Abstract
While most of the recent improvements in multiple sequence alignment accuracy are due to better use of vertical information, which include the incorporation of consistency-based pairwise alignments and the use of profile alignments, we observe that it is possible to further improve accuracy by taking into account alignment of neighboring residues when aligning two residues, thus making better use of horizontal information. By modifying existing multiple alignment algorithms to make use of horizontal information, we show that this strategy is able to consistently improve over existing algorithms on a few sets of benchmark alignments that are commonly used to measure alignment accuracy, and the average improvements in accuracy can be as much as 1–3% on protein sequence alignment and 5–10% on DNA/RNA sequence alignment. Unlike previous algorithms, consistent average improvements can be obtained across all identity levels.
INTRODUCTION
The construction of multiple sequence alignments is among the most important techniques to perform biological sequence analysis, with important applications to many areas of computational biology. The most popular strategy to construct multiple sequence alignments is by employing a progressive alignment algorithm, in which each sequence is treated initially as an alignment and the next two most similar alignments are repeatedly combined until a single multiple alignment is obtained (1–7). This is often followed by iterative refinements that improve the accuracy of the final alignment (3,4,6–8).
There are many recent efforts that lead to significant improvement of alignment accuracy, including the incorporation of consistency-based pairwise alignments that improve the quality of the initial pairwise alignments by aligning through other sequences to increase their agreement with the final multiple alignment (2,4–7), the use of maximal expected accuracy alignment (4–6), the incorporation of secondary structure predictions (7,9–11), the use of local structural information (12–14), and the incorporation of additional sequences from database search (9,10,15,16).
While most of these algorithms are able to significantly improve alignment accuracy by making better use of vertical information, either by incorporating consistency-based pairwise alignments or by using profiles in which each column of an alignment is modeled independently, we observe that most of these algorithms do not make use of horizontal information when constructing alignments, and it may be useful to take into account alignment of neighboring residues when aligning two residues.
There are a few previous approaches that use neighboring information to obtain significant performance improvements in other applications. Spang et al. (17) obtained a jumping alignment that is suitable for remote homology detection between a given sequence and a multiple alignment by aligning each position in the given sequence to one of the sequences in the multiple alignment while penalizing each vertical jump between horizontal moves. Panchenko et al. (18) used average conservation scores across spatial neighboring sites in the local structural environment to improve functional site prediction, while Capra and Singh (19) used conservation scores from neighboring residues to improve the prediction of functionally important residues in aligned sequences.
To incorporate horizontal information in alignments, we develop a window-based method that adjusts the pairwise score of a residue pair between two sequences (or a column pair between two sub-alignments) by incorporating the scores of neighboring residue pairs (or column pairs). This method can be applied to any multiple alignment algorithm that uses pairwise scores during the construction of a multiple alignment.
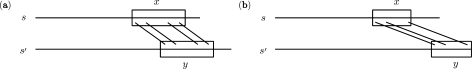
Since conserved residues in core regions tend to be clustered together (19,20), this strategy reduces the differences among neighboring scores within these regions, and can potentially lead to better gap placements by encouraging higher concentrations of consecutively aligned residues and more extensive grouping of consecutive indels, which is especially helpful when the similarity within a core region has large fluctuations. Figure 1 illustrates an example in which the strategy removes one incorrect long gap within an alignment that arises from a short fragment of sequence similarities which do not agree in secondary structure.
Figure 1.
Illustration of the beginning portion of the alignment of sequences 1smvA and 4sbvC from PREFAB (3) by different algorithms. (a) Alignment by MUSCLE (3). (b) Alignment by our algorithm NRAlign that modifies MUSCLE, which agrees with the reference structural alignment in PREFAB, where SS is the secondary structure assignment from DSSP (21), with L denoting loop and E denoting extended strand.
We test the strategy by modifying existing multiple alignment algorithms to make use of horizontal information and show that consistent average improvements can be obtained for these algorithms on all sets of benchmark alignments that we have tested. By using a statistical test that pairs the alignments before and after algorithm modification, we show that highly statistically significant improvements are obtained not just in relative accuracy but also in paired accuracy. We also verify that better gap placements are achieved by comparing the distributions of gaps and the lengths of alignments before and after algorithm modification.
METHODS
Incorporating horizontal information into pairwise scores
Given a residue (or column) at position x in the first sequence (or sub-alignment) s=s1···sm, a residue (or column) at position y in the second sequence (or sub-alignment) s′ = s′1··· s′n}, and a parameter ω, define the window that includes at most ω positions to the left and to the right of (x, y) by the following set of valid offsets in the neighborhood of (x, y) (Figure 2):
We use the following equation to incorporate the scores of the neighboring pairs at position (x + i,y + i) over all offsets i in Nω(x,y) into the score of the given pair (x, y):
| 1 |
where Sold is the original score, Snew is the adjusted score, and β is a parameter that specifies the weight of the neighboring scores during the adjustment. For each alignment of two sequences (or two sub-alignments), this step takes O(ωl2) time, where l is the maximum sequence (or sub-alignment) length.
Figure 2.
Illustration of the window on two sequences s and s′ with ω = 2. (a) The offsets in Nω(x,y) = {−2,−1,1,2} are included. (b) Since y + 1 is the last position in s′, only one position is used to the right of (x,y) and the offsets in Nω(x,y) = {−2,−1,1} are included.
We apply this strategy to TCoffee 5.31 (2) without using structural information, which is among the first multiple alignment algorithms that utilize consistency-based pairwise alignments, to MUSCLE 3.6 (3), which is among the most efficient multiple alignment algorithms that also have high accuracy, to ProbCons 1.10 (4), which is among the first multiple alignment algorithms that utilize the maximal expected accuracy alignment based on a pair-HMM model, and to MUMMALS 1.01 (5), which uses secondary structure information during pair-HMM training to further improve alignment accuracy. Except for MUMMALS, we test both the protein and DNA/RNA versions of each algorithm. For ProbCons, the DNA/RNA version ProbConsRNA was obtained from parameter training on BRAliBase II (22).
In each case, we evaluate the accuracy of each of the modified algorithms (called NRAlign) against each of the original algorithms while using the same parameter setting across different benchmark alignments for each modified algorithm (Table 1), with values of ω in the DNA/RNA version being three times as large as the protein version. Horizontal information is incorporated into each of the algorithms either during the computation of consistency-based pairwise alignments or during the progressive alignment step.
Table 1.
