x. [DOI] [PubMed] [Google Scholar]
- 12.Singh RK, Liburd J, Wardle SJ, Haniford DB. The nucleoid binding protein H-NS acts as an anti-channeling factor to favor intermolecular Tn10 transposition and dissemination. J. Mol. Biol. 2008;376:950–962. doi: 10.1016/j.jmb.2007.12.035. [DOI] [PubMed] [Google Scholar]
- 13.Fang FC, Rimsky S. New insights into transcriptional regulation by H-NS. Curr. Opin. Microbiol. 2008;11:113–120. doi: 10.1016/j.mib.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rimsky S, Zuber F, Buckle M, Buc H. A molecular mechanism for the repression of transcription by the H-NS protein. Mol. Microbiol. 2001;42:1311–1323. doi: 10.1046/j.1365-2958.2001.02706.x. [DOI] [PubMed] [Google Scholar]
- 15.Dame RT, Wyman C, Goosen N. Structural basis for preferential binding of H-NS to curved DNA. Biochimie. 2001;83:231–234. doi: 10.1016/s0300-9084(00)01213-x. [DOI] [PubMed] [Google Scholar]
- 16.Lavoie BD, Chan BS, Allison RG, Chaconas G. Structural aspects of a higher order nucleoprotein complex: induction of an altered DNA structure at the Mu-host junction of the Mu type 1 transpososome. Embo J. 1991;10:3051–3059. doi: 10.1002/j.1460-2075.1991.tb07856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savilahti H, Rice PA, Mizuuchi K. The phage Mu transpososome core: DNA requirements for assembly and function. Embo J. 1995;14:4893–4903. doi: 10.1002/j.1460-2075.1995.tb00170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, Namgoong SY, Zhang X, Harshey RM. Kinetic and structural probing of the precleavage synaptic complex (type 0) formed during phage Mu transposition. Action of metal ions and reagents specific to single-stranded DNA. J. Biol. Chem. 1996;271:9619–9626. doi: 10.1074/jbc.271.16.9619. [DOI] [PubMed] [Google Scholar]
- 19.York D, Reznikoff WS. DNA binding and phasing analyses of Tn5 transposase and a monomeric variant. Nucleic Acids Res. 1997;25:2153–2160. doi: 10.1093/nar/25.11.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhasin A, Goryshin IY, Steiniger-White M, York D, Reznikoff WS. Characterization of a Tn5 pre-cleavage synaptic complex. J. Mol. Biol. 2000;302:49–63. doi: 10.1006/jmbi.2000.4048. [DOI] [PubMed] [Google Scholar]
- 21.Reznikoff WS. The Tn5 transposon. Annu. Rev. Microbiol. 1993;47:945–963. doi: 10.1146/annurev.mi.47.100193.004501. [DOI] [PubMed] [Google Scholar]
- 22.Jilk RA, York D, Reznikoff WS. The organization of the outside end of transposon Tn5. J. Bacteriol. 1996;178:1671–1679. doi: 10.1128/jb.178.6.1671-1679.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhasin A, Goryshin IY, Reznikoff WS. Hairpin formation in Tn5 transposition. J. Biol. Chem. 1999;274:37021–37029. doi: 10.1074/jbc.274.52.37021. [DOI] [PubMed] [Google Scholar]
- 24.Kennedy AK, Guhathakurta A, Kleckner N, Haniford DB. Tn10 transposition via a DNA hairpin intermediate. Cell. 1998;95:125–134. doi: 10.1016/s0092-8674(00)81788-2. [DOI] [PubMed] [Google Scholar]
- 25.Krebs MP, Reznikoff WS. Transcriptional and translational initiation sites of IS50. Control of transposase and inhibitor expression. J. Mol. Biol. 1986;192:781–791. doi: 10.1016/0022-2836(86)90028-8. [DOI] [PubMed] [Google Scholar]
- 26.Weinreich MD, Gasch A, Reznikoff WS. Evidence that the cis preference of the Tn5 transposase is caused by nonproductive multimerization. Genes Dev. 1994;8:2363–2374. doi: 10.1101/gad.8.19.2363. [DOI] [PubMed] [Google Scholar]
