Abstract
It has previously been shown that transcription greatly enhances recombination in mammalian cells. However, the proteins involved in catalysing this process and the recombination pathways involved in transcription-associated recombination (TAR) are still unknown. It is well established that both the BRCA2 protein and the RAD51 paralog protein XRCC2 are required for homologous recombination. Here, we show that the BRCA2 protein is also required for TAR, while the XRCC2 protein is not involved. Expression of the XRCC2 gene in XRCC2 mutated irs1 cells restores the defect in homologous recombination repair of an I-SceI-induced DNA double-strand break, while TAR is unaffected. Interestingly, the XRCC2-deficient irs1 cells are also proficient in recombination induced at slowed replication forks, suggesting that TAR is mechanistically linked with this recombination pathway. In conclusion, we show that TAR depends on BRCA2 but is independent of XRCC2, and that this recombination pathway is separate from that used to repair a two-ended DNA double-strand break.
INTRODUCTION
Transcription-associated recombination (TAR) is conserved in all cellular organisms investigated, from bacteria to mammals (1). The mechanisms and underlying causes of TAR are still largely unknown, as are the proteins catalysing this process. Studies in Saccharomyces cerevisiae has shown that a number of factor influence TAR; for instance DNA damage appears to have a synergistic effect with transcription on the levels of recombination (2). It has also been shown in S. cerevisiae that RNA polymerase I-dependent transcription can result in collision with the replication machinery leading to replication fork blockage and recombination in rDNA repeat copies (3). RNA polymerase II transcription can also impair the replication fork progression, which results in an increased recombination (4). A similar increase in recombination levels can be observed in S. cerevisiae strains with impaired transcription elongation owing to mutations in the THO/TREX complex (5,6). The THO/TREX complex is active at the interface between transcription and messenger ribonucleoprotein (mRNP) metabolism, and as a consequence of the mRNP biogenesis defect in THO/TREX mutants an impaired replication fork progression could be observed, which appear to stimulate TAR (7). Nickoloff and Reynolds (8) were first to show that transcription stimulated homologous recombination in mammalian cells, using heteroallelic neomycin genes. TAR in mammalian cells has been shown to also be associated with replication and restricted to cells in the S phase of the cell cycle (9). The available data thus suggest that TAR is likely involved in bypassing an active RNA polymerase, but details of the recombination mechanisms and proteins involved in the process remain elusive.
Homologous recombination (HR) can be induced by DNA double-strand breaks (DSBs) or at replication forks (10–12). There are two major pathways by which DSBs can be repaired; non-homologous end-joining (NHEJ) or homologous recombination (HR). NHEJ is a fast and error-prone repair pathway that involves ligation of free ends (13), while homologous recombination is slow and error-free if an intact DNA molecule, usually the sister chromatid, is used in the repair (14,15). Several proteins, e.g. BRCA2, RAD51 and five RAD51 paralogs, have been shown to be essential for efficient homologous recombination repair of DSBs in mammalian cells (16–19), and mutations in these genes often results in cellular sensitivity to ionizing radiation. Chinese hamster cells sensitive to ionizing radiation have been isolated (20) and it was later found that one of the five RAD51 paralog genes, XRCC2, correct the ionizing radiation sensitive hamster cell line irs1 (21). The precise function of XRCC2 is unknown, but it has been shown to associate with three other RAD51 paralogs; RAD51B, RAD51C and RAD51D (22). This complex is called BCDX2 and has been shown to bind single-stranded DNA as well as single-stranded regions of DNA and to facilitate RAD51 filament formation (22). Cell lines deficient in XRCC2 are viable, but exhibit chromosomal aberrations, sensitivity to cross-linking agents, a mild sensitivity to γ-radiation (20) and defective RAD51 foci formation (18).
Here, we investigated the connection between homologous recombination and TAR using well-characterized Chinese hamster cell lines, irs1 and V-C8, deficient in XRCC2 and BRCA2, respectively (21,23). We find that BRCA2 defective cells are deficient in TAR, showing that TAR employs homologous recombination proteins. Surprisingly, the XRCC2 defective irs1 cells are still able to carry out TAR, in spite of being defective in homologous recombination of a DSB induced by ionizing radiation or a restriction endonuclease. This genetically separates TAR from DSB-induced homologous recombination. Furthermore, we show that XRCC2-deficient irs1 cells are also proficient in thymidine-induced recombination, which altogether suggest that TAR is employing a similar recombination mechanism to bypass replication blocks as for lesions produced by thymidine treatment.
