Abstract
Single-stranded DNA-binding proteins (SSB) form a class of proteins that bind preferentially single-stranded DNA with high affinity. They are involved in DNA metabolism in all organisms and serve a vital role in replication, recombination and repair of DNA. In this report, we identify human mitochondrial SSB (HmtSSB) as a novel protein-binding partner of tumour suppressor p53, in mitochondria. It binds to the transactivation domain (residues 1–61) of p53 via an extended binding interface, with dissociation constant of 12.7 (± 0.7) μM. Unlike most binding partners reported to date, HmtSSB interacts with both TAD1 (residues 1–40) and TAD2 (residues 41–61) subdomains of p53. HmtSSB enhances intrinsic 3′-5′ exonuclease activity of p53, particularly in hydrolysing 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG) present at 3′-end of DNA. Taken together, our data suggest that p53 is involved in DNA repair within mitochondria during oxidative stress. In addition, we characterize HmtSSB binding to ssDNA and p53 N-terminal domain using various biophysical measurements and we propose binding models for both.
INTRODUCTION
Single-stranded DNA-binding proteins (SSBs) comprise a group of proteins that bind preferentially to single-stranded DNA (ssDNA) with high affinity. SSBs do not exhibit direct catalytic functions, but are key players in DNA metabolism in all organisms (1). SSBs serve a vital role in replication (2), recombination (3) and repair of DNA (4) by binding to intermediate ssDNA products, thereby protecting them from refolding or nucleolytic attacks. Several groups of SSBs can be classified based on sequence comparisons. Viral proteins [e.g. T4 gene 32 protein (5), gene 5 protein from filamentous phages (6)] differ considerably from prokaryotic SSBs [e.g. SSBs from Escherichia coli (7)] and several F-plasmids from E. coli (8). Eukaryotic SSBs are subdivided into two categories, nuclear and mitochondrial SSBs. Nuclear SSBs (RPA) have been found in all eukaryotic cells examined (9). These proteins are essential for replication of chromosomal DNA (10). Moreover, they interact specifically with other proteins involved in DNA metabolism. These proteins, however, do not have any similarities in sequence and oligomeric structure to the bacterial SSBs. Mitochondrial SSBs (mtSSB) have been isolated from various organisms. These eukaryotic proteins include a number of conserved residues that share with E. coli SSBs in their amino-terminal regions (11–13), but are divergent otherwise. The putative biological role of mtSSB is to stabilize single-stranded regions of mitochondrial DNA (mtDNA) in the displacement loop (D-loop) structures and other replicative intermediates (14,15). Recent biochemical data indicate that TWINKLE, a DNA helicase at mtDNA replication fork, is specifically stimulated by mtSSB (16). Further, mtSSB augments polymerization rate (17), DNA processivity (18,19), 3′-5′ exonuclease activity (20), fidelity (18,21) and primer utilization (17,18) of mitochondrial DNA polymerase gamma (Polγ). Together with mtSSB, Polγ and TWINKLE form highly processive replication machinery, capable of generating DNA products of 16 kb (human mtDNA has a length of 16 568 bp) (22,23). In addition, mtSSB is shown to facilitate packaging mtDNA into bacterial nucleoid-like structures (24), is required to maintain the copy number of mtDNA (25,26) and modulates the level transcription mediated by mitochondrial polymerases (27). Moreover, expression of mtSSB is regulated in response to signalling pathways that control mitochondrial biogenesis in mammalian cells (26).
A recent report demonstrated that the tumour suppressor p53 plays a novel role in maintaining mitochondrial genetic stability through its ability to translocate to mitochondria and interact physically with mtDNA and the catalytic subunit of DNA polymerase gamma (PolγA), in response to mtDNA damage induced by exogenous and endogenous insults including reactive oxygen species (ROS) (28). Also known as ‘guardian of the genome’, p53 is a transcription factor involved in cell-cycle regulation, apoptotic cell death, DNA repair and cellular senescence (29). Being in the centre of a huge network of proteins that allow integration of various signals, p53 is activated upon cellular stress such as DNA damage and the presence of oncogenes. Therefore, interaction of p53 and PolγA is crucial in avoiding accumulation of mtDNA mutations and preventing mtDNA depletions (28), which are frequently observed in human cancers (30), ageing tissues (31,32), mitochondrial diseases (33) and neurodegenerative diseases (34).
In this present report, we showed that tumour suppressor p53 interacts physically with human mtSSB (HmtSSB) in vitro via its transactivation domain. HmtSSB is a novel binding partner of p53 in mitochondria and a component of mitochondrial DNA replisome. This interaction is biologically relevant as HmtSSB enhances moderately the intrinsic 3′-5′ exonuclease activity of p53, particularly in hydrolysing 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG) that is a well-known marker of oxidative stress (35). We propose, therefore, that p53 is implicated in DNA repair in mitochondria during oxidative stress.
MATERIALS AND METHODS
Nucleic acids
Single-stranded DNAs used for DNA binding of HmtSSB had the following sequences: 5′-[Fluorescein] GTTTTCCCAGTCACGAC-3′ (denoted as 17-mer), 5′-[Fluorescein] AATATGGTTTGTTATAAAGAGTAAAGATTTC-3′ (31-mer) and 5′-[Fluorescein] GCCCTGATCACGGTACTCGGTTTTTTTTTTTTTTTTTTTTGGCTCCTCTAGACTCGACCG-3′ (60-mer). The following hairpin DNAs were used to test exonuclease activity of p53: 5′-[Fluorescein] CCATCTAAGCAGACTCACGAATTCACCTAGTTGTTCTAGGTGAAG-3′ (hairpinG) and 5′-[Fluorescein] CCATCTAAGCAGACTCACGAATTCACCTAGTTGTTCTAGGTGAAGG-3′ (hairpin2G). The following 8-oxodG-containing hairpin DNAs were used to examine exonuclease activity of PolγA, PolγAB2 and p53: 5′-[Fluorescein] CCATCTAAGCAGACTCACGACTTCACCTAGTTGTTCTAGGTGAA[8-oxodG]-3′ (8-oxoG:C) and 5′-[Fluorescein] CCATCTAAGCAGACTCACGAATTCACCTAGTTGTTCTAGGTGAA[8-oxodG]-3′ (8-oxoG:A). All DNAs were HLPC-purified and were purchased from either Sigma Genosys or Eurogentec.
Cloning, expression and purification of HmtSSB, PolγA, PolγB, p53 and N-terminal domain of p53 (p53N)
HmtSSB
Spodoptera frugiperda Sf9 cells were maintained and propagated in suspension in TNM-FH insect media (Sigma–Aldrich) containing 10% (v/v) fetal calf serum (FCS) and 1% (v/v) lipid, at 27°C. Protein expression was performed by growing 400 ml of Sf9 cells to a density of 1 × 106 cells/ml in suspension. The cells were infected with 10 plague-forming units/cell of recombinant baculovirus. Autographa californica nuclear polyhedrosis virus, recombinant for HmtSSB expression (mitochondrial form of the protein without the import signal, residues 17–148), was a generous gift from Prof. Maria Falkenberg (Karolinska Institute, Stockholm, Sweden). Infected cells were harvested 72 h post-transfection. Cells were frozen in liquid nitrogen and thawed at 4°C in buffer A (50 mM Tris–HCl pH 7.6, 0.5 mM EDTA, 50 mM NaCl, 1 mM DTT) supplemented with DNase, RNase and protease inhibitor. Cells were lysed by sonication. Upon centrifugation, supernatant was loaded onto HiTrap SP FF column (GE Healthcare) pre-equilibrated with buffer A. The flow-through was subsequently re-loaded onto HiTrap Heparin HP column (GE Healthcare) pre-equilibrated with buffer B [25 mM Tris–HCl pH 7.6, 0.5 mM EDTA, 1 mM DTT, 10% (v/v) glycerol]. After washing with 5 column volumes (CV) of buffer B, protein was eluted with linear gradient of NaCl (0–1 M) in buffer B. Fractions containing HmtSSB were pooled, concentrated and loaded onto HiLoad 26/60 Superdex 200 column (GE Healthcare) pre-equilibrated with buffer C [25 mM Tris–HCl pH 7.6, 0.5 mM EDTA, 200 mM NaCl, 1 mM DTT, 10% (v/v) glycerol]. All chromatographic steps were performed at 4°C. HmtSSB concentration was estimated using an extinction coefficient of 19 940 M–1 cm–1.
PolγA
The gene encoding PolγA (mitochondrial form of the protein without the import signal, residues 26–1240) was cloned into pFastBac HT A vector (Invitrogen) using restriction sites BamHI and EcoRI. Recombinant baculovirus was produced and amplified in S. frugiperda Sf9 cells according to manufacturer's instructions. PolγA was expressed and purified from High Five cells (Invitrogen) infected with the recombinant baculovirus. Cells were grown in suspension culture in Ex-Cell 405 medium (Sigma–Aldrich) supplemented with 5% (v/v) FCS and 1% (v/v) lipid. Cells were harvested on the third day of infection and lysed by sonication in buffer A [50 mM Tris–HCl pH 8, 300 mM NaCl, 10 mM imidazole, 0.1% (v/v) 2-mercaptoethanol] supplemented with DNase, RNase and protease inhibitor. After centrifugation, we loaded supernatant onto HisTrap HP column (GE Healthcare) pre-equilibrated with buffer A. After washing with 5 CV of buffer A, protein was eluted with linear gradient of imidazole (0.01–1 M) in buffer A. His-tag was cleaved with TEV protease overnight. His-tag-removed protein was diluted with buffer B [25 mM Tris–HCl pH 8, 0.5 mM EDTA, 1 mM DTT, 10% (v/v) glycerol] to adjust NaCl concentration to below 100 mM and loaded onto HiTrap Heparin HP column (GE Healthcare) pre-equilibrated with buffer B. Upon washing with 5 CV of buffer B, protein was eluted with linear gradient of NaCl (0–1 M) in buffer B. Fractions containing PolγA were pooled, concentrated and loaded onto HiLoad 26/60 Superdex 200 column (GE Healthcare) pre-equilibrated with buffer C [25 mM Tris–HCl pH 8, 0.5 mM EDTA, 100 mM NaCl, 1 mM DTT, 10% (v/v) glycerol]. All chromatographic steps were performed at 4°C. PolγA concentration was estimated using an extinction coefficient of 241 670 M–1 cm–1.
