Abstract
The Z-DNA conformation preferentially occurs at alternating purine-pyrimidine repeats, and is specifically recognized by Zα domains identified in several Z-DNA-binding proteins. The binding of Zα to foreign or chromosomal DNA in various sequence contexts is known to influence various biological functions, including the DNA-mediated innate immune response and transcriptional modulation of gene expression. For these reasons, understanding its binding mode and the conformational diversity of Zα bound Z-DNAs is of considerable importance. However, structural studies of Zα bound Z-DNA have been mostly limited to standard CG-repeat DNAs. Here, we have solved the crystal structures of three representative non-CG repeat DNAs, d(CACGTG)2, d(CGTACG)2 and d(CGGCCG)2 complexed to hZαADAR1 and compared those structures with that of hZαADAR1/d(CGCGCG)2 and the Zα-free Z-DNAs. hZαADAR1 bound to each of the three Z-DNAs showed a well conserved binding mode with very limited structural deviation irrespective of the DNA sequence, although varying numbers of residues were in contact with Z-DNA. Z-DNAs display less structural alterations in the Zα-bound state than in their free form, thereby suggesting that conformational diversities of Z-DNAs are restrained by the binding pocket of Zα. These data suggest that Z-DNAs are recognized by Zα through common conformational features regardless of the sequence and structural alterations.
INTRODUCTION
DNA can adopt various secondary conformations other than the classical right-handed B-DNA structure under certain specific physiological conditions (1). Left-handed Z-DNA has been studied in detail by numerous methods over the last two decades since its first crystal structure was solved (2). Z-DNA conformations in many different sequences have been a challenge to crystallize. Nonetheless, several structures have been determined by X-ray crystallographic study (3). Many characteristic features of the Z-DNA structure have been identified from the accumulated structural data. The overall shapes of most Z-DNA structures share common features that are similar to those seen in the first crystallized Z-DNA structure of d(CGCGCG)2 (2). Z-DNA has a zig-zag sugar phosphate backbone and is longer and thinner than B-DNA. Its nucleotides form in a dinucleotide repeat in which they alternate with syn and anti conformations. Z-DNA is favored in alternating pyrimidine-purine (APP) sequences (4). The alternating CG-repeat sequence is the most favorable energetically for Z-DNA formation (5). Networks of water molecules hydrate Z-DNA, forming hydrogen bonds with atoms in both the backbone and base. However, some Z-DNAs form with different structural features, especially those with sequences without APP or including A-T base pairs (6–8). When A-T base pairs are introduced, the overall Z-DNA structure becomes partially distorted due to disruption of the hydration spine (6). In addition, the non-APP base pairs have been found to be highly buckled when compared with other base pairs in Z-DNA (7,8). Up to now, however, there have been only a limited number of structural studies of Z-DNAs containing non-APP or A-T base pairs. Most studies have been carried out in the presence of excess cations or using dsDNAs modified by methylation or bromation. Thus, study of Z-DNA under low salt conditions or in the presence of Z-DNA-binding domains (Zα) remains to be explored, and this may provide insight into the effect that sequence variability has on Z-DNA conformation under physiological conditions.
The protein Zα domain was first identified from human ADAR1 (double-stranded RNA adenosine deaminase) and subsequently found in other proteins (DLM1, E3L and PKZ). These domains provide a unique opportunity to explore Z-DNA and its various roles in biological systems (9–13). The Zα domains are highly specific for the Z-conformation of nucleic acids, including dsDNA, dsRNA as well as DNA–RNA hybrids. They have binding affinities in the low-nanomolar range. In the crystal structures of double-stranded d(CGCGCG)2 complexed with hZαADAR1, mZαDLM1, yabZαE3L or hZβDLM1 (10,11,14–16), a close resemblance was found between the bound and unbound states of the Z-DNA structure of d(CGCGCG)2 (2). It is now known that Z-DNA is found in the genome as an active transcription modifier that functions by modulating chromatin structure (17,18). The Z-DNA conformation is not limited only to APP sequences, but can appear in many other nucleotide sequences. Recent functional studies of the Z-DNA-binding domains in vaccinia E3L showed that it acts as a transcription modulator of several apoptosis-related genes (19). More recently, the Z-DNA-binding protein DLM1 has been found to act in the innate immune system as a cytosolic receptor for dsDNA, recognizing foreign pathogenic DNA (20). These results support the idea that diverse sequences of Z-DNA can be recognized by Z-DNA-binding domains and that their binding is essential for cellular processes. To understand the binding mode of Zα to Z-DNAs in various sequence contexts, it is necessary to investigate structural features of Z-DNAs bound to Zα and to compare their structures with Z-DNAs stabilized by base-modification and/or positively charged ions. In this regard, we undertook a study to solve Z-DNA structures with non-CG-repeat sequences stabilized by the same Zα domain. Here we report the co-crystal structures of three non-CG-repeat Z-DNAs containing either A-T base pairs or non-APP sequence bound to hZαADAR1. These complexes reveal how the structural diversity of Z-DNA caused by non-CG-repeat sequences is recognized and stabilized by the Z-DNA-binding domain.
