Abstract
UHRF1 (ubiquitin-like, containing PHD and RING finger domains 1) is a multi-domain protein associated with cellular proliferation and epigenetic regulation. The UHRF1 binds to methylated CpG dinucleotides and recruits transcriptional repressors DNA methyltransferase 1 (DNMT1) and histone deacetylase 1 (HDAC1) through its distinct domains. However, the molecular basis of UHRF1-mediated transcriptional regulation via chromatin modifications is yet to be fully understood. Here we show that UHRF1 binds histone lysine methyltransferase G9a, and both are co-localized in the nucleus in a cell-cycle-dependent manner. Concurrent with the cell-cycle progression, gradual deposition of UHRF1 and G9a was observed, which mirrored H3K9me2 accumulation on chromatin. Murine Uhrf1-null embryonic stem (ES) cells displayed a reduced amount of G9a and H3K9me2 on chromatin. UHRF1 recruited and cooperated with G9a to inhibit the p21 promoter activity, which correlated with the elevated p21 protein level in both human UHRF1 siRNA-transfected HeLa cells and murine Uhrf1-null ES cells. Furthermore, endogenous p21 promoter remained bound to UHRF1, G9a, DNMT1 and HDAC1, and knockdown of UHRF1 impaired the association of all three chromatin modifiers with the promoter. Thus, our results suggest that UHRF1 may serve as a focal point of transcriptional regulation mediated by G9a and other chromatin modification enzymes.
INTRODUCTION
The Uhrf1 gene encodes a member of RING-finger E3 ubiquitin ligase that is overexpressed in cancers (1). Mammalian UHRF1, previously known as ICBP90 (inverted CCAAT box binding protein of 90 kDa) in human (2) and Np95 (nuclear protein of 95 kDa) in mouse (3), possesses a UBI (ubiquitin-like), PHD (plant homeodomain), SRA (SET and RING associated) and RING (really interesting new gene) domains. These four distinct domains of the protein serve different functions. The UBI domain exhibits a typical α/β ubiquitin fold along with surface lysine residues similar to those of ubiquitin molecule. The PHD domain is placed between the UBI and SRA domains. Both PHD and SRA domains participate in di- and trimethyl histone H3K9 binding (4). Although the PHD domain determines the binding specificity, SRA domain promotes binding activity. Furthermore, both domains are essential for heterochromatic localization of human UHRF1, and down-regulation of UHRF1 in both human and mouse cells resulted in disrupted distribution of H3K9me3 and Hp1α, two known heterochromatic marks on the mammalian genome (4). The SRA domain of mouse UHRF1 was also shown to bind histones (5), and depletion of UHRF1 in murine cells resulted in hyperacetylated histone H4 and increased transcription of major satellites, demonstrating a role of UHRF1 in pericentromeric heterochromatin formation (6). In relevance to this observation, a recent study demonstrated that the PHD domain of mouse UHRF1 plays a role in large-scale reorganization of pericentromeric heterochromatin (7). Apart from binding to histones, the SRA domain of UHRF1 can bind to methyl-CpG dinucleotides with a preference for hemimethylated CpG sites (8,9). Similarly, in Arabidopsis, a UHRF1 homolog VIM1 has methyl cytosine binding properties via its SRA domain and plays a crucial role in maintenance of heterochromatin (10). The RING domain of UHRF1 was found to be essential for its E3 ubiquitin ligase activity for histones (4,5). Deletion of the RING domain was shown to sensitize cells to the effects of chemotherapeutics such as etoposide and cis-platinum (11).
Chromatin modifications exert a significant impact on gene expression. Several chromatin-modifying enzymes have been identified and known to catalyze specific modifications including methylation, acetylation, phosphorylation and ubiquitination (12). Enzymes that remove the modifications have been also identified, reflecting the dynamic situations in chromatin structure and function. Most of these enzymes have an intrinsic affinity to specific target substrates; however, such preference is not considered to be sufficient to carry out efficient and specific chromatin modifications to accommodate dynamic changes in chromatin structure occurring during normal cell growth. In fact, additional nuclear proteins have been identified and shown to direct these various chromatin-modifying enzymes to specific targets to facilitate timely and efficient modifications on chromatin. Among these additional factors, UHRF1 was found to form a complex with HDAC1 and bind to methylated promoter regions of tumor suppressor genes such as p16 and p14 in cancer cells (8). Recently, UHRF1 was also shown to bind mammalian DNA methyltransferase DNMT1 (9,13,14). The interaction between DNMT1 and UHRF1 facilitates and maintains DNA methylation in both human and mouse genomes (9,14). This interaction was shown to maintain global and local DNA methylation and demonstrated to repress the transcription of retrotransposons and imprinted genes. The loss of UHRF1 resulted in 75% reduction in genomic methylation in mouse embryonic stem (ES) cells (15).
The UHRF1, a known cell-cycle regulator and transcriptional activator of topoisomerase IIα expression, was also shown to be a regulator for retinoblastoma (Rb) expression (2,16). Overexpression of UHRF1 resulted in down-regulation of pRb, and the UHRF1 can form complexes with pRb in human cells, indicating that the protein complexes may affect pRb-regulated promoters. Indeed, the presence of a macromolecular complex of pRb2/p130 on estrogen receptor-α (ER-α) promoter correlated with DNA methylation status of the gene, and the same study suggested that pRb2/p130 could cooperate with UHRF1 and DNA methyltransferases in maintaining a specific methylation pattern of ER-α gene (17). Further experimental evidence of UHRF1-associated nuclear proteins involved in DNA repair and chromatin modification was recently documented by mass-spectometric analyses of UHRF1 pull-down complexes (14). The UHRF1-associated protein complexes suggest that UHRF1 is involved in regulation of local and global epigenome. Therefore, UHRF1 appears to be a transcriptional regulator via regulating DNA methylation and/or recruitment of transcriptional repressors.
Despite significant progress toward understanding the role of UHRF1 in gene expression, the precise role of UHRF1 in transcriptional gene regulation is not fully understood. There are both speculations and evidences of UHRF1 being a focal point of chromatin modification due to its close association with DNMT1 and HDAC1. Here, we demonstrate that UHRF1 recruits G9a, an essential component of histone H3K9 modification. This cooperative binding of UHRF1 and G9a was observed on the endogenous p21 promoter to repress the gene in cancer cells. Therefore, a new mechanism of UHRF1-based gene silencing via histone methyltransferase recruitment is discussed.
MATERIALS AND METHODS
Cell culture, transfections and RNA interference
HeLa, COS-7 and HEK293 cells were obtained from American Type Culture Collection (ATCC) and maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (FBS) (Hyclone). Mouse ES cells (a generous gift from Haruhiko Koseki and Masahiro Muto), E14 and mUhrf1−/− (19–4), were cultured on 0.1% gelatin (StemCell Technologies Inc) in Glasgow's Minimal Essential Medium supplemented with 50 U/ml mouse Leukemia Inhibitory Factor (LIF) (Chemicon), 10% FBS, 1X non-essential amino acids (Invitrogen), 1 mM sodium pyruvate and 55 μM β-mercaptoethanol. All media (Invitrogen) were supplemented with 2 mM l-glutamine and 1% antibiotic solution (ATCC). DNA transfections for luciferase assays were performed using FuGENE 6 (Roche Applied Science) according to the manufacturer's instructions. For siRNA transfections, hUHRF1 (Invitrogen), G9a (New England Biolabs), DNMT1 (New England Biolabs) and control Litmus (New England Biolabs) siRNAs were transfected into HeLa cells twice to a final concentration of 20–100 nM for 4 days using RNAiFECT reagent (Qiagen). Cell synchronization at G1/S was performed essentially following the double thymidine block protocol (18) except that aphidicolin (3 μg/ml) instead of thymidine (2 mM) was used for the first block.