PolγB
pRUN vector harbouring gene encoding PolγB (mitochondrial form of the protein without the import signal, residues 26–486) tagged with 10×His at N-terminal was a generous gift from Dr Ian J. Holt (MRC Dunn Human Nutrition Unit, Cambridge, UK). For protein expression, plasmid was freshly transformed into E. coli C41 (DE3). Cells were grown in 2×TY media supplemented with 100 μg/ml ampicillin, at 37°C. When OD600 reached 0.5–0.6, 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to induce protein expression and temperature was lowered to 23°C. After 24 h, cells were harvested and pellets were frozen in liquid nitrogen. PolγB was purified through a sequence of HisTrap HP column (GE Healthcare), HiTrap Heparin HP column (GE Healthcare) and HiLoad 26/60 Superdex 200 column (GE Healthcare), as for PolγA with the exception that His-tag was not cleaved in PolγB. PolγB concentration was estimated using an extinction coefficient of 71 390 M–1 cm–1.
p53
A superstable mutant of human p53 containing the following mutations in the core domain: M133L/V203A/N239Y/N268D was used to increase protein expression level and sample stability (36,37). Protein expression and purification were performed as described previously (38).
N-terminal domain of p53 (p53N)
A gene fragment, corresponding to residues 1–93 of human p53, was PCR amplified and cloned into pET24a-HLTV vector. For protein expression, plasmid was freshly transformed into E. coli C41 (DE3). Cells were grown in 2×TY media supplemented with 50 μg/ml kanamycin, at 37°C. When OD600 reached 0.5–0.6, 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to induce protein expression and temperature was lowered to 25°C. After 24 h, cells were harvested and pellets were frozen in liquid nitrogen. Cells were thawed at 4°C in buffer A [50 mM KPi pH 8, 300 mM NaCl, 10 mM imidazole, 1% (v/v) 2-mercaptoethanol] supplemented with DNase, RNase and protease inhibitor and were lysed by high-pressure homogenization (EmulsiFlex-C5, Avestin). After centrifugation, we loaded supernatant onto HisTrap HP column (GE Healthcare) pre-equilibrated with buffer A. After washing with 5 CV of buffer A, protein was eluted with linear gradient of imidazole (0.01–1 M) in buffer A. His-tag+lipoyl domain was cleaved with TEV protease overnight. The sample was reloaded onto a second nickel column, after a desalting step to remove imidazole. The flow-through was diluted with buffer B [50 mM KPi pH 8, 5 mM DTT, 10% (v/v) glycerol] to adjust NaCl concentration to ∼60 mM and loaded onto HiTrap Q HP column (GE Healthcare) pre-equilibrated with buffer B. Upon washing with 5 CV of buffer B, protein was eluted with 1 M NaCl in buffer B. Fractions containing p53N were pooled and loaded onto HiLoad 26/60 Superdex 75 column (GE Healthcare) pre-equilibrated with buffer C [25 mM KPi pH 7.2, 150 mM KCl, 5 mM DTT, 10% (v/v) glycerol]. All chromatographic steps were performed at 4°C. p53N concentration was estimated using an extinction coefficient of 16 500 M–1 cm–1. 15N-labelled sample for NMR was produced in M9 media using 15NH4Cl as nitrogen source.
Analytical ultracentrifugation (AUC)
Equilibrium sedimentation experiments were performed on a Beckman XL-I ultracentrifuge by using Ti-60 rotor and 6-sector cells at speeds of 18 000, 25 000 and 35 000 r.p.m. Data were collected at 15°C, following absorbance at 230 nm, 280 nm and interference. The sample volume was 110 μl and concentrations tested were 5–100 μM. Buffer conditions were 25 mM Tris–HCl pH 7.4 and 150 mM NaCl. Samples were considered to be at equilibrium as judged by a comparison of the several scans at each speed. Data were processed and analysed by using UltraSpin software, which is available from our website (www.mrc-cpe.cam.ac.uk).
Differential scanning calorimetry (DSC)
Calorimetric measurements were performed by using a VP-DSC microcalorimeter (MicroCal), equipped with an AutoSampler. Scans were obtained at a protein concentration of 80 μM, in buffer containing 25 mM HEPES pH 7.4 and 150 mM NaCl. A scan rate of 125°C/h was used, over a temperature range from 2–110°C. The reversibility of the transition was checked by cooling and reheating the same sample. Results from the DSC measurements were analysed with the Origin 7.0 software from MicroCal using the routines of the software provided with the instrument (39).
DNA binding or p53N binding of HmtSSB
The binding of HmtSSB to either ssDNA or p53N was performed by using fluorescence anisotropy (40,41). Measurements were recorded on FluoroMax-3 spectrofluorimeter (Jobin-Yvon-Horiba) equipped with a Hamilton Microlab Titrator, at 15°C in buffer containing 25 mM Tris–HCl pH 7.4, 150 mM NaCl, and 5 mM DTT. HmtSSB protein was titrated into a cuvette containing either 1.2-ml fluorescein-labelled ssDNA (initial concentration 1 nM) or AlexaFluor 546-labelled p53N (initial concentration 100 nM), as described previously (40,41). Excitation/emission wavelengths for fluorescein and AlexaFluor 546 were 480/530 nm and 540/569 nm, respectively. Fluorescence anisotropy was calculated from the fluorescence intensities (42) and binding analysis was performed using either a simple one-state binding isotherm or Hill's equation (40,41).
Intrinsic fluorescence measurements
Intrinsic fluorescence measurements were performed with a Cary Eclipse fluorescence spectrophotometer (Varian). Scans were recorded using excitation wavelength of 280 nm and emission wavelength ranged from 300 to 400 nm. Slit widths for both excitation and emission were kept at 5 nm, with a photomultiplier voltage of 800 V. Concentrations of HmtSSB and/or ssDNA were 500 nM, in buffer containing 25 mM HEPES pH 7.4 and 150 mM NaCl.
NMR experiments
Spectra were acquired with a Bruker Avance 700-MHz spectrometer equipped with a CryoProbe and single-axis gradients. HSQC spectra (43) were recorded with protein in a buffer of 50 mM MES pH 6.8, 100 mM NaCl, 5 mM DTT and 5% (v/v) D2O at 25°C. In both control (15N-labelled p53N) and sample (15N-labelled p53N + HmtSSB) runs, concentrations of p53N and HmtSSB were ∼100 μM. All spectra were externally referenced based on the position of the water peak.
DNA analysis on denaturing polyacrylamide gel (PAGE)
All polymerization and exonuclease reactions were conducted in buffer containing 20 mM Tris–HCl pH 7.5, 10 mM MgCl2, 2 mM ATP, 0.1 mg/ml BSA, 1 mM DTT and 10% (v/v) glycerol, at 37°C. A fifty-micro-litre reaction mix was quenched by adding 15 μl loading dye [8 M urea, 50 mM EDTA pH 7.4, 0.01% (w/v) xylene cyanol FF]. Samples were heated in boiling water for 5 min, snap-cooled in ice water, and analysed on 15% denaturing PAGE [15% (v/v) acrylamide:bis solution (37.5:1), 7 M urea, 1 × TBE] in TBE buffer. Gels were run at constant power of 10 W for 60–70 min, at room temperature. Gel images were acquired with Typhoon scanner (GE Healthcare) and analysed with ImageQuant TL software (GE Healthcare).
Confocal immunofluorescence microscopy
Human 143B TK- osteosarcoma cells were grown and maintained in Dulbecco's Modified Eagle's Medium (DMEM+GlutaMax-1, Invitrogen), supplemented with 10% FCS. Cells were sub-cultured at a ratio of 1:8 in standard 6-well plate and cells were grown on glass coverslips overnight. Media were replaced with either fresh DMEM media, DMEM supplemented with rotenone (100 nM/500 nM) or DMEM supplemented with H2O2 (50 μM/250 μM). After 24 or 48 h, cells were washed two times with serum-free DMEM, followed by incubation with MitoTracker Orange (1 μM in serum-free DMEM) for 15 min at room temperature. After mitochondria staining, cells were washed three times with serum-free DMEM and once with PBS. Cells were fixed for 20 min with 4% (w/v) paraformaldehyde in PBS, and permeabilized by washing three times with PBS + 0.1% (v/v) Triton X-100 (TX100). For blocking, the coverslips were incubated for 30 min with PBS + 0.1% (v/v) TX100 + 1% (w/v) BSA and incubated for 1 h with primary antibodies (mouse anti-DNA, diluted 1:20 in blocking solution and/or rabbit anti-p53, diluted 1:200 in blocking solution; PROGEN Biotechnik GmbH). Coverslips were then washed three times with PBS + 0.1% TX100 and incubated for 1 h with secondary antibodies (AlexaFluor 647 goat anti-mouse IgG and/or AlexaFluor 488 goat anti-rabbit IgG, both diluted 1:1000 in blocking solution; Molecular Probes). Coverslips were again washed three times with PBS + 0.1% TX100, air-dried and mounted in Fluoromount-G (SouthernBiotech). All steps were performed in the dark. Images were collected on a BioRad Radiance confocal microscope.
RESULTS
HmtSSB was over-expressed in insect cells (S. frugiperda, Sf9). The advantage of using baculoviral expression system in this case was to avoid cross-contamination from constitutively expressed E. coli SSB, since both HmtSSB and E. coli SSB have analogous physiochemical properties (44). We characterized the oligomeric structure, thermostability and DNA-binding properties of insect cell-expressed HmtSSB as a prelude to the study of physical/functional interactions between HmtSSB and p53.
HmtSSB is a thermostable tetramer
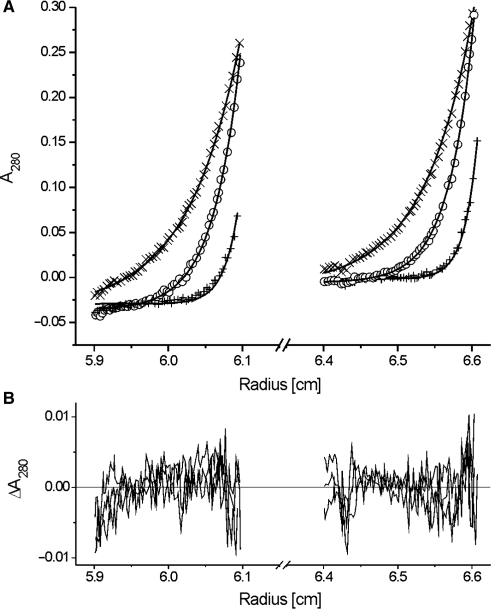
In order to analyse the DNA-binding and p53N-binding of HmtSSB, it was first necessary to determine the oligomerization state of insect cell-expressed HmtSSB in solution. Using equilibrium sedimentation AUC, we showed that HmtSSB existed as homotetramers in solution for all concentrations tested (5, 10, 25, 50, 75 and 100 μM). The data were fitted to a single exponential model (Figure 1), using partial specific volume of 0.7340 for HmtSSB and solvent density of 1.006 (both values were calculated using Sednterp software). An apparent molecular weight of 59.2 (±0.3) kDa was obtained, which is 3.9 times the size of a monomer (15.2 kDa). Therefore, HmtSSB (each monomer of the mitochondrial form has 132 amino acids, tetrameric size = 60.8 kDa) shares the same oligomeric structure as E. coli SSB (each monomer has 178 amino acids, tetrameric size = 75.9 kDa) (7), and differs considerably from heterotrimeric replication protein A (RPA, a nuclear SSB) (9).
