Abstract
DEAD box helicases are involved in nearly all aspects of RNA metabolism. They share a common helicase core, and may comprise additional domains that contribute to RNA binding. The Thermus thermophilus helicase Hera is the first dimeric DEAD box helicase. Crystal structures of Hera fragments reveal a bipartite C-terminal domain with a novel dimerization motif and an RNA-binding module. We provide a first glimpse on the additional RNA-binding module outside the Hera helicase core. The dimerization and RNA-binding domains are connected to the C-terminal RecA domain by a hinge region that confers exceptional flexibility onto the helicase, allowing for different juxtapositions of the RecA-domains in the dimer. Combination of the previously determined N-terminal Hera structure with the C-terminal Hera structures allows generation of a model for the entire Hera dimer, where two helicase cores can work in conjunction on large RNA substrates.
INTRODUCTION
Helicases couple the energy from ATP hydrolysis to structural rearrangements of their nucleic acid substrates. Based on conserved sequence motifs, helicases are divided into superfamilies SF1, SF2 and SF3 (1). RNA helicases of the DEAD box family belong to SF2 and are involved in virtually all processes of RNA metabolism. DEAD box helicases share a helicase core domain that contains all conserved helicase motifs (2). This core domain is subdivided into two RecA-domains (3,4) that are flexibly connected. Additional domains may contribute to substrate binding, confer binding specificity or mediate interactions with other proteins. The N-terminal domain contains many conserved motifs involved in nucleotide-binding, such as the Q-motif, the Walker A motif and the DEAD box. The C-terminal RecA-domain contains motifs IV–VI that are essential for RNA binding. RNA helicases undergo large conformational changes during their catalytic cycle (5). In the closed, ATP-bound state the N- and C-terminal RecA-domains associate to form a continuous binding site for RNA and allow motifs from both domains to form an interaction network.
Closed conformations of RNA helicases were trapped in crystalline form in few instances, such as the exon junction complex (6,7) and the Drosophila melanogaster Vasa RNA helicase (8), which were determined in complex with an ATP analogue and a single stranded RNA oligonucleotide. Information on the open helicase core conformations was gained from structures of eIF4A (9), an uncharacterized Methanococcus jannaschii DEAD (mjDeaD) box helicase (10), UAP56 (11,12), Dhh1p (13) and DDX3X (14). Since these structures comprise the helicase core only, the location of any appendage domains is currently unknown. Most of the helicase core structures show a monomer, but there is some evidence from analysis of crystal contacts that the N-terminal domains of Hera (15), mjDeaD (10) and UAP56 (11,12) may dimerize in solution.
In the heat resistant RNA-dependent ATPase Hera from Thermus thermophilus, the helicase core is followed by a C-terminal extension that mediates interaction with 23S ribosomal RNA fragments and RNase P RNA (16,17). Hera is the first DEAD box helicase that forms a stable dimer in the absence of ligands, which sets it apart from other RNA helicases and raises the question as to how dimerization might be linked to RNA binding and helicase function. N-terminal truncations mapped the dimerization domain to the C-terminal half of Hera (17).
We determined three crystal structures of the C-terminal part of Hera from T. thermophilus (termed Hera_208–419) on the way to a model of the complete Hera helicase. The structures display a number of unique features. First, Hera forms a dimer in the crystal, which was also confirmed in solution studies. Second, the first 50 residues of the C-terminal domain form the dimerization motif, which adopts a novel fold. It is loosely packed and imparts a high degree of flexibility on the Hera dimer. Comparison of the three independent structures shows that this flexibility allows for drastic changes in the juxtaposition of the two helicase cores within the dimer. Third, while the C-terminal 90 residues could not be built into the electron density, their localization and extent are clearly visible and provide a first glimpse on this additional RNA-binding site outside the Hera helicase core. Thus, the C-terminal extension of Hera is a bipartite structure of a dimerization and an RNA-binding module.
MATERIALS AND METHODS
Protein production, purification, size exclusion chromatography and crystallization
Hera was produced and purified as described (17) but failed to crystallize. Two C-terminally truncated constructs, Hera_208–510 and Hera_208–419, were then generated and purified using the same protocol as for authentic Hera. These constructs eluted as dimers from a calibrated S200 gel permeation column (GE Healthcare). Analytical size exclusion chromatography was performed on a S200 10/300 GL column (GE Healthcare) in 50 mM Tris/HCl, pH 7.5, 500 mM NaCl. 10 μM of Hera_1–510, Hera_208–419, or a mixture were incubated at 65°C for 10 min or 20 min to allow for monomer exchange. For crystallization, 5–10 mg/ml of protein was mixed 1:2 with reservoir and incubated in a microbatch setup. Tetragonal crystals of Hera_208–510 grew at 4°C from reservoir containing 0.1 M MES/NaOH pH 6.5, 15–18% ethylene glycol, 0.3–0.5 M NaI, 3–6% PEG20.000 and are of space group P41212 with one molecule per asymmetric unit. Two orthorhombic crystal forms of Hera_208–419 grew from reservoir containing 0.1 M Tris/HCl pH 8.5, 0.2 M (NH4)2SO4, 23% PEG3350 and 0.1 M Tris/HCl pH 7.0, 0.2 M (NH4)2SO4, 25% PEG3350, 5–10% (w/v) glucose or sucrose. Both crystal forms belong to space group P212121 and contain two molecules per asymmetric unit. Further, details for purification, crystallization and phasing of the Hera fragments will be given elsewhere (18).