MATERIALS AND METHODS
Expression and purification
hZαADAR1 (residues 133–209) from human ADAR1 was expressed and purified as described previously (21). In brief, hZαADAR1 was purified through sequential chromatographic steps involving HiTrap metal affinity column (GE Healthcare, Piscataway, NJ), thrombin digestion for the removal of N-terminal his-tag and a Resource S column (GE Healthcare, Piscataway, NJ). The purity and concentration of hZαADAR1 were estimated by SDS–PAGE and the Bradford method, respectively. DNAs were purchased (IDT, Coralville, IA) and purified as described previously (22).
Crystallization
The DNA used for crystallization all had 6nt, plus a 5′ T overhang which acts as a stabilizing capping residue. For crystallization, hZαADAR1 was mixed with dsDNA [(d(TCGCCCG)::d(TCGGGCG), d(TCACGTG)2, d(TCGTACG)2 or d(TCGGCCG)2)] at a 0.66 mM : 0.33 mM molar ratio in 5 mM HEPES-NaOH pH 7.5 containing 20 mM NaCl, and incubated at 303 K for at least 2 h. All crystallization experiments were performed using the hanging drop vapor diffusion method with a VDX plate at 295 K. Among the four dsDNA used in crystallization, only d(TCGCCCG)::d(TCGGGCG) was successfully co-crystallized with hZαADAR1 when ammonium sulfate was used as precipitant. For crystallization of the other three complexes, crystals of hZαADAR1/d(TCGCCCG)::d(TCGGGCG) were used as seeds, and initial crystals were again used as seeds for further crystallization. Diffraction quality crystals were obtained within a month using 2.2 M ammonium sulfate and 10% glycerol in the reservoir solution.
Data collection and structure determination
Preliminary X-ray diffraction analyses were performed at beamline BL6A of PAL (Pohang, Korea). X-ray diffraction data of frozen crystals were collected at 100 K with a MAR CCD165 detector at the BL41-XU beamline of Spring-8 (Harima, Japan). Crystals were frozen either in liquid nitrogen directly or by using paraton as a cryoprotectant. Diffraction data were processed and integrated using HKL2000 (23). The unit cell parameters, space group and other data collection statistics are summarized in Supplementary Table 1. The crystal structure of hZαADAR1/d(TCGCGCG)2 (PDB ID 1QBJ) was used for the initial model of the other complex structures. Refinement and model building were performed by CNS (24) and O (25), respectively. The refinement statistics are summarized in Supplementary Table 1. The crystal structure of hZαADAR1/d(TCGCCCG)::d(TCGGGCG) was not refined because of ambiguity in base assignment. All figures were drawn using Molscript, Raster3D and Pymol (26,27, http://www.pymol.org). The structural superposition was performed using the LSQKAB CCP4 program (28).
RESULTS AND DISCUSSION
Structure determination
The crystal structures of hZαADAR1 complexed with three non-CG-repeat dsDNAs, d(TCACGTG)2, d(TCGTACG)2 and d(TCGGCCG)2 were determined at resolutions of 2.2, 2.5 and 2.7 Å, respectively (Supplementary Table 1). The simulated annealed omit maps contoured at 3σ near the altered sequences confirmed that current structural information represents each non-CG-repeat DNA (Supplementary Figure 1). In all three structures, the deoxythymidine overhangs at the 5′ end of the oligonucleotides were not modeled due to their weak electron densities, and therefore, are not mentioned in current study. There are three hZαADAR1 domains, assigned as chains A, B and C, and three DNA strands, chains D, E and F, in one asymmetric unit. Chains D and E form the Z-DNA duplex, and chain F forms a duplex with chain F in another asymmetric unit that is related by crystallographic 2-fold symmetry. Each DNA strand is bound to one hZαADAR1 domain. The protein/DNA complex made up of the chain C and chain F pair was used for structural analyses unless specified otherwise because its average temperature factor is the lowest among the three complexes in the same asymmetric unit.