Figure 1.
(A) Equilibrium sedimentation AUC of HmtSSB (5 μM) at speeds of 18 000 r.p.m. (times), 25 000 r.p.m. (open circle) and 35 000 r.p.m (plus). Data were fitted to a single-exponential model (smooth line). Experiments were conducted at 15°C in 25 mM Tris–HCl pH 7.4 and 150 mM NaCl. (B) Residuals of curve fits.
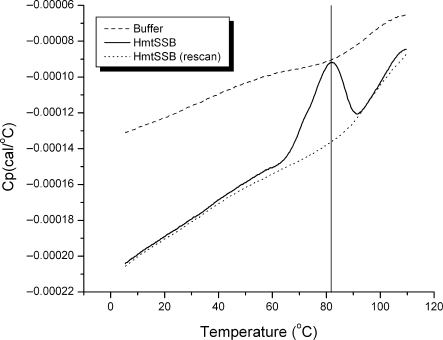
Figure 2 shows the DSC thermogram for HmtSSB (80 μM) at pH 7.4 (25 mM HEPES buffer). The thermogram was asymmetrical, in agreement with the multimeric nature of the protein. Apparent Tm of HmtSSB is ∼82°C. Similar to E. coli SSB (45,46) and murine mtSSB (47), HmtSSB is a thermostable protein. Thermogram of HmtSSB did not show any characteristic signs of heavily aggregated protein, after heat denaturation. Further, HmtSSB denatured irreversibly, as judged from the rescan thermogram.
Figure 2.
DSC thermograms of HmtSSB (80 μM) in buffer containing 25 mM HEPES pH 7.4 and 150 mM NaCl (solid line), of buffer (dashed line) and of sample rescan (dotted line).
HmtSSB binds ssDNA tightly and cooperatively
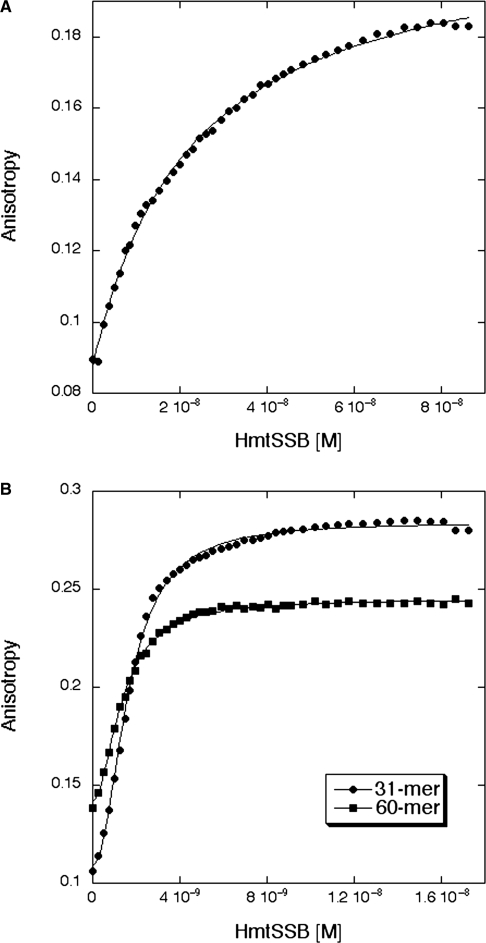
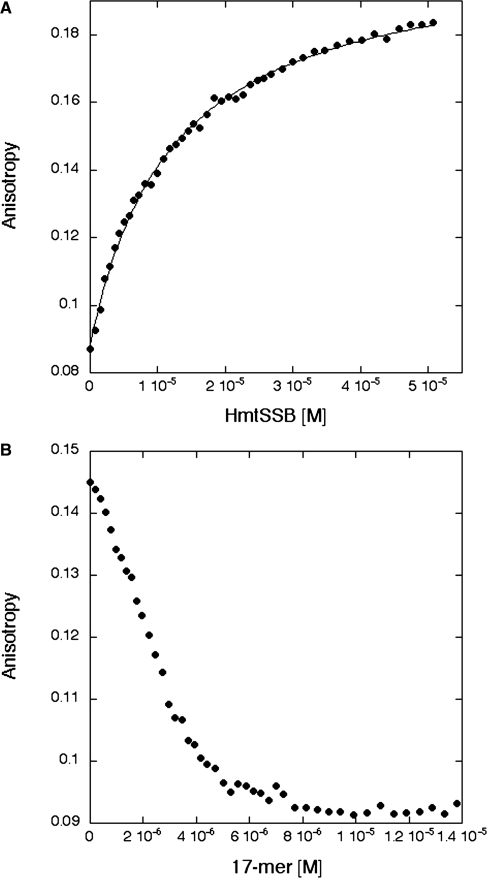
Using fluorescence anisotropy, we performed solution-based DNA-binding experiments to measure the affinity of tetrameric HmtSSB to ssDNAs. In contrast to radioactivity based electrophoretic mobility shift assay (EMSA) that is commonly employed to study DNA-binding of SSBs qualitatively (19,47), fluorescence anisotropy allows quantitative measurement of DNA affinity. Three ssDNAs of different lengths (17-mer, 31-mer and 60-mer) were used and their sequences were optimized to minimize DNA secondary structure formation (Supplementary Figure S1A). Increasing concentrations of HmtSSB were titrated automatically to a sample of fluorescently labelled DNA (1 nM) and DNA-binding experiments were conducted at 150 mM (physiological) ionic strength. The data from 17-mer experiments fitted well to a simple one-state model (Figure 3A), yielding dissociation constant (Kd) of 19.2 (±3.1) nM (Table 1). The same model could not be applied to fit data from 31-mer and 60-mer experiments. These data, however, were fitted well to the Hill equation indicating cooperativity in binding longer ssDNAs (Figure 3B). Upon curve fitting, we obtained Kd and Hill constant of 1.5 (±0.3) nM and 1.9 (±0.2), respectively, for 31-mer (Table 1). In experiments involving 60-mer ssDNA, similar Kd (1.4 ± 0.4 nM) and Hill constant (2.0 ± 0.3) were obtained (Table 1). Thus, HmtSSB binds ∼13-fold tighter to longer ssDNA (31-mer and 60-mer) than to shorter ssDNA (17-mer). Interestingly, strong cooperativity was also observed for 17-mer, when DNA concentration was increased significantly above Kd value (data not shown). All dissociation constants reported in Table 1 were expressed in monomer HmtSSB concentrations for the convenience of comparing affinities from various studies, estimated using different approaches, particularly EMSA.
Figure 3.
Binding titration profiles of HmtSSB with fluorescein-labelled single-stranded DNAs. (A) HmtSSB binding to 17-mer. (B) HmtSSB binding to 31-mer (filled diamond) and 60-mer (filled square). Experiments were conducted at 15°C in 25 mM Tris–HCl pH 7.4, 150 mM NaCl and 5 mM DTT.
Table 1.
Binding of ssDNAs (17-mer, 31-mer and 60-mer) and p53N (residues 1–93) to HmtSSB, measured using fluorescence anisotropy
| DNA/p53N | Simple one-state model | Hill equationa |
Number of titrations repeated | |
|---|---|---|---|---|
| Kd (M) | Kd (M) | Hill constant | ||
| 17-mer | 19.2 ± 3.1 (10−9) | 19.4 ± 4.0 (10−9) | 1.1 ± 0.3 | 4 |
| 31-mer | Poor fit | 1.5 ± 0.3 (10−9) | 1.9 ± 0.2 | 3 |
| 60-mer | Poor fit | 1.4 ± 0.4 (10−9) | 2.0 ± 0.3 | 4 |
| p53N | 12.7 ± 0.7 (10−6) | 14.2 ± 0.3 (10−6) | 0.9 ± 0.0 | 3 |
DNAs and p53N were labelled with 5′-flurescein or AlexaFluor 546, respectively. Data were fitted to either a simple one-state model or Hill equation. All dissociation constants were expressed as HmtSSB monomer concentrations.
aHill equation: 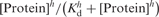 where h is Hill constant.
where h is Hill constant.
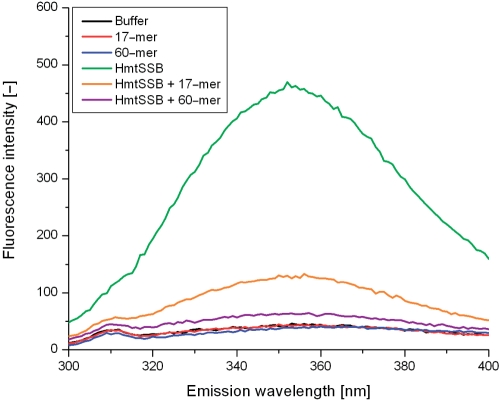
HmtSSB contains two tryptophan residues at positions 65 and 84 (correspond to positions 49 and 68 in the mitochondrial form of the protein). From sequence comparison, these residues are homologous to the tryptophan residues 41 and 55 of E. coli SSB (NCBI accession code P0AGE0) (48). Figure 4 shows the intrinsic fluorescence spectra of HmtSSB, in the absence and presence of ssDNA (17-mer or 60-mer). Excitation at 280 nm gave an emission maximum at 352 nm, implying that these tryptophan residues are solvent exposed. Upon binding 17-mer, the fluorescence emission of HmtSSB was reduced by ∼70%. When longer ssDNA was used (60-mer), there was ∼90% loss in fluorescence emission. These results corroborated previous observations on E. coli SSB, that Trp 41 and Trp 55 are involved in stabilizing protein-nucleic acid complexes via stacking interactions with the nucleotide bases of ssDNA (48–52). Consistent with the spectroscopic data, crystal structure of E. coli SSB bound to two 35-mer ssDNA provides clear evidence that Trp 41, Trp 54 and Phe 61 make extensive interactions with ssDNA (53).
Figure 4.
Intrinsic fluorescence emission spectra of HmtSSB in the absence (green) and presence of 17-mer (orange) and 60-mer (purple), at excitation wavelength of 280 nm. Experiments were conducted at 25°C in 25 mM HEPES pH 7.4 and 150 mM. Concentrations of both HmtSSB and single-stranded DNAs were 500 nM. Control spectra without HmtSSB were also recorded: buffer (black), 17-mer in buffer (red) and 60-mer in buffer (red).