Data collection, structure determination and refinement
Data were collected at 100K and reduced with the HKL programs (19) or with XDS (20) and SADABS (Bruker). The structure was determined by MAD phasing using data from a tetragonal crystal of Hera_208–510 containing a single selenium site per asymmetric unit (18). Model building was initiated from the coordinates of PDB-ID 1hv8, which were manually fit into the electron density map from the MAD experiment after density modification and phase extension. After rebuilding, the tetragonal structure could not be satisfactorily refined, probably due to the anisotropy and disorder, which was observed for >20 datasets collected from tetragonal crystals. In some cases merohedral twinning was present with the true space group being P212121 (a ≈ b) and a dimer in the asymmetric unit (data not shown). Although MAD-phasing using merohedrally twinned data is possible (21), the dataset used here was not twinned. Electron density is present for the C-terminal RNA-binding domain (RBD; Hera residues 420–510), which enables placement of this domain (see Results section). The orthorhombic structures were determined using the tetragonal structure as the starting model for molecular replacement with PHASER (22). Models were built in COOT (23) and refined with BUSTER-TNT (24) or PHENIX (25). 5% of reflections were reserved for Rfree cross-validation in all structures (26). Statistics are summarized in Table 1. Surface complementarity coefficients (27) and solvent accessible surface areas were calculated with the programs SC and AREAIMOL, respectively, as implemented in CCP4 (28), using a 1.7 Å radius probe. Possible hydrogen bonds and van der Waals contacts were detected with CONTACSYM (29) using default parameters. Inter-helical angles were calculated using HELIXANG (28). Figures were created with Bobscript (30) and rendered with Raster3D (31), or PyMol (www.pymol.org).
Table 1.
Data collection, phasing and refinement statistics
| Dataset | 3EAR—form 1, partial | 3EAS—form 1, complete, asymmetric | 3EAQ—form 2, complete, symmetric |
|---|---|---|---|
| Data collection resolution range, (Å)a | 46.0–2.3 (2.36–2.30) | 46.1–2.8 (2.88–2.80) | 46.9–2.3 (2.34–2.30) |
| 100% criterion (Å)b | 2.3 | 2.8 | 2.3 |
| Space group | P212121 | P212121 | P212121 |
| Cell dimensions (Å) | a = 41.6, b = 67.7, c = 183.8 | a = 46.1, b = 70.8, c = 181.2 | a = 62.6, b = 70.8, c = 101.9 |
| Unique reflections | 23 505 (1375) | 15 368 (1089) | 20 794 (772) |
| Multiplicity | 6.3 (6.3) | 6.3 (6.3) | 12.8 (10.3) |
| Completeness (%) | 97.9 (94.8) | 99.9 (100) | 100 (100) |
| Mosaicity (°) | 0.45 | 0.32 | 0.28 |
| Rsym (%)c | 4.8 (66.1) | 7.0 (79.0) | 10.6 (95.0) |
| Average I/σ(I) | 14.7 (1.9) | 10.1 (1.4) | 12.9 (1.7) |
| Refinement resolution range (Å) | 45.9–2.3 | 45.3–2.8 | 46.9–2.3 |
| Rcryst/Rfree (%)d | 23.1/25.5 | 22.8/31.4 | 21.8/26.9 |
| Number of residues/waters | 256/29 | 415/0 | 416/7 |
| Coordinate error (Å)e | 0.34 | 0.58 | 0.29 |
| r.m.s.d. bonds/angles (Å, °) | 0.006/0.92 | 0.008/1.15 | 0.007/1.07 |
| Ramachandran plot (%)f | 93.2/6.8/0 | 92.7/7.3/0 | 92.4/7.3/0.3 |
aValues in parentheses correspond to the highest resolution shell.
bThe 100% criterion was calculated using SFTOOLS and represents the resolution in Å of a 100% complete hypothetical data set with the same number of reflections as the measured data.
cRsym = 100 ∑h∑i|Ii(h) – <I(h)>|/∑h∑iIi(h), where Ii(h) is the i-th measurement of reflection h and <I(h)> is the average value of the reflection intensity.
dRcryst = ∑|Fo| – |Fc|/∑|Fo|, where Fo and Fc are the structure factor amplitudes from the data and the model, respectively. Rfree is Rcryst with 5% of test set structure factors.
eBased on maximum likelihood.
fCalculated using PROCHECK (53). Numbers reflect the percentage amino-acid residues of the core, allowed and generous allowed regions, respectively.
RESULTS
Full-length Hera from T. thermophilus was purified in a nucleotide-free form that binds and hydrolyzes ATP and is an active RNA helicase (17). Crystallization of the full-length Hera has been reported several years ago (16), but was irreproducible in our hands. In a divide et impera approach we cloned the genes coding for N- and C-terminal fragments (residues 1–207, 208–510 and 208–419) to determine their crystal structures separately and prepare a model for the full-length Hera. The structure of the N-terminal domain in complex with AMP was determined earlier (15). Here, we report three significantly different structures of the C-terminal Hera fragment 208–419 in two orthorhombic crystal forms. A third, tetragonal crystal form of Hera_208–510 was used for Se-Met MAD phasing (see Methods section) but the model built into the experimental electron density maps could not be refined satisfactorily (18). However, this model was of sufficient quality for molecular replacement of the orthorhombic Hera_208–419 crystal forms. The asymmetric unit of both forms contains two Hera_208–419 molecules that form dimers. Dataset 1 of crystal form 1 has one of the two RecA domains disordered, which was omitted from the model. This structure is referred to as ‘partial’ throughout. Another dataset of crystal form 1 and data from crystal form 2 allowed tracing of complete Hera_208–419 dimers, called dimer I and dimer II, respectively. The structures were refined to resolutions of 2.8 Å (dimer I) and 2.3 Å (partial structure and dimer II). Data collection and refinement statistics are summarized in Table 1.
