Abstract
Ribosome-inactivating proteins (RIPs) inhibit protein synthesis by enzymatically depurinating a specific adenine residue at the sarcin-ricin loop of the 28S rRNA, which thereby prevents the binding of elongation factors to the GTPase activation centre of the ribosome. Here, we present the 2.2 Å crystal structure of trichosanthin (TCS) complexed to the peptide SDDDMGFGLFD, which corresponds to the conserved C-terminal elongation factor binding domain of the ribosomal P protein. The N-terminal region of this peptide interacts with Lys173, Arg174 and Lys177 in TCS, while the C-terminal region is inserted into a hydrophobic pocket. The interaction with the P protein contributes to the ribosome-inactivating activity of TCS. This 11-mer C-terminal P peptide can be docked with selected important plant and bacterial RIPs, indicating that a similar interaction may also occur with other RIPs.
INTRODUCTION
The stalk of the eukaryotic ribosome is composed of a pentameric complex of acidic ribosomal proteins, P0(P1)2(P2)2, which forms part of the GTPase-associated centre that binds elongation factors during protein synthesis (1). In humans, P0, P1 and P2 share a conserved flexible C-terminal tail that is rich in acidic residues (Figure 1A). The last 11 residues of these proteins, SD 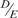 DMGFGLFD, are also highly conserved among all eukaryotic ribosomal stalk proteins. It has been argued that the existence of multiple copies of this conserved C-terminal sequence of stalk proteins, which protrude outward from the ribosomes to the cytoplasm, functions to fetch the elongation factors and draw them into the GTPase-associated centre (1,2).
DMGFGLFD, are also highly conserved among all eukaryotic ribosomal stalk proteins. It has been argued that the existence of multiple copies of this conserved C-terminal sequence of stalk proteins, which protrude outward from the ribosomes to the cytoplasm, functions to fetch the elongation factors and draw them into the GTPase-associated centre (1,2).
Figure 1.
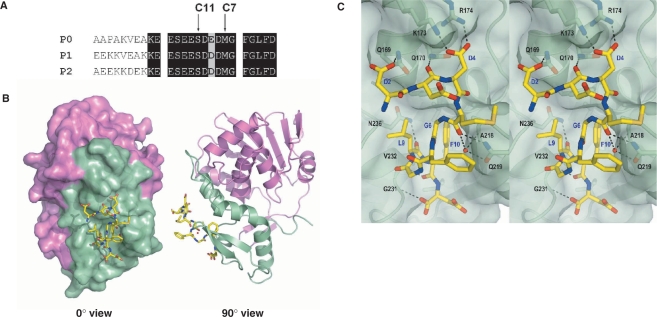
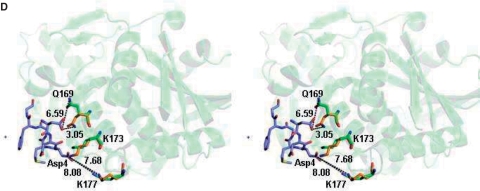
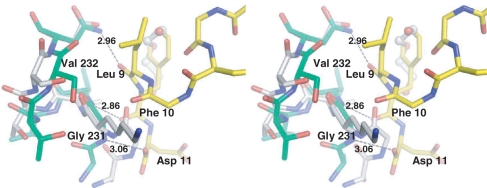
(A) Sequence alignment of the C-terminal residues of mammalian P proteins. The last 17 aa of the P proteins are nearly identical. Positions of the last 11 and 7 aa of the P proteins are marked. (B) The c11-P motif (SDDDMGFGLFD) (yellow) of eukaryotic stalk proteins binds to a pocket in the C-terminal domain of TCS. The N-terminal and the C-terminal domains of TCS are colour-coded violet and green, respectively. (C) A stereo diagram showing the interaction between c11-P (yellow) and TCS (green). Intermolecular hydrogen bonds and salt-bridges are indicated as dashed lines. Residue numbers of TCS and c11-P are in black and blue, respectively. (D) A stereo diagram showing the movement of TCS side chains upon interacting with c11-P. Compared to the wild-type TCS (green), the side chain of Gln169 in TCS–c11-P (orange) flips toward Asp2 of c11-P and forms a hydrogen bond with that residue; the side chain of Lys173 is located between OD1 and OD2 of Asp4; the side chain of Lys177 moves from 8.08 Å to 7.68 Å toward Asp4 of the peptide.
Trichosanthin (TCS) is a type I ribosome-inactivating protein (RIPs) from the medicinal plant Trichosanthes kirilowii Maxim. It has anti-tumour and anti-HIV properties (3) and inactivates ribosomes by enzymatically depurinating an invariant adenine residue (A-4324 in the rat sequence) at the α-sarcin/ricin loop (SRL) of the 28S rRNA in eukaryotic ribosomes. This modification inhibits protein synthesis by altering the conformation of the rRNA and affecting the binding of elongation factors to the GTPase-associated centre. Through systematic deletion studies and scanning charge-to-alanine mutagenesis, we have mapped the TCS interacting site to a conserved 11 mer motif (c11-P), SDDDMGFGLFD, at the C-terminus of the P protein. This interaction is required for the full activity of TCS and is mediated presumably by the interaction of K173, R174 and K177 in the C-terminal domain of TCS with the conserved DDD residues in the 11-mer motif of the P protein, as the substitution of these three aspartate residues with alanine in the P2 protein abolishes the interaction between TCS and P2 (4). Recently, it has also been shown that this conserved 11-mer motif interacts directly with Shiga-like toxin (SLT-1A) and ricin A (RTA). Additionally, a 17-mer peptide of this conserved C-terminal region of the P protein is able to protect ribosomes from SLT-1A in an in vitro protein synthesis assay (5), thus showing that a similar interaction may also occur with other RIPs.