HmtSSB binds N-terminal domain of p53 (p53N)
Upon DNA damage, the tumour suppressor p53 is activated and orchestrates a cellular response by transcriptional regulation of genes or by interacting with other proteins involved in cell-cycle arrest, apoptosis, senescence and DNA repair. One of these protein partners is RPA, a nuclear SSB, which binds p53 transactivation domain (54,55). Accumulating evidence indicates that p53 is also able to translocate to mitochondria and interacts physically with mitochondrial proteins, such as PolγA (28) and mitochondrial transcription factor A (TFAM) (56). On the basis of these previous findings, we hypothesized that HmtSSB could interact with p53 and possibly via its transactivation domain. By using fluorescence anisotropy, we assayed HmtSSB for binding to an AlexaFluor 546-labelled p53N (residues 1–93) at physiological ionic strength and our hypothesis was indeed verified. Data were fitted to a simple one-state model, yielding a Kd of 12.7 (±0.7) μM (Figure 5A). The binding mode of p53N to HmtSSB and/or binding site on HmtSSB is plausibly identical to a short ssDNA, in view of the acidic nature of p53N (pI = 3.47). To test this assumption, we performed a competition assay. Non-fluorescently-labelled 17-mer ssDNAs were titrated into a mixture of 100 nM AlexaFluor 546-labelled p53N and 20 μM HmtSSB. The decrease in anisotropy indicated displacement of p53N from HmtSSB (Figure 5B). HmtSSB bound 17-mer ∼661-fold tighter than it did p53N. Nevertheless, anisotropy dropped to a minimum level at 17-mer concentration of ∼8 μM suggesting the possibility that HmtSSB could bind p53N and 17-mer simultaneously. This is not surprising as four possible binding sites exist in one tetrameric HmtSSB molecule. In the case of nuclear SSB (RPA), p53N binding site also overlaps with ssDNA-binding region (55). We did not measure the intrinsic fluorescence of HmtSSB upon binding p53N, as three solvent-exposed tryptophans (Trp 23, Trp 53 and Trp 91) exist in p53N.
Figure 5.
(A) Binding titration profile of HmtSSB with AlexaFluor 546-labelled p53N (residues 1-93). (B) Competition assay in which non-fluorescently labelled 17-mer was titrated into a mixture of 100 nM AlexaFluor 546-labelled p53N and 20 μM HmtSSB. The decrease in fluorescence anisotropy indicated displacements of p53N from HmtSSB. Experiments were conducted at 15°C in 25 mM Tris–HCl pH 7.4, 150 mM NaCl and 5 mM DTT.
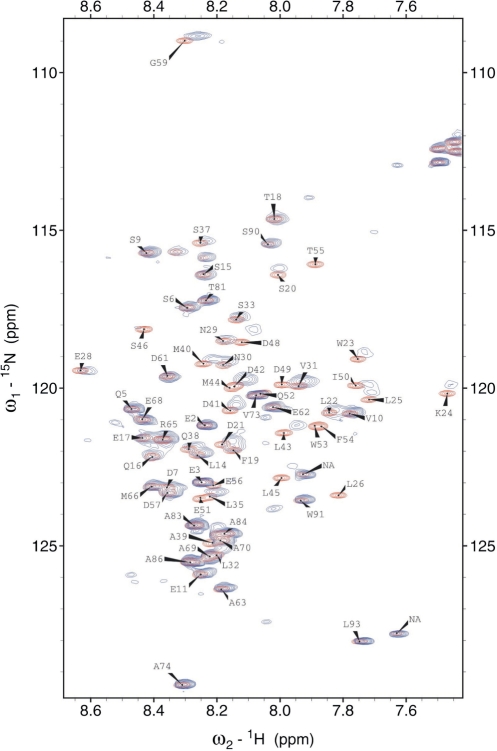
To localize the binding interface on p53N, we recorded NMR HSQC spectra of 15N-labelled p53N (100 μM) in the absence and presence of unlabelled HmtSSB (100 μM). In Figure 6, free 15N-labelled p53N spectrum (red) was overlaid with the HmtSSB-bound spectrum (blue). p53N interacts clearly with HmtSSB through an extended interface. Large chemical shift perturbations were mainly localized in three regions: region I (residues 20–28), region II (residues 35–40) and region III (41–59). Resonances at Leu 26, Trp 53, Phe 54, Thr 55 completely disappeared in the bound spectrum, signifying the importance of hydrophobic interaction in stabilizing p53N-HmtSSB complexes. The p53 transactivation domain is subdivided into two loosely defined subdomains (57,58), TAD1 (residues 1–40) and TAD2 (residues 41–61). Most p53N-binding proteins reported to date, in general, bind to either one of the two transactivation subdomains. Proteins like MDM2 (59), MDM4 (60), TBP (61), TAFII31 (62), TAFII40 (63) and TAFII60 (63) all interact with TAD1, whereas RPA (54,55) and p62/Tfb1 subunit of TFIIH (64,65) bind to TAD2. We found that HmtSSB interacts simultaneous with TADI and TAD2, similar to the transcriptional coactivator p300 (66).
Figure 6.
NMR HSQC spectra of 1H15N-labelled p53N (residues 1–93, 100 μM) in the absence (red) and presence of 100 μM of HmtSSB (blue). Spectra were recorded at 25°C in 50 mM MES pH 6.8, 100 mM NaCl, 5 mM DTT and 5% (v/v) D2O.
PolγA, but not PolγAB2, is capable of hydrolysing 8-oxodG
Being at the site of cellular respiration (oxidative phosphorylation), mtDNA is particularly vulnerable to reactive oxygen species (ROS) that are by-products of normal metabolism. The levels of 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG), an indicator of oxidative DNA damage, were 4-fold higher in mtDNA compared with nuclear DNA in heart tissue obtained from eight mammalian species with varying life spans (3.5 to 46 yr) (67). Further, there was a significant inverse correlation between 8-oxodG levels in mtDNA and maximum life span among the different species (67). 8-oxodG is capable of base-pairing with both 2′-deoxycytidine (dC) and 2′-deoxyadenosine (dA) (68). The 8-oxodG:dC pair mimics natural dG:dC pair, whereas 8-oxodG:dA pair results in GC→TA transversions (69,70). Transversions usually cause non-synonymous amino acid substitutions, if not repaired (71).
The interaction between p53 and SSB (e.g. RPA) has been implicated in DNA repair (72). Since p53 was shown to translocate to mitochondria upon oxidative stress, we decided to examine the exonuclease activities of Polγ and p53 using oxidized DNAs. In this experiment, we designed two DNAs containing 8-oxodG at 3′-termini (Supplementary Figure S1B). The sequences of DNAs were optimized so that the DNAs form hairpins spontaneously, with double-stranded stem at 3′-end and single-stranded arm at 5′-end. These DNAs mimic singly primed template, useful for studying 3′-mismatch hydrolysis and template-directed polymerization. An obvious advantage of using such DNAs is keeping ‘primer’ and ‘template’ at right stoichiometric ratio. One of our two DNAs had 8-oxodG:dC pair at 3′-end (denoted as 8-oxoG:C), while 8-oxodG:dA pair was present at 3′-end of the other DNA (8-oxoG:A).
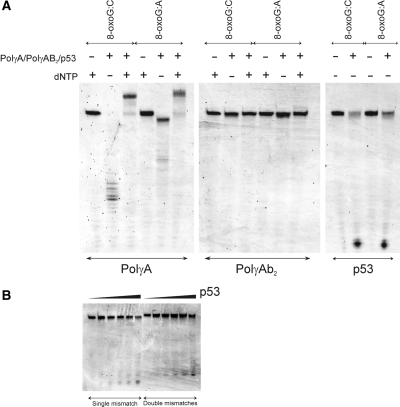
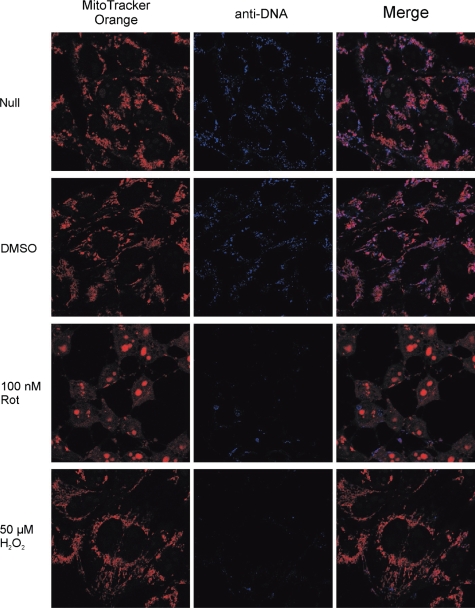
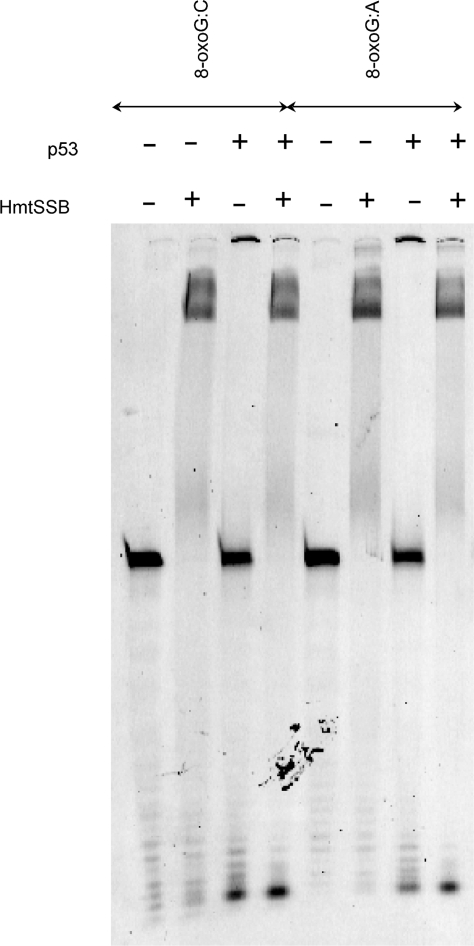
Polγ, exists as a heterotrimer in mitochondria (PolγAB2), comprising a catalytic subunit (PolγA) and two accessory subunits (PolγB). PolγA and PolγB were expressed in insect cells and E. coli, respectively. Both proteins were functional, as verified using various biochemical and biophysical assays (data not shown). PolγA could effectively hydrolyse 3′-end 8-oxodG in both 8-oxoG:C and 8-oxoG:A in the absence of dNTP (Figure 7A). An obvious difference in comparing the degradation patterns of 8-oxoG:C and 8-oxoG:A was the extent of inward digestion. In 8-oxoG:C, exonuclease activity of PolγA proceeded further, generating smaller DNAs. When encountering 8-oxodG:dA mismatch, exonuclease activity of PolγA paused at position after 8-oxodG was removed from 3′-end. The result was in good agreement with another set of experiments, in which non-oxidized DNAs were used (data not shown). Exonuclease activity of PolγA was ‘progressive’ when incubated with 3′-matching DNA and ‘stalling’ when mixed with 3′-mismatching DNA, in the absence of dNTP. In the presence of dNTP, PolγA catalysed polymerization in both 8-oxoG:C and 8-oxoG:A. Surprisingly, both exonuclease and polymerization activities of PolγA were inhibited after complexing with PolγB when oxidized DNAs were used (Figure 7A), which was not observed in non-oxidized DNAs (data not shown). We speculated that mtDNA replication was hindered, upon prolonged oxidative stress exposure. We verified this assumption using confocal microscopic analysis. Human 143B TK- osteosarcoma cells were incubated with rotenone (100 nM/500 nM) and H2O2 (50 μM/250 μM) for 24 and 48 h. Rotenone, a complex I inhibitor, is known to increase mitochondrial ROS by interfering with the respiratory activity (73). After drug treatment, mitochondria were stained with MitoTracker Orange and mtDNA was detected with combination of mouse anti-DNA and AlexaFluor 647 goat anti-mouse IgG. 24-h incubation did not result in evident differences (Supplementary Figure S2), even at 500 nM rotenone and 250 μM H2O2. A drastic mtDNA reduction was noticed after 48 h, at both 100 nM rotenone and 50 μM H2O2 (Figure 8). The same experiment was repeated three times independently. We confirmed that the mtDNA reduction was neither experimental error nor cell-staining artefact.