Architecture of the Hera monomer
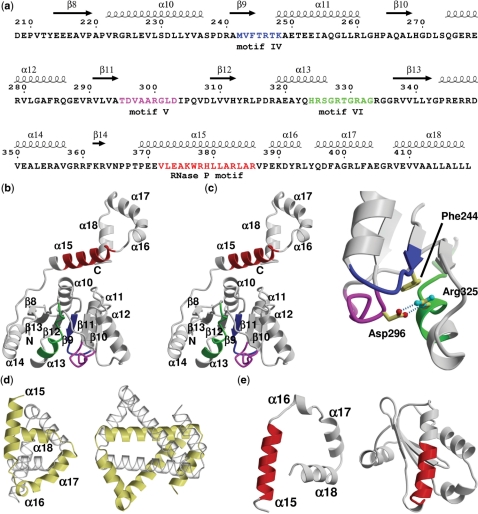
Hera_208–419 folds into a RecA domain connected to a dimerization domain that in authentic Hera is followed by a C-terminal RBD. The RecA-domain comprises a central seven-stranded parallel β-sheet flanked by two and four α-helices on either side (Figure 1a and b). A DALI search (www2.ebi.ac.uk/dali/) for structural homologues results in >50 unique structures with a Z-score > 10, among them the Bacillus subtilis DEAD box helicase YxiN (Z-score 23.9, r.m.s.d. 1.8 Å over 151 residues) and the M. jannaschii DEAD box helicase (Z-score 23.0, r.m.s.d. 1.7 Å over 149 residues), but also the more distantly related helicase domains in D. melanogaster Vasa (Z-score 20.1, r.m.s.d. 2.2 Å over 146 residues; (8), the human exon junction complex (Z-score 21.2, r.m.s.d. 2.1 Å over 154 residues; (6,7), and hepatitis C virus NS3 (Z-score 13.0, r.m.s.d. 2.5 Å over 116 residues; (32), for which structures of nucleic acid complexes exist. The conserved helicase motifs IV–VI in the RecA domain of Hera are located close in space and interact with each other. A key residue in this network is Arg335 of motif VI that bridges motifs IV and VI. Phe244 in motif IV interacts with Arg325 via π–cation interactions, while Asp296 of motif V both neutralizes the positive charge of Arg325 and aligns this side-chain for interaction with Phe244 by two hydrogen bonds (Figure 1c). The helicase motifs thus form a platform for the second RecA domain of the helicase core (33).
Figure 1.
Hera structure sequence relationship and architecture of the Hera_208–419 monomer. (a) Sequence of Hera_208–419 with secondary structure elements indicated on the top. The numbering of sequence and secondary structure elements corresponds to the full-length Hera. Conserved helicase motifs IV–VI are colored blue, magenta and green, respectively. The putative RNase P motif is colored in red. (b) Stereo ribbon diagram of the Hera_208–419 monomer. The secondary structure elements, helicase and RNase P motifs from (a) are indicated. (c) Close-up showing the interactions of helicase motifs IV–VI. Asp296 of motif V connects to Arg325 motif VI via two hydrogen bonds (dashed lines). Arg325 stacks on Phe244 of motif IV. The view is rotated by 180° around the y-axis compared to (b). (d) Comparison of the left-handed super-helices in Hera_208–419 (left) and the hypothetical H. pylori protein HP242 (right). The corresponding monomers are colored identically in yellow and transparent grey. (e) The RNase P motifs in Hera (left) and in the protein component of the T. maritima RNase P (PDB-ID 1nz0; right) are predominantly α-helical but located in a very different structural context. The structures are shown with their RNase P motifs aligned (red).
The RecA domain is followed by a α-helical extension (residues 370–419) that folds into a single turn of a left-handed super-helix. A BLAST search for this region (www.ncbi.nlm.nih.gov/BLAST) revealed no significant sequence homology to any other protein. A DALI search for structural homologues using this domain as a query resulted in only one remotely similar structure with a Z-score of 3.8, where scores <2.5 are defined as structurally dissimilar. The topology of HP0242, a Helicobacter pylori protein of unknown function [PDB-IDs 2ouf; MCSG, unpublished and 2bo3; (34)], is similar to Hera, also forming a left-handed super-helix. However, although both molecules use this domain to form dimers, the differences in length and angles of the helix segments lead to a large r.m.s.d. value of 3.7 Å over 50 residues for these structures, which should therefore be considered unrelated (Figure 1d). As a result, we conclude that this dimerization motif constitutes a novel fold that is currently unique to Hera.
The RNase P motif in Hera
Sequence alignments have shown that the region encompassing residues 372–386 exhibits weak homology to the signature motif of the RNase P-protein component (16). A connection of Hera and RNase P with respect to RNA recognition via the RNase P motif would be exciting, and indeed we have recently shown RNase P RNA to be a specific substrate of Hera (17). However, comparison of the crystal structures of Hera_208–419 and the RNase P-protein from Thermotoga maritima [PDB-ID 1nz0, (35)] clearly shows that both proteins are dissimilar. Although both RNase P motifs form α-helices (though not over the entire sequence in T. maritima RNase P), the structural context is very different (Figure 1e). Hera is all α-helical in its C-terminal region while the RNase P-protein adopts a mixed α/β-fold. A common function might exist for the RNase P motifs in RNA binding due to the presence of conserved arginines [Arg378, Arg383 and Arg386 in Hera (16)]. However, a Hera mutant where these arginines are replaced by alanines is still able to bind RNA, rendering the RNase P motif dispensable for RNA binding (17). It thus appears that the sequence homology between Hera and RNase P is coincidental.
The Hera dimer
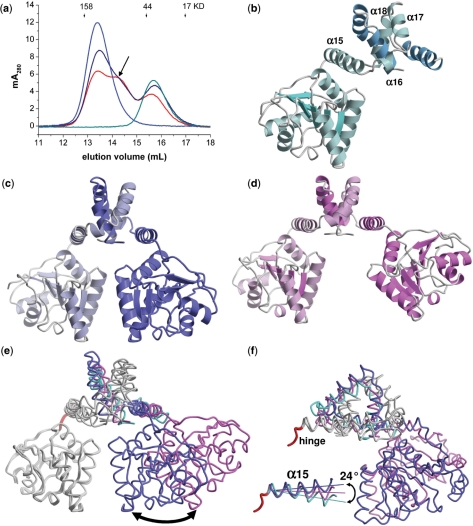
Full-length Hera forms a dimer in solution as judged by gel permeation chromatography. Hera constructs encompassing residues 1–510, 208–510, 208–419 and 370–510 are apparent dimers in solution (17), putting the dimerization domain within the region encompassing residues 370–419. To prove the dimeric nature of Hera in solution, gel permeation chromatography was performed on mixtures of Hera_1–510 (full-length, 56 kDa per monomer) and Hera_208–419 (24 kDa per monomer). Provided rapid monomer exchange, the appearance of a heterodimeric species with intermediate molecular weight is expected. When Hera_1–510 and Hera_208–419 were mixed, only two peaks corresponding to the individual proteins were observed in the chromatogram, indicating slow (or no) monomer exchange (data not shown). After heating the mixture to 65°C for 10 min, however, a third peak corresponding to a heterodimer appeared (Figure 2a). The population of this species increased further when the incubation time was increased to 20 min, consistent with slow exchange kinetics even at elevated temperatures. Controls with the equally treated isolated proteins yielded only one peak at the expected molecular weight of dimers, confirming that the additional peak in the mixture is not an artifact due to incubation at 65°C. The dimeric nature of Hera is confirmed by the crystal structure of the partial dimer (Figure 2b), and further corroborated by the crystal structures of the complete dimers (Figure 2c and d).