MATERIALS AND METHODS
Cell culture
All cell lines used in this study are derived from V79 Chinese hamster cells, with an additional mutation in BRCA2 (V-C8 and V-C8TofZM4), XRCC2 (irs1TofZM14 and irs1TofZM15) or with a partial duplication in the hprt gene S8TofZM5 (Table 1). The cells were cultured in Dulbecco's Modified Eagle Medium with 10% fetal bovine serum and 1% PEST (penicillin–streptomycin). To select for the desired clones and to keep the inserted genes in the cells, 0.25 mg/ml of zeocin and 120 U/ml hygromycin were added to the medium. In order to keep the transcription of the recombination substrate off during growth, 1 μg/ml of doxycycline was also added to the medium. The cells were cultured in an incubator at 37°C with 5% CO2.
Table 1.
Genotype and origin of Chinese hamster cell lines used in this study
| Cell line | Genotype | Defect/modification | Origin | References |
|---|---|---|---|---|
| irs1TofZM14,15 | XRCC2− | XRCC2−, deficient in homologous recombination, carrying TAR substrate | V79 | (37) |
| S8TofZM5 | WT | Wild-type Chinese hamster, partical duplication in hprt gene, carrying TAR substrate | V79 | (38) |
| V-C8 | BRCA2− | BRCA2−, deficient in homologous recombination | V79 | (23) |
| V-C8TofZM4 | BRCA2− | BRCA2−, deficient in homologous recombination, carrying TAR substrate | V79 | (23) |
| V79 | WT | Wild-type Chinese hamster | Lung | (23) |
Transfection
The Chinese hamster cell lines were stably transfected with a recombination substrate using electroporation. The plasmids were purified from Escherichia coli. Fifteen micrograms of plasmid DNA were used for each transfection. The plasmid DNA was diluted in 50 μl dH2O and mixed with 7.5 × 106 cells in an electroporation cuvette. The cells were transfected using the voltage 2.5 kV/cm and the capacitance 25 μF. The electroporated cells were aliqotated in the amounts 20, 50 and 100 μl on Petri-dishes and incubated for 2 days before the appropriate selective agent was added. After the colonies had formed, 30 colonies of varying sizes were picked and transferred to culture plates for further cultivation. Transient transfection was performed using Lipofectamine 2000 from Invitrogen according to the manufacturer's protocol.
Luciferase assay
For measuring luciferase activity individual clones were trypsinised and counted and 2 × 105 cells from each clone were incubated with and without doxycycline for 24 h prior to lysis. After the incubation the cells were rinsed twice with Dulbecco's PBS w/o Ca and Mg. All PBS was removed and 500 μl of lysis buffer was added to each well. The plates were shaken for 20 min in order to lyse the cells and the luciferase activity was measured using an illuminator. Clones exhibiting low background and high inducibility in the luciferase assay were selected for further experiments.
Recombination assay
For measuring recombination 1 × 106 cells were seeded onto four Petri-dishes. To two of the dishes, 10 μl (1 mg/ml) of doxycycline was added to repress expression from the Pbi-1 promoter. In the case of I-SceI-induced recombination, the cells were transiently transfected using Lipofectamine after 24 h. After an additional 24 h the cells were plated on cloning and selection dishes to determine survival and recombination, respectively. On the cloning dishes, 500 cells were seeded and on the selection dishes 300 000 cells were seeded. After 24 h, 100 μl of G418 was added. When colonies had formed (10 days for cloning and 15 days for selection), the cells were stained, using methylene blue in methanol (4 g/l), and counted. The recombination frequency was calculated as the number of recombinants formed in relation to survival. Thus, differential cloning efficiencies are compensated for in the recombination assay. Recombination assays while over-expressing wild-type XRCC2 was also performed. For these assays, 20 μg of XRCC2 plasmid (24) was used for each 10-cm Petri-dish. One microgram of I-SceI plasmid was used for each Petri-dish in order to induce a DSB in the recombination substrate. Transient transfections were performed using Lipofectamine 2000 according to the manufacturer's protocol.
