Abstract
Covalent modifications of nucleic acids play an important role in regulating their functions. Among these modifications, (cytosine-5) DNA methylation is best known for its role in the epigenetic regulation of gene expression. Post-transcriptional RNA modification is a characteristic feature of noncoding RNAs, and has been described for rRNAs, tRNAs and miRNAs. (Cytosine-5) RNA methylation has been detected in stable and long-lived RNA molecules, but its function is still unclear, mainly due to technical limitations. In order to facilitate the analysis of RNA methylation patterns we have established a protocol for the chemical deamination of cytosines in RNA, followed by PCR-based amplification of cDNA and DNA sequencing. Using tRNAs and rRNAs as examples we show that cytosine methylation can be reproducibly and quantitatively detected by bisulfite sequencing. The combination of this method with deep sequencing allowed the analysis of a large number of RNA molecules. These results establish a versatile method for the identification and characterization of RNA methylation patterns, which will be useful for defining the biological function of RNA methylation.
INTRODUCTION
Covalent modifications of nucleic acids are abundant but not very well understood. These modifications are thought to play an important role in regulating the functions of DNA and various cellular RNA species. Some DNA modifications have been analyzed in considerable detail and the characterization of (cytosine-5) DNA methylation has been crucial for developing the molecular basis of epigenetic gene regulation (1). Compared to DNA modifications, RNA modifications are more prevalent and chemically more complex (2), but the current knowledge of the function of RNA modifications is very limited. Modifications have been identified in rRNA, mRNA, miRNAs as well as in other small RNAs, such as tRNAs, which display complex patterns of modifications. Among these RNAs, (cytosine-5) RNA methylation has been described in rRNA and tRNA molecules (2). Both RNA species are stable and long-lived. Whether 5-methylcytosine (m5C) in RNA acts as an inheritable modification, comparable to DNA methylation, has not been tested, mainly because the functional characterization of RNA methylation has been hindered by the lack of molecular methods for RNA methylation analysis.
Methods to detect 5-methylcytosine in DNA were developed a long time ago (3,4). In general, methylation at position 5 of pyrimidines is difficult to detect by conventional molecular methodology since it does not interfere with the base-pairing properties of the nucleoside, thereby excluding hybridization-based detection methods. The most robust and reliable method for the detection of m5C in DNA is based on the selective chemical deamination of cytosine (but not m5C) to uracil by bisulfite treatment (5). This reaction (Figure 1A) is based on the low reactivity of m5C compared to cytosine residues with HSO3-ions in neutral or acidic solutions. Cytidine and some of its derivatives form adducts, which are relatively unstable. Rapid decomposition at alkaline pH leads to chemical deamination of the pyrimidine ring and concomitant conversion to uracil or derivatives of uracil. The conversion of cytosine (but not m5C) residues can be detected by PCR-based methods. DNA sequencing of PCR amplicons and/or of cloned PCR products (DNA bisulfite sequencing) allows the characterization of DNA methylation patterns in their native sequence context and has been of fundamental importance for understanding the human epigenome (6,7).
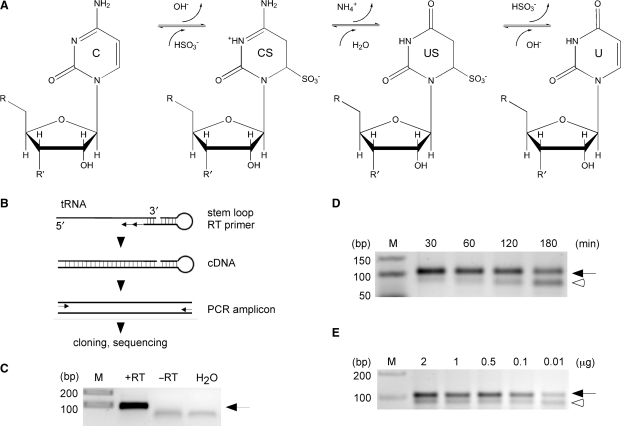
Figure 1.
Establishment of RNA bisulfite sequencing. (A) Schematic diagram of the bisulfite conversion reaction. C, cytidine; CS cytidine sulfonate; US, uridine sulfonate; U, uridine. (B) Outline of the basic strategy to analyze tRNA for m5C methylation. Bisulfite-treated tRNAs are reverse transcribed using a tRNA 3′-sequence-specific stem–loop primer, amplified with primers binding only to deaminated sequences at the 5′ end, followed by standard cloning and sequencing. (C) As an example, in vitro transcribed tRNAAsp served as template for cDNA synthesis. RT, reverse transcriptase; arrow, tRNA amplicon. (D) Increasing deamination times lead to degradation of tRNAAsp. Equal amounts of cellular RNA (1 μg) were subjected to deamination for the time indicated, followed by cDNA synthesis and PCR amplification. (E) Dilution series of total RNA subjected to bisulfite treatment, followed by cDNA synthesis and PCR amplification of tRNAAsp. Ten nanograms of cellular RNA are sufficient to amplify bisulfite-treated tRNAAsp (32 amplification cycles). Arrow, tRNA amplicon; open arrowhead, aberrant PCR amplicon.
