Abstract
In eukaryotic cells translation initiation occurs through two alternative mechanisms, a cap-dependent operating in the majority of mRNAs, and a 5′-end-independent driven by internal ribosome entry site (IRES) elements, specific for a subset of mRNAs. IRES elements recruit the translation machinery to an internal position in the mRNA through a mechanism involving the IRES structure and several trans-acting factors. Here, we identified Gemin5 protein bound to the foot-and-mouth disease virus (FMDV) and hepatitis C virus (HCV) IRES using two independent approaches, riboproteomic analysis and immunoprecipitation of photocroslinked factors. Functional analysis performed in Gemin5 shRNA-depleted cells, or in in vitro translation reactions, revealed an unanticipated role of Gemin5 in translation control as a down-regulator of cap-dependent and IRES-driven translation initiation. Consistent with this, pull-down assays showed that Gemin5 forms part of two distinct complexes, a specific IRES-ribonucleoprotein complex and an IRES-independent protein complex containing eIF4E. Thus, beyond its role in snRNPs biogenesis, Gemin5 also functions as a modulator of translation activity.
INTRODUCTION
Translational control constitutes a major step in gene expression regulation. Initiation of translation in eukaryotes is the rate-limiting step of protein synthesis and involves a set of specialized proteins known as initiation factors (eIFs) that recruit the small ribosome subunit to the m7GTP residue (or cap), located at the 5′-end of most mRNAs (1). The cap-dependent initiation complex scans along the 5′ untranslated regions (UTR) until an AUG codon placed in the appropriate context is recognized by the translation machinery to start protein synthesis (2). Not surprisingly, the 5′ UTR of mRNAs, in concerted action with the 3′ UTR, play a key role in this process serving as platforms for the formation of macromolecular complexes controlling translation initiation.
In contrast to the general cap-dependent mechanism of translation initiation, some cytoplasmic RNA viruses, such as picornaviruses, initiate translation internally (3,4), independently of the 5′-end and bypassing stable RNA structures present on their 5′ UTRs and proteins bound to it. Internal initiation of translation in eukaryotic cells is mediated by cis-acting RNA structures termed internal ribosome entry site (IRES) elements that recruit the translational machinery to an internal position in the mRNA of some viruses (5) and a specific subset of cellular mRNAs (6,7). However, because of the lack of conservation at the level of primary sequence, RNA structure and transacting factor requirements, the general principles governing internal initiation of translation are still poorly understood.
Picornavirus IRES activity relies on the interaction with cellular proteins including translation eIFs and IRES trans-acting factors (ITAFs) (8). Hepatitis C virus (HCV) RNA also initiates translation using a cap-independent mechanism (9,10). Despite performing a similar function, HCV and picornavirus IRES elements differ in primary sequence, RNA structure and ITAFs requirement (11).
Most ITAFs are RNA-binding proteins that regulate RNA life-spam as part of macromolecular complexes operating either in the nucleus or in the cytoplasm of the cell (12). Some of these factors travel with the RNA and are responsible for the establishment and regulation of complex RNA–protein networks that determine the fate of the target mRNA (13).
To gain insights into the mechanism of internal translation initiation, we compared the proteins associated with two unrelated viral IRES elements. We report that two ITAFs with an apparent mobility of 170 kDa correspond to eIF3a and Gemin5. The interaction with foot-and-mouth disease virus (FMDV) and HCV IRES elements is direct and sequence-specific. We also found that Gemin5 forms an RNA-independent protein complex with eIF4E. Functional analysis addressing the role of Gemin5 showed that this IRES-binding factor down-regulated IRES activity. Furthermore, Gemin5 also negatively affected cap-dependent initiation, suggesting that this protein acts as a general down-regulator of translation.
MATERIALS AND METHODS
Constructs
Plasmids expressing different domains of FMDV and HCV IRES elements have been already described (14).
The dicistronic constructs CMVpBIC and CMVp156, bearing either the FMDV or the HCV IRES upstream of the luciferase coding region, were constructed by transferring the MluI-blunt NotI fragment from pBIC-AvrII-Not (15) into the NheI-blunt NotI pCDNA3 vector.
The Gemin5 (pSUPER-GFP-Gemin5) or the polypyrimidine (PTB) silencing plasmids (pSUPER-GFP-PTB) [target sequences GCAUAGUGGUGAUAAUUGA (16) or AACTTCCATCATTCCAGAGAA (17), respectively], were generated according to the manufacturer instructions (OligoEngine). The control shRNA expressed from pSUPER-GFP-TM (18) (target sequence AATTCTCCGAACGTGTCACGT) had no homology to any mammalian gene.
RNA synthesis
T7 transcription was performed at 37°C for 1 h using the Megashortscript kit (Ambion) as recommended by the manufacturer. When needed, transcripts were uniformly labeled using α32P-CTP (400 Ci/mmol). DNA template was digested with 1 U of RQ1 DNase (Promega) and unincorporated α32P-CTP eliminated by exclusion chromatography (19). RNA was extracted with phenol–chloroform, ethanol precipitated and resuspended in TE to a concentration of 0.03 pmol/µl (∼4 × 105 cpm/µl). RNA integrity was examined in 6% acrylamide 7 M urea denaturing gel electrophoresis (20).
Bicistronic RNAs of the form cloranfenicol acetyl transferase (CAT)—IRES—luciferase (LUC) corresponding to the FMDV or the HCV IRES were produced in vitro as described (21,22).
RNA affinity
Transcripts (2.5 µg) encompassing domain 5 (d5), domain 3 (d3), the entire IRES of FMDV, or domain III (dIII) of HCV IRES, with extension of 15 adenines at their 3′-end were incubated with oligo-dT dynabeads (Dynal) in binding buffer (25 µl) [10 mM Tris (pH 7.5), 100 mM KCl, 2 mM MgCl2], at 4°C for 30 min on a rotating wheel as described (23). Unbound RNA was removed and the beads–RNA complex washed twice with binding buffer. The beads–RNA complexes were then incubated with protein extracts in the presence of unspecific RNA competitor, at 4°C for 1 h. Unbound proteins were eliminated by washing with binding buffer, followed by two washes with 10 mM Tris (pH 7.5), 100 mM KCl, 0.5 mM MgCl2. The retained products were then fractionated in SDS–PAGE, and the bands of interest processed for mass spectrometry analysis.