- 27.Weinreich MD, Mahnke-Braam L, Reznikoff WS. A functional analysis of the Tn5 transposase. Identification of domains required for DNA binding and multimerization. J. Mol. Biol. 1994;241:166–177. doi: 10.1006/jmbi.1994.1486. [DOI] [PubMed] [Google Scholar]
- 28.Mahnke Braam LA, Goryshin IY, Reznikoff WS. A mechanism for Tn5 inhibition. carboxyl-terminal dimerization. J. Biol. Chem. 1999;274:86–92. doi: 10.1074/jbc.274.1.86. [DOI] [PubMed] [Google Scholar]
- 29.Reznikoff WS. Tn5 transposition and its regulation. Cell. 1982;31:307–308. doi: 10.1016/0092-8674(82)90123-4. [DOI] [PubMed] [Google Scholar]
- 30.York D, Reznikoff WS. Purification and biochemical analyses of a monomeric form of Tn5 transposase. Nucleic Acids Res. 1996;24:3790–3796. doi: 10.1093/nar/24.19.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jilk RA, Makris JC, Borchardt L, Reznikoff WS. Implications of Tn5-associated adjacent deletions. J. Bacteriol. 1993;175:1264–1271. doi: 10.1128/jb.175.5.1264-1271.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steiniger-White M, Reznikoff WS. The C-terminal alpha helix of Tn5 transposase is required for synaptic complex formation. J. Biol. Chem. 2000;275:23127–23133. doi: 10.1074/jbc.M003411200. [DOI] [PubMed] [Google Scholar]
- 33.Zhou M, Bhasin A, Reznikoff WS. Molecular genetic analysis of transposase-end DNA sequence recognition: cooperativity of three adjacent base-pairs in specific interaction with a mutant Tn5 transposase. J. Mol. Biol. 1998;276:913–925. doi: 10.1006/jmbi.1997.1579. [DOI] [PubMed] [Google Scholar]
- 34.Goryshin IY, Reznikoff WS. Tn5 in vitro transposition. J. Biol. Chem. 1998;273:7367–7374. doi: 10.1074/jbc.273.13.7367. [DOI] [PubMed] [Google Scholar]
- 35.Wiegand TW, Reznikoff WS. Characterization of two hypertransposing Tn5 mutants. J. Bacteriol. 1992;174:1229–1239. doi: 10.1128/jb.174.4.1229-1239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou M, Reznikoff WS. Tn5 transposase mutants that alter DNA binding specificity. J. Mol. Biol. 1997;271:362–373. doi: 10.1006/jmbi.1997.1188. [DOI] [PubMed] [Google Scholar]
- 37.Davies DR, Goryshin IY, Reznikoff WS, Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- 38.Lovell S, Goryshin IY, Reznikoff WR, Rayment I. Two-metal active site binding of a Tn5 transposase synaptic complex. Nat. Struct. Biol. 2002;9:278–281. doi: 10.1038/nsb778. [DOI] [PubMed] [Google Scholar]
- 39.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 40.Foster TJ, Davis MA, Roberts DE, Takeshita K, Kleckner N. Genetic organization of transposon Tn10. Cell. 1981;23:201–213. doi: 10.1016/0092-8674(81)90285-3. [DOI] [PubMed] [Google Scholar]
- 41.Rosner JL. Formation, induction, and curing of bacteriophage P1 lysogens. Virology. 1972;48:679–689. doi: 10.1016/0042-6822(72)90152-3. [DOI] [PubMed] [Google Scholar]
- 42.Zhang A, Rimsky S, Reaban ME, Buc H, Belfort M. Escherichia coli protein analogs StpA and H-NS: regulatory loops, similar and disparate effects on nucleic acid dynamics. Embo J. 1996;15:1340–1349. [PMC free article] [PubMed] [Google Scholar]
- 43.Naumann TA, Reznikoff WS. Trans catalysis in Tn5 transposition. Proc. Natl Acad. Sci. USA. 2000;97:8944–8949. doi: 10.1073/pnas.160107997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Way JC, Davis MA, Morisato D, Roberts DE, Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984;32:369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- 45.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 47.Whitfield CR, Wardle SJ, Haniford DB. Formation, characterization and partial purification of a Tn5 strand transfer complex. J. Mol. Biol. 2006;364:290–301. doi: 10.1016/j.jmb.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 48.Zhang A, Derbyshire V, Salvo JL, Belfort M. Escherichia coli protein StpA stimulates self-splicing by promoting RNA assembly in vitro. RNA. 1995;1:783–793. [PMC free article] [PubMed] [Google Scholar]