Figure 7.
(A) Exonuclease activities of PolγA, PolγAB2 and full-length p53 using two hairpin DNAs containing 8-oxodG at 3′-termini (denoted as 8-oxoG:C and 8-oxoG:A). (B) Exonuclease activities of full-length p53 using two hairpin DNAs containing one dG:dA mismatch (left) or two dG:dA mismatches (right) at 3′-termini. Experiments were conducted at 37°C in 20 mM Tris–HCl pH 7.5, 10 mM MgCl2, 2 mM ATP, 0.1 mg/ml BSA, 1 mM DTT and 10% (v/v) glycerol. DNA samples were analysed on 15% denaturing PAGE in 1 × TBE.
Figure 8.
Confocal microscopic analyses of human 143B TK- cells after incubating with 100 nM rotenone or 50 μM H2O2 for 48 h. Mitochondria were stained with MitoTracker Orange and mtDNA were probed with combination of mouse anti-DNA and AlexaFluor 647 goat anti-mouse IgG.
Intrinsic 3′-5′ exonuclease activity of p53 is capable of excising 8-oxodG
Other than its transcriptional activity, p53 possesses intrinsic 3′-5′ exonuclease activity (74–79). Using two non-oxidized DNAs (hairpinG with one dG:dA mismatch at 3′-end and hairpin2G with two consecutive dG:dA mismatches at 3′-end, see Supplementary Figure S1C), we confirmed that full-length p53 is a weak 3′-5′ exonuclease in the presence of Mg2+ ions (Figure 7B). Same DNAs were tested with PolγA. Both exonuclease and polymerase activities of PolγA were positive (Supplementary Figure S3). We excluded the possibility that the observed p53 exonuclease activity was due to contaminating proteins co-purified with p53. C-terminal truncated p53 or DNA-binding domain of p53, purified using exact purification scheme, did not show noticeable exonuclease activity (data not shown). This concern was also addressed by other authors and same conclusion was drawn (74). Interestingly, p53 was capable of hydrolyzing 3′-end 8-oxodG in 8-oxoG:C and 8-oxoG:A DNAs, both in a ‘progressive’ manner (Figure 7A).
HmtSSB enhances exonuclease activity of p53 moderately
We next checked the effect of HmtSSB on the exonuclease activity of p53, as HmtSSB was previously demonstrated to augment 3′-end mismatch hydrolysis of Polγ (20). Figure 9 exemplified that 3′-end 8-oxodG hydrolysis of p53 was enhanced moderately by addition of HmtSSB, particularly in ‘mismatching’ 8-oxoG:A DNA. The difference in DNA hydrolysis was not due to unequal DNA sample loading to denaturing PAGE. Experiment was repeated and same result was observed. We reckoned that p53-HmtSSB complex had higher affinity towards non-specific DNA, in comparison to p53 alone. This might account for enhanced p53 exonuclease activity.
Figure 9.
Exonuclease activities of full-length p53 using two hairpin DNAs containing 8-oxodG at 3′-termini (denoted as 8-oxoG:C and 8-oxoG:A), in the absence and presence of HmtSSB. Experiments were conducted at 37°C in 20 mM Tris–HCl pH 7.5, 10 mM MgCl2, 2 mM ATP, 0.1 mg/ml BSA, 1 mM DTT and 10% (v/v) glycerol. DNA samples were analysed on 15%denaturing PAGE in 1 × TBE.
DISCUSSION
The binding modes of ssDNA and p53N to HmtSSB
Escherichia coli SSB exhibits two types of cooperative ssDNA-binding behaviours (80). At low ionic strength (<10 mM NaCl) (81,82) and high protein:DNA ratio (83–85), E. coli SSB displays ‘unlimited’ cooperative binding to long ssDNA, resulting in long protein clusters (83,85). At high ionic strength (>200 mM NaCl or 3 mM MgCl2) (81,82) and low protein:DNA ratio (83–85), ‘limited’ cooperativity is favoured in which the protein does not form long clusters along ssDNA. Two binding modes have previously been defined for E. coli SSB, referred as (SSB)35 and (SSB)65 (83,85). The difference between these two modes lies in the number of nucleotides occluded per bound tetramer [35 ± 2 for (SSB)35 and 65 ± 3 for (SSB)65]. In (SSB)35, two of the four E. coli SSB monomers are in contact with ssDNA, while in (SSB)65, all four E. coli monomers are contacting ssDNA (81,82,86). Therefore, ‘unlimited’ cooperativity is thought to be associated with (SSB)35 mode (80,83,85) whereas (SSB)65 promotes ‘limited’ cooperativity (80,83,84).
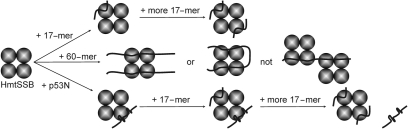
In the present study, we reported ssDNA- and p53N-binding of human mitochondrial SSB, at physiological ionic strength (150 mM NaCl). Based on the data from fluorescence anisotropy and intrinsic fluorescence measurements, we proposed the binding models depicted in Figure 10. For the 17-mer, we expected a (DNA)1:(tetramer)1 ratio since binding data fitted well to a simple one-state model (Figure 3A). When DNA concentration was increased, we observed cooperative behaviour (data not shown) which could be explained by additional 17-mer bound to the same tetramer. In either (DNA)1:(tetramer)1 or (DNA)2:(tetramer)1, not all monomers are in contact with ssDNA because of the limited length of ssDNA. This is consistent with the 30% intrinsic fluorescence retained upon binding 17-mer (Figure 4). As such, the (SSB)35 mode is dominant in binding short ssDNA. For 31-mer and 60-mer, we suggested (DNA)2:(tetramer)1 based on four pieces of evidence: Firstly, binding data fitted well to the Hill equation yielding a Hill constant of 2 (Figure 3B). Secondly, only ∼10% of intrinsic fluorescence was retained upon binding 60-mer suggesting that most monomers are contacting ssDNA (Figure 4). Thirdly, each ssDNA was bound to two monomers and this explains higher affinity (∼13-fold) of HmtSSB for longer ssDNA. Fourthly, the crystal structure of E. coli SSB is bound to two 35-mers indicating (DNA)2:(tetramer)1 (53). Thus, the (SSB)65 binding mode is favoured in binding long ssDNA (35-mer and 60-mer). It has previously been demonstrated that two murine mtSSB tetramers can bind simultaneously to an 80-mer, when the complex is formed in buffer containing 10 mM Tris–HCl pH 8.0, 2 mM DTT and 200 μg/ml BSA (19). We believe that this binding mode was impossible for human mtSSB, based on two compelling reasons. Firstly, HmtSSB aggregated upon dialysis into NaCl-free buffer (data not shown). Other authors have also reported that HmtSSB was insoluble at salt concentrations lower than 50 mM NaCl (44). Secondly, we performed equilibrium sedimentation AUC using a mixture of fluorescein-labelled 60-mer (5 μM) and HmtSSB (107 μM). Data were collected by monitoring fluorescein absorbance at 492 nm, at 12 000, 17 000 and 24 000 r.p.m. Upon data fitting to a single exponential model (Supplementary Figure 4) using partial specific volume of 0.6443 for (DNA)2:(tetramer)1 complex and solvent density of 1.006, we found an apparent molecular weight of 81.8 (± 0.9) kDa. The discrepancy between this value and calculated molecular weight (Mw) for (DNA)2:(tetramer)1 complex (98.6 kDa) results from difficulty in estimating partial specific volume for protein–DNA complex. Certainly, no high-molecular-weight species [Mw of (DNA)1:(tetramer)2 complex is calculated to be 145.5 kDa] was observed. Nonetheless, we could not exclude the possibility that a 60-mer could wrap around the outside of the entire tetramer to give a (DNA)1:(tetramer)1 complex. The calculated Mw of (DNA)1:(tetramer)1 complex (79.7 kDa) was also closed to the apparent Mw from AUC (91.6 ± 1.1 kDa), when data were fitted using partial specific volume of 0.6785 for (DNA)1:(tetramer)1 complex. In the case of p53N-binding, (p53N)1:(tetramer)1 complex was proposed since binding data were well-fitted to a simple one-state binding model. Moreover, it is plausible that p53N and ssDNA can bind simultaneously to HmtSSB tetramer which accounts for the enhanced 3′-5′ exonuclease activity of p53 (Figure 9).
Figure 10.
Schematic models of 17-mer/HmtSSB, 60-mer/HmtSSB and p53N/HmtSSB interactions. HmtSSB exists as homotetramer in solution. 17-mer and p53N bind to HmtSSB in (DNA)1:(tetramer)1 and (p53N)1:(tetramer)1 ratios. 60-mer binds cooperatively to HmtSSB, in either (DNA)2:(tetramer)1 or (DNA)1:(tetramer)1. In 17-mer/HmtSSB or p53N/HmtSSB interactions, not all HmtSSB monomers are in contact with binding partners, whereas in 60-mer/HmtSSB, all monomers are contacting binding partners.
p53 translocation to mitochondria
p53 is a predominantly nuclear protein, lacking a mitochondrial targeting sequence as predicted using Mitoprot software (http://ihg2.helmholtz-muenchen.de/ihg/mitoprot.html). It has been shown that the cytoplasm contained a separate and distinct p53 pool that was the major source for p53 translocation to the mitochondria upon its stress-induced stabilization (87). Albeit not a p53-shuttler, Mdm2 was required for monoubiquitylation of p53 and thus its translocation to mitochondria (87). Upon arrival at the mitochondria, p53 underwent rapid deubiquitylation by mitochondrial HAUSP via a stress-induced mitochondrial p53-HAUSP complex (87). This generated apoptotically active and non-ubiquitylated mitochondrial p53 (87). Nonetheless, contrasting data exist suggesting that mitochondrial p53 originated from nucleus and utilized p53 ubiquitylation for mitochondrial targeting (88). The mechanism by which p53 is translocated to mitochondria remains to be clarified.