Parameter settings for the modified version of each algorithm that uses horizontal information
| Protein |
DNA/RNA |
||||||
|---|---|---|---|---|---|---|---|
| TCoffee | MUSCLE | ProbCons | MUMMALS | TCoffee | MUSCLE | ProbConsRNA | |
| ω | 3 | 2 | 5 | 1 | 9 | 6 | 15 |
| β | 0.7 | 1.0 | 1.0 | 0.8 | 0.7 | 1.0 | 1.0 |
Modification of TCoffee
The TCoffee algorithm consists of the following steps: construct a library of pairwise alignments from the input sequences by using global alignments from ClustalW (1) and local alignments from Lalign (23), assign a weight to each pair of aligned residues in the library according to sequence identity, apply library extension to all the weights in the library to obtain an extended library that utilizes consistency-based information by using a triplet approach, and perform progressive alignment according to a guide tree by aligning two groups of pre-aligned sequences using the average scores between column pairs in the extended library. In NRAlign, we apply Equation (1) to adjust the average extended library scores between column pairs before each progressive alignment step.
Modification of MUSCLE
The MUSCLE algorithm consists of the following steps: compute the k-mer distance for each pair of input sequences to produce an initial tree and perform progressive alignment according to the tree by utilizing log-expectation scores between two aligned columns to obtain an initial multiple alignment, re-estimate the tree using Kimura distances (24) computed from the multiple alignment and perform progressive alignment according to the new tree, then perform iterative refinements to obtain the final alignment. In NRAlign, we apply Equation (1) to adjust the log-expectation scores before each progressive alignment step.
Modification of ProbCons
The ProbCons algorithm consists of the following steps: compute the posterior probability matrix for each pair of input sequences according to the pair-HMM model, compute maximal expected accuracy alignment for each sequence pair by dynamic programming, re-estimate the match quality score matrix for each sequence pair by performing probabilistic consistency transformation, construct a guide tree according to the maximal expected accuracy alignments, perform progressive alignment according to the guide tree by using the transformed scores, and perform iterative refinements to obtain the final alignment. In NRAlign, we apply Equation (1) to adjust the match quality scores for each sequence pair before consistency transformation is performed.
Modification of MUMMALS
The MUMMALS algorithm consists of the following steps: compute the k-mer distance for each pair of input sequences to produce an initial tree and perform progressive alignment according to the tree to obtain an initial multiple alignment, re-estimate the tree using sequence identities computed from the multiple alignment, perform a two-stage progressive alignment in which highly similar sequences are first aligned by using weighted sum-of-pairs BLOSUM62 scores (25), and a representative is chosen from each pre-aligned group to perform progressive consistency-based multiple alignment based on transformed pairwise maximal expected accuracy alignments that are obtained from a pair-HMM model that also includes secondary structure states, then merge the pre-aligned groups according to the alignment of the representatives to obtain the final alignment. In NRAlign, we apply Equation (1) to adjust the scores between column pairs before each progressive alignment step.
Availability
NRAlign is available for download at http://faculty.cs.tamu.edu/shsze/nralign.
RESULTS
Benchmark alignments
To evaluate the accuracy of NRAlign on multiple protein sequence alignment, we use benchmark multiple alignments from BAliBASE 3.0 (26), which contains manually refined structural alignments that are subdivided into five categories, from HOMSTRAD (27), which contains a collection of manually edited structure-based alignments, from PREFAB 4.0 (3), which contains structural alignments of two sequences and automatically generated alignments that are obtained from adding high scoring hits of the two sequences from database search, and from SABmark 1.65 (13), which contains alignments that are derived from the SCOP classification (28).
To evaluate the accuracy of NRAlign on multiple DNA/RNA sequence alignment, we use benchmark multiple alignments from BRAliBase II (22), which contains alignments of non-coding RNA sequences of Group II introns, 5S rRNA, SRP, tRNA and U5 splicesomal RNA from the Rfam database (29), and DNA PREFAB (30), which contains alignments of DNA sequences that are obtained from database search of protein sequences from PREFAB 4.0.
Two reference-dependent scores are used to evaluate the accuracy of each algorithm, including the sum-of-pairs score (SPS), which measures the percentage of residue pairs that are aligned correctly in the reference alignment, and the column score (CS), which measures the percentage of entire columns that are aligned correctly (31). For BAliBASE, PREFAB and DNA PREFAB, evaluations are made only on the core regions that are specified in the reference alignments. For PREFAB, SABmark and DNA PREFAB, the reference alignments are based on sequence pairs and the CS score is not used. For PREFAB and DNA PREFAB, the Q score (3) is computed on the original input sequence pair, which has the same meaning as the SPS score. For SABmark, reference alignments are specified for each sequence pair, and the fD score, which is a sensitivity score that has the same meaning as the SPS score, and an additional fM score, which is a specificity score that measures the percentage of residue pairs that are aligned correctly in the test alignment, are computed by averaging the scores over all sequence pairs for each multiple alignment (13).
In addition to reference-dependent scores, four reference-independent scores are used in the presence of known 3D structures to evaluate the structural agreement between aligned protein sequence pairs, including the Dali Z-score (32), which computes a structural similarity score as a weighted sum of similarities of intramolecular distances between residues in aligned columns normalized according to alignments of random structure pairs [see Equations 2–4 in (32)], the GDT_TS score (33,34), which computes the average of the maximum number of aligned residue pairs that can be superimposed within four different distance thresholds of 1, 2, 4 and 8 Å [see the Equation in (34)], and two LiveBench contact scores ContactA and ContactB (35,36), which compute an overlap score that is the lower of two contact scores, one for each structure, computed based on intramolecular distances between residues in aligned columns that are separated by at least five residues [see Equation 2 in (36)], with ContactA normalized for each residue and ContactB normalized over the entire contact map.
To compute the GDT_TS score, multiple superpositions of aligned residue pairs are needed that optimize the individual score components, and the software from (37) is used with the set of aligned residue pairs as input while omitting the final normalization step. Following the procedure in (5), each score is further weighted and normalized against the reverse alignment that represents a random model. For SABmark, three-dimensional coordinates are extracted from the given PDB files (38), and the scores for each multiple alignment are computed by averaging the scores over all sequence pairs.
For RNA sequence alignment, the structure conservation index (SCI) in (39) is used, which is a reference-independent score that computes the ratio of the consensus RNA folding minimum free energy of an alignment to the average of the RNA folding minimum free energy of each individual sequence in the alignment [see the Equation in (39)].
To evaluate whether the use of NRAlign leads to significant improvements, we use the Wilcoxon matched-pairs signed-ranks test (40) over subsets that are large enough with P = 0.05 as significance cutoff, in which the alignments before and after algorithm modification are paired to evaluate whether the improvements are consistent not just in relative accuracy but also in paired accuracy.
Accuracy of NRAlign on protein sequence alignment
Table 2 shows accuracy comparisons on full length protein sequences in BAliBASE 3.0. Among all the subsets that are large enough, NRAlign always performed at least as well as the original algorithm. Except for MUSCLE, the improvements of NRAlign were more statistically significant in the CS score when compared to the SPS score, and this is especially evident on TCoffee. The improvements in the CS score were >2% in references 1V2, 2, 3 and 4 over MUSCLE and in reference 1V2 over MUMMALS, >4% in reference 5 over MUMMALS, and >1% in the entire set over MUSCLE and MUMMALS.