Plasmid construction and antibodies
GFP fusions of full-length human UHRF1 (GFP-hUHRF1) and mouse UHRF1 (GFP-mUHRF1) were described previously (9). All GST-hUHRF1 constructs were generated by PCR-based cloning procedures, using the GFP-hUHRF1 as a template and cloning the resulting products into EcoRI/XhoI sites of pGEX5X-1 (GE Healthcare Life Sciences). DsRed-G9a and all GST-G9a constructs were previously described (19). To obtain purified hUHRF1 for GST pull-down assays, the full-length hUHRF1 fragment was cloned into NdeI/XhoI sites of pET-28a (Novagen). A 5X Gal4-cdc2 luciferase reporter (pG5-cdc2-luc) was constructed by inserting the cdc2 promoter fragment (−912 to +33) from pcdc2-luc (20) into NheI/BglII sites of pG5luc (Promega). To generate a Gal4 DNA-binding domain (Gal4DBD) fusion of hUHRF1 (pG4-hUHRF1), the full-length hUHRF1 fragment was cloned into SalI/XbaI sites of pBIND (Promega). The EGFP-fused N-terminal deletion mutant of G9a (EGFP-NΔG9a) was previously described (21). The DsRed-NΔG9a was generated by PCR amplification of coding sequence for G9a lacking the N-terminal 394 amino acids and subcloning the PCR product into EcoRI/BamHI sites of pDsRed2-C1 (Clontech). Antibodies (Ab) used for immunoprecipitation and western analyses were as follows: anti-GFP Ab (Roche Applied Science), anti-hUHRF1 Ab (BD Biosciences), anti-G9a Ab (Sigma), anti-human p21 Ab (Cell Signaling Technology), anti-mouse p21 Ab (Abcam), anti-dimethyl histone H3 (Lys9) Ab (Millipore), anti-histone H3 Ab (Cell Signaling Technology), anti-phospho-histone H3 (Ser10) Ab (Cell Signaling Technology), anti-actin Ab (Sigma) and anti-DNMT1 Ab (New England Biolabs).
Coimmunoprecipitation and western blot analysis
After 48 h of transfection, COS-7 cells were washed with PBS once and lysed in ice-cold RIPA buffer with proteinase inhibitor cocktail (Sigma). Cleared cell lysates (1.2 mg) were pre-incubated with BSA-blocked protein G-magnetic beads (New England Biolabs) for 1 h at 4°C to reduce non-specific binding of proteins to the beads. After brief spin, the precleared cell lysates were incubated with 2 μg of indicated antibodies for 2 h at 4°C before precipitation of the immune complexes with protein G-magnetic beads for 1 h. Immunoprecipitates were analyzed using western blot as described previously (19). For coimmunoprecipitation of endogenous proteins, HEK293 cells were synchronized by serum starvation for 20 h and the subsequent release into 10% FBS-containing medium for 15 h before cell harvest. Immunoprecipitation was performed following the same procedure described earlier.
Purification of GST-fusion proteins and GST pull-down assays
Purification of GST-fusion proteins and pull-down assays were described previously (22). Purified hUHRF1 protein was obtained by bacterial expression of 6xHis-tagged hUHRF1 and Ni-sepharose chromatography. G9a was expressed and purified from baculovirus-infected Sf9 cells (New England Biolabs).
Cytochemistry
COS-7 cells were cotransfected with DsRed-G9a and GFP-hUHRF1 or GFP-mUHRF1 plasmids with TransPass D2 reagent (New England Biolabs) for 48 h. Fluorescence microscopy was performed as described previously (19). Cells were visualized with a Zeiss 200M microscope with a 63× oil objective lens at 488 nm for GFP-hUHRF1 and GFP-mUHRF1 proteins, 568 nm for DsRed-G9a detection and 460 nm for DNA staining with Hoechst 33342.
Chromatin isolation and chromatin immunoprecipitation (ChIP)
Chromatin isolation procedure was described previously (9). The chromatin in buffer C (1% SDS, 10 mM EDTA, 50 mM Tris–HCl, pH 8.0) was used for either western-blot analyses or ChIP. For ChIP, the chromatin fractions were diluted 10-fold in ChIP dilution buffer (16.7 mM Tris–HCl, pH 8.0, 167 mM NaCl, 1.2 mM EDTA, 1.1% Triton X-100, 0.01% SDS) and subjected to brief sonication for 20 s followed by centrifugation at 4000 × g for 5 min. The cleared chromatin fractions (30–40 μg based on DNA per IP) were used for immunoprecipitation with 2 μg of indicated antibodies overnight at 4°C. The immune complexes were precipitated by pre-blocked protein G-magnetic beads (New England Biolabs) for 2 h and subjected to sequential washes with low salt buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.1% deoxycholate, 0.1% SDS) three times and high salt buffer (50 mM Tris–HCl, pH 8.0, 500 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.1% deoxycholate, 0.1% SDS) once. DNA was eluted in 1% SDS elution buffer (1% SDS, 10 mM EDTA, 50 mM Tris–HCl, pH 8.0) and de-crosslinked at 65°C overnight. DNA was purified by using QIAquick PCR purification kit (Qiagen) and eluted in 50 μl of TE buffer. The recovered DNA was analyzed by conventional PCR or quantitative PCR (Q-PCR) by using specific primers to the proximal and distal regions of p21 promoter (23). Before each IP, 5% input chromatin was taken and used to normalize the Q-PCR data as % input. The relative promoter occupancy after knockdown (KD) experiments was calculated by dividing the % input of each experimental group by that of control KD (CTL KD).
Luciferase assay
COS-7 cells were cotransfected with pG5-cdc2-luc reporter plus various combinations of pG4-hUHRF1 and GFP-G9a plasmids in six-well plates as indicated in the figure legend (Figures 4 and 6). Similarly, the p21 promoter–luciferase construct [pGL2–p21 (24), a kind gift from Jane B. Trepel] was cotransfected with different combinations of EGFP-hUHRF1 and EGFP-G9a plasmids. Empty vector DNA was included in the transfections to ensure that the same amounts of the DNA are introduced to the cells, and the pCMV-Gluc control plasmid (New England Biolabs) was used to normalize the transfection efficiency. After 48 h transfection, firefly luciferase activity was determined by Luciferase Assay System (Promega) and normalized by Gaussia luciferase activity measured by Gaussia Luciferase Assay Kit (New England Biolabs). Each transfection was performed in duplicate and repeated at least three times.
Quantitative RT–PCR
Total RNA was isolated from siRNA-transfected HeLa cells with RNeasy Micro Kit (Qiagen) and used for cDNA synthesis by DyNAmo SYBR Green 2-step qRT-PCR Kit (Finnzymes). The Q-PCR was performed by a Bio-Rad iCycler using iQ SYBR Green Supermix (Bio-Rad). The amount of p21 mRNA from each sample was normalized by the amount of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) as an internal control. The primer sequences for p21 are 5′-ATGGAACTTCGACTTTGTCACC-3′ and 5′-AGGCACAAGGGTACAAGACAGT-3′ (220 bp). The primer sequences for GAPDH were described previously (20). Each quantitative RT-PCR was performed in triplicate using four independent sets of cDNA.
Cell-proliferation assay
Wild-type (+/+) and mUhrf1-null (−/−) ES cells were seeded on six-well plates at 1 × 105 cells per well, and cell growth was monitored for 3 days by counting cells from six replicates for each group. For BrdU (bromodeoxyuridine)-incorporation assay, cells were plated in a 96-well format at 1000 cells/well. After 24 h incubation, BrdU labeling was performed for 2 h and determined by using cell proliferation ELISA kit (Roche) according to the manufacturer's instruction.