Mitochondrial p53 was demonstrated to be highly efficient in inducing the release of soluble (e.g. cyto C, Smac) and non-soluble (e.g. AIF, EndoG; both tethered to the inner mitochondrial membrane) apoptogenic factors by severely disrupting the integrity of both outer as well as inner mitochondrial membranes (89). This action was associated with p53-induced oligomerization of Bax, Bak and VDAC, and the formation of stress-induced endogenous p53-cyclophilin D complex located at the inner membrane (89). Therefore, mitochondrial membrane permeabilization triggered by p53 is a highly plausible mechanism by which p53 enters mitochondrial matrix where p53 binding partners (e.g. PolγA, TFAM, HmtSSB) are located. Alternatively, p53 could possibly be imported via mitochondrial translocon since p53 has been shown to interact with two key mitochondrial import proteins, mtHsp70 and Hsp60 (88). Recent findings further documented that many nuclear proteins were localized in mitochondrial matrix and directly involved in mitochondrial functions, without harbouring typical N-terminal mitochondrial leader sequence. These include nuclear triiodothyronine receptor (c-ErbAα1) (90), retinoic acid X receptor [(RXR)α] (90) and estrogen receptor (ERα and β) (91).
In mammalian cells, mtDNA replication takes place preferentially within a subset of mitochondria clustered within a perinuclear domain (92). Interestingly, HmtSSB protein is also not distributed uniformly within all mitochondria but is more abundant within perinuclear regions (26). In other words, nuclear p53 is located in close proximity to mitochondrial protein partners (e.g. PolγA and HmtSSB).
Biological implication of p53-HmtSSB interaction
p53 function is directly related to its transcriptional activity, mediated through sequence-specific binding to several DNA targets in the genome. The consensus DNA-binding site comprises two decameric repeats of the general form PuPuPuC(A/T)(A/T)GPyPyPy separated by 0−13 bp (93,94). In addition to its DNA binding capacity, the p53 core domain has an intrinsic 3′-5′ exonuclease activity (75) that is modulated by its C-terminus (74,95). This exonuclease activity has been suggested to be involved in DNA damage repair (96). We showed that full-length p53 is a weak 3′-5′ exonuclease (Figure 7). When tetramerization domain and C-terminal regulatory domain were truncated, p53 is devoid of exonuclease activity (data not shown). Unlike PolγAB2, p53 is capable of hydrolysing 3′-end 8-oxodG and this activity is enhanced moderately by addition of HmtSSB (Figure 9). A possible explanation for the increased mismatch hydrolysis is that HmtSSB augments affinity of p53 towards non-specific DNA.
Although p53/HmtSSB interaction is relatively weak (Kd = 12.7 ± 0.7 μM), concentration of HmtSSB within mitochondria is high (97,98). In human HeLa cells, HmtSSB is 3000-fold more abundant than mtDNA (97). Depending on physiological conditions, the size of mitochondria varies from 0.5 μm to 10 μm. Each mitochondrion contains about 2–10 copies of mtDNA. Taken together and assuming spherical shape of mitochondrion, the calculated HmtSSB concentration ranges from 19 nM to 762 μM. It is important to note that mitochondrial p53 isolated from ML1, HCT116 and RKO cells, after short term genotoxic stress, were phosphorylated on Ser 6, Ser 9, Ser 15, Ser 20, Ser 37 and Ser 46, even though phosphorylation/acetylation are not determining factors for p53's translocation to mitochondria (99). These serine residues were located within TAD1 (residues 1–40) and TAD2 (residues 41–61) regions of p53, the identified binding interface between p53 and HmtSSB. Since phosphorylation increases negative charge on p53 N-terminal, we believe that the affinity between p53/HmtSSB in vivo is higher than value obtained from in vitro measurement.
Remarkably, when the human mitochondrial genome (NC 001807) was inspected for putative p53-recognition sequences, a total of seven candidate sequences were detected: 1553, 1809, 2397 and 2903 positioned within the 12S and 16S rDNA region downstream of the heavy strand promoter; 4637 situated within NADH dehydrogenase subunit 2 gene; 12 230 positioned between NADH dehydrogenase subunit 4 and 5 genes; and 16 190 lying within the regulatory D-loop downstream of the light strand promoter (the numbers indicate the positions of the first nucleotide of the recognition sequence) (100). Of these, recognition sequence 1553 conferred responsivity to p53 and p53′s relatives p73α and β (100). It was, therefore, suggested that mitochondrial p53 might bind directly to mtDNA and perhaps be involved in the regulation of mitochondrial transcription/replication. In addition, the presence of mtDNA likely enhances further p53/HmtSSB interaction. This is made possible since p53/mtDNA interaction (via DNA-binding core domain and possibly C-terminal regulatory domain) and p53/HmtSSB interaction (via N-terminal transactivation domain) involve different functional domains of p53. Likewise for HmtSSB, simultaneous HmtSSB/p53 and HmtSSB/mtDNA interactions is feasible due to its multimeric nature. Our data has shown that p53 is implicated in maintaining mitochondrial genome stability. In addition to p53, we believe other proteins are recruited for mtDNA repair during oxidative stress since p53 is a weak exonuclease compared to PolγA. This explains drastic mtDNA loss in 143B cells, after a 48-h incubation with oxidizing agents (Figure 8). In sum, we propose that mitochondrial p53 plays dual roles: an initiator of a cascade of apoptosis program and a mtDNA repair factor upon cellular stress.
CONCLUSION
In the present study, we identified HmtSSB as a novel binding partner of tumour suppressor p53 in mitochondria. Biophysical measurements, in combination with biochemical assays, provide a systematic platform to characterize the physical and functional interactions between these two proteins in vitro. The validation of this binding in vivo would, of course, require additional cell-based experiments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The European Union FP6 Proteomage (to A.R.F. and T.S.W.); and MRC Career Development Fellowship (to T.S.W.). Funding for open access charge: Medical Research Council UK.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Christopher M. Johnson and Robert Sade for useful discussions, Caroline M. Blair and Fiona Sait for technical assistance. We acknowledge Prof. Maria Falkenberg (Karolinska Institute, Stockholm, Sweden) for providing genes encoding HmtSSB and PolγA, and Dr Ian J. Holt (MRC Dunn Human Nutrition Unit, Cambridge, United Kingdom) for PolγB gene.
REFERENCES
- 1.Kornberg A, Baker TA. DNA Replication. 2nd. New York: W. H. Freeman and Company; 1992. [Google Scholar]
- 2.Sigal N, Delius H, Kornberg T, Gefter ML, Alberts B. A DNA-unwinding protein isolated from Escherichia coli: its interaction with DNA and with DNA polymerases. Proc. Natl Acad. Sci. USA. 1972;69:3537–3541. doi: 10.1073/pnas.69.12.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coverley D, Kenny MK, Lane DP, Wood RD. A role for the human single-stranded DNA binding protein HSSB/RPA in an early stage of nucleotide excision repair. Nucleic Acids Res. 1992;20:3873–3880. doi: 10.1093/nar/20.15.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delius H, Mantell NJ, Alberts B. Characterization by electron microscopy of the complex formed between T4 bacteriophage gene 32-protein and DNA. J. Mol. Biol. 1972;67:341–350. doi: 10.1016/0022-2836(72)90454-8. [DOI] [PubMed] [Google Scholar]
- 6.Wen JD, Gray DM. Ff gene 5 single-stranded DNA-binding protein assembles on nucleotides constrained by a DNA hairpin. Biochemistry. 2004;43:2622–2634. doi: 10.1021/bi030177g. [DOI] [PubMed] [Google Scholar]
- 7.Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol. Rev. 1990;54:342–380. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruvolo PP, Keating KM, Williams KR, Chase JW. Single-stranded DNA binding proteins (SSBs) from prokaryotic transmissible plasmids. Proteins. 1991;9:120–134. doi: 10.1002/prot.340090206. [DOI] [PubMed] [Google Scholar]
- 9.Iftode C, Daniely Y, Borowiec JA. Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 1999;34:141–180. doi: 10.1080/10409239991209255. [DOI] [PubMed] [Google Scholar]
- 10.Zou Y, Liu Y, Wu X, Shell SM. Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J. Cell Physiol. 2006;208:267–273. doi: 10.1002/jcp.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghrir R, Lecaer JP, Dufresne C, Gueride M. Primary structure of the two variants of Xenopus laevis mtSSB, a mitochondrial DNA binding protein. Arch. Biochem. Biophys. 1991;291:395–400. doi: 10.1016/0003-9861(91)90152-9. [DOI] [PubMed] [Google Scholar]
- 12.Stroumbakis ND, Li Z, Tolias PP. RNA- and single-stranded DNA-binding (SSB) proteins expressed during Drosophila melanogaster oogenesis: a homolog of bacterial and eukaryotic mitochondrial SSBs. Gene. 1994;143:171–177. doi: 10.1016/0378-1119(94)90093-0. [DOI] [PubMed] [Google Scholar]
- 13.Tiranti V, Rocchi M, DiDonato S, Zeviani M. Cloning of human and rat cDNAs encoding the mitochondrial single-stranded DNA-binding protein (SSB) Gene. 1993;126:219–225. doi: 10.1016/0378-1119(93)90370-i. [DOI] [PubMed] [Google Scholar]
- 14.Van Tuyle GC, Pavco PA. The rat liver mitochondrial DNA-protein complex: displaced single strands of replicative intermediates are protein coated. J. Cell Biol. 1985;100:251–257. doi: 10.1083/jcb.100.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clayton DA. Replication of animal mitochondrial DNA. Cell. 1982;28:693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- 16.Korhonen JA, Gaspari M, Falkenberg M. TWINKLE Has 5′ -> 3′ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J. Biol. Chem. 2003;278:48627–48632. doi: 10.1074/jbc.M306981200. [DOI] [PubMed] [Google Scholar]