In gel digestion and mass spectrometry
To prepare samples for ESI-Q-TOF, slices from Coomassie blue stained gels were subjected to in-gel digestion (24) using a ProGest Investigator robot (Genomic Solutions, Ann Arbor, MI). Peptides were separated using an UltiMate nanoLC (LC Packings, Amsterdam) equipped with a PepMap C18 trap and column. The eluent was sprayed into a Q-Star Pulsar XL tandem mass spectrometer (Applied Biosystems, Foster City, CA). The MS/MS data file generated was analyzed using the Mascot search engine against MSDB. In all the cases analyzed, the experimental Mr values were in good agreement with the theoretical values for identified proteins. Sequence, score Mascot and M/H+ of matched peptides are given in Supplementary Tables S1 and S2.
RNA–protein photocrosslinking
For cell extract preparation, BHK-21, MDBK, IBRS-2, HeLa, or HEK293T cells were grown to 100% confluence in 10 cm dishes in 5% calf serum supplemented DMEM, washed twice with cold phosphate buffer saline (PBS), scraped, collected by centrifugation and processed as described (25). Uniformly radiolabeled probes (0.03 pmol, ∼4 × 105 cpm) were incubated with S10 cell extracts (40 µg protein) and UV-irradiated as described (14). To increase the specificity of binding in the UV-crosslinking assay, yeast tRNA (1 µg/µl, ∼100-fold molar excess) or 10–1000-fold molar excess of total cytoplasmic RNA was added to the reaction. Following RNase treatment, samples were subjected to SDS–PAGE and 32P-labeled proteins were visualized by autoradiography of dried gels.
Immunodetection
Immunoprecipitation of RNA–protein complexes was performed using antibodies recognizing eIF3, PTB, PABP1, Gemin5, DHX9 or PA2G4 (14,17,18,26–29). Antibodies recognizing Gemin5 (Santa Cruz), hnRNP U (Immuquest), eIF4E (BD Transduction Laboratories), tubulin (Sigma), and appropriate peroxidase conjugated secondary antibodies (Pierce) were used according to the manufacturer instructions.
m7GTP affinity
HEK293T or HeLa cell extracts, photocrosslinked with the indicated RNAs, were subjected to m7GTP–sepharose (Sigma) chromatography, using 10 µl of resin at 4°C for 1 h. Unbound proteins were eliminated by five washes with RIPA buffer [50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1% NaDOC], and once with TBS [20 mM Tris–HCl (pH 7.5), 140 mM NaCl]. Proteins retained in the resin were analyzed in SDS–PAGE followed by Western blotting.
RNA interference
For gene silencing, HEK293T cells were transfected with pSUPER-GFP-Gemin5, pSUPER-GFP-PTB or pSUPER-GFP-TM. In parallel to verification of protein depletion by Western blot, the effect of protein depletion on IRES activity was assessed by cotransfecting CMVpBIC (FMDV IRES) or CMVp156 (HCV IRES) plasmids together with the shRNA expressing constructs. Relative IRES activity of bicistronic mRNAs expressed from the CMV promoter was quantified as the expression of luciferase (Luc) normalized to cloranfenicol acetyltransferase (CAT), as described (30). Each experiment was repeated at least three times.
The relative amounts of reporter bicistronic RNAs present in shRNA transfected cells were measured by real time RT–PCR using oligonucleotides specific for the FMDV IRES (5′-GCCTGTCACCAGTGTGTGGGTACCAG-3′ and 5′-GGGGTAACACTGAATTCTGTGTTTGGCTCCACG-3′) or the HCV IRES (5′-CGGCGAAGCTTGTTACGTTTGGTTTTTC-3′ and 5′-CCAGGAATTCCTCCCGGGATGCCTGATAG-3′), respectively. Total RNA from cytoplasmic extracts was extracted using the Tripure reagent (Roche) and RT–PCR was carried out with the SuperScriptII RT (Invitrogen) using 50 ng of RNA as template and the reporter specific primer pBIC-as (5′-GGCCTTTCTTTATGTTTTTGGCG-3′) (30). The LightCycler system with the FastStart DNA Master Green I (Roche) was used. The data were generated from duplicates of three independent experiments.
In vitro translation
Gemin5 transcript synthesized in vitro (300 ng) was translated in 70% rabbit reticulocyte lysate (RRL) (Promega) supplemented with 10 µCi of 35S-methionine 20 min prior to addition of either of the bicistronic RNA (200 ng) bearing the FMDV or HCV IRES as described (21).
RESULTS
Identification of novel IRES transacting factors
To identify new ITAFs and seek for similarities or differences in IRES function, we conducted a proteomic analysis of complexes formed on IRES sequences from two RNA viruses, FMDV and HCV (11). In the case of FMDV IRES, we focused our attention on domain 5 (d5), which in contrast to a control RNA, specifically associated with many of the proteins detected with the entire IRES (Figure 1A and Supplementary Figure S1A). FMDV d5 consists of a stem-loop followed by an unpaired region upstream of the first start codon of the viral RNA (31,32) (Supplementary Figure S1B). Likewise, domain III (dIII) of the HCV IRES (Supplementary Figure S1C) was used in this study, as it is involved in the interaction with eIF3 and the 40S ribosomal subunit (10).