Overall structures
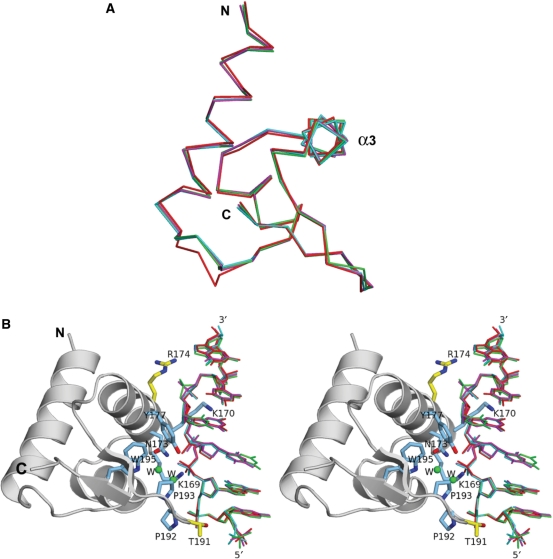
Overall, the hZαADAR1 domains bound to the three different Z-DNAs used in this study have almost identical structures to that of hZαADAR1 bound to d(CGCGCG)2 (14). hZαADAR1 has an α/β topology containing a three-helix bundle flanked on one side by a twisted antiparallel β sheet. The root mean square deviations (RMSDs) between hZαADAR1 bound to d(CGCGCG)2 and hZαADAR1 bound to d(CACGTG)2, d(CGTACG)2 or d(CGGCCG)2 are 0.52, 0.19 and 0.19 Å, respectively, when calculated using the 64 Cα atoms of hZαADAR1 (Figure 1A). The three Z-DNAs bound to hZαADAR1 are all in the Z-conformation with alternating anti-and syn-glycosidic bonds regardless of their sequence (Figure 1B). With the exception of purines G3 and pyrimidines C4 of d(CGGCCG)2 that adopt the anti and syn conformations, respectively, all other purines and pyrimidines are in the syn and anti conformations, respectively (Figure 1B). The calculated double-stranded DNA parameters are within the range of a Z conformation of DNA (Supplementary Table 2). For Z-DNAs complexed with hZαADAR1, the RMSD values between d(CGCGCG)2 and other DNAs, d(CACGTG)2, d(CGTACG)2 and d(CGGCCG)2 were 0.28, 0.33 and 0.56 Å, respectively, when calculated with 63 DNA backbone atoms of chain F for each DNA structure (Figure 1B and Supplementary Table 3).
Figure 1.
Structural comparison of the hZαADAR1 domain and the Z-DNAs in several complexes. (A) Overlapping Cα traces of the hZαADAR1 domains when complexed to d(CGCGCG)2 [cyan], (NDB ID PH0001), d(CACGTG) 2 [red], d(CGTACG)2 [magenta] and d(CGGCCG)2 [green]. Sixty-four Cα atoms of chain C in each structure were used for the superposition. The N- and C-termini and α3 are labeled. (B) Four Z-DNA strands, CACGTG (red), CGTACG (magenta), CGGCCG (green) and CGCGCG (cyan), bound to hZαADAR1 were compared by superimposing four Zα/Z-DNA complexes using 64 Cα atoms of hZαADAR1 domains. In order to show the relative orientation of the Z-DNA and Zα, chain C of the hZαADAR1/d(CGCGCG)2 complex was drawn in a ribbon diagram. The amino acid residues and water molecules involved in DNA binding are drawn as sky blue stick models and green balls, respectively. Arg174 and Thr191 are marked by yellow stick models to differentiate them from other core resides. These residues are mostly disordered in the current study, but are well defined and bind to DNA in the crystal structure of the hZαADAR1/d(CGCGCG)2 complex.
Interactions between hZαADAR1 and non-CG-repeat Z-DNAs
The structural analyses of hZαADAR1, mZαDLM1 and yabZαE3L bound to the Z conformation of d(CGCGCG)2 revealed well-conserved interactions with the Zα domains (10,11,14). Zα domains with a winged helix–turn–helix motif bind to Z-DNA in a conformation-specific manner. The residues in the recognition helix (α3) and in the wing play critical roles in binding Z-DNA through direct or water-mediated hydrogen bonds and van der Waals interactions (Figure 2). On Z-DNA, one continuous surface composed of the sugar-phosphate backbone of Z-DNA is mostly involved in protein contact.
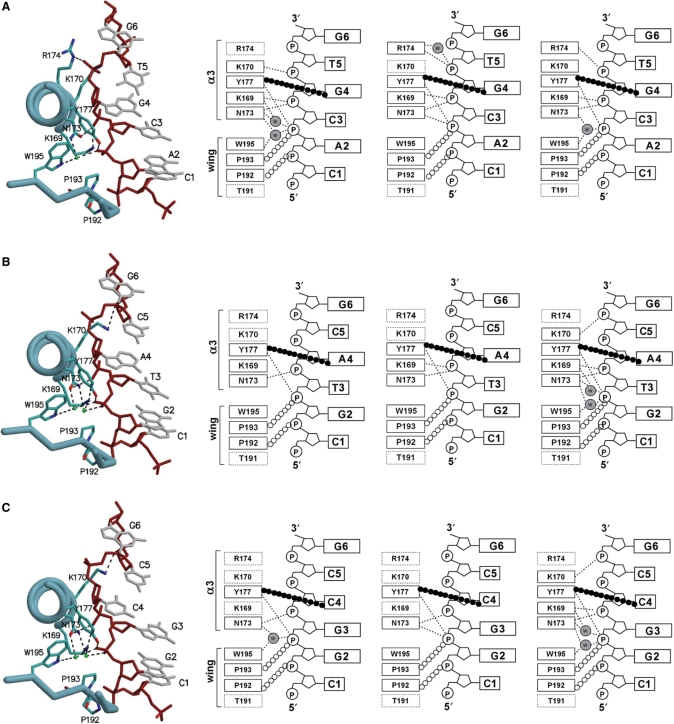
Figure 2.