RAD51 foci
To score for Rad51 foci, cells were treated with ionizing radiation or thymidine or left untreated as a control. For cells that were transiently transfected with a plasmid containing XRCC2, transfection was performed as described previously. Irradiated cells were treated with 5 Gy and left in an incubator for 5 h in order for foci to appear. For the thymidine treatment, the cells were treated with 2 mM of thymidine for 24 h. For Rad51 foci detection, 5 × 104 cells per slide were seeded onto sterilized cover slides. The next day the slides were either irradiated or treated with thymidine. After the treatment the cells were fixed with 3% paraformaldehyde containing 0.1% Triton X-100. The cells were then rinsed with PBS containing 0.1% TX-100 2 × 10 min. After rinsing the cells were permabilised with PBS + 0.3% TX-100 for 10 min. The cells were then blocked with PBS + 3% BSA for 40 min. After blocking, the primary antibody, h92 Santa Cruz anti RAD51 polyclonal rabbit diluted 1 : 1000 in PBS + 3% BSA was added and the cells were left at 4°C over night. The next day, the cells were rinsed with PBS containing 0.1% TX-100. After rinsing the cells were permabilized with PBS + 0.3% TX-100 for 10 min. After permabilization, the secondary antibody was added; donkey anti rabbit alexa 555 from Invitrogen, diluted 1 : 500 in PBS + 3% BSA. The slides were the incubated in the dark for 1 h. The cells were then rinsed with PBS containing 0.1% TX-100 2 × 10 min and permabilized with PBS + 0.3% TX-100 for 10 min. They were then rinsed with PBS for 5 min and stained with ToPro from Invitrogen for 30 min. The slides were then rinsed with PBS for 5 min and mounted with anti fade kit and sealed with nail polish. The slides were coded to make scoring impartial. Cells with more than 10 foci were scored as positive and 200 cells per slide were scored.
Western blot
To confirm that transient transfection with wild-type XRCC2 increased the levels of the XRCC2 protein in the cells, western blots were performed. An antibody against XRCC2 diluted 1 : 200 was used. An antibody against α-tubulin diluted 1 : 1000 was used as a loading control. The proteins were extracted using lysis buffer and the protein concentration was measured by using the Coomassie (Bradford) Protein Assay Kit from Pierce. A standard western blot protocol was used and the proteins were separated on a NuPage Novex 10% Bis–Tris gel from Invitrogen and then blotted onto a PVDF membrane. The proteins were visualized using SuperSignal Western blotting kit from Pierce.
RESULTS
BRCA2 is required for TAR
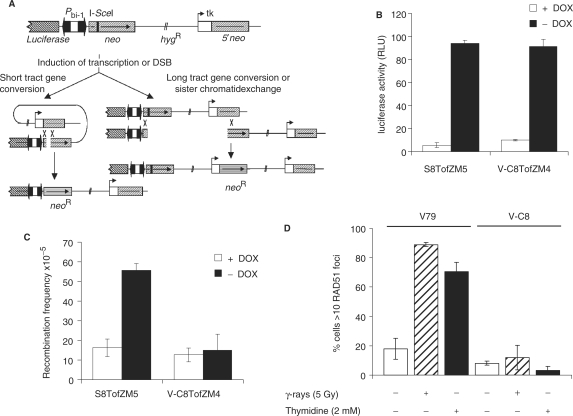
We have previously constructed a substrate to investigate the effect of transcription on homologous recombination (9). Briefly, the construct contains two non-functional neomycin resistance genes; one neo repeat has a 18-bp long recognition site for I-SceI inserted, which introduce a stop codon that truncates the gene product, and the second neo repeat has a 3′ deletion and is called 5′neo (Figure 1A). A functional neoR gene can be regained either through gene conversion (GC) or sister chromatid exchange (SCE), but not through single-strand annealing. The truncated neo repeat is under the control of a bi-directional inducible promoter based on a Tet-Off gene expression system. Using this approach, the levels of transcription over this neo repeat can be measured using the expression levels of the luciferase gene, simultaneously expressed in the opposite direction in the vector (9).
Figure 1.