Figure 2.
The Hera_208–419 dimer. (a) Size exclusion chromatography of Hera_1–510 (blue), Hera_208–419 (cyan) and a mixture of both proteins after incubation at 65°C for 10 min (dark blue) or 20 min (red). Monomer exchange produces a heterodimer with intermediate molecular weight (arrow), confirming the dimeric nature of Hera in solution. (b)–(d) Ribbon representations of the partial dimer, dimer I and dimer II. The monomers are colored in slightly different hues. The helices of the dimerization domain in (b) are labeled. (e) Superposition of (b)–(d) onto the first RecA-domain shows the different poses of the second RecA-domain (arrow: ca. 20 Å). The colors match those in (b)–(d). (f) View rotated 90° about the y-axis. Inset: magnification of the hinge region (red), showing the changes in the directions of the α15 helix.
The overall fold of the Hera monomer is replicated in the Hera dimers. Both dimer structures adopt a ∧-shape with the helicase motifs IV–VI facing towards a flat surface that constitutes part of the interface with the N-terminal RecA domain in the closed conformation (Figures 1b and 4). The arrangement of the second RecA domains and the dimer interfaces are very different when the dimer structures are compared to each other, and thus indicate pronounced flexibility of Hera also in solution. Whereas the overall structure of dimer II is almost symmetric, with a pseudo-twofold axis perpendicular to the dimerization domain (Figure 2c), dimer I is highly asymmetric (Figure 2d). Superposition of the first RecA domains for dimers I and II shows a large displacement of the second RecA domain. The two Hera conformations are characterized by a 34° rotation, which places equivalent residues up to 25 Å apart from each other (Figure 2e and f). One cause for this flexibility in Hera is a hinge region (red in Figure 2e and f) formed by residues Pro366–Pro369, which allows for different directions of the adjacent helix α15. Comparison of the three Hera_208–419 structures shows the α15 inter-helical angles to change in steps of 12° (inset in Figure 2f). The hinge allows the C-terminal ends of helix α15 to vary by up to 9 Å (at Ala385), with a concomitant amplification in the displacement of the RecA domain of the second monomer. This mobility can be further augmented by equivalent hinge movements in the second monomer. The hinge around Pro366–Pro369 is, however, not the sole origin of flexibility in the Hera dimer, as the dimer interface also shows considerable flexibility.
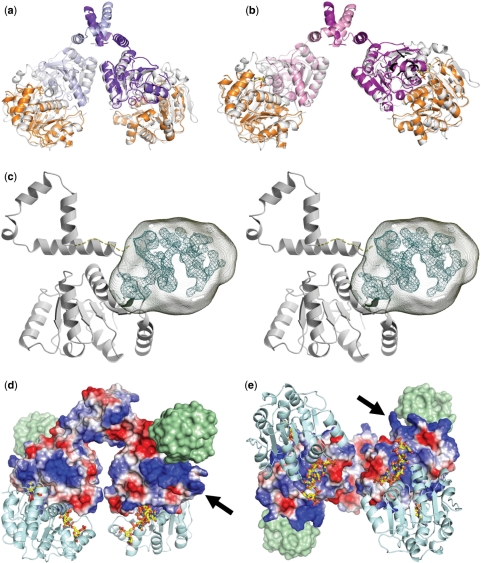
Figure 4.
Model for the dimeric Hera helicase and location of the RBD. (a, b) Construction of the Hera_1–419 dimers by superposition of the isolated Hera_N (PDB-ID 1gxs) and Hera_208–419 structures onto the Vasa-RNA complex. The view and color code for Hera_208–419 are the same as in Figure 2. Hera_N is colored orange, and the template Vasa is colored in white. (c) Stereo view of the location of the Hera RBD (residues 420–510) in crystals of Hera_208–510. Electron density from the MAD-phased tetragonal crystals is contoured at the 1σ level. This density is not explained by residues 208–419 and must thus belong to residues 420–510. The approximate volume for these residues is drawn as a transparent hull. The Hera_208–419 monomer is taken from dimer II and shown in gray. The connection between the Hera_208–419 C-terminus and the RBD is shown as a dashed line. (d) Electrostatic potential calculated for dimer II showing a distinct positively polarized patch (arrow) close to the RBD (location shown as a green surface). (e) View rotated 90° about the x-axis to emphasize the close proximity of the RNA-binding sites. The nucleotide AMP was taken from the Hera_N structure, the RNA from Vasa. Both are shown as stick models.