The protein–DNA interactions in three different Zα-non-alternating CG ZDNA complexes. (A) hZαADAR1/d(CACGTG)2, (B) hZαADAR1/d(CGTACG)2 and (C) hZαADAR1/d(CGGCCG)2. One representative Zα/DNA complex (chains C and F) of the three complexes in one asymmetric unit is drawn as a tubular ribbon diagram (left). The three complexes in the asymmetric unit are then shown as schematic diagrams (left to right). These are chains A and D, chains B and E, and chains C and F, respectively. In the schematic diagrams, the amino acids identified as the DNA-contacting residues in the structure of the hZαADAR1/d(CGCGCG)2 complex are marked by boxes. Disordered residues are indicated by dotted boxes. Hydrogen bonds are represented by dotted lines and van der Waals contacts by open circles. The CH–π interaction between the conserved Tyr residue and syn deoxynucleoside is indicated by filled circles. Waters are shown as gray circles. In the tubular ribbon diagrams, the same residues used in the schematic diagrams are drawn as stick models and labeled. The DNA backbones and labeled bases are shown as red and gray stick models, respectively. Water molecules are shown as green spheres. Hydrogen bonds are drawn as dashed lines.
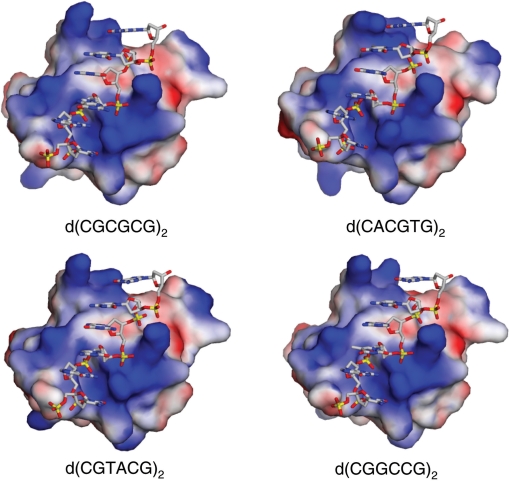
Similarly, in the crystal structures of hZαADAR1 bound to the three non-CG-repeat dsDNAs the residues located in α3 and the wing mostly make contact with the Z-DNA backbones either directly or through water-mediated interactions (Figure 2). When the DNA-binding surfaces of the hZαADAR1 domains bound to the four Z-DNAs are compared, their curvatures and surface charge distributions are nearly identical (Figure 3). These results, together with the limited structural alterations found in hZαADAR1 domains, strongly demonstrate that the overall binding mode of hZαADAR1 to Z-DNA are well conserved regardless of the sequence of the bound Z-DNA.
Figure 3.
Surface charge distributions of the hZαADAR1 domains complexed with various Z-DNAs, as viewed along the DNA binding cleft. DNA-binding surfaces of the hZαADAR1 domains bound to d(CGCGCG)2 (upper left), d(CACGTG)2 (upper right), d(CGTACG)2 (bottom left) and d(CGGCCG)2 (bottom right) are drawn, and their surface charge distributions are displayed. Arg174 and Thr174 were not used for the surface charge calculations since they are not well defined in most structures. The red and blue areas represent the negatively and positively charged surfaces, respectively. DNA backbones are shown in stick models with phosphate atoms in yellow, oxygen in red, nitrogen in blue and carbon in gray.
The number and type of hZαADAR1 residues contacting bases on each DNA vary, mainly because some of the important residues that were identified as the DNA-contacting residues in hZαADAR1/d(CGCGCG)2 (14) are disordered (Figure 2). However, it does not appears that variations in protein/DNA contacts are related to the sequence of the Z-DNA, since the number of contacting residues is not the same, even among the three hZαADAR1 domains in one asymmetric unit of each ZαADAR1/DNA crystal. For example, in most structures, the roles of Arg174 and Thr191 are neither well defined nor involved in DNA contact. In hZαADAR1/d(CGCGCG)2, Arg174 was shown to form a direct hydrogen bond to a phosphate atom and a water-mediated hydrogen bond to the ribose ring (14). However, in seven of the nine hZαADAR1 domains used in this study, the amine groups of Arg174 are not modeled due to their weak electron densities (Figure 2). In the case of Thr191, no DNA interaction has been found. Similarly, Lys169 and Lys170 are only in contact with DNA in some structures (Figure 2). In contrast, Asn173, Tyr177, Pro192, Pro193 and Trp195 of hZαADAR1 contribute to the recognition of Z-DNA in all cases. Asn173 and Tyr177, located in the recognition helix (α3), recognize the phosphate backbone of Z-DNA through direct or water-mediated hydrogen bonds as observed in hZαADAR1/d(CGCGCG)2 (14). Likewise, Pro192 and Pro193 in the wing interact with Z-DNA through van der Waals interactions. In some cases, Trp195 makes a water-mediated hydrogen bond to a phosphate, but its major role seems to be supporting Tyr177 via the hydrophobic edge-to-face interaction which stabilizes the interactions between Tyr177 and the bases in the N4 position (Figure 2). Moreover, some coordinated waters that are well defined and mediate hydrogen bonds between protein and DNA in the crystal structure of hZαADAR1/d(CGCGCG)2 are not found in some structures.