Bisulfite sequencing
Bisulfite conversion of genomic DNA (2 μg) was performed by using EpiTect Bisulfite kit (Qiagen) following the manufacturer's instruction. The primers for p21 promoter region (−398 to +11) are as follows: 5′-TTTTTGTTTGTTAGAGTGGGTTAG-3′ and 5′- ACAACTACTCACACCTCAACTAA-3′. The PCR products were ligated into pCR2.1-TOPO by using the TOPO TA cloning system (Invitrogen), and at least 20 separate clones per group were sequenced.
RESULTS
hUHRF1 interacts with G9a in vivo and in vitro
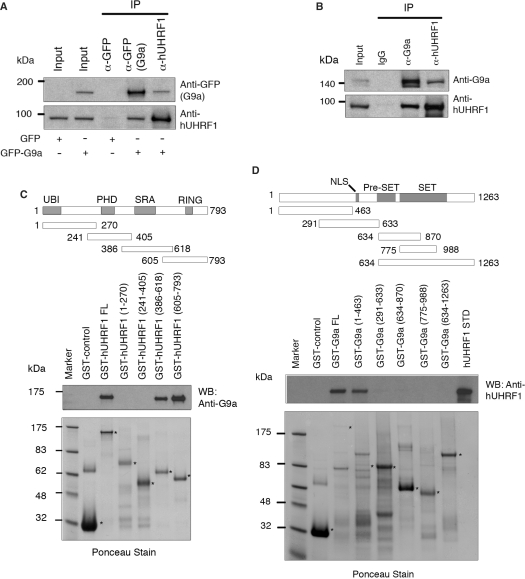
Human UHRF1 (hUHRF1) was shown to form a complex with HDAC1 (8). Both hUHRF1 and mUHRF1 (mouse UHRF1) were found to interact with DNMT1 and contribute to maintenance DNA methylation (9,14). These observations led to a possibility that UHRF1 may have an extended role for recruiting a broad range of chromatin modification enzymes during epigenetic regulation. To establish possible interactions between hUHRF1 and G9a, COS-7 cells were transfected with an expression vector encoding GFP-fused full-length G9a (GFP-G9a) to overcome a low abundance of G9a relative to hUHRF1. The cell extracts were used for immunoprecipitation with either anti-GFP antibody or anti-hUHRF1 antibody, and the immunoprecipitates were analyzed by western blot with specific antibodies to detect the in vivo association of the GFP-G9a and endogenous hUHRF1 proteins. Both proteins were detected by reciprocal immunoprecipitation when cells were transfected with GFP-G9a as opposed to transfection with GFP alone, indicating the presence of specific association between G9a and hUHRF1 (Figure 1A). In a similar experiment, we were able to detect the coimmunoprecipitation of the endogenous G9a and hUHRF1 in synchronized HEK293 cells (Figure 1B). We also performed GST pull-down assays to determine whether there are direct physical interactions between the two proteins in vitro and to map the regions on the proteins involved in the interactions. Various GST-fusion fragments of hUHRF1 and G9a were generated and incubated with purified G9a or hUHRF1. The hUHRF1 was found to directly interact with G9a predominantly through its C-terminus covering the SRA and RING domains with a stronger binding affinity of the RING domain-containing fragment to G9a (Figure 1C). A similar approach was taken to identify the region of the G9a involved in a direct interaction with hUHRF1, and revealed that G9a interacts with hUHRF1 through its N-terminus (Figure 1D). Taken together, we demonstrated that hUHRF1 interacts with G9a in vivo and in vitro.
Figure 1.
Physical interaction of hUHRF1 and G9a. (A) Coimmunoprecipitation of hUHRF1 and G9a in cell extracts from COS-7 cells transfected with GFP control or GFP-G9a. The antibodies used for immunoprecipitation (IP) are indicated at the top of the panel. Western-blot analysis of immunoprecipitates was performed with antibodies as indicated. The input shows 2% of each lysate. (B) Coimmunoprecipitation of endogenous hUHRF1 and G9a in HEK293 cell extracts. HEK293 cells were synchronized to late S phase by serum starvation for 20 h and the subsequent release into 10% FBS-containing medium for 15 h before cell harvest. Antibodies used for immunoprecipitation and western blot are shown. The input represents 2% of each lysate. (C) Direct binding of hUHRF1 to G9a and mapping of the G9a-binding region on hUHRF1 using GST fusions of hUHRF1 fragments. Various domains of hUHRF1 are indicated along with a schematic presentation of the GST fusion constructs marked with amino acid numbers. The blot from GST pull-down assay was probed with anti-G9a antibody and stained with Ponceau solution to visualize the transferred proteins. Positions of fusion proteins are marked with asterisks. (D) Mapping of hUHRF1-binding region on G9a. Schematic diagram of the various GST-fusion constructs of G9a is shown with amino acid numbers. Western blot with anti-hUHRF1 antibody is shown along with the corresponding Ponceau-stained membrane.
UHRF1 colocalizes with G9a in vivo
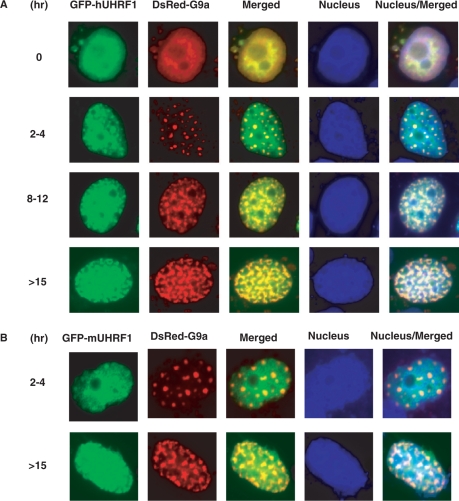
The observation that hUHRF1 interacts with G9a prompted us to examine the subnuclear localization of the two proteins by cotransfecting COS-7 cells with GFP-hUHRF1 and DsRed-G9a. The transfected cells were synchronized by treatment of aphidicolin, released from G1/S border and followed for the indicated time points to monitor the localization of the two proteins. Although both proteins were present in a diffused pattern throughout the nucleus at 0 h time point (G1/S border), they formed more distinct foci along the progression of the S phase, which is more obvious for G9a represented by red spots (Figure 2A). Most of major distinct foci of the hUHRF1 and G9a were found to be colocalized as revealed by yellow spots in the merged images (Figure 2A). This colocalization study was repeated with GFP-mUHRF1 (mouse UHRF1) and DsRed-G9a, essentially giving similar results and confirming that both human and mouse UHRF1 can associate with G9a in the nucleus (Figure 2B). Furthermore, deletion of N-terminal hUHRF1-interacting region on G9a (DsRed-N▵G9a) substantially impaired colocalization with GFP-fused hUHRF1 in COS-7 cells (Figure S1).
Figure 2.
Colocalization of UHRF1 and G9a. (A) Subnuclear localization of GFP-hUHRF1 and DsRed-G9a transiently expressed in COS-7 cells. Cells were synchronized with aphidicolin and released from G1 arrest for a given number of hours through S phase. Nuclei were visualized with Hoechst stain. (B) Colocalization of GFP-mUHRF1 and DsRed-G9a in COS-7 cells at given hours of release from synchronization.