The dimerization domain and flexibility in the Hera dimer
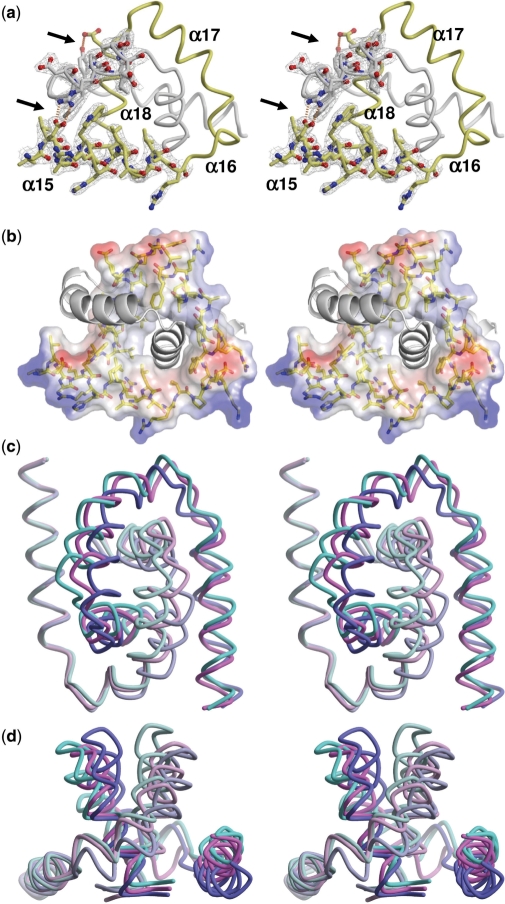
Two left-handed helical regions formed by Hera residues 370–419 associate to form a pseudo-symmetric dimer that has well-defined electron density (Figure 3a). No other regions seem to be contributing to Hera dimerization, in accord with the solution studies. The predominantly hydrophobic dimer interface is both extensive and of high complementarity. While the partial structure buries a surface of 3300 Å2, more than 5300 Å2 are solvent inaccessible in dimer I and dimer II (Table 2). The average surface complementarity coefficient Sc is ∼0.75, indicating a very good fit of the monomers, which is also mirrored in the large number of inter-subunit contacts. The interface is dominated by van der Waals interactions: around 200 are present, many of them centered at residues Trp377, Tyr392, Tyr395, Phe398 and helix α18 formed by residues Val408-Leu419 (see Supplementary Table 1 for a complete listing). The parallel orientation of helices α18 forms the hydrophobic core of the dimer interface (Figure 3b) and at first sight resembles a leucine zipper. The inter-helical angle in the prototypic GCN4 leucine zipper is ∼24°. In contrast, the α18 inter-helical angles in the partial structure, dimer I and dimer II are 52°, 54° and 58°, respectively, excluding a leucine-zipper type interaction. The r.m.s. differences in the α18 inter-helical angles also point to considerable flexibility in the dimer interface. There are a few polar contacts in the interface, most notably between the side-chains of Glu374/Arg401 and Tyr392/Glu409 (Figure 3a), but these are not present in all complexes. Indeed, a closer inspection of the interface contacts shows strong differences, both when comparing the different structures among each other and when comparing reciprocal interactions of the chains within one dimer. While the total number of contacts and buried surface areas in dimers I and II are roughly the same, there are fewer contacts and less buried surface in the partial structure, although the dimerization domain is complete. This discrepancy points to a less compact packing in the partial structure, indicating substantial plasticity in how the interface is organized.
Figure 3.
The dimer interface. (a) The σA-weighted 2Fo–Fc electron density map of Hera structure dimer II is contoured at 1.0σ. Three inter-subunit hydrogen bonds are shown as dashed orange lines and indicated by arrows. (b) Stereo view of the electrostatic potential calculation of one monomer shows the predominantly hydrophobic dimer interface made up by helices α15–α18. (c) Variation in the dimerization core shown by overlay of the three Hera_208–419 structures onto helix α15. The color code of this stereo image adheres to Figure 2. (d) View perpendicular to (c).
Table 2.
Overview of the Hera_208–419 dimer interfaces
| Structure | Partial | Dimer I | Dimer II |
|---|---|---|---|
| BSA (Å2)a | 3355 | 5330 | 5361 |
| Scb | 0.736 | 0.771 | 0.751 |
| Number of H-bonds/salt bridgesc | 0/4 | 1/4 | 1/2 |
| Number of vdWc | 199 | 221 | 228 |
| Angle between α18 (°) | 54 | 52 | 58 |
aTotal buried surface area (BSA) in the dimer calculated with a probe radius of 1.7Å.
bA surface complementarity coefficient of one would denote perfect complementarity.
cNumber of hydrogen bonds, salt bridges and van der Waals interactions in the dimer.
Superposition of the dimerization domains onto helix α15 reveals that this domain can rotate about an axis parallel to the principal axis of helix α15. The extent of the rotation can be as large as 17°, resulting in maximum displacements of the apical atoms of the domain by up to 7 Å (Cα atoms of Ala404; Figure 3c and d). In conclusion, the overall structural plasticity of the Hera dimer is the sum of hinge movements and rigid body rotations of segments in the dimer interface. Due to this flexibility monomeric Hera may exist at elevated temperatures, but the observed slow monomer exchange underscores a high stability of the dimer even at physiologic temperatures for T. thermophilus.
Construction of the Hera helicase core from its N- and C-terminal domains
The Vasa helicase was crystallized in a closed conformation of its two RecA-domains in complex with an ATP analogue and a U10 ssRNA (8), and represents one extreme of the conformational space accessible to DEAD box helicases. The question now arises, how the structures of the dimeric Hera_208–419 and the N-terminal Hera domain would compare to Vasa and if a complete model for the Hera helicase core can be constructed without violating stereochemistry. The RecA-domains of Hera and Vasa superimpose closely with r.m.s.d. values of ∼1.5 Å over 196 (N-terminal) and 125 residues (C-terminal), respectively. Indeed, Vasa-based alignment of the Hera dimer I and dimer II structures results in two feasible models for the complete Hera helicase core including the dimerization domain (Figure 4a and b). There are no stereochemical hindrances between the modeled RecA-domains that could not be rectified by slight conformational adjustments of loops and side-chains. Inspection of the RNA-binding site in Vasa shows that many of the side-chains contacting RNA have their Hera analogues in close proximity. Thus, not only is the construction of the artificial Hera helicase core stereochemically feasible, but it also makes biochemical sense and allows drawing conclusions on RNA binding by Hera (see below). Very similar models can be generated using the hepatitis C virus NS3 helicase (32) and the human exon junction complex (6,7) as templates. A possible limitation of these Hera helicase core models is the missing C-terminal domain. These additional modules are implicated in specific RNA binding (16,17,36–40) or in contributing to high affinity RNA binding by providing unspecific interactions with RNA (41–44). Their relative orientation with respect to the helicase core is currently unknown.