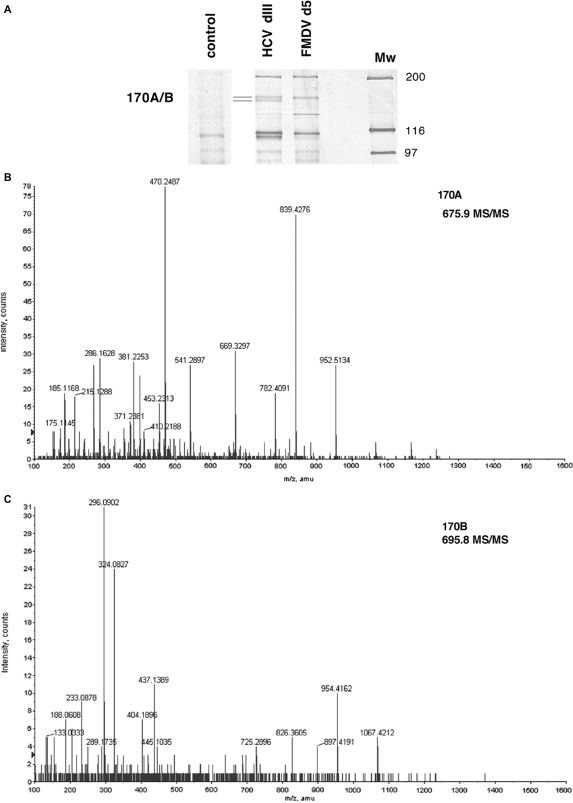
Figure 1.
Identification of proteins interacting with the FMDV and HCV IRES. (A) RNA affinity chromatography. Silver stained 7% SDS–PAGE loaded with proteins bound to FMDV d5, HCV dIII IRES, or a control RNA sequences. (B) Mass spectrometry analysis of the p170 interacting with the HCV IRES. A detail of the 675.9 Da peptide MS/MS spectrum, matched to the sequence LATLLGLQAPPTR of eIF3a. (C) Detail of the 695.8 Da peptide MS/MS spectrum, matched to the sequence CYLGATSAYDAAK of Gemin5.
Figure 1A shows the pattern of high molecular weight proteins bound to either FMDV d5 or HCV dIII transcripts in RNA affinity pull-down assays. The sequence of the polypeptides present in the 170A/B kDa doublet obtained by ESI-Q-TOF revealed that these two bands correspond to the eIF3a subunit of eIF3 (170A, Supplementary Table S1 and Figure 1B) and Gemin5 (170B, Figure 1C and Supplementary Table S2). Identification of eIF3a is consistent with the role of eIF3 in the assembly of the 48S initiation complex on HCV IRES (9), as well as with the direct interaction of the eIF3b/c subunit (p116/110) to HCV IRES domain III (14). Gemin5 is the RNA-binding factor of the survival motor neuron (SMN) complex (16). This complex assembles the Sm proteins on small nuclear RNAs (snRNAs) and plays a critical role in the biogenesis of small nuclear ribonucleoproteins (snRNP), the essential components of the mRNA splicing machinery (33). Hence, identification of Gemin5 as a new FMDV and HCV IRES-binding factor suggested an unanticipated role of this protein in translational control.
p170A/B are ubiquitous polypeptides that bind directly to IRES elements
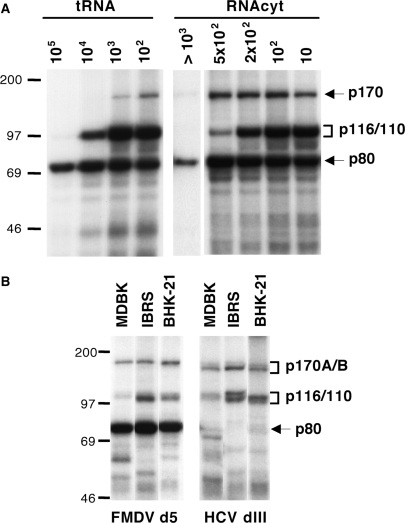
The RNA affinity method described in Figure 1A does not reveal which of the proteins pulled-down directly interacts with the RNA. To address this, we performed photocrosslinking assays using 32P-labeled RNAs incubated with soluble cell extracts, as this method only reveals factors covalently linked to the probe. Three polypeptides interact with FMDV IRES domain 5 in the presence of a large excess of tRNA: p170, p116/110 and p80 (Figure 2A). Addition of total cytoplasmic RNA (RNAcyt) to the binding mixture increased the intensity of p170 without affecting other factors (Figure 2A). The p80 and p116/110 polypeptides have been previously identified as eIF4B and eIF3b/c (14). Thus, in this report we focused on the characterization of the p170 band.
Figure 2.
Effect of unspecific competitors on the IRES–protein interaction pattern. (A) Photocrosslinking RNA–protein pattern obtained with radiolabeled domain 5 of the FMDV IRES, using BHK-21 S10 extracts in the presence of increasing amounts of tRNA or total cytoplasmic RNA (RNAcyt). (B) UV-crosslinking RNA–protein pattern of radiolabeled FMDV d5 and HCV dIII in the presence of 5 × 102 molar excess cytoplasmic RNA. Autoradiography of proteins resolved in 8% SDS–PAGE.
Since FMDV and HCV IRES elements function in several mammalian cell types (34,35), we compared the binding of the p170 doublet using cell extracts from various cell lines in the presence of cytoplasmic RNA. While the HCV probe was preferentially crosslinked to the lower p170 band, a more intense crosslink to the upper p170 band was detected with FMDV RNA. In both cases, the p170A/B-IRES crosslink was observed with all cell extracts tested, indicating that this doublet contains ubiquitous proteins able to bind directly to FMDV and HCV IRES elements.