Sodium bisulfite has also been applied to treat RNA in approaches to investigate amino-acylation and the three-dimensional structure of tRNAs (8,9). However, the general use of bisulfite deamination for RNA analysis was never seriously considered, since the harsh reaction conditions (high pH) are detrimental to the stability of phosphodiester bonds in RNA. Previous attempts using bisulfite treatment of RNAs for m5C mapping in tRNAHis (10) did not permit to interrogate the methylation status of individual RNA sequences in a manner comparable to DNA bisulfite sequencing. In order to determine m5C in the sequence context of RNAs on a general basis, we developed a bisulfite deamination protocol for RNA. A PCR based assay allows for cDNA amplification from low quantities of cellular RNAs and the analysis of clonal methylation patterns in their native sequence context. We show here that this approach can be used to precisely and quantitatively determine methylation patterns of various known RNA molecules in a rapid and cost-efficient way and thus represents a versatile method for the characterization of RNA methylation patterns.
MATERIALS AND METHODS
Materials
Unmodified tRNAAsp was generated as described previously (11). Drosophila Dnmt2 mutants were generated by P-element mediated transposon mutagenesis, as described elsewhere (12).
Isolation of RNA
RNA was isolated using Trizol (Invitrogen). Fractionation to enrich for tRNA was performed by fractionating total RNA (Trizol) to 15% denaturing urea–PAGE. RNAs were visualized using SYBR-Gold and tRNAs were excised under UV light. RNAs were extracted using NH4Cl and precipitated.
Bisulfite treatment
RNAs were digested with DNAse (Ambion). For bisulfite treatment of RNAs the EpiTect Bisulfite Kit (Qiagen) was used. Conversions were carried out in a reaction volume of 70 µl. One microgram total RNA, dissolved in 20 µl RNase-free water was mixed with 42.5 µl bisulfite mix and 17.5 µl DNA protect buffer (both provided with the kit). RNA was denatured at 70°C for 5–10 min, followed by a reaction period of 1 h at 60°C. For tRNA and 16S rRNA the reaction was submitted to a 3× cycle of the above described parameters. The cycle number was increased to 6× for the treatment of human 28S rRNA. The RNA was isolated from the bisulfite reaction mix with Micro Bio-Spin 6 columns (Bio-Rad). tRNAs were treated with 0.5 M Tris–HCl, pH 9 at 37°C for 1 h. The isolated RNA was ethanol precipitated (3 vol ethanol, 1/10 vol 2 M (NH4)OAc, 20 µg GlycoBlue) overnight at −80°C.
cDNA synthesis
Reverse transcription reactions were carried out with the SuperScript III Reverse Transcriptase kit (Invitrogen), following the manufacturers instructions. The concentration of dNTPs was chosen depending on the structure of the template to be between 0.5 and 2.5 mM for each dNTP. 250 ng random primers (random hexamers) or 1 μM specific primers (or stem–loop primers) were used in the reverse transcription reaction. The amount of RNA used was in the range between 10 and 500 ng. After mixing RNA, dNTPs and primers, the reaction was heated to 65°C for 5 min and immediately cooled on ice. 5× First-Strand buffer, DTT, RNaseOUT and SuperScript RT (200 U/µl) were added (as recommended by the manufacturer). The reaction was incubated for 10 min at 25°C, 50 min at 50°C and 15 min at 70°C.
Conventional cloning and sequencing
cDNA was PCR amplified using primers specific for deaminated sequences (Table 1). PCR products were separated from unincorporated primers using low melting agarose, followed by phenol–chloroform extraction (products <100 bp) and ethanol precipitation or by Gel Extraction Kit (Qiagen) processing for products >100 bp. PCR amplicons were cloned using the TOPO TA Cloning Kit for Sequencing (Invitrogen). The reaction product was used to transform chemically competent SURE 2 Supercompetent cells (Stratagene), according to the manufacturers instructions.
Table 1.