Table 2.
Average SPS and CS scores (in %) on full length protein sequences in BAliBASE 3.0
| TCoffee |
MUSCLE |
ProbCons |
MUMMALS |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SPS | ||||||||||||
| 1V1 {38} | 53.81 | 54.21 | 56.21 | 56.98 | 64.46 | 64.48 | 64.41 | 64.23 | ||||
| 1V2 {44} | 91.55 | 91.98 | 90.62 | 91.50 | 93.50 | 93.65 | 93.53 | 94.00 | ||||
| 1 (V1–V2) {82} | 74.06 | 74.48 | 0.02 | 74.67 | 75.50 | 0.003 | 80.05 | 80.13 | – | 80.03 | 80.20 | – |
| 2 {41} | 89.04 | 88.82 | 88.08 | 88.24 | 89.93 | 89.94 | 89.18 | 89.39 | ||||
| 3 {30} | 71.09 | 71.19 | 75.01 | 76.27 | 78.62 | 78.30 | 80.76 | 80.79 | ||||
| 4 {49} | 82.21 | 82.37 | 84.83 | 85.64 | 87.43 | 87.25 | 83.69 | 83.97 | ||||
| 5 {16} | 81.94 | 80.98 | 82.69 | 82.83 | 87.69 | 87.87 | 86.33 | 87.40 | ||||
| All (1–5) {218} | 78.88 | 78.97 | 0.04 | 80.11 | 80.82 | 0.006 | 83.93 | 83.89 | – | 83.14 | 83.39 | – |
| CS | ||||||||||||
| 1V1 {38} | 31.34 | 32.21 | 35.63 | 33.95 | 40.45 | 41.00 | 41.61 | 41.39 | ||||
| 1V2 {44} | 81.64 | 82.68 | 80.75 | 82.93 | 85.52 | 85.77 | 83.98 | 86.41 | ||||
| 1 (V1–V2) {82} | 58.33 | 59.29 | 1×10−4 | 59.84 | 60.23 | 0.01 | 64.63 | 65.02 | 0.02 | 64.34 | 65.55 | – |
| 2 {41} | 37.85 | 38.88 | 35.27 | 37.61 | 40.63 | 40.49 | 42.83 | 43.46 | ||||
| 3 {30} | 36.00 | 36.83 | 40.57 | 42.73 | 54.37 | 54.80 | 49.40 | 49.57 | ||||
| 4 {49} | 48.20 | 48.78 | 47.37 | 49.67 | 53.67 | 53.14 | 48.55 | 49.76 | ||||
| 5 {16} | 50.63 | 49.31 | 47.94 | 44.94 | 57.38 | 57.31 | 52.88 | 57.00 | ||||
| All (1–5) {218} | 48.56 | 49.27 | 7×10−9 | 48.89 | 50.07 | 0.002 | 55.71 | 55.77 | 0.04 | 53.85 | 55.02 | 0.001 |
Reference 1 contains alignments of sequences that are subdivided into two subsets 1V1 (<20% identity) and 1V2 (20–40% identity). Reference 2 contains alignments that include orphan sequences. Reference 3 contains alignments of clusters of sequences from different families. Reference 4 contains alignments of sequences with large terminal extensions, while reference 5 contains alignments of sequences with internal insertions. The number in braces denotes the number of alignments in each subset. For each algorithm, the first number shows the accuracy of the original algorithm (TCoffee, MUSCLE, ProbCons, MUMMALS) that does not use horizontal information. The second number shows the accuracy of the modified algorithm NRAlign that makes use of horizontal information, with the higher accuracy value in bold. The third number shows the P-value, with – indicating insignificant differences. Since many of the subsets are small, P-values are computed only for reference 1 and for the entire set.
Table 3 shows accuracy comparisons on HOMSTRAD. Except for 70–100% protein sequence identity where the improvements of NRAlign were statistically significant only over MUMMALS, all improvements at other identity levels were statistically significant (except for 0–20% over MUMMALS). The improvements were especially statistically significant when the identity is moderately low (20–40%), while the overall improvements were highly statistically significant over all algorithms.
Table 3.
Average SPS and CS scores (in %) on HOMSTRAD
| TCoffee |
MUSCLE |
ProbCons |
MUMMALS |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SPS | ||||||||||||
| 0–20% {156} | 46.68 | 47.21 | 0.005 | 48.08 | 50.18 | 4 × 10−4 | 49.67 | 50.67 | 6 × 10−8 | 54.39 | 54.44 | – |
| 20–40% {459} | 79.19 | 79.71 | 4 × 10−13 | 78.86 | 80.11 | 1 × 10−10 | 80.55 | 81.44 | 3 × 10−22 | 82.67 | 82.71 | 4 × 10−4 |
| 40–70% {348} | 94.48 | 94.80 | 2 × 10−11 | 94.45 | 94.77 | 1 × 10−4 | 94.75 | 95.19 | 7 × 10−12 | 95.04 | 95.14 | 9 × 10−4 |
| 70–100% {69} | 99.10 | 99.16 | – | 99.02 | 99.07 | – | 99.10 | 99.08 | – | 98.94 | 99.14 | 0.005 |
| All {1032} | 80.76 | 81.19 | 2 × 10−22 | 80.82 | 81.80 | 6 × 10−16 | 81.91 | 82.60 | 6 × 10−38 | 83.65 | 83.72 | 5 × 10−8 |
| CS | ||||||||||||
| 0–20% {156} | 39.97 | 40.64 | 2 × 10−4 | 41.77 | 43.70 | 0.003 | 43.12 | 44.15 | 3 × 10−7 | 47.94 | 48.00 | – |
| 20–40% {459} | 72.97 | 73.76 | 9 × 10−17 | 73.01 | 74.61 | 2 × 10−11 | 74.67 | 75.80 | 5 × 10−24 | 77.31 | 77.43 | 0.001 |
| 40–70% {348} | 91.79 | 92.33 | 2 × 10−13 | 91.90 | 92.28 | 8 × 10−5 | 92.20 | 92.84 | 6 × 10−13 | 92.61 | 92.77 | 3 × 10−5 |
| 70–100% {69} | 99.03 | 99.10 | – | 98.98 | 99.03 | – | 99.06 | 99.02 | – | 98.87 | 99.08 | 0.007 |
| All {1032} | 76.07 | 76.71 | 1 × 10−30 | 76.39 | 77.53 | 8 × 10−16 | 77.44 | 78.32 | 6 × 10−40 | 79.47 | 79.60 | 4 × 10−8 |
Each subset includes all protein sequence alignments with average pairwise identity within the specified range. For each algorithm, the higher accuracy value is in bold.