- 17.Thommes P, Farr CL, Marton RF, Kaguni LS, Cotterill S. Mitochondrial single-stranded DNA-binding protein from Drosophila embryos. Physical and biochemical characterization. J. Biol. Chem. 1995;270:21137–21143. doi: 10.1074/jbc.270.36.21137. [DOI] [PubMed] [Google Scholar]
- 18.Williams AJ, Kaguni LS. Stimulation of Drosophila mitochondrial DNA polymerase by single-stranded DNA-binding protein. J. Biol. Chem. 1995;270:860–865. doi: 10.1074/jbc.270.2.860. [DOI] [PubMed] [Google Scholar]
- 19.Mikhailov VS, Bogenhagen DF. Effects of Xenopus laevis mitochondrial single-stranded DNA-binding protein on primer-template binding and 3′–>5′ exonuclease activity of DNA polymerase gamma. J. Biol. Chem. 1996;271:18939–18946. doi: 10.1074/jbc.271.31.18939. [DOI] [PubMed] [Google Scholar]
- 20.Farr CL, Wang Y, Kaguni LS. Functional interactions of mitochondrial DNA polymerase and single-stranded DNA-binding protein. Template-primer DNA binding and initiation and elongation of DNA strand synthesis. J. Biol. Chem. 1999;274:14779–14785. doi: 10.1074/jbc.274.21.14779. [DOI] [PubMed] [Google Scholar]
- 21.Hoke GD, Pavco PA, Ledwith BJ, Van Tuyle GC. Structural and functional studies of the rat mitochondrial single strand DNA binding protein P16. Arch. Biochem. Biophys. 1990;282:116–124. doi: 10.1016/0003-9861(90)90094-f. [DOI] [PubMed] [Google Scholar]
- 22.Korhonen JA, Pham XH, Pellegrini M, Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 2004;23:2423–2429. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 24.Bogenhagen DF, Wang Y, Shen EL, Kobayashi R. Protein components of mitochondrial DNA nucleoids in higher eukaryotes. Mol. Cell Proteomics. 2003;2:1205–1216. doi: 10.1074/mcp.M300035-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Farr CL, Matsushima Y, Lagina A.T., III, Luo N, Kaguni LS. Physiological and biochemical defects in functional interactions of mitochondrial DNA polymerase and DNA-binding mutants of single-stranded DNA-binding protein. J. Biol. Chem. 2004;279:17047–17053. doi: 10.1074/jbc.M400283200. [DOI] [PubMed] [Google Scholar]
- 26.Schultz RA, Swoap SJ, McDaniel LD, Zhang B, Koon EC, Garry DJ, Li K, Williams RS. Differential expression of mitochondrial DNA replication factors in mammalian tissues. J. Biol. Chem. 1998;273:3447–3451. doi: 10.1074/jbc.273.6.3447. [DOI] [PubMed] [Google Scholar]
- 27.Barat-Gueride M, Dufresne C, Rickwood D. Effect of DNA conformation on the transcription of mitochondrial DNA. Eur. J. Biochem. 1989;183:297–302. doi: 10.1111/j.1432-1033.1989.tb14928.x. [DOI] [PubMed] [Google Scholar]
- 28.Achanta G, Sasaki R, Feng L, Carew JS, Lu W, Pelicano H, Keating MJ, Huang P. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. EMBO J. 2005;24:3482–3492. doi: 10.1038/sj.emboj.7600819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Romer L, Klein C, Dehner A, Kessler H, Buchner J. p53–a natural cancer killer: structural insights and therapeutic concepts. Angew. Chem. Int. Ed. Engl. 2006;45:6440–6460. doi: 10.1002/anie.200600611. [DOI] [PubMed] [Google Scholar]
- 30.Higuchi M. Regulation of mitochondrial DNA content and cancer. Mitochondrion. 2007;7:53–57. doi: 10.1016/j.mito.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conley KE, Marcinek DJ, Villarin J. Mitochondrial dysfunction and age. Curr. Opin. Clin. Nutr. Metab. Care. 2007;10:688–692. doi: 10.1097/MCO.0b013e3282f0dbfb. [DOI] [PubMed] [Google Scholar]
- 32.Terzioglu M, Larsson NG. Mitochondrial dysfunction in mammalian ageing. Novartis Found. Symp. 2007;287 doi: 10.1002/9780470725207.ch14. 197–208; discussion 208–113. [DOI] [PubMed] [Google Scholar]
- 33.Copeland WC. Inherited mitochondrial diseases of DNA replication. Annu. Rev. Med. 2008;59:131–146. doi: 10.1146/annurev.med.59.053006.104646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baron M, Kudin AP, Kunz WS. Mitochondrial dysfunction in neurodegenerative disorders. Biochem. Soc. Trans. 2007;35:1228–1231. doi: 10.1042/BST0351228. [DOI] [PubMed] [Google Scholar]
- 35.Loft S, Poulsen HE. Markers of oxidative damage to DNA: antioxidants and molecular damage. Methods Enzymol. 1999;300:166–184. doi: 10.1016/s0076-6879(99)00124-x. [DOI] [PubMed] [Google Scholar]
- 36.Joerger AC, Allen MD, Fersht AR. Crystal structure of a superstable mutant of human p53 core domain. Insights into the mechanism of rescuing oncogenic mutations. J. Biol. Chem. 2004;279:1291–1296. doi: 10.1074/jbc.M309732200. [DOI] [PubMed] [Google Scholar]
- 37.Nikolova PV, Henckel J, Lane DP, Fersht AR. Semirational design of active tumor suppressor p53 DNA binding domain with enhanced stability. Proc. Natl Acad. Sci. USA. 1998;95:14675–14680. doi: 10.1073/pnas.95.25.14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Veprintsev DB, Freund SM, Andreeva A, Rutledge SE, Tidow H, Canadillas JM, Blair CM, Fersht AR. Core domain interactions in full-length p53 in solution. Proc. Natl Acad. Sci. USA. 2006;103:2115–2119. doi: 10.1073/pnas.0511130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plotnikov V, Rochalski A, Brandts M, Brandts JF, Williston S, Frasca V, Lin LN. An autosampling differential scanning calorimeter instrument for studying molecular interactions. Assay Drug Dev. Technol. 2002;1:83–90. doi: 10.1089/154065802761001338. [DOI] [PubMed] [Google Scholar]
- 40.Weinberg RL, Veprintsev DB, Bycroft M, Fersht AR. Comparative binding of p53 to its promoter and DNA recognition elements. J. Mol. Biol. 2005;348:589–596. doi: 10.1016/j.jmb.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 41.Weinberg RL, Veprintsev DB, Fersht AR. Cooperative binding of tetrameric p53 to DNA. J. Mol. Biol. 2004;341:1145–1159. doi: 10.1016/j.jmb.2004.06.071. [DOI] [PubMed] [Google Scholar]
- 42.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3rd. New York: Springer; 2006. [Google Scholar]
- 43.Bodenhausen G, Ruben DJ. Natural abundance N-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 1980;69:185–189. [Google Scholar]
- 44.Curth U, Urbanke C, Greipel J, Gerberding H, Tiranti V, Zeviani M. Single-stranded-DNA-binding proteins from human mitochondria and Escherichia coli have analogous physicochemical properties. Eur. J. Biochem. 1994;221:435–443. doi: 10.1111/j.1432-1033.1994.tb18756.x. [DOI] [PubMed] [Google Scholar]
- 45.Meyer RR, Glassberg J, Kornberg A. An Escherichia coli mutant defective in single-strand binding protein is defective in DNA replication. Proc. Natl Acad. Sci. USA. 1979;76:1702–1705. doi: 10.1073/pnas.76.4.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyer RR, Glassberg J, Scott JV, Kornberg A. A temperature-sensitive single-stranded DNA-binding protein from Escherichia coli. J. Biol. Chem. 1980;255:2897–2901. [PubMed] [Google Scholar]
- 47.Li K, Williams RS. Tetramerization and single-stranded DNA binding properties of native and mutated forms of murine mitochondrial single-stranded DNA-binding proteins. J. Biol. Chem. 1997;272:8686–8694. doi: 10.1074/jbc.272.13.8686. [DOI] [PubMed] [Google Scholar]
- 48.Khamis MI, Casas-Finet JR, Maki AH, Murphy JB, Chase JW. Investigation of the role of individual tryptophan residues in the binding of Escherichia coli single-stranded DNA binding protein to single-stranded polynucleotides. A study by optical detection of magnetic resonance and site-selected mutagenesis. J. Biol. Chem. 1987;262:10938–10945. [PubMed] [Google Scholar]
- 49.Casas-Finet JR, Khamis MI, Maki AH, Chase JW. Tryptophan 54 and phenylalanine 60 are involved synergistically in the binding of E. coli SSB protein to single-stranded polynucleotides. FEBS Lett. 1987;220:347–352. doi: 10.1016/0014-5793(87)80844-x. [DOI] [PubMed] [Google Scholar]
- 50.Zang LH, Maki AH, Murphy JB, Chase JW. Triplet state sublevel kinetics of tryptophan 54 in the complex of Escherichia coli single-stranded DNA binding protein with single-stranded poly(deoxythymidylic) acid. Biophys. J. 1987;52:867–872. doi: 10.1016/S0006-3495(87)83280-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsao DH, Casas-Finet JR, Maki AH, Chase JW. Triplet state properties of tryptophan residues in complexes of mutated Escherichia coli single-stranded DNA binding proteins with single-stranded polynucleotides. Biophys. J. 1989;55:927–936. doi: 10.1016/S0006-3495(89)82891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khamis MI, Casas-Finet JR, Maki AH, Murphy JB, Chase JW. Role of tryptophan 54 in the binding of E. coli single-stranded DNA-binding protein to single-stranded polynucleotides. FEBS Lett. 1987;211:155–159. doi: 10.1016/0014-5793(87)81427-8. [DOI] [PubMed] [Google Scholar]
- 53.Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat. Struct. Biol. 2000;7:648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 54.Bochkareva E, Kaustov L, Ayed A, Yi GS, Lu Y, Pineda-Lucena A, Liao JC, Okorokov AL, Milner J, Arrowsmith CH, et al. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc. Natl Acad. Sci. USA. 2005;102:15412–15417. doi: 10.1073/pnas.0504614102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vise PD, Baral B, Latos AJ, Daughdrill GW. NMR chemical shift and relaxation measurements provide evidence for the coupled folding and binding of the p53 transactivation domain. Nucleic Acids Res. 2005;33:2061–2077. doi: 10.1093/nar/gki336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshida Y, Izumi H, Torigoe T, Ishiguchi H, Itoh H, Kang D, Kohno K. P53 physically interacts with mitochondrial transcription factor A and differentially regulates binding to damaged DNA. Cancer Res. 2003;63:3729–3734. [PubMed] [Google Scholar]
- 57.Chang J, Kim DH, Lee SW, Choi KY, Sung YC. Transactivation ability of p53 transcriptional activation domain is directly related to the binding affinity to TATA-binding protein. J. Biol. Chem. 1995;270:25014–25019. doi: 10.1074/jbc.270.42.25014. [DOI] [PubMed] [Google Scholar]