Primer sequences
| Reverse transcription primers | ||
| Stem–loop primer (nondeaminated sequences) | ||
| HD250 | D.m.tRNAAsp | 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTGGCTCCC-3′ |
| Stem–loop primer (deaminated sequences) | ||
| HD361 | D.m. tRNAAsp | 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTGGCTCCCAA-3′ |
| HD385 | H.s. tRNAAsp | 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTGGCTCCCCA-3′ |
| HD312 | tRNALys | 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTAACACCCAACA-3′ |
| Non-stem–loop primer (deaminated sequences) | ||
| HD366* | tRNAVal | 5′-CCCAAAATCAAACCAAAA-3′ |
| HD407 | E. coli 16SC967 | 5′-CTCTAAAAACTTCCATAA-3′ |
| HD409 | E. coli 16SC1407 | 5′-TAAACACCCTCCCAAAAA-3′ |
| HD411 | H.s. 28SC4417 | 5′-ATCTAAACCCAACTCACA-3′ |
| PCR primers | ||
| Forward primer (non-deaminated sequences) | ||
| HD254 | D.m. tRNAAsp | 5′-TCCTCGATAGTATAG-3′ |
| Forward primers (deaminated sequences) | ||
| HD267 | D.m. tRNAAsp | 5′-AGTATAGTGGTGAGTATT-3′ |
| HD360 | tRNALys | 5′-GTTTGGTTAGTTTAGtT-3′ |
| HD319 | H.s. tRNAAsp | 5′-TAGTATAGTGGTAGTATT-3′ |
| HD367* | tRNAVal | 5′-GTTTTTGTGGTGTAGTG-3′ |
| HD406 | E. coli 16SC967 | 5′-ATGAATTGATGGGGGTTT-3′ |
| HD408 | E. coli 16SC1407 | 5′-TGAATATGTTTTTGGGTT-3′ |
| HD410 | H.s. 28SC4417 | 5′-TTTTAAGTAGGAGGTGTT-3′ |
| Reverse primers | ||
| Universal stem–loop primer (nondeaminated and deaminated sequences) | ||
| HD258 | tRNAAsp tRNALys | 5′-CACGACACCAGTTGA-3′ |
| Non-stem–loop primer (deaminated sequences) | ||
| HD366* | tRNAVal | 5′-CCCAAAATCAAACCAAAA-3′ |
| HD407 | E. coli 16SC967 | 5′-CTCTAAAAACTTCCATAA-3′ |
| HD409 | E. coli 16SC1407 | 5′-TAAACACCCTCCCAAAAA-3′ |
| HD411 | H.s. 28SC4417 | 5′-ATCTAAACCCAACTCACA-3′ |
| 454 deep-sequencing PCR primers | ||
| Forward primer (deaminated sequences) | ||
| HD346 | tRNALys | 5′-GCCTCCCTCGCGCCATCAGCACAGTTTGGTTAGTTTAGTT-3′ |
| HD377* | tRNAVal | 5′-GCCTCCCTCGCGCCATCAGTCTCGTTTTTGTGGTGTAGTG-3′ |
| Reverse primer | ||
| HD366* | tRNAVal | 5′-CCCAAAATCAAACCAAAA-3′ |
| HD258 | tRNALys | 5′-CACGACACCAGTTGA-3′ |
Primers with an asterisk amplify both tRNAVal,3 and tRNAVal,4 (tRNAs can be distinguished by sequence analysis).
Underlined sequences represent hybridization of RT primer with RNA.
Bold letters denote bar-code sequences for deep sequencing.
Deep sequencing of tRNA-derived cDNAs
cDNA was PCR amplified (28 cycles) using primers specific for deaminated sequences (Table 1). One-tenth of the PCR products was again amplified (10–15 cycles) using primers specific for deaminated sequences plus the 5′-primer containing the 454 linker sequence (bar-coded). PCR products were separated from unincorporated primers using low melting agarose, followed by phenol–chloroform extraction (products <100 bp) and ethanol precipitation or Gel Extraction kit (Qiagen) processing for products >100 bp. PCR amplicons were quantified using NanoDrop and 1011 molecules per bar-coded PCR amplicon were processed for 454 deep sequencing according to the Roche 454 sequencing manual. Deep-sequencing data were analyzed for sequence specific cytosine content using the GS Amplicon Variant Analyzer (Roche).
RESULTS
Establishment of RNA bisulfite sequencing
Recently, the cytosine-5 methyltransferase Dnmt2 has been shown to catalyze the methylation of tRNAAsp at position C38 in various organisms, including Drosophila melanogaster (13). We took advantage of this finding to develop an RNA bisulfite sequencing protocol. Total RNA preparations from Drosophila were subjected to bisulfite conversion (Figure 1A) using a low-temperature protocol, followed by desalting and removal of aminoacylation. In order to specifically amplify tRNAAsp from a complex mixture of RNAs, a stem–loop primer for reverse transcription (RT) of tRNAAsp was designed (Figure 1B). Stem–loop primers are also being used to amplify other short RNAs such as miRNAs (14). Using in vitro transcribed tRNAAsp from D. melanogaster as a template we established that the tRNAAsp specific stem–loop RT primer allows the synthesis of cDNA that can be amplified by PCR using sequence-specific primers (Figure 1C).