Gemin5 binds directly to IRES elements as a complex with other factors
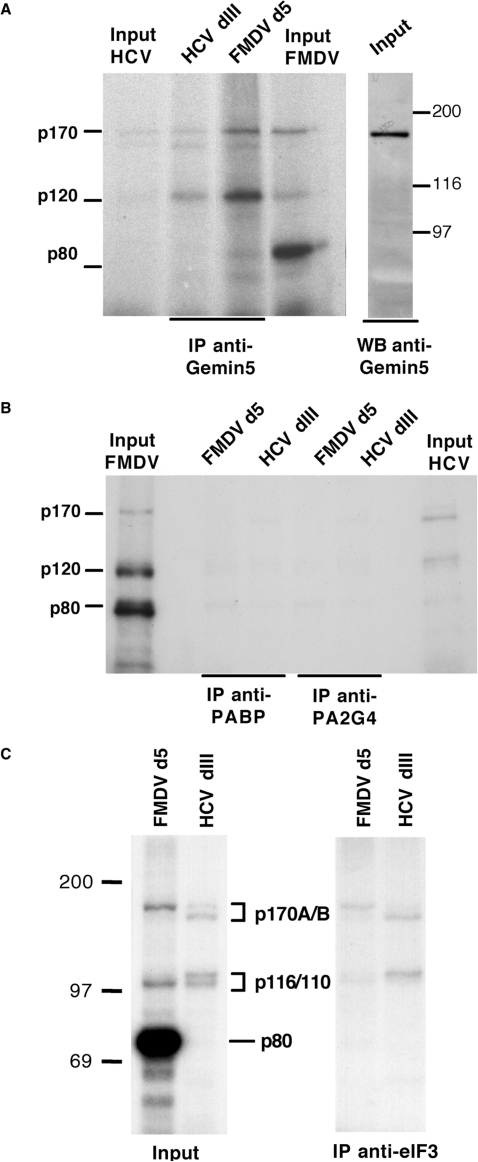
To verify if the p170 products detected in photocrosslinking studies were the same peptides identified by mass spectrometry, we conducted immunoprecipitation of the proteins crosslinked to radiolabeled RNA probes. Recognition of p170 photocrosslinked to the HCV and FMDV IRES by anti-Gemin5 antibodies confirmed its identity (Figure 3A). Strikingly, despite the specific recognition of a single polypeptide in Western blot assays (right panel in Figure 3A), a p170 doublet bound to both FMDV and HCV IRES probes was coimmunoprecipitated with anti-Gemin5 antibody, together with an additional p120 band (Figure 3A). In contrast, and in support of the specificity of the immunoprecipitation observed in Figure 3A, antibodies recognizing other IRES-interacting proteins, such as the poly(A)-binding protein 1 (PABP1) or the proliferation-associated 2G4 (PA2G4), also termed ErbB3-binding protein 1 (EbP1) (23,36), failed to coimmunoprecipitate p170 photocrosslinked to these RNAs (Figure 3B). A similar result was obtained using anti-PTB or anti-DEAH-box polypeptide 9 (DHX9) antibodies (data not shown).
Figure 3.
Gemin5 binds directly to FMDV and HCV IRES as part of a protein complex. (A) Anti-Gemin5 immunoprecipitation of S10 extracts from BHK-21 cells photocrosslinked to radiolabeled FMDV d5 or HCV dIII (left) and Western blot analysis of S10 extracts using anti-Gemin5 antibodies (right panel). (B) Immunoprecipitation of S10 extracts prepared from HeLa cells photocrosslinked to radiolabeled FMDV d5 or HCV dIII with anti-PABP and anti-PA2G4 antibodies. (C) Immunoprecipitation of S10 extracts prepared from BHK-21 cells photocrosslinked to radiolabeled FMDV d5 or HCV dIII UV-crosslinked proteins using anti-eIF3 serum. In all cases the input corresponds to 5% of the immunoprecipitation sample.
Confirming the identification of eIF3a in the proteomic analysis, p170 interacting with both FMDV d5 and HCV dIII IRES regions were immunoprecipitated using anti-eIF3 sera (Figure 3C). Note that p170 coimmunoprecipitated with the p116/110 doublet corresponding to eIF3b,c (14). Lack of p80 detection again verified the specificity of the immunoprecipitation. Interestingly, and similarly to the results obtained with the Gemin5 antibody, a p170 kDa doublet was pull-down with the eIF3 antibody (Figure 3C). Collectively, these results indicate that Gemin5 forms part of a protein complex bound to the IRES elements of FMDV and HCV viral RNAs that contain a specific subset of IRES-binding proteins including eIF3a,b,c.
Gemin5 functions as a down-regulator of translation
To analyze the role of Gemin5 in IRES-driven translation, we used two different experimental approaches. In the first one, we tested the effect of Gemin5 protein in in vitro translation assays programmed with bicistronic RNAs. In the second one, we used gene silencing to test the effect of Gemin5 depletion in the translation efficiency of bicistronic constructs transiently transfected in cells.
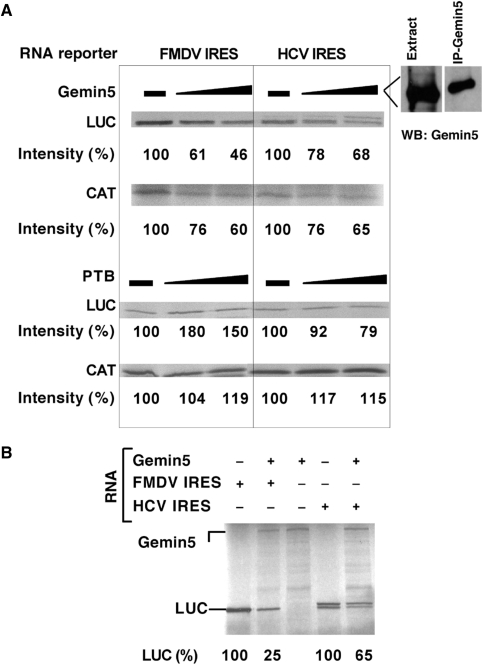
Addition of immunoprecipitated Gemin5 protein from BHK-21 cells to the in vitro translation reaction resulted in lower IRES activity (Figure 4A, upper panels), regardless of the IRES used to drive luciferase synthesis, FMDV or HCV. In addition, a moderate decrease of cap-dependent CAT synthesis was observed (Figure 4A, lower panels). As a control, addition of purified PTB did not induce a decrease in translation efficiency, and as expected (37), a stimulatory effect on FMDV IRES-driven translation was observed. Furthermore, expression of Gemin5 in reticulocyte lysates prior to the addition of the IRES-containing RNA decreased FMDV and HCV IRES efficiency (Figure 4B). Coexpression of an unrelated protein, or addition of a control unrelated protein did not inhibit translation (data not shown). Altogether, our results therefore indicate that Gemin5 is a novel IRES-binding factor that causes a reduction in translation capacity of eukaryotic mRNAs.