BRCA2 is required for transcription-associated recombination. (A) Transcription-associated recombination is monitored between a mutated neoR and a truncated neoR gene, which after homologous recombination can revert to a functional neoR gene that can be selected with G418. Recombination can be induced either by enhanced transcription after removal of doxycycline (DOX) or after induction of a DSB at the I-SceI site following transient transfection with the pCMV3xnlsI-SceI vector. A functional neomycin resistance gene can be regained following short tract gene conversion or long tract gene conversion or sister chromatid exchange (see ref. 9 for details). (B) The inducibility at the bidirectional Pbi-1 promoter is determined by measuring luciferase activity in S8TofZM5, VC-8TofZM4, both with stably integrated copies of both the pTetoffZeo and pBI-LMscI vectors (9). (C) The recombination frequency was monitored as reversion to a functional neomycin resistant gene following removal of doxycyclin, which increases transcription at the recombination substrate. Transcription enhanced recombination in wild-type S8TofZM5 (P < 0.01 in t-test), but not in BRCA2 defective V-C8TofZM4 (P > 0.05 in t-test). (D) The number of cells with RAD51 foci was scored in BRCA2 defective V-C8 and parental V79 cell line 5 h after irradiation with 5 Gy or following 24 h 2 mM thymidine treatments. The numbers of cells with foci increased in wild-type cells following both treatments (P < 0.01 in t-test), but not in BRCA2 defective V-C8 cells following any treatment (P > 0.05 in t-test). The average and standard deviation of three independent experiments is depictured in all experiments.
Here, we transfected wild-type, XRCC2 and BRCA2 mutated Chinese hamster cells with both the Tet-regulatory vector pTetOffZeo and the recombination construct pBI-LMScI and selected for individual clones resistant to zeocin and hygromycin, to obtain cells with both vectors stably integrated into the genome. Withdrawal of doxycycline from the media resulted in increased luciferase activity in both wild-type (S8TofZM5) and BRCA2 defective (V-C8TofZM4) cells (Figure 1B), showing that BRCA2 does not influence the inducibility of transcription on the recombination substrate.
Transcription on the recombination substrate enhanced recombination levels in S8TofZM5 cells 3.7-fold (Figure 1C), which is in agreement with what was reported earlier for other wild-type hamster cells (9). However, transcription did not induce recombination in BRCA2 defective V-C8TofZM4 cells (Figure 1C). This is expected as the BRCA2 protein is vital for RAD51-mediated recombination (17), and in particular the gene conversion events produced by transcription (9,25). Here, we confirm the homologous recombination defect in BRCA2 defective V-C8 cells and show that RAD51 foci cannot form following either ionizing radiation [which has been shown before (16)] or following thymidine treatments (Figure 1D).
Transcription associated recombination is independent of XRCC2
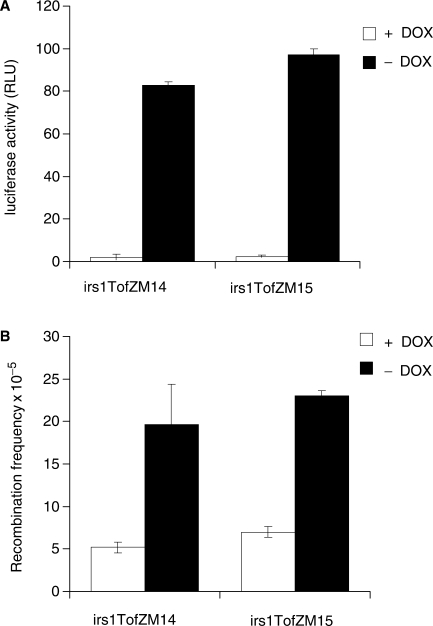
We also studied TAR in XRCC2 mutated irs1 cells carrying the Tet-regulatory vector pTetOffZeo and the recombination construct pBI-LMScI. Transcription was efficiently increased over the recombination substrate following removal of doxycycline from the media, as determined by measuring the luciferase activity (Figure 2A). Surprisingly, both the irs1TofZM14 and irs1TofZM15 clones showed a 3.8- and 3.3-fold increase in TAR following removal of doxycycline, respectively (Figure 2B), indicating a functional recombination pathway. This was unexpected as the XRCC2-defective irs1 clone earlier has been shown to be defective in homology directed repair of DSBs (26,27) and to be defective in RAD51 foci formation following ionizing radiation treatment (18,19).
Figure 2.
XRCC2 defective irs1 cells are proficient in transcription-associated recombination. (A) The inducibility at the bidirectional Pbi-1 promoter is determined by measuring luciferase activity in irs1TofZM14 and irs1TofZM15 cells, both with stably integrated copies of both the pTetoffZeo and pBI-LMscI vectors (9). (B) The recombination frequency was monitored as reversion to a functional neomycin resistance gene following removal of doxycycline, which increases transcription at the recombination substrate. Transcription enhanced recombination in both XRCC2 defective clones irs1TofZM14 and irs1TofZM15 (P < 0.01 in t-test). The average and standard deviation of three independent experiments are depictured.