The decrease in protein/DNA interactions in the three complex structures compared to hZαADAR1/d(CGCGCG)2 can be explained in part by the decrease in diffraction resolution. However, it seems that protein–DNA interactions are not all affected by diffraction resolution since well-defined interactions are consistently observed in all three structures despite their resolution differences. For example, the electron density of the well-defined hydrogen bonds between Tyr177 and the phosphate group of G3 are very clear even in the case of hZαADAR1/d(CGGCCG)2 whose structure was determined at 2.7 Å resolution (Supplementary Figure 2). Conversely, it is suspected that some of the unseen protein/DNA interactions from these three Z-DNA structures are neither strong nor essential for Z-DNA recognition. From these results, it can be hypothesized that some of the residues previously identified as Z-DNA binders are not indispensable for Z-DNA binding but that they have auxiliary roles in DNA binding and probably produce tighter binding. However, we cannot rule out the possibility that crystal packing forces may have affected or destabilized some of the interactions between hZαADAR1 and Z-DNA.
In the Zα/d(CGCGCG)2 structure as well as in the current crystal structure of hZαADAR1/d(CATGCG)2, the tyrosine residue of the recognition helix (α3) has a unique role in recognizing the C-8 carbon of the syn deoxyguanosine at the forth position (G4) via a CH–π interaction (10,11,14; Figure 2A). In the case of Zα/d(CGTACG)2, the deoxyadenosine (A4) also adopts the syn conformation, which was very similar to that observed in previous studies (Figure 2B). More interesting is the observation that hZαADAR1 stabilizes the syn conformation of deoxycytidine at the forth position (C4) of d(CGGCCG)2 (Figure 2C). Generally, the syn conformation is not favored for pyrimidine nucleotides unless they are modified (3,4). However in the crystal structure of hZαADAR1/d(CGGCCG)2, the syn conformation of deoxycytidine is stabilized by the CH–π interaction between the C-5 and C-6 carbons of C4 and the π orbital of the tyrosine ring (Figure 2C). These data show that the binding mode of hZαADAR1 observed for d(CGCGCG)2 is also a template for non-CG-repeat Z-DNAs containing non-APP repeat sequences or A-T base pairs. As with the earlier structure, there are no sequence-specific interactions between Zα and Z-DNA. These results reinforce the idea that the binding of Zα to Z-DNA is sequence-independent but conformation specific.
Structural variations between free and hZαADAR1-bound Z-DNAs
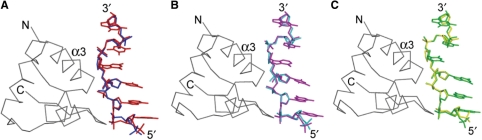
The crystal structures of d(CGCGCG)2, d(CACGTG)2, d(m5CGTAm5CG)2 and d(m5CGGCm5CG)2 were used to represent Z-DNAs free of protein binding (Supplementary Table 3; 2,8,29,30), and they have been compared with the hZαADAR1-bound Z-DNA structure. In order to stabilize and crystallize Z-DNA with non-APP or A-T base pairs, base modification is used or it is necessary to add cations such as metal ions or polyamines. The 5 position of cytosine was methylated for the crystallization of Z-form d(CGTACG)2 and d(CGGCCG)2 (8,29), and spermine was added for the crystallization of Z-form d(CGCGCG)2 and d(CACGTA)2 (2,30). When the protein-free and hZαADAR1-bound Z-DNAs were compared by superimposing each DNA pair using 63 DNA backbone atoms, RMSDs of d(CGCGCG)2, d(CACGTG)2, d(CGTACG)2 and d(CGGCCG)2 pairs were 0.89, 0.75, 0.57 and 0.61 Å, respectively (Figure 4 and Supplementary Table 3). Regardless of their sequence, the main structural differences between the free and bound Z-DNAs were found in the helical rise: when bound to hZαADAR1 it increased in the N3pN4 step, but decreased in the N2pN3 and N4pN5 steps (Supplementary Table 2). However, the distance changes in the N1pN2 and N5pN6 steps occurred regardless of Zα binding (Supplementary Table 2).
Figure 4.