Figure 4.
Role of Gemin5 in cap- and IRES-dependent translation in vitro. (A) Increasing amounts of Gemin5 protein, purified by immunoprecipitation from BHK-21 cells, was added to RRL programmed with equal amounts of bicistronic RNAs bearing the FMDV or the HCV IRES in the intercistronic region. The intensity of 35S-labeled Luciferase (luc) (IRES-dependent translation) and cloranfenicol acetyl transferase (Cat) (5′-end-dependent translation) proteins is indicated below each lane. Likewise, recombinant PTB (43) was included in a parallel assay. (B) Equal amounts of bicistronic RNAs, synthesized in vitro, were added to RRL after 20 min of Gemin5 RNA incubation, prepared from plasmid Gemin5 V5-His (27,39). 35S-labeled proteins were resolved in 8% SDS–PAGE.
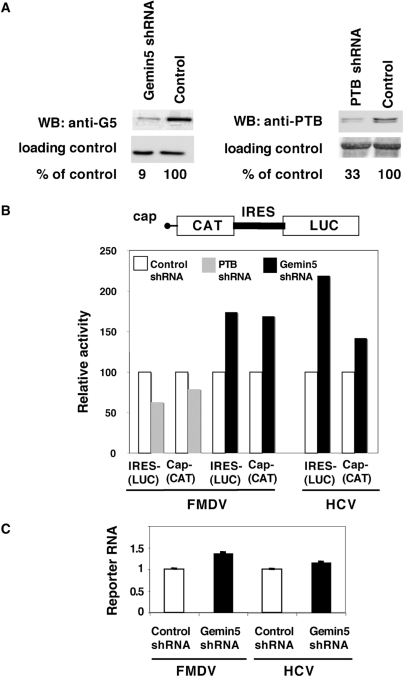
Next, to confirm the role of Gemin5 in translation regulation and expand this result to an in vivo system, we measured IRES activity in cells depleted of Gemin5. Transient expression of short hairpins RNA (shRNA) in HEK293T cells led to a 90% decrease in Gemin5 levels (Figure 5A). As a positive control, we also expressed shRNA against PTB, a RNA-binding protein required for picornavirus IRES activity (38).
Figure 5.
Role of Gemin5 in IRES activity assessed by gene silencing. (A) Western blot analysis of HEK293T cell extracts expressing shRNAs targeted to Gemin5, PTB, or a shRNA control with no target sequence in mammalian mRNAs. Tubulin was used as loading control. (B) IRES- and cap-dependent translation in Gemin5-depleted cells upon transfection with equal amounts of the bicistronic plasmids (see diagram at the top). PTB-depleted cells were used as positive control. Each experiment was repeated at least three times. Activity was calculated as the % of the values observed in the control shRNA. (C) Real time RT–PCR analysis of reporter RNAs in control and Gemin5 shRNAs silenced cells. The relative amount of RNA was determined by duplicate assays of three independent experiments.
Compared to cells expressing a negative control shRNA, Gemin5 depletion in cells transfected with a bicistronic plasmid containing FMDV IRES, led to an increase in IRES-dependent luciferase synthesis, as well as cap-dependent CAT synthesis (Figure 5B). This result is fully consistent with the in vitro translation assays shown in Figure 4. Similar results were obtained in Gemin5 depleted cells expressing a bicistronic plasmid containing the HCV IRES. In contrast, PTB depletion reduced FMDV IRES activity ∼2-fold (Figure 5B), consistent with PTB protein positively influencing FMDV IRES function (37). As shown in Figure 5C, no differences in the amount of reporter RNAs were detected by real time RT–PCR in Gemin5 shRNA silenced cells relative to that observed in control shRNA cells, discarding an effect of Gemin5 in RNA stability. These results indicate that a reduction in Gemin5 levels favor both cap-dependent and IRES-dependent translation, and therefore argue for a general translation down-regulatory role of Gemin5 in translation.
Gemin5 also forms part of an IRES-independent protein complex
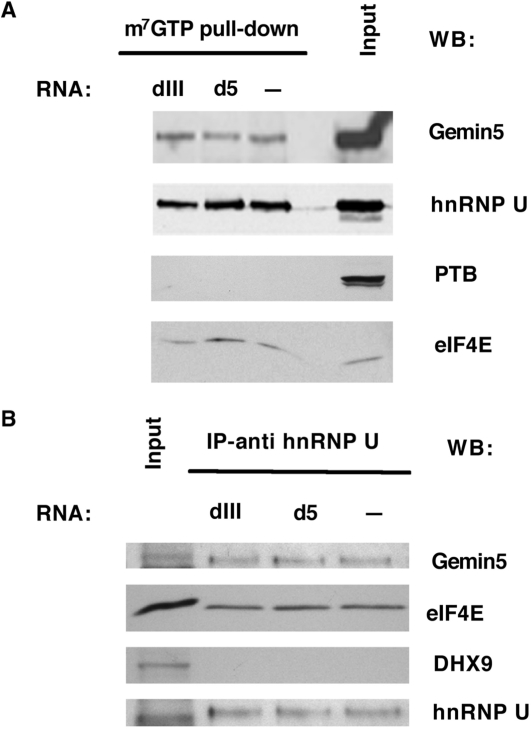
Gemin5 has been reported to interact with the cap-binding factor eIF4E (39) which can be retained on m7GTP–sepharose resins. In an attempt to gain insights into Gemin5-associated factors, we used m7GTP–sepharose to bind eIF4E and thus, eIF4E-interacting proteins. Western blot analysis of the input and pull-down samples demonstrated that the resin was able to retain most of the eIF4E protein present in the extract (Figure 6A, bottom panel). Immunodetection of Gemin5 revealed its ability to associate with eIF4E, independently of HCV and FMDV RNAs. These samples also contained the heterogeneous nuclear ribonucleoprotein U (hnRNP U), in agreement with previous data showing the association of hnRNPs to the SMN complex (40). However, the IRES-binding factor PTB (hnRNP I) was not pulled down in these assays (Figure 6A). As these samples were treated with RNase A prior to its incubation with the resin, it is likely that this analysis detects protein–protein interactions not mediated by RNA.