Sodium bisulfite mediated deamination of cytosines in DNA is a function of temperature and time (15) and efficient melting of the DNA double-strand structure is crucial for efficient deamination. Bisulfite treatment of RNA under high temperature (95°C) and for extended time periods has been attempted previously (10,16). These conditions cause rapid degradation of RNA, thus reducing the number of molecules available for cDNA synthesis. We reasoned that RNA denaturation could be achieved at lower temperatures, because most RNA structures are defined by regions of single-strandedness followed by relatively short double-stranded (stem) structures and single-stranded loops. In order to develop a bisulfite deamination protocol for tRNAAsp, we tested various temperatures and times to optimize the conversion of unmethylated cytosines in total RNA or gel-purified tRNA fractions. First, we established the optimal deamination temperature that permitted robust PCR amplification with bisulfite-specific primers to be at 60°C (data not shown). At this temperature, RNA could be bisulfite-treated for as long as 180 min, without major loss of the PCR amplicon of the expected size (Figure 1D). This protocol allowed PCR amplification of the correct fragment from as little as 10 ng of total RNA as starting material (Figure 1E). Longer treatment times and lower amounts of starting material resulted in poorer results, as evidenced by the appearance of additional PCR products (Figure 1D and E; open arrowheads).
In order to quantitatively determine the deamination efficiency at various time points we treated total RNA from Drosophila embryos with bisulfite for 30, 60, 120, 180 and 300 min, and sequenced five independently cloned tRNAAsp molecules for every time point. The results showed that deamination rates increased from 67% after 30 min, to 79% after 60 min, to 91% after 120 min, to 99% after 180 min and to 100% after 300 min (Figure 2). Deamination rates >95% are generally considered to be sufficient for the generation of high-quality results (17). Because this benchmark was exceeded after 180 min of bisulfite treatment, these conditions were used as basis for further experiments. It is important to notice that the low levels (<5%) of randomly distributed nonconverted cytosines after 180 min of bisulfite treatment (deamination artifacts) can be used as indicators for the complexity of the RNA-derived cDNA library.
Figure 2.
In vivo RNA bisulfite sequencing of tRNAAsp from total RNA (1 μg) after various deamination times. Five clones were sequenced for each time point and deamination rates (DR) were calculated based on the number of all cytosines (minus known m5C residues at positions C38 and C48). Black boxes indicate methylated cytosine residues, white boxes indicate unmethylated cytosine residues. Each PCR amplicon queried 17 cytosines in tRNAAsp (C = 17). Numbers below boxes indicate the cytosine positions in the primary RNA sequence.
To validate the bisulfite sequencing protocol for the analysis of the in vivo methylation pattern of tRNAAsp we analyzed total RNA preparations from wild-type and from Dnmt2 mutant embryos. Dnmt2 has been shown to specifically methylate C38 of tRNAAsp (13), which predicted that the C38 methylation signal detected in tRNAAsp from wild-type tissue should be lost in Dnmt2 mutant tissue. Indeed, bisulfite sequencing of multiple independently cloned tRNAAsp sequences revealed 70% C38 methylation in wild-type tissue (Figure 3A), compared to 6% in Dnmt2 mutants (Figure 3B). Our data thus confirm the Dnmt2-dependent methylation of tRNAAsp at C38 in Drosophila (13). C48 was methylated both in wild-type and in Dnmt2 mutant samples, which is consistent with the notion that C48 is methylated by the tRNA methylase Trm4p (18). These results establish RNA bisulfite sequencing as a reliable method for the direct and quantitative analysis of in vivo tRNAAsp methylation.
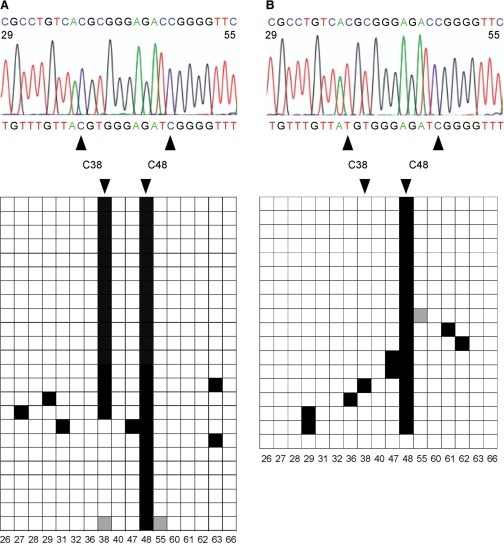
Figure 3.
Validation of RNA bisulfite sequencing by the detection of tRNAAsp methylation changes in Dnmt2 mutant tissues. (A) Bisulfite sequencing analysis of tRNAAsp from wild-type D. melanogaster embryos. A representative sequencing trace showing the deaminated tRNAAsp sequence (nucleotides 29–55 containing nine cytosines) with the nondeaminated sequence for comparison (above). The diagram shows the methylation status of individual cytosines (horizontal, C=17) in several independent clones (vertical, N = 24). Black boxes indicate methylated cytosine residues, white boxes indicate unmethylated cytosine residues, gray boxes indicate cytosine residues with unknown methylation status (due to bad sequence reads). Numbers below boxes indicate the cytosine positions in the primary RNA sequence. The deamination rate was calculated to be 98%. C38 (methylated by the Dnmt2 methyltransferase) and C48 (methylated by the Trm4 methyltransferase) were methylated in 70% and 100% of the sequences, respectively. (B) Bisulfite sequencing analysis of tRNAAsp from Dnmt2 mutant D. melanogaster embryos. The deamination rate was calculated to be 97%. C38 and C48 were methylated in 6% and 94% of the sequences (N = 18), respectively.