Table 4 shows accuracy comparisons on PREFAB 4.0. When only the original input protein sequence pair are aligned, the accuracy improvement characteristics of NRAlign were similar to those of HOMSTRAD, except that the improvements of NRAlign were statistically significant also for 70–100% identity over ProbCons. The improvements were more statistically significant in this case than in the case when the full set of at most 50 protein sequences are aligned, although using the full set of sequences gives better accuracy for each of the original and modified algorithms on divergent sequences (0–40% identity).
Table 4.
Average Q scores (in %) on PREFAB 4.0
| TCoffee |
MUSCLE |
ProbCons |
MUMMALS |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Q(2) | ||||||||||||
| 0–20% {887} | 37.92 | 38.27 | 1 × 10−5 | 38.22 | 39.69 | 8 × 10−8 | 38.95 | 40.17 | 7 × 10−31 | 43.59 | 43.62 | 0.005 |
| 20–40% {588} | 82.60 | 82.92 | 4 × 10−8 | 81.75 | 83.87 | 1 × 10−29 | 82.84 | 84.30 | 4 × 10−39 | 85.39 | 85.45 | 2 × 10−4 |
| 40–70% {112} | 96.37 | 96.51 | 0.005 | 96.24 | 96.58 | 0.01 | 96.41 | 96.83 | 5 × 10−6 | 96.59 | 96.75 | 5 × 10−4 |
| 70–100% {95} | 97.94 | 98.04 | – | 97.97 | 97.91 | – | 97.76 | 98.05 | 3 × 10−4 | 97.75 | 97.93 | 0.03 |
| All {1682} | 60.82 | 61.13 | 1 × 10−12 | 60.68 | 62.21 | 7 × 10−29 | 61.44 | 62.64 | 3 × 10−71 | 64.79 | 64.85 | 5 × 10−8 |
| Q(50) | ||||||||||||
| 0–20% {887} | 49.67 | 50.00 | 6 × 10−6 | 50.71 | 50.95 | – | 55.63 | 55.72 | 0.02 | 57.68 | 57.91 | 0.02 |
| 20–40% {588} | 83.94 | 84.20 | 8 × 10−7 | 85.09 | 85.13 | – | 87.24 | 87.38 | 3 × 10−7 | 87.24 | 87.30 | 0.02 |
| 40–70% {112} | 95.99 | 95.55 | 0.02* | 94.72 | 96.46 | – | 95.39 | 95.48 | 0.004 | 95.34 | 95.41 | – |
| 70–100% {95} | 97.97 | 98.04 | – | 97.50 | 97.69 | – | 97.26 | 97.40 | 0.001 | 96.68 | 97.04 | 0.005 |
| All {1682} | 67.46 | 67.70 | 2 × 10−9 | 68.30 | 68.57 | – | 71.68 | 71.79 | 1 × 10−7 | 72.73 | 72.89 | 5 × 10−4 |
Each subset includes all structure pairs with protein sequence identity within the specified range, with * indicating worse accuracy in P-value. The Q(2) scores are obtained from aligning only the original input protein sequence pair, while the Q(50) scores are obtained from aligning the full set of protein sequences (at most 50) that also include random hits from database search and evaluations are made on the original input sequence pair. For each algorithm, the higher accuracy value is in bold.
Table 5 shows accuracy comparisons on the Twilight and Superfamily subsets of SABmark 1.65. Unlike previous algorithms that have improvements mostly on divergent protein sequences, the improvements of NRAlign were more statistically significant on the Superfamily subset than on the more divergent Twilight subset. Similar to the results in (5), there are strong correlations between the reference-dependent and reference-independent results, which indicate that the improvements are not only at the protein sequence level but also at the structural level.
Table 5.
Average fD and fM scores and average normalized Dali Z-score, GDT_TS score, and ContactA and ContactB scores (in %) on the Twilight and Superfamily subsets of SABmark 1.65
| TCoffee |
MUSCLE |
ProbCons |
MUMMALS |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Twilight {205} | ||||||||||||
| fD | 24.07 | 23.99 | – | 24.07 | 25.29 | 0.008 | 29.26 | 29.72 | 0.01 | 31.57 | 31.63 | 0.04 |
| fM | 18.08 | 18.08 | – | 16.47 | 16.84 | – | 21.00 | 21.02 | – | 22.87 | 22.97 | 0.009 |
| Dali Z-score | 11.10 | 11.19 | 0.02 | 13.14 | 13.68 | 0.02 | 13.88 | 14.32 | 3 × 10−5 | 15.32 | 15.38 | 0.03 |
| GDT_TS | 10.67 | 10.78 | 0.007 | 12.45 | 12.91 | 0.03 | 13.38 | 13.68 | 5 × 10−4 | 14.52 | 14.54 | – |
| ContactA | 6.72 | 6.76 | – | 7.62 | 7.95 | 0.03 | 8.67 | 8.87 | 0.01 | 9.41 | 9.45 | – |
| ContactB | 8.98 | 9.03 | – | 10.06 | 10.47 | – | 12.10 | 12.37 | 0.01 | 12.59 | 12.61 | – |
| Superfamily {422} | ||||||||||||
| fD | 52.91 | 53.30 | 2 × 10−5 | 53.12 | 53.91 | 0.008 | 57.06 | 57.30 | 8 × 10−8 | 59.50 | 59.65 | 0.004 |
| fM | 41.30 | 41.52 | 5 × 10−4 | 39.87 | 40.26 | 0.04 | 43.57 | 43.61 | 0.03 | 45.15 | 45.25 | 0.01 |
| Dali Z-score | 33.09 | 33.25 | 0.04 | 35.34 | 35.85 | 0.002 | 35.84 | 36.26 | 9 × 10−21 | 37.79 | 37.87 | 0.001 |
| GDT_TS | 31.07 | 31.23 | 0.01 | 32.98 | 33.47 | 5 × 10−4 | 33.67 | 33.92 | 1 × 10−17 | 35.05 | 35.11 | 0.01 |
| ContactA | 23.07 | 23.14 | – | 24.23 | 24.54 | 0.001 | 25.29 | 25.45 | 2 × 10−9 | 26.41 | 26.45 | – |
| ContactB | 28.91 | 28.94 | – | 30.30 | 30.59 | 0.007 | 32.10 | 32.22 | 1 × 10−4 | 33.11 | 33.17 | 0.04 |
The Twilight subset contains protein sequence alignments that represent a SCOP fold (⩽25% identity), while the Superfamily subset contains protein sequence alignments that represent a SCOP superfamily (⩽50% identity). Four cases are omitted in the Twilight subset and three cases are omitted in the Superfamily subset since no high quality reference alignments are available. For each algorithm, the higher accuracy value is in bold.