Structural comparisons of free- and hZαADAR1-bound Z-DNAs. Superposition of the free d(CACGTG)2 (blue) with Zα-bound d(CACGTG)2 (red) (A), the free d(m5CGTAm5CG)2 (cyan) with Zα-bound d(CGTACG)2 (magenta) (B) and the free d(m5CGGCm5CG)2 (yellow) with Zα-bound d(CGGCCG)2 (green) (C). For the structural overlap, 63 DNA backbone atoms of chain F of each ZαADAR1-bound Z-DNA and chain A of each free Z-DNA were used. The Cα trace of hZαADAR1 in each complex is drawn in gray. The N- and C- termini, recognition helix (α3) of hZαADAR1 and the 5′ and 3′ ends of the DNA are labeled.
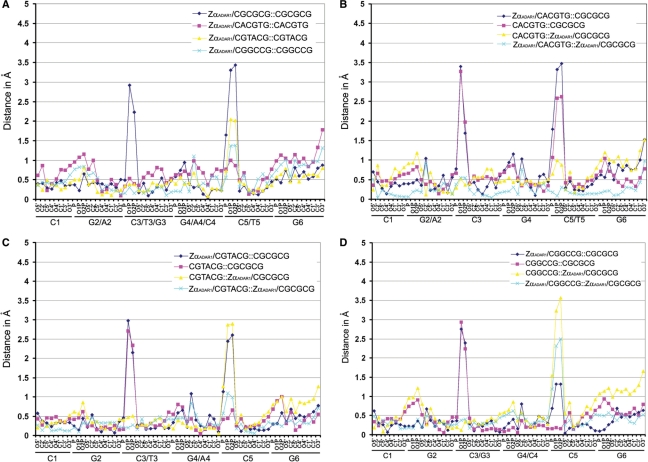
Structural variations between protein-free and hZαADAR1-bound Z-DNAs were compared directly by plotting the distance between two corresponding atoms in Z-DNAs along the DNA backbone atoms (Figure 5A). In this manner, the structures of non-CG-repeat Z-DNAs and CG-repeat Z-DNA were also analyzed (Figure 5B–D). Structural deviations are plotted for all comparisons of protein-free and Zα-bound Z-DNA sequences. The largest structural variation between protein-free and hZαADAR1-bound Z-DNAs was detected in the two phosphates at the N2pN3 and N4pN5 phosphodiester steps, respectively. These results were expected because the ZI conformation of the GpC phosphodiester step is preferred when Z-DNA forms a complex with hZαADAR1 due to the specific interaction between the phosphate groups of DNA and the charged residues of Zα, whereas protein-free Z-DNA can have two alternative conformations, ZI and ZII (14,31). The difference between two corresponding atoms of the compared Z-DNAs is >2.5 Å when the protein-free Z-DNA is in the ZII conformation, and it is <0.5 Å when the protein-free Z-DNA is in the ZI conformation (Figure 5A). It is interesting that the current structures revealed that hZαADAR1-bound Z-DNAs do not absolutely have the ZI conformation. Because ionic interactions between the phosphates at the N5 position and the charged residues, Lys170 and Arg174, are absent or weak in d(CGTACG)2 and d(CGGCCG)2, the phosphodiester conformation of the N4pN5 step is not a typical ZI conformation (Figures 1B, 5C and D). As a result, in both Z-DNAs, the phosphates at the N5 position still show structural deviation from that of d(CGCGCG)2 in hZαADAR1-bound structures (sky blue lines in Figure 5C and D). Specifically, the N4pN5 phosphodiester step of d(CGGCCG)2 adopts an intermediate conformation between ZI and ZII in the Zα/DNA complex.
Figure 5.
Structural deviation of each DNA backbone atom, comparing free Z-DNA and ZαADAR1-bound Z-DNA in both non-CG-repeat and CG-repeat Z-DNAs. The distances in angstrom between two backbone atoms in the same position of each paired DNA were plotted against the DNA backbone atoms. The structures were compared in the same way as shown in Figure 4. (A) The backbone atoms of Z-DNAs with CGCGCG, CACGTG, CGTACG and CGGCCG sequences in free- and hZαADAR1-bound forms were compared. (B) CACGTG, (C) CGTACG and (D) CGGCCG DNAs in both free and hZαADAR1-bound forms were compared with free- and hZαADAR1-bound CG-repeat Z-DNA (CGCGCG).
It is well known that there is no significant sequence-dependent conformational alteration in the Z-DNA backbone structure, although base-packing parameters vary in a sequence-dependent manner (2,3,8,30–32). However, when corresponding atoms of each DNA pair are compared, differences near all of the phosphate groups are notable although the extent varies depending on their positions (Figure 5B–D). However, the differences near the phosphate groups of each DNA pair are reduced when the DNAs are bound to hZαADAR1 (Supplementary Table 3 and Figure 5) except in the case of N3pN4 in comparing d(CGCGCG)2 and d(CGGCCG)2. The reduction in structural alteration of the phosphate groups of Zα-bound DNA is probably due to structural restraint enforced by Zα binding. These results strongly support the idea that the preformed binding pocket of Zα functions as a mold in recruiting various Z-DNAs, and it freezes the backbone conformations of Z-DNAs. As a result, structural variations of Z-DNAs that are caused mostly by conformational heterogeneity near the phosphate groups are reduced upon their binding into the Zα cleft.