rRNA bisulfite sequencing
Having demonstrated the feasibility of RNA bisulfite sequencing for tRNA methylation analysis we next asked whether other cellular RNAs could also be analyzed. A prominent RNA species which has been reported to contain 5-methylcytosine is ribosomal RNA (2). 5-methylcytosine is found in rRNA from eubacteria to eukaryotes, with distinct rRNAs being methylated at different positions. It has been shown that 16S as well as 23S rRNA of Escherichia coli contains 5-methylcytosine (16,19). 5-methylcytosine has also been detected in ribosomal RNAs of eukaryotic organisms and 28S rRNA of Xenopus laevis and Homo sapiens is methylated on at least two different cytosines (20).
Using total RNA preparations from E. coli we performed RNA bisulfite sequencing on 16S rRNA. cDNA was synthesized using sequence-specific RT primers (supplemented with random hexamers), and two regions harboring the mapped m5C sites at position C967 and C1407 were amplified by PCR, cloned and sequenced. The results confirm C967 and C1407 methylation in 100% of the sequenced clones (Figure 4A and B). Interestingly, we detected an additional C at position 1402 that was maintained after bisulfite treatment in 88% of the sequenced clones (Figure 4B). C1402 has been shown previously to contain N4, 2′-O-dimethylcytidine (m4mC, Figure 4C), a cytdine that contains a methyl group at the exocyclic N4 position of the base as well as a methyl group at the 2′-position of the ribose (16,21). Our data thus indicate that m4mC is protected against bisulfite-mediated deamination, since it lacks one proton at the nitrogen atom (Figure 1A), and will therefore be scored as methylated in a bisulfite sequencing analysis. Together, these results demonstrate that RNA bisulfite sequencing can be applied to detect m5C in rRNA and show that at least one additional cytosine modification, m4mC, could also be analyzed.
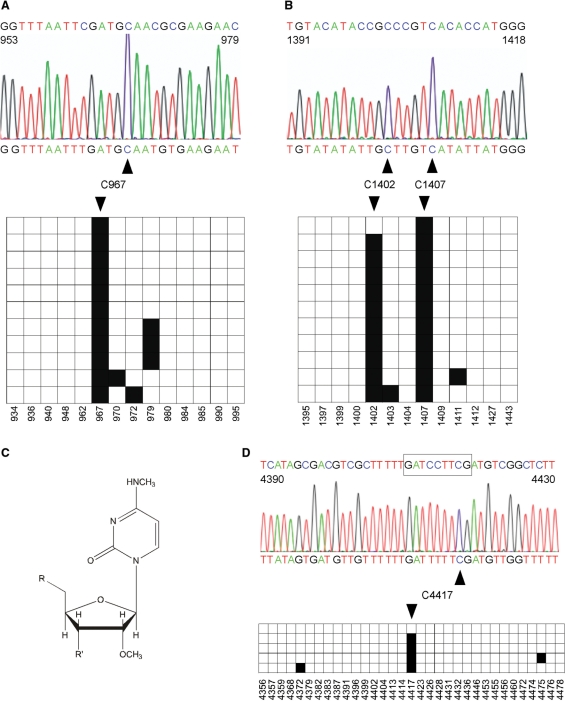
Figure 4.
RNA bisulfite sequencing detects cytosine methylation in ribosomal RNAs. Black boxes indicate methylated cytosine residues, white boxes indicate unmethylated cytosine residues. (A) A representative sequencing trace showing the deaminated 16S rRNA from E. coli (nucleotides 953–979 containing five cytosines) with the nondeaminated sequence for comparison (above). The diagram shows the methylation status of individual cytosines (horizontal, C = 14) in several independent clones (vertical, N = 11). The deamination rate was calculated to be 96%. C967 (methylated by the RsmB/Fmu methyltransferase) was methylated in 100% of the sequences. (B) Parallel analysis of nucleotides 1391–1418 from E. coli 16S rRNA. C1407 (methylated by the RsmF/YebU methyltransferase) was methylated in 100% of the sequences. Interestingly, C1402 containing m4mC, N4,2′-O-dimethylcytidine, was scored as methylated in 88% of the sequences. The diagram shows the methylation status of individual cytosines (horizontal, C = 13) in several independent clones (vertical, N = 11). The deamination rate was calculated to be 98%. (C) Chemical structure of N4,2′-O-dimethylcytidine which can be detected by RNA bisulfite sequencing at position C1402 in 16S rRNA. (D) Precision mapping of m5C in human 28S rRNA. A representative sequencing trace showing the deaminated 28S rRNA from the human HCT116 cell line (nucleotides 4390–4430) with the nondeaminated sequence for comparison (above). The boxed sequence highlights the region to which m5C was localized previously (20). The diagram shows the methylation status of individual cytosines (horizontal, C = 33) in five independent clones. The deamination rate was calculated to be 99%. Bisulfite sequencing uncovers the exact position of m5C at nucleotide 4417. Numbers below boxes indicate the cytosine positions in the primary RNA sequence.