When comparisons were made on the improvements among the different algorithms, we found that MUMMALS was the hardest to improve on HOMSTRAD and on PREFAB when using the original input protein sequence pair for moderate to low identity. ProbCons was the hardest to improve on BAliBASE, the easiest to improve on HOMSTRAD except for 70–100% identity and on PREFAB when using the original input protein sequence pair, while the improvements on TCoffee and MUSCLE varied across different benchmarks. This is in contrast with the better accuracy of ProbCons and MUMMALS over TCoffee and MUSCLE for moderate to low identity. The improvement characteristics were especially different on PREFAB depending on whether the original input protein sequence pair or the full set of protein sequences are aligned, when it was easier to improve on MUMMALS than on MUSCLE in the latter case.
Accuracy of NRAlign on DNA/RNA sequence alignment
Table 6 shows accuracy comparisons on Data-set 1 of BRAliBase II. The improvements of NRAlign were more statistically significant in the reference-independent SCI score when compared to the SPS and CS scores, where the improvements in the SCI score were >3% for moderate to low RNA sequence identity (0–75%) and were statistically significant for high RNA sequence identity (75–100%) over all algorithms. This is especially evident on TCoffee, where the improvements in the SCI score were >12% for moderate to low identity (0–75%) and >9% in the entire set. In the SPS and CS scores, except for 75–100% identity where the improvements of NRAlign were statistically significant only over ProbConsRNA in the CS score, all improvements at other identity levels were statistically significant (except for 55–75% over MUSCLE in the SPS score), with improvements of >3% over TCoffee.
Table 6.
Average SPS, CS and SCI scores (in %) on Data-set 1 of BRAliBase II
| TCoffee |
MUSCLE |
ProbConsRNA |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| SPS | |||||||||
| 0–55% {96} | 57.87 | 63.01 | 2 × 10−11 | 65.10 | 67.65 | 0.01 | 73.20 | 74.86 | 1 × 10−5 |
| 55–75% {218} | 80.07 | 83.41 | 5 × 10−22 | 83.62 | 84.35 | – | 86.08 | 87.06 | 4 × 10−8 |
| 75–100% {167} | 95.01 | 95.23 | – | 95.28 | 95.28 | – | 96.05 | 96.19 | – |
| All {481} | 80.83 | 83.44 | 9 × 10−32 | 83.97 | 84.81 | 0.004 | 86.97 | 87.80 | 1 × 10−12 |
| CS | |||||||||
| 0–55% {96} | 36.42 | 41.68 | 2 × 10−7 | 45.83 | 48.73 | 0.02 | 56.32 | 57.87 | 0.005 |
| 55–75% {218} | 65.29 | 70.56 | 7 × 10−23 | 71.03 | 72.30 | 0.02 | 74.57 | 75.97 | 2 × 10−6 |
| 75–100% {167} | 89.90 | 90.46 | – | 90.73 | 90.76 | – | 91.94 | 92.24 | 0.03 |
| All {481} | 68.07 | 71.70 | 5 × 10−28 | 72.84 | 74.00 | 0.002 | 76.96 | 78.01 | 2 × 10−8 |
| SCI | |||||||||
| 0–55% {96} | 31.84 | 46.13 | 3 × 10−14 | 50.80 | 55.22 | 2 × 10−4 | 57.33 | 61.63 | 3 × 10−5 |
| 55–75% {218} | 54.17 | 66.92 | 2 × 10−27 | 66.26 | 69.88 | 3 × 10−4 | 67.07 | 71.22 | 2 × 10−17 |
| 75–100% {167} | 87.58 | 88.75 | 0.03 | 89.30 | 89.99 | 0.03 | 89.23 | 90.09 | 1 × 10−4 |
| All {481} | 61.31 | 70.35 | 2 × 10−39 | 71.17 | 73.93 | 2 × 10−7 | 72.82 | 75.85 | 8 × 10−23 |
Each subset includes all alignments of five RNA sequences with average pairwise identity within the specified range. For each algorithm, the higher accuracy value is in bold.
Table 7 shows accuracy comparisons on the mdsa_all set of DNA PREFAB. Except for 70–100% DNA sequence identity over MUSCLE, all the improvements of NRAlign were statistically significant. The improvements were especially statistically significant for moderate to low identity (20–70%) and for the entire set, with improvements of >7% over MUSCLE and >4% for 40–70% identity over all algorithms.
Table 7.
Average Q scores (in %) on the mdsa_all set of DNA PREFAB
| Q | TCoffee |
MUSCLE |
ProbConsRNA |
||||||
|---|---|---|---|---|---|---|---|---|---|
| 0–20% {123} | 2.75 | 3.14 | 0.002 | 3.85 | 6.33 | 4 × 10−4 | 2.90 | 3.49 | 4 × 10−4 |
| 20–40% {1030} | 12.80 | 14.51 | 1 × 10−77 | 15.93 | 23.09 | 3 × 10−65 | 16.13 | 19.88 | 2 × 10−94 |
| 40–70% {436} | 51.78 | 56.47 | 6 × 10−69 | 60.17 | 73.19 | 1 × 10−64 | 59.38 | 66.43 | 2 × 10−69 |
| 70–100% {87} | 96.74 | 97.03 | 2 × 10−5 | 97.05 | 97.03 | – | 96.74 | 97.09 | 2 × 10−6 |
| All {1676} | 26.56 | 28.88 | 7 × 10−153 | 30.76 | 38.73 | 1 × 10−131 | 30.60 | 34.80 | 8 × 10−171 |
Each subset includes all pairs with DNA sequence identity within the specified range. For each algorithm, the higher accuracy value is in bold.
When comparisons were made on the improvements among the different algorithms, we found that MUSCLE was the hardest to improve on BRAliBase and the easiest to improve on accuracy in DNA PREFAB, while TCoffee was the easiest to improve on BRAliBase. This is in contrast with the better accuracy of MUSCLE and ProbConsRNA over TCoffee.
Overall accuracy of NRAlign
In all the subsets that we have assessed, NRAlign always performed at least as well as the original algorithm (except for one case). The overall improvements were highly statistically significant in most cases, even when the average improvements in accuracy can sometimes be small, and the improvements were much more evident on DNA/RNA sequence alignment than on protein sequence alignment. Unlike previous algorithms that have improvements mostly on divergent sequences, consistent average improvements can be obtained across all identity levels, and it is not always the case that the most improvements were obtained on highly divergent sequences.
Supplementary Tables S1 to S6 show the percentage of cases in which each of the scores in Tables 2–7 respectively becomes better and worse on each set of benchmark alignments when comparing the results of NRAlign to the original algorithm. The percentage of cases that become better was almost always larger than the percentage of cases that become worse even when the identity is very high, and the degree of relative improvement was reflected by the corresponding P-value in Tables 2–7, with less cases becoming better and less cases becoming worse simultaneously as identity increases in most situations.