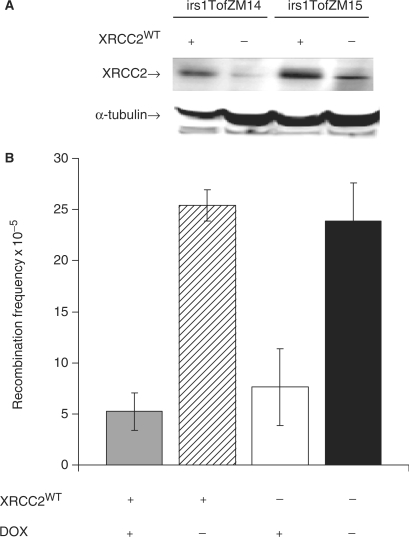
To fully test if XRCC2 is not involved in TAR, we transiently transfected irs1TofZM14 cells with the wild-type XRCC2 in the pIRESneo2 vector and determined expression using western blot (Figure 3A). We found that expression of XRCC2 did not influence TAR levels (Figure 3B), confirming that the XRCC2 protein or XRCC2 protein level have no function in TAR.
Figure 3.
XRCC2 does not influence transcription-associated recombination. (A) irs1TofZM14 and irs1TofZM15 cells were transiently transfected with a plasmid expressing wild-type XRCC2 (XRCC2WT). The expression levels were verified using western blot. (B) The influence of XRCC2 expression on transcription-associated recombination was monitored in irs1TofZM14 cells as the reversion to G418 resistant clones among 105 surviving cells. The average and standard deviation of three independent experiments are depictured.
TAR is mechanistically distinct from homologous recombination repair of DSBs
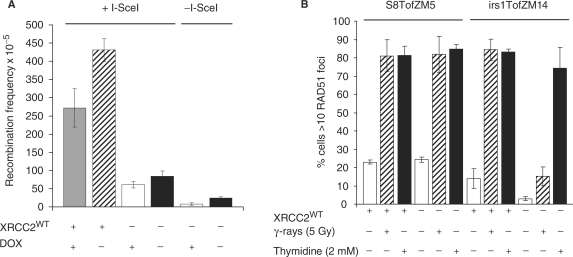
The irs1 cells used here have previously been reported to be defective in DSB-induced homologous recombination repair of an I-SceI-induced DSB (27). We found that transfection of the irs1 cell line with an XRCC2 expression vector did not influence TAR, which could be explained by XRCC2 not being involved in TAR, or alternatively, that the XRCC2 expressed in the irs1TofZM14 is non-functional. As our recombination construct contains an I-SceI site, we tested if the XRCC2 expressed in irs1TofZM14 cells could complement the reported deficiency in homologous recombination. To test this, we transiently transfected irs1TofZM14 cells with both the pCMV3xnlsI-SceI vector and/or XRCC2 (24) with or without doxycycline. We found that the XRCC2 vector efficiently reverts the defect in homology directed DSB repair in irs1TofZM14 cells (Figure 4A); showing that the over-expressed XRCC2 protein is functional. Furthermore, we find that the effect of TAR is additive, which is in agreement with earlier findings that transcription does not influence the homology directed repair of a DSB (9,28,29). Interestingly, we find that homologous recombination is induced by an I-SceI-induced DSB also in non-complimented irs1TofZM14 cells (P < 0.05), while the original report showed no induction of recombination in the same irs1 cells (27). The data presented here suggests that irs1 cells are defective in homologous recombination repair of I-SceI-induced DSBs and are at the same time proficient in TAR. Thus, there might be a sub-pathway of homologous recombination repair in irs1 cells that is still functional. It was previously reported that irs1 cells are proficient in thymidine-induced RAD51 foci, but defective in hydroxyurea and γ-ray-induced RAD51 foci (30). Thymidine is an agent that depletes only the (dCTP) pool in cells, as a consequence of a negative feedback mechanism of the R1 subunit of ribonucleotide reductase that follows high (dTTP) levels (31). The consequence is that the progression of the replication fork is slowed down during replication, which results in a unique replication lesion, with no DSBs, that is separate from those produced by hydroxyurea. Hydroxyurea directly inhibits ribonucleotide reductase, depleting all (dNTP) pools resulting in stalled replication forks and DSB formation (32). Thymidine efficiently induces homologous recombination at replication forks (32), which likely involves a template switching mechanism (33). Here, we tested if also the irs1TofZM14 cells are proficient in thymidine-induced RAD51 foci as this may explain the proficiency in TAR. We found that the irs1TofZM14 cells were defective in RAD51 foci induced by ionizing radiation as compared to wild-type cells and proficient in RAD51 foci formation induced by thymidine (Figure 4B), which altogether suggests that the irs1TofZM14 cells are still proficient in a sub-pathway of homologous recombination repair at replication forks. The deficiency in RAD51 foci formation following ionizing radiation was reverted by transient transfection with a plasmid containing wt XRCC2, while not altering the number of RAD51 foci following thymidine treatment, confirming that the XRCC2 deficiency is responsible for the lack of RAD51 foci following ionizing radiation in irs1TofZM14 cells.