Z-DNAs containing non-APP sequences or A-T base pair(s) have intrinsic structural instability and an increased solvent-exposed surface since base pairs are buckled out of the base-pair plane and protrude into the major groove (3,8,30,31). These structural features are still observed in hZαADAR1-bound Z-DNAs (Supplementary Table 2). For example, d(CGTACG)2 and d(CGGCCG)2 reveal a huge buckle and decreased stacking interactions, whether bound to Zα or not (Figure 4 and Supplementary Table 2). Therefore, it is thought that the base packing pattern and structural instability of non-CG-repeat Z-DNAs are always maintained, whereas the phosphate backbone conformations are restrained by the Z-DNA-binding pocket of hZαADAR1. Overall, additional structural instability of the Z conformation caused by non-CG-repeat sequences did not significantly affect the maintenance of the Z confirmation of DNAs in their complexes with Zα. These results suggest that more energy is gained upon Zα binding than is needed to overcome the structural instability of non-CG-repeat Z-DNAs.
CONCLUSIONS
Our structural data strongly support the idea that hZαADAR1 recognizes Z-DNA in a conformation-specific manner, but not in a sequence-specific manner. The structures of hZαADAR1 complexed with three different non-CG-repeat double-stranded Z-DNAs (with non-APP sequences or A-T base pairs) reveal that hZαADAR1 binds and stabilizes the Z conformation of DNA via a similar binding mode to that of the Zα/d(CGCGCG)2 complex, regardless of sequence context. Most notably, the CH–π interaction was observed between Tyr177 and the cytosine in the syn conformation. While structures of phosphate backbones in free and hZαADAR1-bound states display some variation near each phosphate group, hZαADAR1 does not exert a profound effect on the Z-DNA base pair parameters induced by incorporation of non-APP and A-T base pairs. Irrespective of the heterogeneity in sequence and structure of non-CG-repeat Z-DNAs, Zα recognizes the DNA backbone atoms and imposes structural restraint through the core residues located on the DNA-binding surface. It has been suggested that different Z-DNA-binding proteins can bind to chromosomal or foreign DNAs and take part in essential biological processes. In this context, the Zα domain is expected to recognize and stabilize stretches of DNA in the Z-DNA conformation. Our results provide a molecular basis for understanding how the Zα proteins recognize and stabilize Z-DNAs in various sequence contexts.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Korea Science and Engineering Foundation (KOSEF) through the National Research Laboratory Program funded by the Ministry of Education, Science and Technology (NRL-2006-02287). Funding for open access charges: the National Laboratory Program grant of the Korean government (MEST).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Rich A. DNA comes in many forms. Gene. 1993;135:99–109. doi: 10.1016/0378-1119(93)90054-7. [DOI] [PubMed] [Google Scholar]
- 2.Wang AH, Quigley GJ, Kolpak FJ, Crawford JL, van Boom JH, van der Marel G, Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979;282:680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- 3.Ho PS, Mooers BHM. Z-DNA crystallography. Biopolymers. 1997;44:65–90. doi: 10.1002/(SICI)1097-0282(1997)44:1<65::AID-BIP5>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 4.Rich A, Nordheim A, Wang AH. The chemistry and biology of left-handed Z-DNA. Annu. Rev. Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- 5.Ho PS. The non-B-DNA structure of d(CA/TG)n does not differ from that of Z-DNA. Proc. Natl Acad. Sci. USA. 1994;91:9549–9553. doi: 10.1073/pnas.91.20.9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang AH, Hakoshima T, van der Marel G, van Boom JH, Rich A. AT base pairs are less stable than GC base pairs in Z-DNA: the crystal structure of d(m5CGTAm5CG) Cell. 1984;37:321–331. doi: 10.1016/0092-8674(84)90328-3. [DOI] [PubMed] [Google Scholar]
- 7.Schroth GP, Kagawa TF, Ho PS. Structure and thermodynamics of nonalternating C.G base pairs in Z-DNA: the 1.3-A crystal structure of the asymmetric hexanucleotide d(m5CGGGm5CG).d(m5CGCCm5CG) Biochemistry. 1993;32:13381–13392. doi: 10.1021/bi00212a002. [DOI] [PubMed] [Google Scholar]