DISCUSSION
Characteristics of alignments
Supplementary Tables S7 to S12 show that the number of gaps in an alignment (a string of consecutive indels within a sequence is counted as one gap), the average length of gaps and the length of the alignment had the tendency to become smaller, larger and smaller respectively when comparing the results of NRAlign to the original algorithm, with generally decreasing tendencies as we move from one category to the next in the above order as demonstrated by the P-values. This confirms that better gap placements are achieved to a larger extent through reducing the number of gaps. While each tendency to become smaller, larger and smaller respectively was almost always larger than the opposite tendency to become larger, smaller and larger respectively, each tendency to become smaller, larger and smaller respectively also diminished simultaneously with the opposite tendency to become larger, smaller and larger respectively as identity increases in most situations.
Pairwise alignment versus multiple alignment
The above results on PREFAB show that the improvements of NRAlign were more statistically significant on pairwise alignments. Since the reference alignments for SABmark are based on sequence pairs, we investigate this further by performing pairwise alignments over all protein sequence pairs instead of obtaining a single multiple alignment, and computing the scores for each multiple alignment by averaging the scores over all sequence pairs.
When compared to Table 5, Table 8 shows that the improvements in SABmark were more statistically significant when pairwise alignments are performed, and this is especially evident on the Superfamily subset, although obtaining a single multiple alignment gives better accuracy on both the Twilight and Superfamily subsets for each of the original and modified algorithms of ProbCons and MUMMALS, and on the Superfamily subset for each of the original and modified algorithms of MUSCLE.
Table 8.
Average fD and fM scores and average normalized Dali Z-score, GDT_TS score, and ContactA and ContactB scores (in %) on the Twilight and Superfamily subsets of SABmark 1.65 when pairwise alignments are performed over all protein sequence pairs instead of obtaining a single multiple alignment
| TCoffee |
MUSCLE |
ProbCons |
MUMMALS |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Twilight {205} | ||||||||||||
| fD | 24.88 | 25.06 | 0.005 | 25.30 | 26.50 | 4 × 10−5 | 26.23 | 26.49 | 4 × 10−4 | 29.13 | 29.17 | 0.02 |
| fM | 16.78 | 16.85 | – | 17.05 | 17.68 | 3 × 10−4 | 17.92 | 17.96 | 0.04 | 19.64 | 19.65 | – |
| Dali Z-score | 13.41 | 13.60 | 1 × 10−4 | 13.83 | 14.40 | 3 × 10−5 | 13.46 | 13.74 | 4 × 10−10 | 15.06 | 15.10 | 0.03 |
| GDT_TS | 12.74 | 12.89 | 7 × 10−8 | 13.16 | 13.69 | 2 × 10−7 | 12.88 | 13.10 | 5 × 10−11 | 14.24 | 14.26 | – |
| ContactA | 7.70 | 7.79 | 0.002 | 8.01 | 8.34 | 4 × 10−4 | 8.09 | 8.15 | 5 × 10−4 | 8.93 | 8.94 | – |
| ContactB | 10.17 | 10.29 | 0.01 | 10.75 | 10.98 | – | 10.99 | 10.94 | – | 11.90 | 11.92 | – |
| Superfamily {422} | ||||||||||||
| fD | 50.73 | 51.01 | 1 × 10−13 | 50.79 | 51.79 | 3 × 10−16 | 51.60 | 52.27 | 1 × 10−28 | 54.79 | 54.83 | 3 × 10−6 |
| fM | 38.09 | 38.24 | 5 × 10−9 | 38.16 | 38.85 | 3 × 10−11 | 39.10 | 39.45 | 7 × 10−19 | 41.06 | 41.08 | 5 × 10−5 |
| Dali Z-score | 33.82 | 33.98 | 3 × 10−11 | 33.80 | 34.60 | 2 × 10−19 | 33.56 | 34.23 | 7 × 10−45 | 35.67 | 35.64 | 1 × 10−5 |
| GDT_TS | 31.81 | 31.95 | 2 × 10−13 | 31.84 | 32.52 | 2 × 10−22 | 31.72 | 32.18 | 3 × 10−39 | 33.34 | 33.36 | 1 × 10−5 |
| ContactA | 23.11 | 23.19 | 2 × 10−6 | 23.21 | 23.74 | 9 × 10−20 | 23.29 | 23.63 | 3 × 10−25 | 24.65 | 24.64 | 0.01 |
| ContactB | 28.85 | 28.91 | 0.003 | 29.10 | 29.51 | 3 × 10−9 | 29.28 | 29.53 | 4 × 10−9 | 30.74 | 30.73 | – |
For each algorithm, the higher accuracy value is in bold.
To further investigate the effect of the number of sequences on the accuracy of NRAlign, we group the results on HOMSTRAD according to the number of protein sequences in each alignment. Table 9 shows that except for TCoffee, the improvements on HOMSTRAD were more statistically significant when the number of sequences is small, and the differences are especially evident when comparing pairwise alignments to multiple alignments.
Table 9.
Average SPS and CS scores (in %) on HOMSTRAD
| TCoffee |
MUSCLE |
ProbCons |
MUMMALS |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SPS | ||||||||||||
| 2 seqs {630} | 80.88 | 81.17 | 1 × 10−6 | 80.40 | 81.55 | 1 × 10−11 | 81.65 | 82.42 | 1 × 10−20 | 83.50 | 83.56 | 6 × 10−6 |
| 3 seqs {169} | 80.52 | 81.29 | 2 × 10−9 | 81.26 | 82.52 | 1 × 10−5 | 81.50 | 82.20 | 2 × 10−8 | 83.33 | 83.43 | 0.002 |
| 4–5 seqs {122} | 79.78 | 80.45 | 1 × 10−9 | 80.97 | 81.23 | – | 82.26 | 82.89 | 2 × 10−9 | 83.53 | 83.59 | 0.04 |
| ⩾ 6 seqs {111} | 81.55 | 81.94 | 4 × 10−4 | 82.34 | 82.72 | 0.04 | 83.64 | 83.95 | 1 × 10−7 | 85.15 | 85.27 | – |
| CS | ||||||||||||
| 2 seqs {630} | 80.88 | 81.17 | 1 × 10−6 | 80.40 | 81.55 | 1 × 10−11 | 81.65 | 82.42 | 1 × 10−20 | 83.50 | 83.56 | 6 × 10−6 |
| 3 seqs {169} | 74.51 | 75.50 | 1 × 10−9 | 75.41 | 77.14 | 6 × 10−6 | 75.54 | 76.43 | 1 × 10−6 | 77.92 | 78.06 | 0.007 |
| 4–5 seqs {122} | 68.38 | 69.48 | 1 × 10−10 | 70.17 | 70.69 | – | 71.69 | 72.94 | 2 × 10−10 | 73.58 | 73.74 | 0.02 |
| ⩾6 seqs {111} | 59.59 | 61.23 | 8 × 10−10 | 62.03 | 62.80 | 0.04 | 62.77 | 63.79 | 3 × 10−8 | 65.47 | 65.93 | 0.04 |
Each subset includes all alignments with number of protein sequences within the specified range. For each algorithm, the higher accuracy value is in bold.