Figure 4.
XRCC2 is required for homology directed DSB repair and dispensable in transcription or thymidine-induced recombination. (A) The influence of transcription and XRCC2 on homologous recombination triggered by an I-SceI-induced DSB following transient transfection of the pCMV3xnlsI-SceI vector was monitored in irs1TofZM14 cells as the reversion to G418 resistant clones among 105 surviving cells. (B) RAD51 foci were visualized in wild-type (S8TofZM5) or XRCC2 defective (irs1TofM14) cells 5 h following γ-ray treatment (5 Gy) or following a 24-h thymidine (2 mM) treatment. Cells containing >10 RAD51 foci were counted as positive. The average and standard deviation of three independent experiments are depicted.
DISCUSSION
TAR is a phenomenon present in all investigated cellular organisms but, in spite of this, the proteins involved in catalysing TAR are unknown. Here, we show that BRCA2 is required for TAR, which strongly support that TAR involves RAD51-mediated strand invasion (34). This is an expected finding and therefore, it is more surprising to find that the XRCC2 protein is not required for TAR. The XRCC2 protein was earlier found to be important in catalysing DSB-induced homologous recombination and RAD51 foci formation after ionizing radiation (18,19,27). Here we report that the irs1 cell line is able to catalyse TAR and that overexpression of a wild-type XRCC2 protein does not influence TAR levels, which altogether show that XRCC2 is not involved in TAR. As irs1 cells are functional for TAR, while defective for DSB-induced homologous recombination, these experiments clearly show that the recombination induced following transcription is functionally different than homologous recombination induced by an I-SceI-induced DSB. This finding is of biological importance as the two-ended DSB formed by ionizing radiation or an endonuclease likely is a rare event in vivo. However, the discrepancy of the XRCC2 gene involved in one pathway, but not another, is anticipated as homologous recombination is involved in repairing several different types of lesions (33).
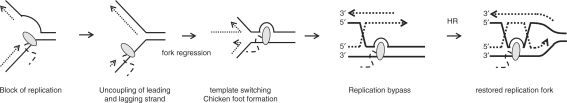
Using the irs1TofZM14 and irs1TofZM15 cell lines, we now have a system to genetically separate TAR from ordinary DSB-induced homologous recombination repair. Interestingly, it was reported that the irs1 cell line is partially proficient for thymidine-induced homologous recombination, while defective for DSB or hydroxyurea-induced homologous recombination (30), which we can confirm to be the case also in the irs1 cells used here. In speculation, it is possible that the thymidine-induced and transcription-induced homologous recombination occur with similar mechanisms in mammalian cells, a mechanism that appears to be largely distinct from DSB-induced homologous recombination. We have shown in several reports that thymidine causes a unique substrate for homologous recombination, which may involve template switching to bypass a lesion (33,34,35,36). The DNA polymerase is about 20 times faster than RNA polymerase II and would need to over take the RNA polymerase not to be slowed down. The model for thymidine-induced recombination may also explain how the RNA polymerase could be bypassed during replication (Figure 5). There is already strong support for TAR being connected with replication in both mammals and yeast (3,9) and it has been shown in yeast that recombination is induced when a replication fork hits an elongating RNA polymerase (4). Also, in mammalian cells TAR is restricted to the S-phase of the cell cycle and relies on a rapidly moving replication fork (9).
Figure 5.