- 8.Eichman BF, Schroth GP, Basham BE, Ho PS. The intrinsic structure and stability of out-of-alternation base pairs in Z-DNA. Nucleic Acids Res. 1999;27:543–550. doi: 10.1093/nar/27.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herbert A, Alfken J, Kim YG, Mian IS, Nishikura K, Rich A. A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proc. Natl Acad. Sci. USA. 1997;94:8421–8426. doi: 10.1073/pnas.94.16.8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz T, Behlke J, Lowenhaupt K, Heinemann U, Rich A. Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nat. Struct. Biol. 2001;8:761–765. doi: 10.1038/nsb0901-761. [DOI] [PubMed] [Google Scholar]
- 11.Ha SC, Lokanath NK, Quyen DV, Wu CA, Lowenhaupt K, Rich A, Kim YG, Kim KK. A poxvirus protein forms a complex with left-handed Z-DNA: crystal structure of a Yatapoxvirus Zalpha bound to DNA. Proc. Natl Acad. Sci. USA. 2004;101:14367–14372. doi: 10.1073/pnas.0405586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu CY, Zhang YB, Huang GP, Zhang QY, Gui JF. Molecular cloning and characterisation of a fish PKR-like gene from cultured CAB cells induced by UV-inactivated virus. Fish Shellfish Immunol. 2004;17:353–366. doi: 10.1016/j.fsi.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Rothenburg S, Deigendesch N, Dittmar K, Koch-Nolte F, Haag F, Lowenhaupt K, Rich A. A PKR-like eukaryotic initiation factor 2alpha kinase from zebrafish contains Z-DNA binding domains instead of dsRNA binding domains. Proc. Natl Acad. Sci. USA. 2005;102:1602–1607. doi: 10.1073/pnas.0408714102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwartz T, Rould MA, Lowenhaupt K, Herbert A, Rich A. Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science. 1999;284:1841–1845. doi: 10.1126/science.284.5421.1841. [DOI] [PubMed] [Google Scholar]
- 15.Ha SC, Lowenhaupt K, Rich A, Kim YG, Kim KK. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature. 2005;437:1183–1186. doi: 10.1038/nature04088. [DOI] [PubMed] [Google Scholar]
- 16.Ha SC, Kim D, Hwang HY, Rich A, Kim YG, Kim KK. The crystal structure of the second Z-DNA binding domain of human DAI (ZBP1) in complex with Z-DNA reveals an unusual binding mode to Z-DNA. Proc. Natl Acad. Sci. USA. 2009 doi: 10.1073/pnas.0810463106. doi:10.1073/pnas.0810463106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu H, Mulholland N, Fu H, Zhao K. Cooperative activity of BRG1 and Z-DNA formation in chromatin remodeling. Mol. Cell. Biol. 2006;26:2550–2559. doi: 10.1128/MCB.26.7.2550-2559.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang G, Vasquez KM. Z-DNA, an active element in the genome. Front. Biosci. 2007;12:4424–4438. doi: 10.2741/2399. [DOI] [PubMed] [Google Scholar]
- 19.Kwon JA, Rich A. Biological function of the vaccinia virus Z-DNA-binding protein E3L: gene transactivation and antiapoptotic activity in HeLa cells. Proc Natl Acad. Sci. USA. 2005;102:12759–12764. doi: 10.1073/pnas.0506011102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 21.Schwartz T, Lowenhaupt K, Kim YG, Li L, Brown B.A., II, Herbert A, Rich A. Proteolytic dissection of Zab, the Z-DNA-binding domain of human ADAR1. J. Biol. Chem. 1999;274:2899–2906. doi: 10.1074/jbc.274.5.2899. [DOI] [PubMed] [Google Scholar]
- 22.Schwartz T, Shafer K, Lowenhaupt K, Hanlon E, Herbert A, Rich A. Crystallization and preliminary studies of the DNA-binding domain Zα from ADAR1 complexed to left-handed DNA. Acta Crystallogr. D Biol. Crystallogr. 1999;55:1362–1364. doi: 10.1107/s090744499900582x. [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 26.Merritt EA. A program for photorealistic molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 27.Kraulis P. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 28.Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 29.Wang AH, Gessner RV, van der Marel GA, van Boom JH, Rich A. Crystal structure of Z-DNA without an alternating purine-pyrimidine sequence. Proc. Natl Acad. Sci. USA. 1985;82:3611–3615. doi: 10.1073/pnas.82.11.3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narayana N, Shamala N, Ganesh KN, Viswamitra MA. Interaction between the Z-type DNA duplex and 1,3-propanediamine: crystal structure of d(CACGTG)2 at 1.2 Å resolution. Biochemistry. 2006;45:1200–1211. doi: 10.1021/bi051569l. [DOI] [PubMed] [Google Scholar]
- 31.Wang AJ, Quigley GJ, Kolpak FJ, van der Marel G, van Boom JH, Rich A. Left-handed double helical DNA: variations in the backbone conformation. Science. 1981;211:171–176. doi: 10.1126/science.7444458. [DOI] [PubMed] [Google Scholar]
- 32.Sadasivan C, Gautham N. Sequence-dependent microheterogeneity of Z-DNA: the crystal and molecular structures of d(CACGCG):d(CGCGTG) and d(CGCACG):d(CGTGCG) J. Mol. Biol. 1995;248:918–930. doi: 10.1006/jmbi.1995.9894. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.