UHRF1 plays a role in chromatin association of G9a
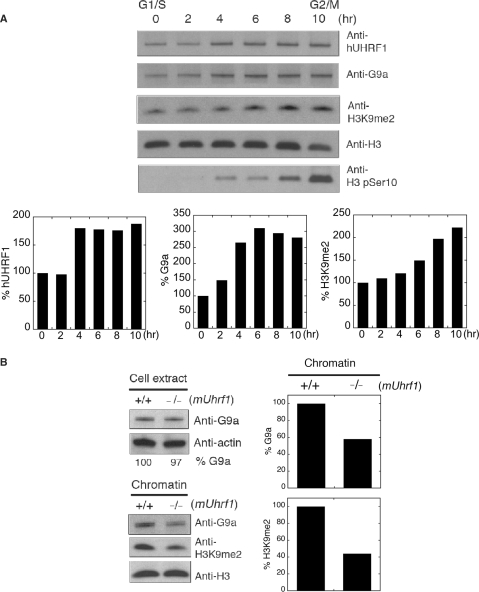
We have previously shown that UHRF1 recruits DNMT1 to chromatin (9). This finding led us to speculate that UHRF1 may assist in the recruitment of G9a onto chromatin by direct physical interaction. To validate this hypothesis, we first examined the chromatin-loading patterns of hUHRF1 and G9a in HeLa cells after synchronization at G1/S border and release from the arrest for given hours. Consistent with the colocalization data (Figure 2A), hUHRF1 and G9a displayed an increase in chromatin association over time (Figure 3A). The progressive chromatin loading patterns of hUHRF1 and G9a were followed by the gradual increase in histone H3-K9 dimethylation (H3K9me2) that is the reaction product of G9a. Next, we investigated if the absence of UHRF1 protein would affect the overall chromatin association of G9a by examining the chromatin fractions isolated from mouse wild-type and mUhrf1-null (−/−) ES cells. The G9a protein levels in cell extracts were similar in both ES cell lines, whereas the chromatin fractions from mUhrf1-null ES cells contained a significantly lower amount of G9a than those from wild-type cells, along with the corresponding decrease in H3K9me2 (Figure 3B). These data suggest that UHRF1 plays a role in chromatin association of G9a.
Figure 3.
UHRF1 affects chromatin association of G9a. (A) Concurrent loading of hUHRF1 and G9a onto chromatin during S phase. HeLa cells were synchronized by double aphidicolin/thymidine block and released to regular medium for hours indicated at the top of the panel. The chromatin fractions at each time point were used for western-blot analysis with antibodies indicated. Anti-H3 and H3 phopho-Ser10 (H3 pSer10) antibodies were used for a loading control and mitosis marker, respectively. Densitometric scans of hUHRF1, G9a and H3K9me2 levels in each chromatin fraction are shown after normalization by the corresponding H3 level. (B) Impaired chromatin association of G9a in the absence of mUHRF1. Cell extracts and chromatin were prepared from wild type (+/+) and mUhrf1-null (−/−) ES (ES) cells and used to detect the presence of G9a and dimethylated H3K9 (H3K9me2) with antibodies indicated. The relative G9a level in cell extracts is shown as percentage G9a at the bottom of the panel by a densitometric analysis after normalization by the actin-loading control. Normalized densitometric scans of G9a and H3K9me2 levels in chromatin are shown in graphs.
hUHRF1 cooperates with G9a to enhance the transcriptional repression
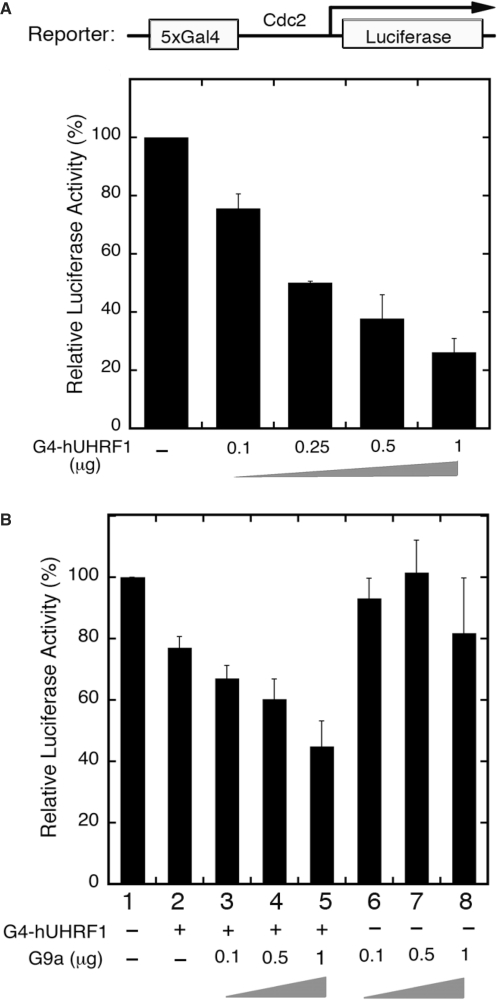
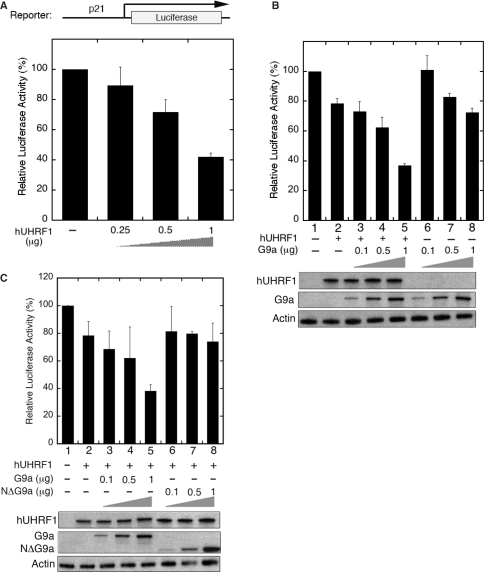
G9a has been shown to be involved in transcriptional repression in euchromatin (23,25–29). We hypothesized that hUHRF1 might recruit G9a to specific promoters in euchromatin to suppress the transcription. To test this possibility, reporter assays were devised using a Gal4-driven luciferase construct (pG5-cdc2-luc) with varied combinations of Gal4DBD-fused hUHRF1 (G4-hUHRF1) and GFP-G9a plasmids. The low basal activity of pG5luc reporter containing a minimal TATA box made it difficult to detect any distinct decrease in luciferase activity possibly mediated by hUHRF1. To allow for a clear detection of transcriptional repression mediated by hUHRF1 and/or G9a, we engineered the pG5luc reporter by replacing the intrinsic minimal promoter with a partial cdc2 promoter fragment containing several Sp1 sites (20), creating the pG5-cdc2-luc plasmid. Transfection with increasing amounts of G4-hUHRF1 alone resulted in progressive repression of the reporter gene expression (Figure 4A). To determine if hUHRF1 can cooperate with G9a to enhance the transcriptional inhibition of the reporter, increasing amounts of GFP-G9a were cotransfected to COS-7 cells with or without a constant amount of G4-hUHRF1. In the absence of G4-hUHRF1, exogenous GFP-G9a had little effects on the repression of the reporter gene transcription (Figure 4B, lanes 6–8) while it significantly suppressed the transcription of the reporter in a dose-dependent manner in the presence of G4-hUHRF1 (Figure 4B, lanes 3–5), suggesting that the G9a-mediated transcriptional inhibition is dependent on the presence of hUHRF1. To determine whether hUHRF1 affects the G9a-mediated transcriptional repression by additional mechanisms other than physical recruitment of G9a to the promoter, we further examined the possibility that hUHRF1 may modulate G9a methyltransferase activity by its interaction with G9a. Using the purified hUHRF1 and G9a that were used for GST pull-down assays (Figure 1C and D), G9a methyltransferase activity was measured in vitro in the presence or absence of hUHRF1, and no significant difference in activity was observed (data not shown). These results provide the evidence supporting that hUHRF1 cooperates with G9a to enhance the transcriptional repression primarily by recruiting G9a to the target promoters.