Model how template switching at replication forks may be used for the DNA polymerase to bypass the transcription machinery. The leading and lagging strand of replication may uncouple as the leading strand of DNA polymerase is stalled by the RNA polymerase. The replication fork may then regress and use template switching to bypass the RNA polymerase. Following the bypass the DNA end is used for replication restart using homologous recombination, which would account for the transcription-associated recombination reported in this study. This model explains how the RNA polymerase may be over taken by the DNA polymerase, as well as explaining how the XRCC2 allele in irs1 cells are proficient for both thymidine and transcription-induced recombination.
In conclusion, we show a differential involvement of the BRCA2 and XRCC2 proteins in catalysing TAR, demonstrating that homology directed DSB repair and TAR employ distinct recombination pathways.
FUNDING
The Swedish Cancer Society; the Swedish Children's Cancer Foundation; the Swedish Research Council; the Lawski Foundation; the Swedish Pain Relief Foundation; Medical Research Council. Funding for open access charge: Swedish Pain Relief Foundation.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank John Thacker, Mark Meuth and Malgorzata Zdzienicka for materials.
REFERENCES
- 1.Aguilera A. The connection between transcription and genomic instability. EMBO J. 2002;21:195–201. doi: 10.1093/emboj/21.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia-Rubio M, Huertas P, Gonzalez-Barrera S, Aguilera A. Recombinogenic effects of DNA-damaging agents are synergistically increased by transcription in Saccharomyces cerevisiae. New insights into transcription-associated recombination. Genetics. 2003;165:457–466. doi: 10.1093/genetics/165.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeuchi Y, Horiuchi T, Kobayashi T. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 2003;17:1497–1506. doi: 10.1101/gad.1085403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prado F, Aguilera A. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J. 2005;24:1267–1276. doi: 10.1038/sj.emboj.7600602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chavez S, Beilharz T, Rondon AG, Erdjument-Bromage H, Tempst P, Svejstrup JQ, Lithgow T, Aguilera A. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J. 2000;19:5824–5834. doi: 10.1093/emboj/19.21.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chavez S, Garcia-Rubio M, Prado F, Aguilera A. Hpr1 is preferentially required for transcription of either long or G+C-rich DNA sequences in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001;21:7054–7064. doi: 10.1128/MCB.21.20.7054-7064.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wellinger RE, Prado F, Aguilera A. Replication fork progression is impaired by transcription in hyperrecombinant yeast cells lacking a functional THO complex. Mol. Cell. Biol. 2006;26:3327–3334. doi: 10.1128/MCB.26.8.3327-3334.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nickoloff JA, Reynolds RJ. Transcription stimulates homologous recombination in mammalian cells. Mol. Cell. Biol. 1990;10:4837–4845. doi: 10.1128/mcb.10.9.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottipati P, Cassel TN, Savolainen L, Helleday T. Transcription-associated recombination is dependent on replication in Mammalian cells. Mol. Cell. Biol. 2008;28:154–164. doi: 10.1128/MCB.00816-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang F, Han M, Romanienko PJ, Jasin M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl Acad. Sci. USA. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnaudeau C, Lundin C, Helleday T. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 2001;307:1235–1245. doi: 10.1006/jmbi.2001.4564. [DOI] [PubMed] [Google Scholar]
- 12.Lundin C, Erixon K, Arnaudeau C, Schultz N, Jenssen D, Meuth M, Helleday T. Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol. Cell. Biol. 2002;22:5869–5878. doi: 10.1128/MCB.22.16.5869-5878.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: From mechanistic understanding to cancer treatment. DNA Repair (Amst) 2007;6:923–935. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Richardson C, Moynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saleh-Gohari N, Helleday T. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 2004;32:3683–3688. doi: 10.1093/nar/gkh703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan SS, Lee SY, Chen G, Song M, Tomlinson GE, Lee EY. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999;59:3547–3551. [PubMed] [Google Scholar]