Localization of the Hera C-terminal RBD
The crystal structures presented here lack the C-terminal 90 residues that presumably form a separate domain, which was shown to be important for substrate RNA binding (17). The first crystals obtained and used for MAD-phasing were grown using a Hera construct encompassing residues 208–510, and thus included the C-terminal domain. Despite the fact that the experimental electron density generated from these crystals was of sufficient quality to trace residues 208–419 and also to use this model for successful molecular replacement of the orthorhombic crystal forms, no satisfactory refinement was obtained. What is obvious, however, from the electron density, is the location of the Hera C-terminal RBD. While we cannot assign secondary structure elements for the RBD, its envelope and location with respect to the remainder of the helicase are very clear. After building of the RecA-and dimerization domains, a globular volume of electron density wedged between these domains remains that must belong to the RBD (Figure 4c). The location of this domain is sensible as the C-terminus of Hera_208–419 points toward the un-modeled density. Leu419 is followed by a low complexity linker sequence (GGAPA) that can traverse the upper part of the RecA-domain towards the region of un-modeled electron density (dashed in Figure 4c).
Using the information of the localization of the RBD a complete model for the full-length dimeric Hera helicase can be assembled (Figure 4d and e). In this model, the RNA-binding sites of the dimers face each other, suggesting a possible coordinated RNA unwinding capability. Calculation of the electrostatic surface potential for the almost symmetric dimer II reveals a positively polarized patch generated by arginines 223, 345, 347 and 348 (arrow in Figure 4d and e). Most helicase cores do not display sequence specificity and, in agreement, the Vasa structure shows many protein–RNA phosphate backbone interactions. The positively polarized region around arginines 223, 345, 347 and 348 may serve as an unspecific binding site for the phosphate backbone of longer RNA substrates. In addition, this electropositive patch is uniquely positioned just underneath the location of the C-terminal RNA-binding domain, which further strengthens the notion that RNA substrates extend to the C-terminal domain via this path.
DISCUSSION
The crystal structure of the Hera_208–419 fragment confirms the location and interactions of the helicase motifs IV–VI involving residues Phe244, Asp296 and Arg325, respectively. The phenylalanine in motif IV was suggested as an anchor for the rigidity of the C-terminal RecA domain of the helicase core by π–cation interactions with an arginine in motif VI. Mutation of the motif IV phenylalanine to leucine or alanine in the SF2 helicase Ded1 results in loss of cooperative ATP/RNA binding (33). In Hera, Asp296 in motif V orients Arg335 by hydrogen bonding for optimal contact with Phe244, putting Arg335 at the interface of motifs V and VI. The same situation is observed in the mjDeaD helicase (10). A multiple sequence alignment of SF2 helicases [PFAM PF00271; pfam.sanger.ac.uk; (9)] shows that the Phe/Arg/Asp residue combination is not mandatory for SF2 helicase function. Instead, the network of interactions between these motifs can be established in the context of very different sequences or conformations with the phenylalanine replaced by hydrophobic and the aspartate replaced by small, often polar residues.
Possible binding of large RNA molecules to Hera
While the helicase core interacts non-specifically with RNA via residues in motif IV that contact the RNA backbone, the C-terminal domains in a few SF2 helicases mediate sequence-specific RNA binding. One well-studied example is the B. subtilis helicase YxiN, which contains a C-terminal domain of ∼80 residues that confers specificity for hairpin 92 of the 23S rRNA (38). This domain folds into an RNA recognition motif that is found in many eukaryotic RNA-binding proteins, but employs a different RNA-binding mode (40). Although the Hera and YxiN RBDs have similar dimensions, they display no sequence homology and most likely adopt different structures, consistent with the impossibility to fit this domain into the electron density of the Hera RBD. Nevertheless, 23S rRNA fragments comprising hairpin 92 have also been identified as specific substrates for Hera, and binding of these RNAs requires the Hera RBD (17). A 32/9mer derived from the 23S rRNA, consisting of hairpin 92 and an adjacent 9-bp helix, is unwound by Hera in an ATP-dependent manner. The Hera model suggests a binding mode in which sequence- or structure-specific RNA binding to the Hera-RBD would serve as an anchor for hairpin structures, bringing the helicase core of Hera into proximity of RNA helices to be unwound. The electropositive patch generated by arginines 223, 345, 347 and 348 is located between the RBD and the helicase core, and could thus contribute to RNA positioning. Such a positioning would be even more important to direct unwinding or remodeling of larger RNA molecules that display a higher degree of conformational freedom. For example, the 379 nucleotide RNase P RNA is a specific Hera substrate and induces the closed conformation of the helicase core. The interaction of Hera with RNase P RNA also requires the RBD (17). The inherent flexibility of the RBD with respect to the dimerization motif, and of the dimerization motif with respect to the C-terminal RecA domain, would afford positioning of RNA with different sizes and structures for unwinding by the helicase core. Such a function is reminiscent of the suggested role of the basic CTDs of the DEAD box helicases Mss116p and CYT-19 (41,44,45). Clearly, a detailed description of the RNA-binding mode in Hera will have to await the structure of the Hera/RNA complex.
Implications of the RNA helicase dimer for RNA unwinding
Helicases in general display a variety of oligomerization states, from ring-shaped hexamers to dimers and monomers. Most DExH box helicases, e.g. the HCV-NS3 helicase (in the absence of the protease domain), function as monomers (46), but an enhanced activity and increased processivity of the dimeric form has been demonstrated (47,48). These observations point towards the existence of a functional dimer, whereas structural studies of NS3 support a monomer (32,49). The DEAD box helicases YxiN and DbpA exist as monomers in solution (50,51). In contrast, the structures of Hera_208–419 from T. thermophilus described here represent first examples for a dedicated dimerization module in a DEAD-box RNA helicase. Based on the large buried surface area and high surface complementarity and the slow monomer exchange even at high temperatures it is likely that Hera forms a stable dimer that is not meant to dissociate during the catalytic cycle. Dimerization is a common strategy for protein stabilization in organisms living at high ambient temperatures (52). However, Hera homologues from the closely related mesophilic Deinococcus geothermalis (Q1J0S9) and the moderately thermophilic D. radiodurans (DR_1624) contain sequences with similarities to the Hera dimerization motif (17), arguing against a purely stabilizing role and in favor of a functional role of the helicase dimer.