Figure 4.
hUHRF1 cooperates with G9a to enhance transcriptional repression. (A) Transcriptional repression of luciferase reporter gene by hUHRF1. COS-7 cells were cotransfected with a Gal4-driven luciferase reporter (2 μg) and increasing amounts of Gal4DBD-hUHRF1 (G4-hUHRF1). Luciferase activity was measured 48 h post-transfection and represented by the means ± SD of duplicate determinations from three independent experiments. (B) Enhanced transcriptional repression by hUHRF1-mediated G9a recruitment. Luciferase activities were measured after 48 h of transfection using COS-7 cells cotransfected with the same reporter in (A), increasing amounts of EGFP-G9a (0.1–1 μg) and with or without a constant amount (0.1 μg) of G4-hUHRF1. The data represent the means ± SD of duplicate determinations from four separate experiments.
hUHRF1 suppresses p21 expression
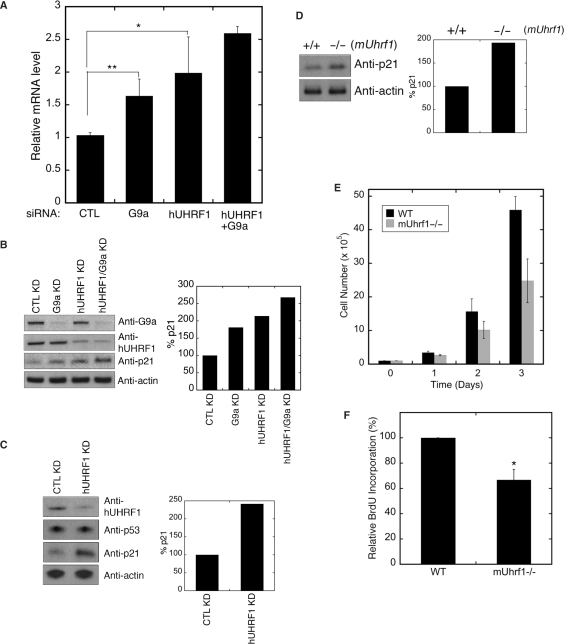
Several studies demonstrated that hUHRF1 protein levels are elevated in tumor tissues (8,11,30), and the protein is required for proliferation in cancer cells (2,8,11). The hUHRF1 was also shown to bind to the methylated promoter of tumor suppressors such as p14 and p16 (8), possibly to suppress the expression of these genes in cancer cells. However, the same study showed that hUHRF1 expression level did not have any correlation with the expression of these cell-cycle regulators in proliferating cells, suggesting that there might be additional hUHRF1 targets involved in cell-cycle regulation to promote proliferation in cancer cells. Previously, it was shown that G9a cooperates with CDP/cut and Gfi1 transcriptional regulators to suppress p21 expression (23,28). On the basis of these prior observations, we hypothesized that p21 promoter may be one of the in vivo targets to which hUHRF1 directs G9a to enhance the repression of the tumor suppressor, thus promoting proliferation in cancer cells. To test this hypothesis, we examined the p21 mRNA and protein levels after the individual KD of hUHRF1 and G9a in HeLa cells by siRNA transfection. The G9a KD resulted in ∼60% increase in p21 mRNA level determined by quantitative RT–PCR, compared to the CTL KD (Figure 5A). Although the fold increase is relatively small (<2-fold), it was a statistically significant change that was reflected by the roughly corresponding change in the protein levels (Figure 5B). As expected, hUHRF1 KD significantly induced the expression of p21 at both mRNA and protein levels (Figure 5A and B). In addition, dual hUHRF1 and G9a KD resulted in additive effects on the induction of p21 expression at both transcript and protein levels (Figure 5A and B). The p21 is a well-characterized target gene of p53-mediated transcriptional regulation (31). However, HeLa cells tested in this study lack functional p53, suggesting that the p21 induction in siRNA-transfected HeLa cells is a p53-independent process. To verify this observation, we performed a similar experiment using COS-7 cell line where the endogenous p53 is abundantly expressed but inactivated by SV40 large T antigen. As shown in Figure 5C, UHRF1 KD did not affect the p53 protein level, yet substantially elevated the p21 expression, demonstrating that the UHRF1 KD-mediated p21 elevation is independent of p53. Then, we further validated the observation that UHRF1 is involved in the repression of p21 expression by examining the amounts of p21 protein in cell extracts of mouse wild-type and mUhrf1-null (−/−) ES cells. As shown in Figure 5D, the p21 protein level was found to be elevated in the mUhrf1(−/−) cells. Furthermore, mUhrf1(−/−) ES cells displayed retarded growth and less BrdU (bromodeoxyuridine) incorporation compared to the wild-type counterpart although it is not clear whether these effects directly resulted from the increased p21 expression in mUhrf1(−/−) ES cells (Figure 5E and F).
Figure 5.
hUHRF1 suppresses p21 expression in cooperation with G9a. (A) Quantitative RT–PCR analysis of p21 expression. Total RNA was isolated from HeLa cells transfected with siRNAs as indicated. The Q-PCR data normalized by GAPDH control are shown as the means ± SD of triplicate determinations from four independent experiments. Statistical significance of the differences among the groups was determined by Student's t-test. *P < 0.05; **P < 0.01. (B) Western blot analysis of p21 expression in siRNA-mediated KD HeLa cells. After each KD, as indicated at the top of the panel, cell extracts were used for detection of p21. The densitometric scan of p21 expression is shown at the right. (C) Western blot analysis of p53 and p21 expression in COS-7 cells after siRNA-mediated KD of hUHRF1. The densitometric scan of p21 expression is shown at the right. (D) Enhanced p21 expression in mUhrf1−/− ES cells. The blot for p21 is shown along with the densitometric analysis. (E) Growth of wild type (+/+) and mUhrf1-null (−/−) ES cells. Cell growth was monitored over 3 days after plating, and the data represent the means ± SD of six replicates. (F) BrdU incorporation of wild-type (+/+) and mUhrf1-null (−/−) ES cells. After 24 h incubation, BrdU labeling was performed for 2 h and determined by a colorimetric assay. The data represent the means ± SD of three separate experiments. Statistical significance of the difference between the groups was determined by Student's t-test. *P < 0.05.
hUHRF1 cooperates with G9a to enhance the transcriptional repression of p21 promoter
We also examined whether hUHRF1 is recruited to the p21 promoter by reporter assays using pGL2–p21. If the p21 promoter is one of the natural promoters targeted by hUHRF1, it should be recruited to the naive promoter without using the Gal4 reporter system that was used in Figure 4. Indeed, exogenous expression of hUHRF1 inhibited the p21 promoter activity in a dose-dependent manner, possibly by recruiting the endogenous epigenetic regulators including G9a to the p21 promoter (Figure 6A). Cotransfection of increasing amounts of G9a expression vector with a constant amount of hUHRF1 plasmid resulted in further repression of the reporter (Figure 6B, lanes 3–5), compared to the case without the exogenous hUHRF1 (Figure 6B, lanes 6–8). The dose-dependent decrease in p21 promoter activity in the absence of exogenous hUHRF1 may have been caused by the recruitment of endogenous hUHRF1 to the promoter (Figure 6B, lanes 6–8). To validate that the UHRF1-mediated p21 promoter repression resulted from the direct interaction between G9a and UHRF1, a G9a mutant lacking the N-terminal UHRF1-interacting region (21) was used for the identical reporter assay in parallel with wild-type G9a. As shown in Figure 6C, the deletion of UHRF1-interaction region on G9a (N▵G9a) impaired UHRF1/G9a-mediated repression of p21 promoter. Taken together, p21 appears to be one of the in vivo targets whose expression is repressed by coordinated actions of UHRF1/G9a.