- 17.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 18.O'Regan P, Wilson C, Townsend S, Thacker J. XRCC2 is a nuclear RAD51-like protein required for damage-dependent RAD51 focus formation without the need for ATP binding. J. Biol. Chem. 2001;276:22148–22153. doi: 10.1074/jbc.M102396200. [DOI] [PubMed] [Google Scholar]
- 19.Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson LH, Takeda S. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones NJ, Cox R, Thacker J. Isolation and cross-sensitivity of X-ray-sensitive mutants of V79-4 hamster cells. Mutat. Res. 1987;183:279–286. doi: 10.1016/0167-8817(87)90011-3. [DOI] [PubMed] [Google Scholar]
- 21.Cartwright R, Tambini CE, Simpson PJ, Thacker J. The XRCC2 DNA repair gene from human and mouse encodes a novel member of the recA/RAD51 family. Nucleic Acids Res. 1998;26:3084–3089. doi: 10.1093/nar/26.13.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davies AA, Masson JY, McIlwraith MJ, Stasiak AZ, Stasiak A, Venkitaraman AR, West SC. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 23.Kraakman-van der Zwet M, Overkamp WJ, van Lange RE, Essers J, van Duijn-Goedhart A, Wiggers I, Swaminathan S, van Buul PP, Errami A, Tan RT, Jaspers NG, et al. Brca2 (XRCC11) deficiency results in radioresistant DNA synthesis and a higher frequency of spontaneous deletions. Mol. Cell. Biol. 2002;22:669–679. doi: 10.1128/MCB.22.2.669-679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohindra A, Bolderson E, Stone J, Wells M, Helleday T, Meuth M. A tumour-derived mutant allele of XRCC2 preferentially suppresses homologous recombination at DNA replication forks. Hum. Mol. Genet. 2004;13:203–212. doi: 10.1093/hmg/ddh022. [DOI] [PubMed] [Google Scholar]
- 25.Saleh-Gohari N, Helleday T. Strand invasion involving short tract gene conversion is specifically suppressed in BRCA2-deficient hamster cells. Oncogene. 2004;23:9136–9141. doi: 10.1038/sj.onc.1208178. [DOI] [PubMed] [Google Scholar]
- 26.Hamilton AA, Thacker J. Gene recombination in X-ray-sensitive hamster cells. Mol. Cell. Biol. 1987;7:1409–1414. doi: 10.1128/mcb.7.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson RD, Liu N, Jasin M. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature. 1999;401:397–399. doi: 10.1038/43932. [DOI] [PubMed] [Google Scholar]
- 28.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 29.Weng YS, Xing D, Clikeman JA, Nickoloff JA. Transcriptional effects on double-strand break-induced gene conversion tracts. Mutat. Res. 2000;461:119–132. doi: 10.1016/s0921-8777(00)00043-4. [DOI] [PubMed] [Google Scholar]
- 30.Liu N, Lim CS. Differential roles of XRCC2 in homologous recombinational repair of stalled replication forks. J. Cell Biochem. 2005;95:942–954. doi: 10.1002/jcb.20457. [DOI] [PubMed] [Google Scholar]
- 31.Bjursell G, Reichard P. Effects of thymidine on deoxyribonucleoside triphosphate pools and deoxyribonucleic acid synthesis in Chinese hamster ovary cells. J. Biol. Chem. 1973;248:3904–3909. [PubMed] [Google Scholar]
- 32.Lundin C, Erixon K, Arnaudeau C, Schultz N, Jenssen D, Meuth M, Helleday T. Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol. Cell. Biol. 2002;22:5869–5978. doi: 10.1128/MCB.22.16.5869-5878.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Helleday T. Pathways for mitotic homologous recombination in mammalian cells. Mutat. Res. 2003;532:103–115. doi: 10.1016/j.mrfmmm.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 34.Davies AA, Masson JY, McIlwraith MJ, Stasiak AZ, Stasiak A, Venkitaraman AR, West SC. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 35.Bolderson E, Scorah J, Helleday T, Smythe C, Meuth M. ATM is required for the cellular response to thymidine induced replication fork stress. Hum. Mol. Genet. 2004;13:2937–2945. doi: 10.1093/hmg/ddh316. [DOI] [PubMed] [Google Scholar]
- 36.Saleh-Gohari N, Bryant HE, Schultz N, Parker KM, Cassel TN, Helleday T. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol. Cell. Biol. 2005;25:7158–7169. doi: 10.1128/MCB.25.16.7158-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deans B, Griffin CS, Maconochie M, Thacker J. Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. EMBO J. 2000;19:6675–6685. doi: 10.1093/emboj/19.24.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Helleday T, Arnaudeau C, Jenssen D. A partial hprt gene duplication generated by non-homologous recombination in V79 Chinese hamster cells is eliminated by homologous recombination. J. Mol. Biol. 1998;279:687–694. doi: 10.1006/jmbi.1998.1809. [DOI] [PubMed] [Google Scholar]