- 58.Walker KK, Levine AJ. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proc. Natl Acad. Sci. USA. 1996;93:15335–15340. doi: 10.1073/pnas.93.26.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 60.Shvarts A, Steegenga WT, Riteco N, van Laar T, Dekker P, Bazuine M, van Ham RC, van der Houven van Oordt W, Hateboer G, van der Eb AJ, et al. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 1996;15:5349–5357. [PMC free article] [PubMed] [Google Scholar]
- 61.Truant R, Xiao H, Ingles CJ, Greenblatt J. Direct interaction between the transcriptional activation domain of human p53 and the TATA box-binding protein. J. Biol. Chem. 1993;268:2284–2287. [PubMed] [Google Scholar]
- 62.Buschmann T, Lin Y, Aithmitti N, Fuchs SY, Lu H, Resnick-Silverman L, Manfredi JJ, Ronai Z, Wu X. Stabilization and activation of p53 by the coactivator protein TAFII31. J. Biol. Chem. 2001;276:13852–13857. doi: 10.1074/jbc.M007955200. [DOI] [PubMed] [Google Scholar]
- 63.Thut CJ, Chen JL, Klemm R, Tjian R. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 64.Di Lello P, Jenkins LM, Jones TN, Nguyen BD, Hara T, Yamaguchi H, Dikeakos JD, Appella E, Legault P, Omichinski JG. Structure of the Tfb1/p53 complex: Insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53. Mol. Cell. 2006;22:731–740. doi: 10.1016/j.molcel.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 65.Xiao H, Pearson A, Coulombe B, Truant R, Zhang S, Regier JL, Triezenberg SJ, Reinberg D, Flores O, Ingles CJ, et al. Binding of basal transcription factor TFIIH to the acidic activation domains of VP16 and p53. Mol. Cell. Biol. 1994;14:7013–7024. doi: 10.1128/mcb.14.10.7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Teufel DP, Freund SM, Bycroft M, Fersht AR. Four domains of p300 each bind tightly to a sequence spanning both transactivation subdomains of p53. Proc. Natl Acad. Sci. USA. 2007;104:7009–7014. doi: 10.1073/pnas.0702010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barja G, Herrero A. Oxidative damage to mitochondrial DNA is inversely related to maximum life span in the heart and brain of mammals. FASEB J. 2000;14:312–318. doi: 10.1096/fasebj.14.2.312. [DOI] [PubMed] [Google Scholar]
- 68.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 69.Klein JC, Bleeker MJ, Saris CP, Roelen HC, Brugghe HF, van den Elst H, van der Marel GA, van Boom JH, Westra JG, Kriek E, et al. Repair and replication of plasmids with site-specific 8-oxodG and 8-AAFdG residues in normal and repair-deficient human cells. Nucleic Acids Res. 1992;20:4437–4443. doi: 10.1093/nar/20.17.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zaccolo M, Williams DM, Brown DM, Gherardi E. An approach to random mutagenesis of DNA using mixtures of triphosphate derivatives of nucleoside analogues. J. Mol. Biol. 1996;255:589–603. doi: 10.1006/jmbi.1996.0049. [DOI] [PubMed] [Google Scholar]
- 71.Wong TS, Roccatano D, Schwaneberg U. Are transversion mutations better? A mutagenesis assistant program analysis on P450 BM-3 heme domain. Biotechnol. J. 2007;2:133–142. doi: 10.1002/biot.200600201. [DOI] [PubMed] [Google Scholar]
- 72.Abramova NA, Russell J, Botchan M, Li R. Interaction between replication protein A and p53 is disrupted after UV damage in a DNA repair-dependent manner. Proc. Natl Acad. Sci. USA. 1997;94:7186–7191. doi: 10.1073/pnas.94.14.7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li N, Ragheb K, Lawler G, Sturgis J, Rajwa B, Melendez JA, Robinson JP. Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J. Biol. Chem. 2003;278:8516–8525. doi: 10.1074/jbc.M210432200. [DOI] [PubMed] [Google Scholar]
- 74.Janus F, Albrechtsen N, Knippschild U, Wiesmuller L, Grosse F, Deppert W. Different regulation of the p53 core domain activities 3′-to-5′ exonuclease and sequence-specific DNA binding. Mol. Cell. Biol. 1999;19:2155–2168. doi: 10.1128/mcb.19.3.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mummenbrauer T, Janus F, Muller B, Wiesmuller L, Deppert W, Grosse F. p53 Protein exhibits 3′-to-5′ exonuclease activity. Cell. 1996;85:1089–1099. doi: 10.1016/s0092-8674(00)81309-4. [DOI] [PubMed] [Google Scholar]
- 76.Bakhanashvili M, Gedelovich R, Grinberg S, Rahav G. Exonucleolytic degradation of RNA by p53 protein in cytoplasm. J. Mol. Med. 2008;86:75–88. doi: 10.1007/s00109-007-0247-5. [DOI] [PubMed] [Google Scholar]
- 77.Bakhanashvili M, Novitsky E, Rubinstein E, Levy I, Rahav G. Excision of nucleoside analogs from DNA by p53 protein, a potential cellular mechanism of resistance to inhibitors of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 2005;49:1576–1579. doi: 10.1128/AAC.49.4.1576-1579.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bakhanashvili M. Exonucleolytic proofreading by p53 protein. Eur. J. Biochem. 2001;268:2047–2054. doi: 10.1046/j.1432-1327.2001.02075.x. [DOI] [PubMed] [Google Scholar]
- 79.Lilling G, Elena N, Sidi Y, Bakhanashvili M. p53-associated 3′–>5′ exonuclease activity in nuclear and cytoplasmic compartments of cells. Oncogene. 2003;22:233–245. doi: 10.1038/sj.onc.1206111. [DOI] [PubMed] [Google Scholar]
- 80.Lohman TM, Ferrari ME. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 81.Lohman TM, Overman LB. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J. Biol. Chem. 1985;260:3594–3603. [PubMed] [Google Scholar]
- 82.Bujalowski W, Lohman TM. Escherichia coli single-strand binding protein forms multiple, distinct complexes with single-stranded DNA. Biochemistry. 1986;25:7799–7802. doi: 10.1021/bi00372a003. [DOI] [PubMed] [Google Scholar]
- 83.Lohman TM, Overman LB, Datta S. Salt-dependent changes in the DNA binding co-operativity of Escherichia coli single strand binding protein. J. Mol. Biol. 1986;187:603–615. doi: 10.1016/0022-2836(86)90338-4. [DOI] [PubMed] [Google Scholar]
- 84.Chrysogelos S, Griffith J. Escherichia coli single-strand binding protein organizes single-stranded DNA in nucleosome-like units. Proc. Natl Acad. Sci. USA. 1982;79:5803–5807. doi: 10.1073/pnas.79.19.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Griffith JD, Harris LD, Register J., III Visualization of SSB-ssDNA complexes active in the assembly of stable RecA-DNA filaments. Cold Spring Harb. Symp. Quant. Biol. 1984;49:553–559. doi: 10.1101/sqb.1984.049.01.062. [DOI] [PubMed] [Google Scholar]
- 86.Bujalowski W, Lohman TM. Negative co-operativity in Escherichia coli single strand binding protein-oligonucleotide interactions. II. Salt, temperature and oligonucleotide length effects. J. Mol. Biol. 1989;207:269–288. doi: 10.1016/0022-2836(89)90455-5. [DOI] [PubMed] [Google Scholar]
- 87.Marchenko ND, Wolff S, Erster S, Becker K, Moll UM. Monoubiquitylation promotes mitochondrial p53 translocation. EMBO J. 2007;26:923–934. doi: 10.1038/sj.emboj.7601560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dumont P, Leu JI, Della Pietra A.C., III, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat. Genet. 2003;33:357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 89.Wolff S, Erster S, Palacios G, Moll UM. p53′s mitochondrial translocation and MOMP action is independent of Puma and Bax and severely disrupts mitochondrial membrane integrity. Cell Res. 2008;18:733–744. doi: 10.1038/cr.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Casas F, Daury L, Grandemange S, Busson M, Seyer P, Hatier R, Carazo A, Cabello G, Wrutniak-Cabello C. Endocrine regulation of mitochondrial activity: involvement of truncated RXRalpha and c-Erb Aalpha1 proteins. FASEB J. 2003;17:426–436. doi: 10.1096/fj.02-0732com. [DOI] [PubMed] [Google Scholar]
- 91.Chen JQ, Delannoy M, Cooke C, Yager JD. Mitochondrial localization of ERalpha and ERbeta in human MCF7 cells. Am. J. Physiol. Endocrinol. Metab. 2004;286:E1011–E1022. doi: 10.1152/ajpendo.00508.2003. [DOI] [PubMed] [Google Scholar]
- 92.Davis AF, Clayton DA. In situ localization of mitochondrial DNA replication in intact mammalian cells. J. Cell. Biol. 1996;135:883–893. doi: 10.1083/jcb.135.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat. Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 94.Funk WD, Pak DT, Karas RH, Wright WE, Shay JW. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol. Cell Biol. 1992;12:2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shakked Z, Yavnilovitch M, Kalb Gilboa AJ, Kessler N, Wolkowicz R, Rotter V, Haran TE. DNA binding and 3′-5′ exonuclease activity in the murine alternatively-spliced p53 protein. Oncogene. 2002;21:5117–5126. doi: 10.1038/sj.onc.1205667. [DOI] [PubMed] [Google Scholar]
- 96.Albrechtsen N, Dornreiter I, Grosse F, Kim E, Wiesmuller L, Deppert W. Maintenance of genomic integrity by p53: complementary roles for activated and non-activated p53. Oncogene. 1999;18:7706–7717. doi: 10.1038/sj.onc.1202952. [DOI] [PubMed] [Google Scholar]
- 97.Takamatsu C, Umeda S, Ohsato T, Ohno T, Abe Y, Fukuoh A, Shinagawa H, Hamasaki N, Kang D. Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA-binding protein. EMBO Rep. 2002;3:451–456. doi: 10.1093/embo-reports/kvf099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang Y, Bogenhagen DF. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J. Biol. Chem. 2006;281:25791–25802. doi: 10.1074/jbc.M604501200. [DOI] [PubMed] [Google Scholar]
- 99.Nemajerova A, Erster S, Moll UM. The post-translational phosphorylation and acetylation modification profile is not the determining factor in targeting endogenous stress-induced p53 to mitochondria. Cell Death Differ. 2005;12:197–200. doi: 10.1038/sj.cdd.4401526. [DOI] [PubMed] [Google Scholar]
- 100.Heyne K, Mannebach S, Wuertz E, Knaup KX, Mahyar-Roemer M, Roemer K. Identification of a putative p53 binding sequence within the human mitochondrial genome. FEBS Lett. 2004;578:198–202. doi: 10.1016/j.febslet.2004.10.099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.