Effect of parameters ω and β
While the same parameters ω and β are used for each modified algorithm across different benchmarks, we found that not only different algorithms have different preferences of ω and β, different benchmarks also have different preferences of ω and β even when the same algorithm is used. Table 10 shows that the effect of varying ω that specifies the maximum number of horizontal positions that are included to the left and to the right was much more pronounced than varying β that specifies the weight of the neighboring scores on HOMSTRAD and BRAliBase, and our chosen parameter setting was not the one that gives the best accuracy. It is possible to further improve accuracy significantly if another parameter setting is chosen that is different across benchmarks, even when no significant differences in accuracy were obtained with our chosen parameter setting.
Table 10.
Average SPS and CS scores (in %) on HOMSTRAD and average SPS, CS and SCI scores (in %) on Data-set 1 of BRAliBase II by varying the parameter ω that specifies the maximum number of horizontal positions that are included to the left and to the right, and the parameter β that specifies the weight of the neighboring scores
| MUMMALS on HOMSTRAD |
MUSCLE on BRAliBase |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ω =1 | ω =3 | ω =5 | ω =7 | ω =9 | ω =3 | ω =6 | ω =9 | ω =12 | ω =15 | ||
| SPS | SPS | ||||||||||
| β =0.2 | 83.72 | 83.72 | 83.64 | 83.49 | 83.18 | β =0.2 | 84.64 | 84.83 | 84.63 | 84.30 | 83.64 |
| β =0.4 | 83.70 | 83.70 | 83.60 | 83.34 | 82.94 | β =0.4 | 84.63 | 84.65 | 84.58 | 84.19 | 83.15 |
| β =0.6 | 83.72 | 83.72 | 83.54 | 83.27 | 82.84 | β =0.6 | 84.54 | 84.64 | 84.42 | 84.12 | 83.04 |
| β =0.8 | 83.72 | 83.72 | 83.52 | 83.24 | 82.78 | β =0.8 | 84.69 | 84.71 | 84.36 | 84.00 | 82.95 |
| β =1.0 | 83.73 | 83.70 | 83.50 | 83.21 | 82.75 | β =1.0 | 84.69 | 84.81 | 84.41 | 83.98 | 82.91 |
| CS | CS | ||||||||||
| β =0.2 | 79.60 | 79.61 | 79.56 | 79.39 | 79.03 | β =0.2 | 73.84 | 74.20 | 73.92 | 73.42 | 72.52 |
| β =0.4 | 79.57 | 79.61 | 79.52 | 79.22 | 78.74 | β =0.4 | 73.92 | 73.93 | 73.85 | 73.31 | 71.81 |
| β =0.6 | 79.59 | 79.64 | 79.44 | 79.13 | 78.62 | β =0.6 | 73.67 | 73.92 | 73.54 | 73.16 | 71.48 |
| β =0.8 | 79.60 | 79.63 | 79.41 | 79.08 | 78.55 | β =0.8 | 74.03 | 74.03 | 73.47 | 73.03 | 71.27 |
| β =1.0 | 79.60 | 79.61 | 79.39 | 79.04 | 78.51 | β =1.0 | 74.03 | 74.00 | 73.57 | 72.90 | 71.22 |
| SCI | |||||||||||
| β =0.2 | 73.20 | 74.19 | 74.29 | 74.19 | 73.26 | ||||||
| β =0.4 | 73.34 | 73.96 | 74.02 | 74.11 | 72.83 | ||||||
| β =0.6 | 73.19 | 73.94 | 73.92 | 74.12 | 72.69 | ||||||
| β =0.8 | 73.28 | 73.79 | 73.64 | 73.97 | 72.41 | ||||||
| β =1.0 | 73.11 | 73.93 | 73.84 | 74.11 | 72.15 | ||||||
For each modified algorithm and each score measure, the highest accuracy value and the values of ω and β that correspond to our chosen parameter setting that is the same across different benchmarks are in bold.
CONCLUSION
We have developed a strategy NRAlign that incorporates horizontal information in alignments and it proves to be useful in all situations. Unlike previous algorithms, consistent average improvements can be obtained that are mostly not dependent on the identity level, even for very high identity. Table 11 shows that NRAlign was at most a few times slower than TCoffee and MUSCLE, and was slightly slower than ProbCons and MUMMALS, which indicates that the window-based adjustment procedure takes up a small part of the computation time of ProbCons, and the use of a small ω = 1 does not add much to the computation time of MUMMALS.
Table 11.
Computation time on HOMSTRAD and on Data-set 1 of BRAliBase II represented as a pair of the form average,maximum in seconds
| HOMSTRAD | TCoffee |
MUSCLE |
ProbCons |
MUMMALS |
||||
|---|---|---|---|---|---|---|---|---|
| 2 seqs {630} | 0.19,1.25 | 0.27,1.33 | 0.03,0.20 | 0.07,0.57 | 0.39,4.67 | 0.42,4.99 | 0.38,4.57 | 0.40,3.92 |
| 3 seqs {169} | 0.38,2.12 | 0.64,4.37 | 0.07,0.52 | 0.21,2.00 | 0.62,5.84 | 0.67,6.38 | 0.67,9.62 | 0.70,13.14 |
| 4–5 seqs {122} | 0.88,3.51 | 1.79,7.71 | 0.14,1.08 | 0.48,2.74 | 1.28,11.35 | 1.40,12.84 | 1.88,11.60 | 1.93,11.89 |
| ⩾6 seqs {111} | 10.44,129.86 | 26.73,348.34 | 0.45,4.66 | 1.57,20.73 | 7.06,147.96 | 8.39,174.65 | 10.77,205.44 | 10.51,209.66 |
| BRAliBase | TCoffee | MUSCLE | ProbConsRNA | |||||
| All {481} | 2.57,9.54 | 12.69,37.63 | 0.05,0.21 | 0.20,1.15 | 0.43,2.42 | 0.62,3.12 | ||
For each algorithm, the first pair shows the running time of the original algorithm and the second pair shows the running time of the modified algorithm NRAlign.
To further improve accuracy, it may be useful to utilize different weights for neighboring scores that are at different distances from the given pair (x, y). In addition to using horizontal information from neighboring scores in sequences, it is also possible to utilize spatial neighboring information in the local structural environment when such information is available and combine the scores from both types of neighbors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation (DBI-0624077). Funding for open access charge: National Science Foundation (DBI-0624077).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the anonymous referees for invaluable comments that significantly improved the paper.