Additionally, we used RNA bisulfite sequencing on total RNA for precision mapping of a m5C residue that had been previously suggested to occur in region 4410–4418 of the human 28S rRNA sequence, but has not yet been mapped to an exact nucleotide (20). RNA bisulfite sequencing uncovered a nondeaminated cytosine at position 4417 in 80% of the clones analyzed (Figure 4D), thus further illustrating the capability of RNA bisulfite sequencing to precisely map m5C residues in RNA.
RNA methylation analysis by deep sequencing
Having demonstrated that RNA bisulfite sequencing could generally be applied to tRNA and rRNA methylation analysis we next sought to extend the use of the method to the analysis of other tRNAs. tRNAs contain modifications that can block reverse transcription (commonly methylation of adenine 58; m1A58). We therefore tested several experimental approaches to facilitate reverse transcription from such tRNA templates. First, we moved the RT primer-binding site closer to the anticodon stem–loop covering m1A58 (patch primer) to exclude the blocking effect of this modification. Second, we performed the RT reaction using increased dNTP concentrations (up to 2.5 mM), which allows stalled RT molecules to overcome the transcriptional block. Third, we primed the RT reaction using random hexamers creating a mixed rather than a tRNA specific cDNA library. These adjustments in the RT reactions allowed us to amplify additional tRNAs, including tRNAVal,3, tRNAVal,4 and tRNALys from bisulfite-treated total RNA preparations. Conventional bisulfite sequencing of cloned tRNAVal,3 sequences (N = 10) suggested scattered low-level RNA methylation (Figure 5A). tRNAVal,4 (N = 6) was partially methylated at C38 (Figure 5A), while all clones of tRNALys (N = 10) were methylated at C48 (Figure 5A). These results indicated that bisulfite sequencing could be used to detect methylated nucleotides in various tRNAs However, querying a limited number of clones by conventional sequencing did not allow definite conclusions about the methylation status of cytosine residues that were spuriously methylated at low levels.
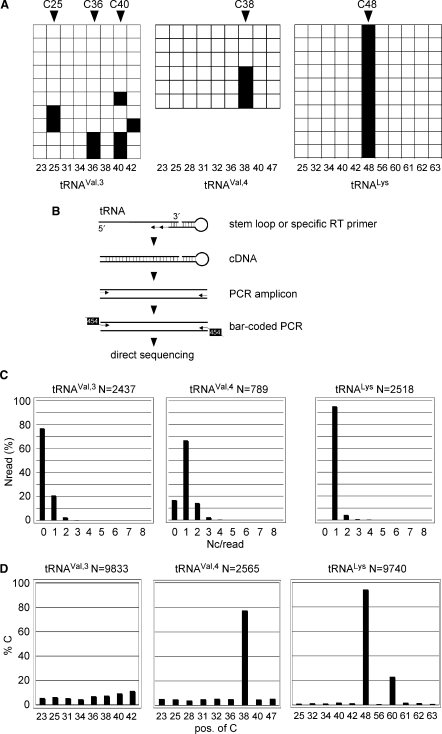
Figure 5.
Deep sequencing of tRNA methylation patterns. (A) Conventional sequencing results. Bisulfite sequencing analysis of tRNAs from wild-type D. melanogaster embryos. The diagram shows the methylation status of individual cytosines (horizontal squares) in several independent clones (vertical squares) in tRNAVal,3, tRNAval,4 and tRNALys. Black boxes indicate methylated cytosine residues, white boxes indicate unmethylated cytosine residues. Numbers below boxes show cytosine position in primary RNA sequence. (B) Outline of the strategy to analyze tRNA for m5C methylation using 454 technology. Bisulfite-treated tRNAs are reverse transcribed using a stem–loop primer or a specific tRNA primer (tRNAVal), followed by PCR amplification step 1 with primers binding only to deaminated sequences at the 5′ end and step 2 with primers introducing the bar-coded 454 sequence. PCR amplicons were sequenced directly. (C) 454 deep-sequencing results. Distribution of nondeaminated cytosines per read (Nc/read) for the illustration of pattern complexity and deamination efficiency. (D) Bisulfite sequencing analysis of tRNAs from wild-type D. melanogaster embryos. Fractions of nondeaminated cytosines are plotted against the position of the cytosine in individual tRNA amplicons, thus revealing the pattern of m5C modification in individual tRNAs.