Figure 6.
hUHRF1 cooperates with G9a to enhance the transcriptional repression of p21 promoter. (A) hUHRF1-mediated transcriptional repression of the p21 promoter-luciferase reporter. COS-7 cells were cotransfected with the reporter (2 μg) and increasing amounts of EGFP-hUHRF1 as indicated at the bottom of the panel. (B) Enhanced transcriptional repression by G9a in the presence of exogenous hUHRF1. Luciferase activities were measured from COS-7 cells cotransfected with the same reporter described earlier and increasing amounts of EGFP-G9a (0.1–1 μg) with or without a constant amount (0.4 μg) of EGFP-hUHRF1. The luciferase assays were performed as described in Figure 4, and the data represent the means ± SD of duplicate determinations from three separate experiments. Western blot analyses of hUHRF1 and G9a expression by anti-GFP antibody are shown for each cotransfection group. (C) Loss of interaction between UHRF1 and G9a abolishes the UHRF1/G9a-mediated repression of p21 promoter. Reporter assays were performed as described in (B), using the wild-type G9a plasmid (EGFP-G9a) and its deletion mutant lacking the N-terminal UHRF1-interacting region (EGFP-NΔG9a). Expression of hUHRF1 and G9a/NΔG9a is shown by western blot analyses with anti-GFP antibody.
hUHRF1 recruits G9a and other chromatin-modifying enzymes to p21 promoter
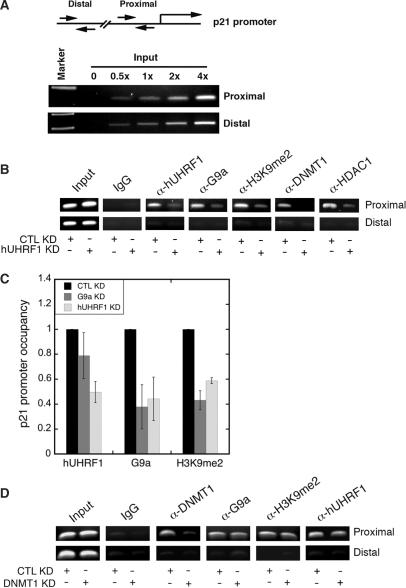
To demonstrate that hUHRF1-mediated G9a recruitment to the endogenous p21 promoter is one of the mechanisms underlying p21 repression in HeLa cells, we transfected HeLa cells with hUHRF1 or control siRNAs and performed ChIP with selected antibodies. The hUHRF1 and G9a were found to associate with the proximal region of the p21 promoter in CTL KD (transfection with control siRNA), suggesting that these proteins are recruited to the p21 promoter under the native conditions (Figure 7B). In agreement with G9a association with the promoter, dimethylation at histone H3K9 (H3K9me2) was detected in CTL KD. Furthermore, DNMT1 was also found to associate with the proximal region of the promoter. As demonstrated by the previous studies, the proximal region of the p21 promoter also displayed the association with HDAC1 (32). None of these enzymes were found on the distal region of the promoter. Then, we examined the effects of hUHRF1 on p21 promoter association of G9a and other chromatin modifiers described earlier. As expected, hUHRF1 KD significantly reduced the amount of hUHRF1 bound to the proximal region of the p21 promoter compared to the CTL KD. The hUHRF1 KD also decreased G9a association with the p21 promoter resulting in reduced H3K9me2 modification on it, suggesting that G9a loading to the promoter is dependent on hUHRF1. We have previously shown that hUHRF1 KD impairs the recruitment of DNMT1 onto chromatin at global levels (9). Consistent with this finding, hUHRF1 KD reduced the DNMT1 association with the p21 promoter. In addition, HDAC1 association was also decreased upon hUHRF1 KD (Figure 7B). Next, we investigated whether G9a and its histone methylation can affect the loading of hUHRF1 to the promoter, because hUHRF1 was recently identified as a methyl K9-specific histone H3-binding protein (4). Therefore, G9a protein level was reduced by siRNA transfection and the relative p21 promoter occupancy of hUHRF1 was examined by quantitative ChIP analysis. As shown in Figure 7C, G9a KD moderately impaired the hUHRF1 association with the promoter (∼20% compared to the CTL KD), whereas hUHRF1 KD more profoundly (∼55%) disrupted the G9a binding to the promoter and reduced the H3K9me2 to a comparable level to that of G9a KD. Previously, we have also shown that siRNA-mediated KD of DNMT1 impairs G9a loading onto chromatin (19). Moreover, there are several studies demonstrating the interdependency between histone and DNA methylation (33–35). To test whether DNMT1 KD can negatively affect G9a recruitment onto the p21 promoter, HeLa cells were transfected with DNMT1 or control siRNAs. After DNMT1 KD, the promoter occupancy of DNMT1 was substantially decreased, whereas G9a association and H3K9me2 modification were not affected much compared to CTL KD (Figure 7D). These results indicate that DNMT1 makes little contribution to the physical recruitment of G9a to p21 promoter. In addition, DNMT1 KD did not appear to affect UHRF1 loading onto the promoter significantly (Figure 7D). Consistent with this observation, bisulfite sequencing of the p21 promoter region (−398 to +11) revealed that the CpGs within the sequence is infrequently methylated except for one CpG dinucleotide at −371 position which was methylated by 50% (Figure S2). Although the functional significance of DNMT1 recruitment onto the p21 promoter is not clear given the infrequent methylation patterns on the p21 promoter, these results suggest that UHRF1 recruitment onto p21 promoter may use additional mechanisms other than CpG methylation.
Figure 7.
hUHRF1 recruits G9a and other chromatin modification enzymes to p21 promoter. (A) Linearity of PCR amplification using primer sets for proximal (−385 to −240) and distal (−4164 to −3959) regions of p21 promoter with increasing amount of input DNA. (B) ChIP analysis of p21 promoter after KD of hUHRF1. HeLa cells were transfected with either control siRNA (CTL KD) or hUHRF1 siRNA (hUHRF1 KD). Using the chromatin isolated from the KD cells, ChIP was performed to detect the proteins or histone modification as indicated at the top of the panel. 5% input is shown. (C) Quantitative ChIP analysis for relative p21 promoter occupancy of hUHRF1, G9a and dimethylated H3K9 (H3K9me2) after KD of hUHRF1 or G9a. Q-PCR data of each group were normalized to its input as % input. The relative p21 promoter occupancy of hUHRF1 or G9a KD samples represents the fold change in percentage input over that of the CTL KD. Error bars indicate standard deviation of three independent experiments. (D) ChIP analysis of p21 promoter after KD of DNMT1. HeLa cells were transfected with either control siRNA (CTL KD) or DNMT1 siRNA (DNMT1 KD). CHIP was performed as described in (B) and 5% input is shown.