REFERENCES
- 1.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 3.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Do CB, Mahabhashyam MS, Brudno M, Batzoglou S. ProbCons: probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pei J, Grishin NV. MUMMALS: multiple sequence alignment improved by using hidden Markov models with local structural information. Nucleic Acids Res. 2006;34:4364–4374. doi: 10.1093/nar/gkl514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roshan U, Livesay DR. Probalign: multiple sequence alignment using partition function posterior probabilities. Bioinformatics. 2006;22:2715–2721. doi: 10.1093/bioinformatics/btl472. [DOI] [PubMed] [Google Scholar]
- 7.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinformatics. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 8.Gotoh O. Significant improvement in accuracy of multiple protein sequence alignments by iterative refinement as assessed by reference to structural alignments. J. Mol. Biol. 1996;264:823–838. doi: 10.1006/jmbi.1996.0679. [DOI] [PubMed] [Google Scholar]
- 9.Zhou H, Zhou Y. SPEM: improving multiple sequence alignment with sequence profiles and predicted secondary structures. Bioinformatics. 2005;21:3615–3621. doi: 10.1093/bioinformatics/bti582. [DOI] [PubMed] [Google Scholar]
- 10.Pei J, Grishin NV. PROMALS: towards accurate multiple sequence alignments of distantly related proteins. Bioinformatics. 2007;23:802–808. doi: 10.1093/bioinformatics/btm017. [DOI] [PubMed] [Google Scholar]
- 11.Wilm A, Higgins DG, Notredame C. R-Coffee: a method for multiple alignment of non-coding RNA. Nucleic Acids Res. 2008;36:e52. doi: 10.1093/nar/gkn174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Sullivan O, Suhre K, Abergel C, Higgins DG, Notredame C. 3DCoffee: combining protein sequences and structures within multiple sequence alignments. J. Mol. Biol. 2004;340:385–395. doi: 10.1016/j.jmb.2004.04.058. [DOI] [PubMed] [Google Scholar]
- 13.Van Walle I, Lasters I, Wyns L. Align-m—a new algorithm for multiple alignment of highly divergent sequences. Bioinformatics. 2004;20:1428–1435. doi: 10.1093/bioinformatics/bth116. [DOI] [PubMed] [Google Scholar]
- 14.Pei J, Kim B-H, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marti-Renom MA, Madhusudhan MS, Sali A. Alignment of protein sequences by their profiles. Protein Sci. 2004;13:1071–1087. doi: 10.1110/ps.03379804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simossis VA, Kleinjung J, Heringa J. Homology-extended sequence alignment. Nucleic Acids Res. 2005;33:816–824. doi: 10.1093/nar/gki233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spang R, Rehmsmeier M, Stoye J. A novel approach to remote homology detection: jumping alignments. J. Comput. Biol. 2002;9:747–760. doi: 10.1089/106652702761034172. [DOI] [PubMed] [Google Scholar]
- 18.Panchenko AR, Kondrashov F, Bryant S. Prediction of functional sites by analysis of sequence and structure conservation. Protein Sci. 2004;13:884–892. doi: 10.1110/ps.03465504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Capra JA, Singh M. Predicting functionally important residues from sequence conservation. Bioinformatics. 2007;23:1875–1882. doi: 10.1093/bioinformatics/btm270. [DOI] [PubMed] [Google Scholar]
- 20.Bartlett GJ, Porter CT, Borkakoti N, Thornton JM. Analysis of catalytic residues in enzyme active sites. J. Mol. Biol. 2002;324:105–121. doi: 10.1016/s0022-2836(02)01036-7. [DOI] [PubMed] [Google Scholar]
- 21.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 22.Gardner PP, Wilm A, Washietl S. A benchmark of multiple sequence alignment programs upon structural RNAs. Nucleic Acids Res. 2005;33:2433–2439. doi: 10.1093/nar/gki541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang X, Miller W. A time-efficient linear-space local similarity algorithm. Adv. Appl. Math. 1991;12:337–357. [Google Scholar]
- 24.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, U.K: Cambridge University Press; 1983. [Google Scholar]
- 25.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson JD, Koehl P, Ripp R, Poch O. BAliBASE 3.0: latest developments of the multiple sequence alignment benchmark. Proteins. 2005;61:127–136. doi: 10.1002/prot.20527. [DOI] [PubMed] [Google Scholar]
- 27.Mizuguchi K, Deane CM, Blundell TL, Overington JP. HOMSTRAD: a database of protein structure alignments for homologous families. Protein Sci. 1998;7:2469–2471. doi: 10.1002/pro.5560071126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 29.Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carroll H, Beckstead W, O'Connor T, Ebbert M, Clement M, Snell Q, McClellan D. DNA reference alignment benchmarks based on tertiary structure of encoded proteins. Bioinformatics. 2007;23:2648–2649. doi: 10.1093/bioinformatics/btm389. [DOI] [PubMed] [Google Scholar]
- 31.Thompson JD, Plewniak F, Poch O. A comprehensive comparison of multiple sequence alignment programs. Nucleic Acids Res. 1999;27:2682–2690. doi: 10.1093/nar/27.13.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holm L, Sander C. Dictionary of recurrent domains in protein structures. Proteins. 1998;33:88–96. doi: 10.1002/(sici)1097-0134(19981001)33:1<88::aid-prot8>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 33.Zemla A, Venclovas Č., Moult J, Fidelis K. Processing and analysis of CASP3 protein structure predictions. Proteins. 1999;(Suppl. 3):22–29. doi: 10.1002/(sici)1097-0134(1999)37:3+<22::aid-prot5>3.3.co;2-n. [DOI] [PubMed] [Google Scholar]
- 34.Venclovas Č., Zemla A, Fidelis K, Moult J. Assessment of progress over the CASP experiments. Proteins. 2003;53:585–595. doi: 10.1002/prot.10530. [DOI] [PubMed] [Google Scholar]
- 35.Rychlewski L, Fischer D, Elofsson A. LiveBench-6: large-scale automated evaluation of protein structure prediction servers. Proteins. 2003;53:542–547. doi: 10.1002/prot.10535. [DOI] [PubMed] [Google Scholar]
- 36.Wallner B, Elofsson A. Can correct protein models be identified? Protein Sci. 2003;12:1073–1086. doi: 10.1110/ps.0236803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 38.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Washietl S, Hofacker IL, Stadler PF. Fast and reliable prediction of noncoding RNAs. Proc. Natl Acad. Sci. USA. 2005;102:2454–2459. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilcoxon F. Probability tables for individual comparisons by ranking methods. Biometrics. 1947;3:119–122. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.