To increase the analytical depth of the RNA bisulfite sequencing method and to precisely quantify tRNA methylation levels, we analyzed the methylation patterns of tRNAVal,3, tRNAVal,4 and tRNALys using deep-sequencing technology on cDNA libraries derived from Drosophila embryos (see scheme in Figure 5B). For tRNAVal,3 we obtained 9833 reads (2437 after elimination of partial reads) querying eight cytosines. For tRNAVal,4 we obtained 2565 reads (789 after elimination of partial reads) querying nine cytosines, and for tRNALys we obtained 9740 reads (2518 after elimination of partial reads) querying 11 cytosines. Analysis of complete reads (entire sequence of tRNA available) and plotting of the number of cytosines per individual read (Nc/read) against the number of reads (Nread) containing these cytosines provided a measure for the overall deamination efficiency (Figure 5C). We calculated the deamination rate to be >95% for all analyzed tRNAs. The clonal diversity of the sequenced library was confirmed by the manual analysis of the distribution of cytosines in sequences with >3 nondeaminated cytosines (data not shown). Figure 5C shows that the majority of tRNAVal,3 molecules is not methylated while most of the tRNAVal,4 and tRNALys molecules are highly methylated at a single site.
Analyzing both complete and partial reads allowed to analyze the methylation levels for individual cytosines in their sequence context. Methylation of individual cytosines varied greatly and ranged from 1 to 94% (Figure 5D). tRNAVal,3 showed low overall methylation levels at all eight cytosine residues analyzed, thus indicating that the tRNA is unmethylated in the analyzed tissue (Figure 5D). In contrast, tRNAVal,4 was found to be highly (77%) methylated at C38 (Figure 5D). Similarly, tRNALys was highly (94%) methylated at C48, and, to a lower extent (22%) at C60 (Figure 5D). These data are in good agreement with published methylation data (22). C60 methylation in tRNALys could not be uncovered by conventional bisulfite sequencing of a few individual clones (Figure 5A) and has not been reported in the literature. This finding provides a strong argument that deep sequencing of RNA molecules might support the detection and characterization of novel methylation sites in RNA. Together, these results illustrate the advantage of deep-sequencing technologies for a comprehensive characterization of RNA cytosine methylation and underscore the capability of RNA bisulfite sequencing to precisely quantify tRNA methylation levels.
DISCUSSION
Sequence-context-specific RNA modifications have been historically mapped by nuclease digest combined with radioactive labeling and chromatographic methods, such as thin-layer chromatography and high-performance liquid chromatography. These methods are comparatively time-consuming and do not readily allow the quantitative analysis of methylation patterns in their native sequence context.
We have established RNA bisulfite sequencing as a generally applicable and cost-efficient method for a clonal and sequence-specific analysis of RNA cytosine methylation. Using small amounts of unfractionated RNA preparations (<100 ng) we were able to detect m5C in several abundant RNA species from various organisms. The bisulfite conversion rate of small RNAs such as tRNAs was generally high (>95%) and comparable to similar studies of DNA methylation patterns (17), which minimized the occurrence of conversion artifacts. Lower conversion rates, as observed during the analysis of larger RNA molecules (e.g. 28S rRNA), which form extended secondary structures, can be overcome by adjustments in the duration of melting and treatment cycles. RNA degradation at low temperatures (60°C) became limiting only with extended treatment times (Figure 1). This problem can be solved by amplifying comparably small PCR fragments. Nevertheless, extended treatment times as well as low amounts of starting material led to the appearance of aberrant PCR products, at the expense of the correctly sized fragment (Figure 1D and E), thus defining the lower limit in the amount of starting material for cDNA synthesis. A major role of cDNA and PCR amplification bias can be excluded by analyzing the diversity of sequencing results and by mapping the occurrence of the <5% nondeaminated cytosines in individual clones. A random distribution of these cytosines is expected if the library contains a representative number of RNA-derived cDNAs, which show differential distribution of low-level deamination artifacts. We anticipate the possibility to analyze m5C patterns in RNA without the need of PCR amplification (23), which would circumvent the PCR bias problems and would allow the mapping and quantification of the methylated RNA transcriptome (23).
Additionally, the detection of bacterial m4mC at position 1402 of 16S rRNA indicates the possibility that bisulfite sequencing can be used to query RNA sequences for any given cytosine modification interfering with the bisulfite deamination process. Among these are 3-methylcytidine (3mC), detected in human 18S and 28S rRNA (24), N4-methylcytidine (4mC), found in mitochondrial 13S rRNA (25) as well as N4-acetylation (ac4C and ac4Cm) of cytidines (26). It will be interesting to experimentally validate RNA bisulfite sequencing for these modifications, which should further expand the application potential of the method.
The presence of RNA modifications, including m5C, is currently only known for abundant RNA species, and the precise physiological importance of cytosine methylation in tRNA and rRNA is mostly unknown. It was demonstrated that the presence of m5C at the wobble position of the anticodon loop in yeast tRNALeu modulates its efficiency as a suppressor in vivo (27). Structural studies have also suggested the importance of m5C for correct spatial organization of the yeast tRNAPhe anticodon stem–loop (28). It is also possible that m5C in long-lived RNA molecules could serve as an inheritable modification in the regulation of RNA-mediated epigenetic inheritance (29). Methylation mapping of RNAs in various experimental and mutant backgrounds will lead to detailed and novel insights into the function of this nucleic acid modification.