Figure 6.
Characterization of Gemin5-interacting proteins. (A) Western blot analysis of HEK293T cell extracts photocrosslinked to unlabeled d5 or dIII RNAs and subsequently bound to m7-GTP resins. (B) Western blot analysis of proteins associated to d5 or dIII unlabeled RNAs following anti-hnRNPU immunoprecipitation of the photocrosslinked ribonucleoprotein complexes assembled with HeLa cell extracts.
Conversely, immunoprecipitation of photocrosslinked RNA complexes with anti-hnRNP U serum revealed the presence of both Gemin5 and eIF4E, whereas other IRES-binding proteins e.g. the RNA helicase DHX9 (also known as RHA) (41) were absent (Figure 6B). This result indicates that the protein complex analyzed here is different from that associated with the SMN complex, which contains RHA (40). Our results therefore show that FMDV and HCV IRES sequences form a Gemin5-containing complex together with a specific set of IRES-binding proteins such as eIF3 polypeptides. Additionally, Gemin5 forms an IRES-independent but eIF4E-dependent complex that also contains hnRNPU. Proteins known to interact with IRES elements, such as PTB or DHX9 are absent in the eIF4E-dependent/Gemin5 complex. Thus, Gemin5 forms part of at least two different macromolecular complexes, beyond the well-described SMN complex.
DISCUSSION
In this study we have identified Gemin5 as a novel IRES-binding factor that contributes to down-regulate translation efficiency. Two independent approaches, riboproteomic analysis and photocrosslinking assays, aided by immunoprecipitation with specific antibodies, led to the identification of a doublet of about 170 kDa as eIF3a and Gemin5 interacting with the IRES element of HCV and FMDV RNA. While identification of eIF3a as an IRES-binding factor confirms previous results (14,42), the finding that Gemin5 is an ITAF was unprecedented. Functional analysis involving gene silencing in transfected cells, as well as co-expression or addition of the protein to in vitro translation assays, underscored a novel function of Gemin5 protein in translation control. The higher cap-dependent and IRES-dependent translation efficiencies in Gemin5-depleted cells, together with the lower efficiency of protein synthesis in vitro upon addition of increasing Gemin5 levels, indicate that Gemin5 acts as a down-regulator of translation.
It is worth noting that the repressor effect of Gemin5 in mRNA translation, although moderate, has been reproduced in different experimental settings. Furthermore, the magnitude of this effect in transfected cells is comparable to that of a well characterized ITAF whose activity stimulates IRES function, such as PTB (43,44). Gemin5 translational down-regulation may become more significant in the context of the highly competitive cytoplasmic environment, a situation that may be further compromised during picornavirus infection (11). Although studies of this protein during infection await further work, it is interesting that Gemin3, which is the RNA helicase of the SMN complex, has been recently found to be a target of picornavirus proteases (45).
Proteins acting as translation repressors often recognize 3′ UTR sequences in their target mRNAs (46). However, and although rare, IRES-binding proteins acting as inhibitors of translation have recently been reported (47). Gemin5 is the RNA-binding factor of the SMN complex (16), which assembles Sm proteins on snRNAs and, thus, plays a critical role in the biogenesis of key components of the mRNA splicing machinery, such as snRNPs (33). Thus, the role of Gemin5 in translation regulation represents an unanticipated and novel finding.
Gemin5 is found mainly in the cell cytoplasm (48). The excess of free Gemin5 is therefore consistent with a more general role of this protein. In this scenario, Gemin5 may recruit factors to mRNAs and thus, regulate their translation. The presence of WD repeats and a coiled-coil motif in Gemin5 (27) could mediate these interactions. Specifically, the eIF4E-Gemin5 interaction, mediated by a YXXXLØ motif, was recently described (39).
The binding of Gemin5 to viral IRES elements is unprecedented. Despite the fact that HCV and FMDV viral RNA are never found in the nuclei of infected cells, complexes formed on IRES elements share components with the splicing machinery. Involvement of splicing factors and hnRNPs in internal initiation has been reported for SRp20 (49), SF2/ASF (50), PTB (hnRNP I) (37,43), hnRNP A1 (51), or hnRNP Q (52). Most of the identified ITAFs are very abundant and shuttle between the nucleus and the cytoplasm.
Collectively, we have shown that Gemin5 acts as a translation regulator, repressing both cap-dependent and IRES-driven translation through two different pathways. The cap-dependent down-regulatory effect of Gemin5 may rely on eIF4E–Gemin5 interaction (39), possibly resulting in the sequestration of this rate limiting initiation factor. Importantly, we have discovered that Gemin5 directly binds to two different viral IRES elements and form a complex with other IRES-binding proteins, such as eIF3a,b,c and hnRNP U, which may explain its IRES-dependent down-regulation ability. Our findings will be instrumental to determine whether Gemin5 also binds and down-regulates translation initiation driven by other IRES elements.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by grant BFU-2005-00948 and by an Institutional grant from Fundación Ramón Areces. Funding for open access charge: Spanish Ministry of Science and Education.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are indebted to J Hershey, G Dreyfuss, M Garcia Blanco, G Squatrito, R Godbout, A Nieto and I Fierro-Monti for their generosity in sharing reagents, to JF Santaren and K Botting for help with the proteomic analysis. We also thank S. Reigadas for early contribution to this work, and B. Desvoyes and C. Gutierrez for helpful suggestions.