DISCUSSION
In mammalian cells, gene expression is regulated via epigenetic mechanisms. Some mechanisms involve covalent modifications on DNA and histone molecules on the chromatin to create either a permissive or a repressive transcriptional environment. Given the emerging evidence supporting the role of UHRF1 as a transcriptional regulator (2,16,17) and its interaction with a histone methyltransferase G9a in this study, we hypothesized that UHRF1 might serve as a transcriptional corepressor along with G9a that has been shown to be involved in transcriptional repression in euchromatin (25). Indeed, UHRF1 alone was capable of repressing reporter gene transcription in a dose-dependent manner, possibly by recruiting the endogenous epigenetic regulators to the target promoters. Consistent with this notion, coexpression of UHRF1 and G9a enhanced the transcriptional repression of the reporter genes, suggesting that the direct physical interaction of UHRF1 and G9a constitutes an effective silencer complex. Among many possible endogenous target genes regulated by the cooperation of UHRF1/G9a, we examined the expression of p21 in HeLa cells where it is poorly expressed in a p53-independent manner. The p21 is a cell-cycle regulator by inhibiting cyclin-dependent kinases and a modulator of apoptosis by interacting with other proteins involved in the regulation of apoptosis (36). Transcription of p21 gene relies on the control of multiple different regulators, of which many are yet to be identified. In this study, UHRF1 appears to be one of the intrinsic regulators of p21 gene expression. We have demonstrated that the endogenous p21 promoter displayed the presence of UHRF1, G9a and other chromatin-modifying enzymes such as DNMT1 and HDAC1 in HeLa cells by ChIP assays. The siRNA-mediated UHRF1 KD significantly reduced the p21 promoter occupancy of all three chromatin-modifying enzymes, placing UHRF1 as a focal point of recruitment of these enzymes and also suggesting possible coordinated efforts of UHRF1, G9a, DNMT1 and HDAC1 for efficient repression of p21. This observation raises an intriguing question on whether all these proteins form a single macromolecular complex or various distinct complexes depending on the specific promoter architecture and cellular context. Gel-filtration chromatography of Jurkat cell lysates containing the endogenously expressed proteins mentioned earlier has revealed that UHRF1 cofractionates with G9a, DNMT1 and HDAC1 to a various extent, indicating the possible formation of the common complexes that are assembled by UHRF1 (data not shown).
A similar coordinated epigenetic repression of p21 was previously reported, involving a transcriptional regulator, Gfi1 (28). The Gfi1 recruits G9a and HDAC1 to p21 promoter and represses its expression in HL-60 cells. Both HDAC1 and G9a were found in the repressive complex assembled by Gfi1, and the KD of Gfi1 elevated the p21 expression by 2–3-fold. We also observed a similar level of p21 induction after UHRF1 KD in HeLa and COS-7 cells. Other repressor proteins can also modulate p21 expression by epigenetic mechanisms. For example, a transcription factor CDP/cut was shown to recruit G9a to the human p21 promoter, and the transcriptional repression function of CDP/cut is mediated through the activity of its associated G9a (23). Polycomb group (PcG) proteins are chromatin modifiers that can transcriptionally silence their target genes and maintain them in the repressed state through cell divisions during development (37). Recently, NSPc1, a transcriptional repressor homologous with PcG protein Bmi-1, was demonstrated to repress p21 expression via direct binding to the retinoid acid response element of its promoter (38).
Furthermore, the murine Uhrf1-null ES cells also displayed a higher level of p21 protein, implicating the role of UHRF1 in transcriptional repression in ES cell environment. An orphan nuclear receptor TLX recruits a set of HDACs to its target genes for transcriptional repression in neural stem cells (39). One of the genes targeted by this repressive complex is p21, suggesting a role very similar to that of UHRF1-G9a repressive complex. Both histone H3K9me2 accumulation via G9a and deacetylation by HDACs are found on many repressed genes in cell lines. As siRNA-mediated KD of G9a or inhibition of HDACs can result in p21 derepression, it is also plausible that HDACs and G9a crosstalk during p21 repression. In fact, a recent study on SHP-mediated regulation of CYP7A1 promoter revealed the functional interplay between SHP-recruited HDACs and G9a in altering the chromatin structure of CYP7A1 promoter (29). These findings suggest multiple mechanisms of p21 gene expression that is modulated by both histone methylation and deacetylation. Moreover, as exemplified earlier, the presence of multiple functional equivalents of UHRF1 that can recruit similar effectors such as G9a and HDAC1 appears to ensure tight repression of p21 in cancer cells. Perhaps, this may constitute the basis for the finding that functional disruption of a single factor in p21 repression only results in relatively minor p21 derepression (∼2–3-fold) as we and other investigators observed.
The UHRF1-mediated p21 repression presents another intriguing idea of reciprocal regulation occurring between UHRF1 and p21 expression in response to different extracellular stimuli, because hUHRF1 expression was shown to be down-regulated by p53/p21-dependent DNA damage check point signal (40). This observation may lead to a hypothesis that, in the presence of DNA damage, the p53/p21-dependent checkpoint response may outweigh the transcriptional repression effects of UHRF1 against p21 resulting in down-regulation of UHRF1 expression, whereas the absence of DNA damage signal allows UHRF1 to keep p21 in a repressed state promoting cell proliferation. The reciprocal down-regulation, involving UHRF1, can be observed in another case where overexpression of hUHRF1 negatively regulates pRb expression (16), whereas pRb interacts with E2F-1 transcription factor and thereby inhibits the expression of the E2F-1 target genes including UHRF1 (8,30). These findings point to the dynamic regulation mode for UHRF1 in cancer cells.
In summary, our data demonstrate a new role of UHRF1 as a transcriptional corepressor in recruitment of histone methyltransferase G9a and other chromatin-modifying enzymes to target promoters, and suggest that UHRF1 acts as a focal point of gene repression mediated by various chromatin modifiers.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: New England Biolabs, Inc.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Jane B. Trepel for kindly providing the pGL2–p21 plasmid. We also thank George R. Feehery and other colleagues of our laboratory for technical assistance and support. We are grateful to Drs D.G. Comb and Rich Roberts at New England Biolabs, Inc. for their support and encouragement. S. E. J. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Bronner C, Achour M, Arima Y, Chataigneau T, Saya H, Schini-Kerth VB. The UHRF family: oncogenes that are drugable targets for cancer therapy in the near future? Pharmacol. Ther. 2007;115:419–434. doi: 10.1016/j.pharmthera.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Hopfner R, Mousli M, Jeltsch JM, Voulgaris A, Lutz Y, Marin C, Bellocq JP, Oudet P, Bronner C. ICBP90, a novel human CCAAT binding protein, involved in the regulation of topoisomerase IIalpha expression. Cancer Res. 2000;60:121–128. [PubMed] [Google Scholar]
- 3.Muto M, Utsuyama M, Horiguchi T, Kubo E, Sado T, Hirokawa K. The characterization of the monoclonal antibody Th-10a, specific for a nuclear protein appearing in the S phase of the cell cycle in normal thymocytes and its unregulated expression in lymphoma cell lines. Cell Prolif. 1995;28:645–657. doi: 10.1111/j.1365-2184.1995.tb00051.x. [DOI] [PubMed] [Google Scholar]