- 49.de la Cruz NB, Weinreich MD, Wiegand TW, Krebs MP, Reznikoff WS. Characterization of the Tn5 transposase and inhibitor proteins: a model for the inhibition of transposition. J. Bacteriol. 1993;175:6932–6938. doi: 10.1128/jb.175.21.6932-6938.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goryshin I, Kil YV, Reznikoff WS. DNA length, bending, and twisting constraints on IS50 transposition. Proc. Natl Acad. Sci. USA. 1994;91:10834–10838. doi: 10.1073/pnas.91.23.10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spurio R, Falconi M, Brandi A, Pon CL, Gualerzi CO. The oligomeric structure of nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for DNA bending. Embo J. 1997;16:1795–1805. doi: 10.1093/emboj/16.7.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stella S, Spurio R, Falconi M, Pon CL, Gualerzi CO. Nature and mechanism of the in vivo oligomerization of nucleoid protein H-NS. Embo J. 2005;24:2896–2905. doi: 10.1038/sj.emboj.7600754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sonnenfield JM, Burns CM, Higgins CF, Hinton JC. The nucleoid-associated protein StpA binds curved DNA, has a greater DNA-binding affinity than H-NS and is present in significant levels in hns mutants. Biochimie. 2001;83:243–249. doi: 10.1016/s0300-9084(01)01232-9. [DOI] [PubMed] [Google Scholar]
- 54.Steiniger M, Adams CD, Marko JF, Reznikoff WS. Defining characteristics of Tn5 Transposase non-specific DNA binding. Nucleic Acids Res. 2006;34:2820–2832. doi: 10.1093/nar/gkl179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adams CD, Schnurr B, Marko JF, Reznikoff WS. Pulling apart catalytically active Tn5 synaptic complexes using magnetic tweezers. J. Mol. Biol. 2007;367:319–327. doi: 10.1016/j.jmb.2006.12.064. [DOI] [PubMed] [Google Scholar]
- 56.Krebs MP, Reznikoff WS. Use of a Tn5 derivative that creates lacZ translational fusions to obtain a transposition mutant. Gene. 1988;63:277–285. doi: 10.1016/0378-1119(88)90531-8. [DOI] [PubMed] [Google Scholar]
- 57.Bischerour J, Chalmers R. Base-flipping dynamics in a DNA hairpin processing reaction. Nucleic Acids Res. 2007;35:2584–2595. doi: 10.1093/nar/gkm186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tippner D, Afflerbach H, Bradaczek C, Wagner R. Evidence for a regulatory function of the histone-like Escherichia coli protein H-NS in ribosomal RNA synthesis. Mol. Microbiol. 1994;11:589–604. doi: 10.1111/j.1365-2958.1994.tb00339.x. [DOI] [PubMed] [Google Scholar]
- 59.Tippner D, Wagner R. Fluorescence analysis of the Escherichia coli transcription regulator H-NS reveals two distinguishable complexes dependent on binding to specific or nonspecific DNA sites. J. Biol. Chem. 1995;270:22243–22247. doi: 10.1074/jbc.270.38.22243. [DOI] [PubMed] [Google Scholar]
- 60.Adams CD, Schnurr B, Skoko D, Marko JF, Reznikoff WS. Tn5 transposase loops DNA in the absence of Tn5 transposon end sequences. Mol. Microbiol. 2006;62:1558–1568. doi: 10.1111/j.1365-2958.2006.05471.x. [DOI] [PubMed] [Google Scholar]
- 61.Steiniger-White M, Bhasin A, Lovell S, Rayment I, Reznikoff WS. Evidence for “unseen” transposase–DNA contacts. J. Mol. Biol. 2002;322:971–982. doi: 10.1016/s0022-2836(02)00877-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.