REFERENCES
- 1.Sonenberg N, Dever TE. Eukaryotic translation initiation factors and regulators. Curr. Opin. Struct. Biol. 2003;13:56–63. doi: 10.1016/s0959-440x(03)00009-5. [DOI] [PubMed] [Google Scholar]
- 2.Pestova TV, Lorsch JR, Hellen CUT. The mechanism of translation initiation in eukaryotes. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translation Control in Biology and Medicine. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2007. pp. 87–128. [Google Scholar]
- 3.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 4.Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez-Salas E. The impact of RNA structure on picornavirus IRES activity. Trends Microbiol. 2008;16:230–237. doi: 10.1016/j.tim.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baird SD, Turcotte M, Korneluk RG, Holcik M. Searching for IRES. RNA. 2006;12:1755–1785. doi: 10.1261/rna.157806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spriggs KA, Bushell M, Mitchell SA, Willis AE. Internal ribosome entry segment-mediated translation during apoptosis: the role of IRES-trans-acting factors. Cell Death Differ. 2005;12:585–591. doi: 10.1038/sj.cdd.4401642. [DOI] [PubMed] [Google Scholar]
- 8.Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI, Hellen CU. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl Acad. Sci. USA. 2001;98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser CS, Doudna JA. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat. Rev. Microbiol. 2007;5:29–38. doi: 10.1038/nrmicro1558. [DOI] [PubMed] [Google Scholar]
- 11.Martinez-Salas E, Pacheco A, Serrano P, Fernandez N. New insights into internal ribosome entry site elements relevant for viral gene expression. J. Gen. Virol. 2008;89:611–626. doi: 10.1099/vir.0.83426-0. [DOI] [PubMed] [Google Scholar]
- 12.Semler BL, Waterman ML. IRES-mediated pathways to polysomes: nuclear versus cytoplasmic routes. Trends Microbiol. 2008;16:1–5. doi: 10.1016/j.tim.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Lopez de Quinto S, Lafuente E, Martinez-Salas E. IRES interaction with translation initiation factors: functional characterization of novel RNA contacts with eIF3, eIF4B, and eIF4GII. RNA. 2001;7:1213–1226. doi: 10.1017/s1355838201010433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez de Quinto S, Saiz M, de la Morena D, Sobrino F, Martinez-Salas E. IRES-driven translation is stimulated separately by the FMDV 3′-NCR and poly(A) sequences. Nucleic Acids Res. 2002;30:4398–4405. doi: 10.1093/nar/gkf569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Battle DJ, Lau CK, Wan L, Deng H, Lotti F, Dreyfuss G. The Gemin5 protein of the SMN complex identifies snRNAs. Mol. Cell. 2006;23:273–279. doi: 10.1016/j.molcel.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 17.Wollerton MC, Gooding C, Wagner EJ, Garcia-Blanco MA, Smith CW. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell. 2004;13:91–100. doi: 10.1016/s1097-2765(03)00502-1. [DOI] [PubMed] [Google Scholar]
- 18.Burgui I, Yanguez E, Sonenberg N, Nieto A. Influenza virus mRNA translation revisited: is the eIF4E cap-binding factor required for viral mRNA translation? J. Virol. 2007;81:12427–12438. doi: 10.1128/JVI.01105-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez-Miragall O, Martinez-Salas E. Structural organization of a viral IRES depends on the integrity of the GNRA motif. RNA. 2003;9:1333–1344. doi: 10.1261/rna.5950603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandez-Miragall O, Ramos R, Ramajo J, Martinez-Salas E. Evidence of reciprocal tertiary interactions between conserved motifs involved in organizing RNA structure essential for internal initiation of translation. RNA. 2006;12:223–234. doi: 10.1261/rna.2153206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez de Quinto S, Martinez-Salas E. Involvement of the aphthovirus RNA region located between the two functional AUGs in start codon selection. Virology. 1999;255:324–336. doi: 10.1006/viro.1999.9598. [DOI] [PubMed] [Google Scholar]
- 22.Lafuente E, Ramos R, Martinez-Salas E. Long-range RNA-RNA interactions between distant regions of the hepatitis C virus internal ribosome entry site element. J. Gen. Virol. 2002;83:1113–1121. doi: 10.1099/0022-1317-83-5-1113. [DOI] [PubMed] [Google Scholar]
- 23.Pacheco A, Reigadas S, Martinez-Salas E. Riboproteomic analysis of polypeptides interacting with the internal ribosome entry site element of foot-and-mouth disease virus. Proteomics. 2008;8:4782–4790. doi: 10.1002/pmic.200800338. [DOI] [PubMed] [Google Scholar]
- 24.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 25.Lopez de Quinto S, Martinez-Salas E. Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA. 2000;6:1380–1392. doi: 10.1017/s1355838200000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez Pulido M, Serrano P, Saiz M, Martinez-Salas E. Foot-and-mouth disease virus infection induces proteolytic cleavage of PTB, eIF3a,b, and PABP RNA-binding proteins. Virology. 2007;364:466–474. doi: 10.1016/j.virol.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 27.Gubitz AK, Mourelatos Z, Abel L, Rappsilber J, Mann M, Dreyfuss G. Gemin5, a novel WD repeat protein component of the SMN complex that binds Sm proteins. J. Biol. Chem. 2002;277:5631–5636. doi: 10.1074/jbc.M109448200. [DOI] [PubMed] [Google Scholar]
- 28.Godbout R, Packer M, Bie W. Overexpression of a DEAD box protein (DDX1) in neuroblastoma and retinoblastoma cell lines. J. Biol. Chem. 1998;273:21161–21168. doi: 10.1074/jbc.273.33.21161. [DOI] [PubMed] [Google Scholar]
- 29.Squatrito M, Mancino M, Donzelli M, Areces LB, Draetta GF. EBP1 is a nucleolar growth-regulating protein that is part of pre-ribosomal ribonucleoprotein complexes. Oncogene. 2004;23:4454–4465. doi: 10.1038/sj.onc.1207579. [DOI] [PubMed] [Google Scholar]