The high structural similarity of the Hera N- and C-terminal RecA-domains compared to the Vasa and eIF4A-III helicases enabled us to construct a model for the closed Hera dimer. Inclusion of the information for the position of the RBDs results in an impressive architecture without any severe stereochemical hindrance (Figure 4d and e). The RNA-binding sites of the helicase cores face towards each other, suggesting that the two subunits could remodel RNA by simultaneously acting on the same RNA molecule, possibly in a coordinated fashion, which may be beneficial for efficient remodeling of large RNA substrates. To answer this question, mechanistic studies of monomeric Hera and structural information of the authentic Hera dimer bound to a large RNA would be essential.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
Deutsche Forschungsgemeinschaft (to M.G.R.); VolkswagenStiftung and the Swiss National Science Foundation (to D.K.). Funding for open access charge: Swiss National Science Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank I. Hertel, K. Gasow and A. Schmidt for excellent technical assistance and M. Linden for purifying full-length Hera. We also thank the colleagues at the SLS and BESSY synchrotrons for beamtime and guidance, and G. Sheldrick for providing a conversion program from XDS to SADABS reflection data format.
REFERENCES
- 1.Koonin EV, Gorbalenya AE. The superfamily of UvrA-related ATPases includes three more subunits of putative ATP-dependent nucleases. Protein Seq. Data Anal. 1992;5:43–45. [PubMed] [Google Scholar]
- 2.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 3.Story RM, Steitz TA. Structure of the recA protein-ADP complex. Nature. 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- 4.Story RM, Weber IT, Steitz TA. The structure of the E. coli recA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 5.Theissen B, Karow AR, Kohler J, Gubaev A, Klostermeier D. Cooperative binding of ATP and RNA induces a closed conformation in a DEAD box RNA helicase. Proc. Natl Acad. Sci. USA. 2008;105:548–553. doi: 10.1073/pnas.0705488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, Pedersen JS, Seraphin B, Le Hir H, Andersen GR. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 7.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-Box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 9.Caruthers JM, Johnson ER, McKay DB. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc. Natl Acad. Sci. USA. 2000;97:13080–13085. doi: 10.1073/pnas.97.24.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Story RM, Li H, Abelson JN. Crystal structure of a DEAD box protein from the hyperthermophile Methanococcus jannaschii. Proc. Natl Acad. Sci. USA. 2001;98:1465–1470. doi: 10.1073/pnas.98.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi H, Cordin O, Minder CM, Linder P, Xu RM. Crystal structure of the human ATP-dependent splicing and export factor UAP56. Proc. Natl Acad. Sci. USA. 2004;101:17628–17633. doi: 10.1073/pnas.0408172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao R, Shen J, Green MR, MacMorris M, Blumenthal T. Crystal structure of UAP56, a DExD/H-box protein involved in pre-mRNA splicing and mRNA export. Structure. 2004;12:1373–1381. doi: 10.1016/j.str.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Cheng Z, Coller J, Parker R, Song H. Crystal structure and functional analysis of DEAD-box protein Dhh1p. RNA. 2005;11:1258–1270. doi: 10.1261/rna.2920905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hogbom M, Collins R, van den Berg S, Jenvert RM, Karlberg T, Kotenyova T, Flores A, Karlsson Hedestam GB, Schiavone LH. Crystal structure of conserved domains 1 and 2 of the human DEAD-box helicase DDX3X in complex with the mononucleotide AMP. J. Mol. Biol. 2007;372:150–159. doi: 10.1016/j.jmb.2007.06.050. [DOI] [PubMed] [Google Scholar]
- 15.Rudolph MG, Heissmann R, Wittmann JG, Klostermeier D. Crystal structure and nucleotide binding of the Thermus thermophilus RNA helicase Hera N-terminal domain. J. Mol. Biol. 2006;361:731–743. doi: 10.1016/j.jmb.2006.06.065. [DOI] [PubMed] [Google Scholar]
- 16.Morlang S, Weglohner W, Franceschi F. Hera from Thermus thermophilus: the first thermostable DEAD-box helicase with an RNase P-protein motif. J. Mol. Biol. 1999;294:795–805. doi: 10.1006/jmbi.1999.3282. [DOI] [PubMed] [Google Scholar]
- 17.Linden MH, Hartmann RK, Klostermeier D. The putative RNase P motif in the DEAD box helicase Hera is dispensable for efficient interaction with RNA and helicase activity. Nucleic Acids Res. 2008;36:5800–5811. doi: 10.1093/nar/gkn581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rudolph MG, Wittmann JG, Klostermeier D. Crystallisation and preliminary characterization of the Thermus thermophilus Hera RNA helicase C-terminal Domain. Acta Cryst. 2008 doi: 10.1107/S1744309108043145. F., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 20.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 1993;26:795–800. [Google Scholar]
- 21.Rudolph MG, Kelker MS, Schneider TR, Yeates TO, Oseroff V, Heidary DK, Jennings PA, Wilson IA. Use of multiple anomalous dispersion to phase highly merohedrally twinned crystals of interleukin-1β. Acta Cryst. 2003;D59:290–298. doi: 10.1107/s0907444902021704. [DOI] [PubMed] [Google Scholar]
- 22.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Cryst. 2007;D63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 24.Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Cryst. 2004;D60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 25.Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, et al. Automated structure solution with the PHENIX suite. Methods Mol. Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 26.Brünger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 27.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 28.CCP4 The collaborative computational project number 4, suite programs for protein crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 29.Sheriff S, Hendrickson WA, Smith JL. Structure of myohemerythrin in the azidomet state at 1.7/1.3 Å resolution. J. Mol. Biol. 1987;197:273–296. doi: 10.1016/0022-2836(87)90124-0. [DOI] [PubMed] [Google Scholar]