- 4.Karagianni P, Amazit L, Qin J, Wong J. ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol. Cell Biol. 2008;28:705–717. doi: 10.1128/MCB.01598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Citterio E, Papait R, Nicassio F, Vecchi M, Gomiero P, Mantovani R, Di Fiore PP, Bonapace IM. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol. Cell Biol. 2004;24:2526–2535. doi: 10.1128/MCB.24.6.2526-2535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papait R, Pistore C, Negri D, Pecoraro D, Cantarini L, Bonapace IM. Np95 is implicated in pericentromeric heterochromatin replication and in major satellite silencing. Mol. Biol Cell. 2007;18:1098–1106. doi: 10.1091/mbc.E06-09-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papait R, Pistore C, Grazini U, Babbio F, Cogliati S, Pecoraro D, Brino L, Morand AL, Dechampesme AM, Spada F, et al. The PHD Domain of Np95 (mUHRF1) is involved in large-scale reorganization of pericentromeric heterochromatin. Mol. Biol Cell. 2008;19:3554–3563. doi: 10.1091/mbc.E07-10-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Unoki M, Nishidate T, Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004;23:7601–7610. doi: 10.1038/sj.onc.1208053. [DOI] [PubMed] [Google Scholar]
- 9.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 10.Woo HR, Pontes O, Pikaard CS, Richards EJ. VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev. 2007;21:267–277. doi: 10.1101/gad.1512007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenkins Y, Markovtsov V, Lang W, Sharma P, Pearsall D, Warner J, Franci C, Huang B, Huang J, Yam GC, et al. Critical role of the ubiquitin ligase activity of UHRF1, a nuclear RING finger protein, in tumor cell growth. Mol. Biol Cell. 2005;16:5621–5629. doi: 10.1091/mbc.E05-03-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Achour M, Jacq X, Ronde P, Alhosin M, Charlot C, Chataigneau T, Jeanblanc M, Macaluso M, Giordano A, Hughes AD, et al. The interaction of the SRA domain of ICBP90 with a novel domain of DNMT1 is involved in the regulation of VEGF gene expression. Oncogene. 2008;27:2187–2197. doi: 10.1038/sj.onc.1210855. [DOI] [PubMed] [Google Scholar]
- 14.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 15.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeanblanc M, Mousli M, Hopfner R, Bathami K, Martinet N, Abbady AQ, Siffert JC, Mathieu E, Muller CD, Bronner C. The retinoblastoma gene and its product are targeted by ICBP90: a key mechanism in the G1/S transition during the cell cycle. Oncogene. 2005;24:7337–7345. doi: 10.1038/sj.onc.1208878. [DOI] [PubMed] [Google Scholar]
- 17.Macaluso M, Montanari M, Noto PB, Gregorio V, Bronner C, Giordano A. Epigenetic modulation of estrogen receptor-alpha by pRb family proteins: a novel mechanism in breast cancer. Cancer Res. 2007;67:7731–7737. doi: 10.1158/0008-5472.CAN-07-1476. [DOI] [PubMed] [Google Scholar]
- 18.Jin P, Hardy S, Morgan DO. Nuclear localization of cyclin B1 controls mitotic entry after DNA damage. J. Cell Biol. 1998;141:875–885. doi: 10.1083/jcb.141.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esteve PO, Chin HG, Smallwood A, Feehery GR, Gangisetty O, Karpf AR, Carey MF, Pradhan S. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20:3089–3103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Gac G, Esteve PO, Ferec C, Pradhan S. DNA damage-induced down-regulation of human Cdc25C and Cdc2 is mediated by cooperation between p53 and maintenance DNA (cytosine-5) methyltransferase 1. J. Biol. Chem. 2006;281:24161–24170. doi: 10.1074/jbc.M603724200. [DOI] [PubMed] [Google Scholar]
- 21.Esteve PO, Patnaik D, Chin HG, Benner J, Teitell MA, Pradhan S. Functional analysis of the N- and C-terminus of mammalian G9a histone H3 methyltransferase. Nucleic Acids Res. 2005;33:3211–3223. doi: 10.1093/nar/gki635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim GD, Ni J, Kelesoglu N, Roberts RJ, Pradhan S. Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. EMBO J. 2002;21:4183–4195. doi: 10.1093/emboj/cdf401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishio H, Walsh MJ. CCAAT displacement protein/cut homolog recruits G9a histone lysine methyltransferase to repress transcription. Proc. Natl Acad. Sci. USA. 2004;101:11257–11262. doi: 10.1073/pnas.0401343101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee SJ, Ha MJ, Lee J, Nguyen P, Choi YH, Pirnia F, Kang WK, Wang XF, Kim SJ, Trepel JB. Inhibition of the 3-hydroxy-3-methylglutaryl-coenzyme A reductase pathway induces p53-independent transcriptional regulation of p21(WAF1/CIP1) in human prostate carcinoma cells. J. Biol. Chem. 1998;273:10618–10623. doi: 10.1074/jbc.273.17.10618. [DOI] [PubMed] [Google Scholar]
- 25.Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boulias K, Talianidis I. Functional role of G9a-induced histone methylation in small heterodimer partner-mediated transcriptional repression. Nucleic Acids Res. 2004;32:6096–6103. doi: 10.1093/nar/gkh947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gyory I, Wu J, Fejer G, Seto E, Wright KL. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat. Immunol. 2004;5:299–308. doi: 10.1038/ni1046. [DOI] [PubMed] [Google Scholar]
- 28.Duan Z, Zarebski A, Montoya-Durango D, Grimes HL, Horwitz M. Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol. Cell Biol. 2005;25:10338–10351. doi: 10.1128/MCB.25.23.10338-10351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang S, Miao J, Xiang L, Ponugoti B, Treuter E, Kemper JK. Coordinated recruitment of histone methyltransferase G9a and other chromatin-modifying enzymes in SHP-mediated regulation of hepatic bile acid metabolism. Mol. Cell Biol. 2007;27:1407–1424. doi: 10.1128/MCB.00944-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mousli M, Hopfner R, Abbady AQ, Monte D, Jeanblanc M, Oudet P, Louis B, Bronner C. ICBP90 belongs to a new family of proteins with an expression that is deregulated in cancer cells. Br. J. Cancer. 2003;89:120–127. doi: 10.1038/sj.bjc.6601068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 32.Ocker M, Schneider-Stock R. Histone deacetylase inhibitors: signalling towards p21cip1/waf1. Int. J. Biochem. Cell Biol. 2007;39:1367–1374. doi: 10.1016/j.biocel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 33.Xin Z, Tachibana M, Guggiari M, Heard E, Shinkai Y, Wagstaff J. Role of histone methyltransferase G9a in CpG methylation of the Prader-Willi syndrome imprinting center. J. Biol. Chem. 2003;278:14996–15000. doi: 10.1074/jbc.M211753200. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen CT, Weisenberger DJ, Velicescu M, Gonzales FA, Lin JC, Liang G, Jones PA. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2'-deoxycytidine. Cancer Res. 2002;62:6456–6461. [PubMed] [Google Scholar]
- 35.Espada J, Ballestar E, Fraga MF, Villar-Garea A, Juarranz A, Stockert JC, Robertson KD, Fuks F, Esteller M. Human DNA methyltransferase 1 is required for maintenance of the histone H3 modification pattern. J. Biol. Chem. 2004;279:37175–37184. doi: 10.1074/jbc.M404842200. [DOI] [PubMed] [Google Scholar]
- 36.Gartel AL. The conflicting roles of the cdk inhibitor p21(CIP1/WAF1) in apoptosis. Leuk. Res. 2005;29:1237–1238. doi: 10.1016/j.leukres.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 37.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 38.Gong Y, Yue J, Wu X, Wang X, Wen J, Lu L, Peng X, Qiang B, Yuan J. NSPc1 is a cell growth regulator that acts as a transcriptional repressor of p21Waf1/Cip1 via the RARE element. Nucleic Acids Res. 2006;34:6158–6169. doi: 10.1093/nar/gkl834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun G, Yu RT, Evans RM, Shi Y. Orphan nuclear receptor TLX recruits histone deacetylases to repress transcription and regulate neural stem cell proliferation. Proc. Natl Acad. Sci. USA. 2007;104:15282–15287. doi: 10.1073/pnas.0704089104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arima Y, Hirota T, Bronner C, Mousli M, Fujiwara T, Niwa S, Ishikawa H, Saya H. Down-regulation of nuclear protein ICBP90 by p53/p21Cip1/WAF1-dependent DNA-damage checkpoint signals contributes to cell cycle arrest at G1/S transition. Genes Cells. 2004;9:131–142. doi: 10.1111/j.1356-9597.2004.00710.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.