FUNDING
The Deutsche Forschungsgemeinschaft (Priority Programme Epigenetics; to M.S. and F.L.). Funding for open access charge: Deutsche Forschungsgemeinschaft DFG.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Mark Helm for stimulating discussions, Madeleine Meusburger for in vitro transcribed tRNAs, Gerald Nyakatura for 454 sequencing and Thomas Horn and Anne Arens for bioinformatics support.
REFERENCES
- 1.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 2.Rozenski J, Crain PF, McCloskey JA. The RNA Modification Database: 1999 update. Nucleic Acids Res. 1999;27:196–197. doi: 10.1093/nar/27.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rein T, DePamphilis ML, Zorbas H. Identifying 5-methylcytosine and related modifications in DNA genomes. Nucleic Acids Res. 1998;26:2255–2264. doi: 10.1093/nar/26.10.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomassin H, Oakeley EJ, Grange T. Identification of 5-methylcytosine in complex genomes. Methods. 1999;19:465–475. doi: 10.1006/meth.1999.0883. [DOI] [PubMed] [Google Scholar]
- 5.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakraburtty K. Effect of sodium bisulfite modification on the arginine acceptance of E. coli tRNA Arg. Nucleic Acids Res. 1975;2:1793–1804. doi: 10.1093/nar/2.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabban EL, Bhanot OS. The effect of bisulfite-induced C to U transitions on aminoacylation of Escherichia coli glycine tRNA. J. Biol. Chem. 1982;257:4796–4805. [PubMed] [Google Scholar]
- 10.Gu W, Hurto RL, Hopper AK, Grayhack EJ, Phizicky EM. Depletion of Saccharomyces cerevisiae tRNA(His) guanylyltransferase Thg1p leads to uncharged tRNAHis with additional m(5)C. Mol. Cell Biol. 2005;25:8191–8201. doi: 10.1128/MCB.25.18.8191-8201.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hengesbach M, Meusburger M, Lyko F, Helm M. Use of DNAzymes for site-specific analysis of ribonucleotide modifications. RNA. 2008;14:180–187. doi: 10.1261/rna.742708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jurkowski TP, Meusburger M, Phalke S, Helm M, Nellen W, Reuter G, Jeltsch A. Human DNMT2 methylates tRNA(Asp) molecules using a DNA methyltransferase-like catalytic mechanism. RNA. 2008;14:1663–1670. doi: 10.1261/rna.970408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 14.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noller HF. Structure of ribosomal RNA. Annu. Rev. Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- 17.Taylor KH, Kramer RS, Davis JW, Guo J, Duff DJ, Xu D, Caldwell CW, Shi H. Ultradeep bisulfite sequencing analysis of DNA methylation patterns in multiple gene promoters by 454 sequencing. Cancer Res. 2007;67:8511–8518. doi: 10.1158/0008-5472.CAN-07-1016. [DOI] [PubMed] [Google Scholar]
- 18.Motorin Y, Grosjean H. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA. 1999;5:1105–1118. doi: 10.1017/s1355838299982201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith JE, Cooperman BS, Mitchell P. Methylation sites in Escherichia coli ribosomal RNA: localization and identification of four new sites of methylation in 23S rRNA. Biochemistry. 1992;31:10825–10834. doi: 10.1021/bi00159a025. [DOI] [PubMed] [Google Scholar]
- 20.Maden BE. Locations of methyl groups in 28 S rRNA of Xenopus laevis and man. Clustering in the conserved core of molecule. J. Mol. Biol. 1988;201:289–314. doi: 10.1016/0022-2836(88)90139-8. [DOI] [PubMed] [Google Scholar]
- 21.Brosius J, Palmer ML, Kennedy PJ, Noller HF. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl Acad. Sci. USA. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sprinzl M, Vassilenko KS. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:D139–D140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 24.Iwanami Y, Brown GM. Methylated bases of ribosomal ribonucleic acid from HeLa cells. Arch. Biochem. Biophys. 1968;126:8–15. doi: 10.1016/0003-9861(68)90553-5. [DOI] [PubMed] [Google Scholar]
- 25.Dubin DT, Taylor RH, Davenport LW. Methylation status of 13S ribosomal RNA from hamster mitochondria: the presence of a novel riboside, N4-methylcytidine. Nucleic Acids Res. 1978;5:4385–4397. doi: 10.1093/nar/5.11.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas G, Gordon J, Rogg H. N4-Acetylcytidine. A previously unidentified labile component of the small subunit of eukaryotic ribosomes. J. Biol. Chem. 1978;253:1101–1105. [PubMed] [Google Scholar]
- 27.Strobel MC, Abelson J. Effect of intron mutations on processing and function of Saccharomyces cerevisiae SUP53 tRNA in vitro and in vivo. Mol. Cell Biol. 1986;6:2663–2673. doi: 10.1128/mcb.6.7.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gustilo EM, Vendeix FA, Agris PF. tRNA's modifications bring order to gene expression. Curr. Opin. Microbiol. 2008;11:134–140. doi: 10.1016/j.mib.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]