- 30.Lopez de Quinto S, Martinez-Salas E. Conserved structural motifs located in distal loops of aphthovirus internal ribosome entry site domain 3 are required for internal initiation of translation. J. Virol. 1997;71:4171–4175. doi: 10.1128/jvi.71.5.4171-4175.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandez-Miragall O, Martinez-Salas E. In vivo footprint of a picornavirus internal ribosome entry site reveals differences in accessibility to specific RNA structural elements. J. Gen. Virol. 2007;88:3053–3062. doi: 10.1099/vir.0.83218-0. [DOI] [PubMed] [Google Scholar]
- 32.Vagnozzi A, Stein DA, Iversen PL, Rieder E. Inhibition of foot-and-mouth disease virus infections in cell cultures with antisense morpholino oligomers. J Virol. 2007;81:11669–11680. doi: 10.1128/JVI.00557-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Will CL, Luhrmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- 34.Borman AM, Le Mercier P, Girard M, Kean KM. Comparison of picornaviral IRES-driven internal initiation of translation in cultured cells of different origins. Nucleic Acids Res. 1997;25:925–932. doi: 10.1093/nar/25.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saiz JC, Lopez de Quinto S, Ibarrola N, Lopez-Labrador FX, Sanchez-Tapias JM, Rodes J, Martinez-Salas E. Internal initiation of translation efficiency in different hepatitis C genotypes isolated from interferon treated patients. Arch. Virol. 1999;144:215–229. doi: 10.1007/s007050050499. [DOI] [PubMed] [Google Scholar]
- 36.Monie TP, Perrin AJ, Birtley JR, Sweeney TR, Karakasiliotis I, Chaudhry Y, Roberts LO, Matthews S, Goodfellow IG, Curry S. Structural insights into the transcriptional and translational roles of Ebp1. EMBO J. 2007;26:3936–3944. doi: 10.1038/sj.emboj.7601817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andreev DE, Fernandez-Miragall O, Ramajo J, Dmitriev SE, Terenin IM, Martinez-Salas E, Shatsky IN. Differential factor requirement to assemble translation initiation complexes at the alternative start codons of foot-and-mouth disease virus RNA. RNA. 2007;13:1366–1374. doi: 10.1261/rna.469707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Florez PM, Sessions OM, Wagner EJ, Gromeier M, Garcia-Blanco MA. The polypyrimidine tract binding protein is required for efficient picornavirus gene expression and propagation. J. Virol. 2005;79:6172–6179. doi: 10.1128/JVI.79.10.6172-6179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fierro-Monti I, Mohammed S, Matthiesen R, Santoro R, Burns JS, Williams DJ, Proud CG, Kassem M, Jensen ON, Roepstorff P. Quantitative proteomics identifies Gemin5, a scaffolding protein involved in ribonucleoprotein assembly, as a novel partner for eukaryotic initiation factor 4E. J. Proteome Res. 2006;5:1367–1378. doi: 10.1021/pr0504539. [DOI] [PubMed] [Google Scholar]
- 40.Yong J, Wan L, Dreyfuss G. Why do cells need an assembly machine for RNA-protein complexes? Trends Cell Biol. 2004;14:226–232. doi: 10.1016/j.tcb.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 41.Fuller-Pace FV. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–4215. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buratti E, Tisminetzky S, Zotti M, Baralle FE. Functional analysis of the interaction between HCV 5'UTR and putative subunits of eukaryotic translation initiation factor eIF3. Nucleic Acids Res. 1998;26:3179–3187. doi: 10.1093/nar/26.13.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaminski A, Hunt SL, Patton JG, Jackson RJ. Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA. 1995;1:924–938. [PMC free article] [PubMed] [Google Scholar]
- 44.Conte MR, Grune T, Ghuman J, Kelly G, Ladas A, Matthews S, Curry S. Structure of tandem RNA recognition motifs from polypyrimidine tract binding protein reveals novel features of the RRM fold. EMBO J. 2000;19:3132–3141. doi: 10.1093/emboj/19.12.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Almstead LL, Sarnow P. Inhibition of U snRNP assembly by a virus-encoded proteinase. Genes Dev. 2007;21:1086–1097. doi: 10.1101/gad.1535607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richter JD. CPEB: a life in translation. Trends Biochem. Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 47.Merrill MK, Gromeier M. The double-stranded RNA binding protein 76:NF45 heterodimer inhibits translation initiation at the rhinovirus type 2 internal ribosome entry site. J. Virol. 2006;80:6936–6942. doi: 10.1128/JVI.00243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hao le T, Fuller HR, Lam le T, Le TT, Burghes AH, Morris GE. Absence of gemin5 from SMN complexes in nuclear Cajal bodies. BMC Cell Biol. 2007;8:28. doi: 10.1186/1471-2121-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bedard KM, Daijogo S, Semler BL. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J. 2007;26:459–467. doi: 10.1038/sj.emboj.7601494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Michlewski G, Sanford JR, Caceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol. Cell. 2008;30:179–189. doi: 10.1016/j.molcel.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 51.Cammas A, Pileur F, Bonnal S, Lewis SM, Leveque N, Holcik M, Vagner S. Cytoplasmic relocalization of heterogeneous nuclear ribonucleoprotein A1 controls translation initiation of specific mRNAs. Mol. Biol. Cell. 2007;18:5048–5059. doi: 10.1091/mbc.E07-06-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim TD, Woo KC, Cho S, Ha DC, Jang SK, Kim KT. Rhythmic control of AANAT translation by hnRNP Q in circadian melatonin production. Genes Dev. 2007;21:797–810. doi: 10.1101/gad.1519507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.