- 30.Esnouf RM. An extensively modified version of MOLSCRIPT that includes greatly enhanced coloring capabilities. J. Mol. Graph. 1997;15:132–134. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 31.Merritt EA, Murphy MEP. Raster3D Version 2.0 - a program for photorealistic molecular graphics. Acta Cryst. 1994;D50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 32.Kim JL, Morgenstern KA, Griffith JP, Dwyer MD, Thomson JA, Murcko MA, Lin C, Caron PR. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 33.Banroques J, Cordin O, Doere M, Linder P, Tanner NK. A conserved phenylalanine of motif IV in superfamily 2 helicases is required for cooperative, ATP-dependent binding of RNA substrates in DEAD-box proteins. Mol. Cell Biol. 2008;28:3359–3371. doi: 10.1128/MCB.01555-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai JY, Chen BT, Cheng HC, Chen HY, Hsaio NW, Lyu PC, Sun YJ. Crystal structure of HP0242, a hypothetical protein from Helicobacter pylori with a novel fold. Proteins. 2006;62:1138–1143. doi: 10.1002/prot.20864. [DOI] [PubMed] [Google Scholar]
- 35.Kazantsev AV, Krivenko AA, Harrington DJ, Carter RJ, Holbrook SR, Adams PD, Pace NR. High-resolution structure of RNase P-protein from Thermotoga maritima. Proc. Natl Acad. Sci. USA. 2003;100:7497–7502. doi: 10.1073/pnas.0932597100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diges CM, Uhlenbeck OC. Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J. 2001;20:5503–5512. doi: 10.1093/emboj/20.19.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsu CA, Kossen K, Uhlenbeck OC. The Escherichia coli DEAD protein DbpA recognizes a small RNA hairpin in 23S rRNA. RNA. 2001;7:702–709. doi: 10.1017/s1355838201010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kossen K, Karginov FV, Uhlenbeck OC. The carboxy-terminal domain of the DExDH protein YxiN is sufficient to confer specificity for 23S rRNA. J. Mol. Biol. 2002;324:625–636. doi: 10.1016/s0022-2836(02)01140-3. [DOI] [PubMed] [Google Scholar]
- 39.Karginov FV, Caruthers JM, Hu Y, McKay DB, Uhlenbeck OC. YxiN is a modular protein combining a DEx(D/H) core and a specific RNA-binding domain. J. Biol. Chem. 2005;280:35499–35505. doi: 10.1074/jbc.M506815200. [DOI] [PubMed] [Google Scholar]
- 40.Wang S, Hu Y, Overgaard MT, Karginov FV, Uhlenbeck OC, McKay DB. The domain of the Bacillus subtilis DEAD-box helicase YxiN that is responsible for specific binding of 23S rRNA has an RNA recognition motif fold. RNA. 2006;12:959–967. doi: 10.1261/rna.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halls C, Mohr S, Del Campo M, Yang Q, Jankowsky E, Lambowitz AM. Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J. Mol. Biol. 2007;365:835–855. doi: 10.1016/j.jmb.2006.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grohman JK, Del Campo M, Bhaskaran H, Tijerina P, Lambowitz AM, Russell R. Probing the mechanisms of DEAD-box proteins as general RNA chaperones: the C-terminal domain of CYT-19 mediates general recognition of RNA. Biochemistry. 2007;46:3013–3022. doi: 10.1021/bi0619472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohr S, Matsuura M, Perlman PS, Lambowitz AM. A DEAD-box protein alone promotes group II intron splicing and reverse splicing by acting as an RNA chaperone. Proc. Natl Acad. Sci. USA. 2006;103:3569–3574. doi: 10.1073/pnas.0600332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang HR, Rowe CE, Mohr S, Jiang Y, Lambowitz AM, Perlman PS. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc. Natl Acad. Sci. USA. 2005;102:163–168. doi: 10.1073/pnas.0407896101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mohr G, Del Campo M, Mohr S, Yang Q, Jia H, Jankowsky E, Lambowitz AM. Function of the C-terminal domain of the DEAD-box protein Mss116p analyzed in vivo and in vitro. J. Mol. Biol. 2008;375:1344–1364. doi: 10.1016/j.jmb.2007.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco I., Jr, Pyle AM, Bustamante C. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Serebrov V, Pyle AM. Periodic cycles of RNA unwinding and pausing by hepatitis C virus NS3 helicase. Nature. 2004;430:476–480. doi: 10.1038/nature02704. [DOI] [PubMed] [Google Scholar]
- 48.Locatelli GA, Spadari S, Maga G. Hepatitis C virus NS3 ATPase/helicase: an ATP switch regulates the cooperativity among the different substrate binding sites. Biochemistry. 2002;41:10332–10342. doi: 10.1021/bi026082g. [DOI] [PubMed] [Google Scholar]
- 49.Yao N, Hesson T, Cable M, Hong Z, Kwong AD, Le HV, Weber PC. Structure of the hepatitis C virus RNA helicase domain. Nat. Struct. Biol. 1997;4:463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]
- 50.Wang S, Overgaard MT, Hu Y, McKay DB. The Bacillus subtilis RNA helicase YxiN is distended in solution. Biophys. J. 2008;94:L01–L03. doi: 10.1529/biophysj.107.120709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Talavera MA, Matthews EE, Eliason WK, Sagi I, Wang J, Henn A, De La Cruz EM. Hydrodynamic characterization of the DEAD-box RNA helicase DbpA. J. Mol. Biol. 2006;355:697–707. doi: 10.1016/j.jmb.2005.10.058. [DOI] [PubMed] [Google Scholar]
- 52.Jaenicke R, Schurig H, Beaucamp N, Ostendorp R. Structure and stability of hyperstable proteins: glycolytic enzymes from hyperthermophilic bacterium Thermotoga maritima. Adv. Protein Chem. 1996;48:181–269. doi: 10.1016/s0065-3233(08)60363-0. [DOI] [PubMed] [Google Scholar]
- 53.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.