Here, we describe the solved crystal structure of the TCS–c11-P complex and reveal the specific interaction between the two moieties. We have found that both the N- and C-terminal regions of c11-P are involved in the interaction and contribute to the full activity of TCS. By docking c11-P to SLT-1A, RTA and saporin (SO6), we further show that a similar interaction may also take place with different RIPs.
MATERIALS AND METHODS
Construction and expression of TCS variants
Variants of TCS were generated by PCR using mutagenic primers and cloned into pET8c for protein expression. The clones were sequenced to ensure that the mutation was correct and that there were no secondary mutations. TCS or its variants were expressed in Escherichia coli BL21 (DE3) pLysS; they were then purified with a 5 ml Hi-Trap CM-Sepharose column followed by a 5 ml Hi-Trap SP-Sepharose column (GE Healthcare) in 20 mM phosphate buffer, pH 6.7.
Assays of biological activities and the interaction with ribosomes
The ability of TCS and its variants to inhibit protein synthesis in a nuclease-untreated rabbit reticulocyte lysate and to depurinate A-4324 of the 28S rRNA were detected as described previously (4). The affinities between TCS and its variants were investigated with pull-down and cross-linking assays. Detailed methods are described in ref. (4).
Crystallisation and data collection of the TCS–c11-P complex, [V232K/N236D]TCS and [K173A/R174A/K177A]TCS
Twenty milligrams TCS per millilitre and 10 mg c11-P peptide (SDDDMGFGLFD) per millilitre were mixed for 6 h before crystallisation. The complex was crystallised in 7% PEG 20 K, 0.1 M MES, pH 6.5. [V232K/N236D]TCS was crystallised in 1 M LiSO4, 2% PEG8000; [K173A/R174A/K177A]TCS was crystallised in 100 mM Na-Acetate, pH 5.7, 100 mM CaCl2 and 20% KCl; both were at a concentration of 20 mg/ml. Diffraction data were collected with an in-house Rigaku MicroMax-007 X-ray generator with a R-AXIS IV++ IP detector at 100 K. Indexing, integration and scaling of all images, as well as intensity data acquisition were performed in MOSFILM (6).
Structure determination and refinement
The structures were solved by the MOLREP molecular replacement program in CCP4 (7,8). TCS complexed with NADPH (PDB code 1TCS) was used as the search model. The electron densities were generated by CNS (9) and visualised by XtalView (10). Refinement was carried out by alternating between CNS and manual building and minimisation.
Modeling of c11-P peptide to selected RIPs
Structures of RTA (pdb: 1RTC) (11), SO6 (pdb: 1QI7) (12), SLT-1A (pdb: 1DM0) (13) and PAP (pdb: 1D6A) (14) were aligned with the TCS–c11-P complex by DALI (15). The c11-P peptide was docked and merged to each of the proteins. The model was manipulated by CooT (16) and global energy was minimised by CNS (9).
RESULTS
Structure of TCS–c11-P
The structure of the TCS–c11-P complex was solved to a resolution of 2.2 Å (Table 1). The asymmetric unit contains two molecules of TCS (chains A and B) and belongs to space group P21. Refinement was first performed on TCS, and then c11-P peptide was added as chain C and chain D. Each molecule of TCS forms a 1:1 complex with c11-P. For chain C, 10 residues (DDDMGFGLFD) could be fitted to the observed electron density. For chain D, the electron density of residues SD was less well defined, because of the subtle difference in the orientation of Gln169 of chain B and resulting in the absence of the hydrogen bond between this residue and Asp2 of chain D. Hence, only residues DDMGFGLFD were fitted. The positions of the residues in chain C and chain D are almost identical. The c11-P peptide binds to a depression at the C-terminal domain of TCS (Figure 1B) and buries a solvent accessible surface area of ∼1100 Å2 (∼650 Å2 for non-polar atoms and ∼450 Å2 for polar atoms). The Cα r.m.s. deviation of the TCS moiety in TCS–c11-P and wild-type TCS is 0.62 Å. The overall structures are similar, except for the positions of the side chains of Gln169, Lys173 and Lys177 (Figure 1D).
Table 1.
Crystal parameters, data collection and refinement statistics
| [K173A/R174A,/K177A]TCS | [V232k/N236D]TCS | TCS/C11-P | |
|---|---|---|---|
| Resolution (Å) | 50–2.4 | 63.76–2.3 | 50–2.2 |
| Space group | P212 121 | P3121 | P21 |
| Unit cell constants | a = 70.03 Å b = 95.84 Å c = 101.83 Å | a = 73.64 Å b = 73.64 Å c = 143.44 Å | a = 62.59 Å b = 43.96 Å c = 92.24 Å |
| Total number of reflections | 145 673 (19 904)a | 76 680 (10 020)b | 61 482 (8864)c |
| Number of unique reflections | 27 383 (3875)a | 20 607 (2898)b | 24 055 (3741)c |
| Mean(I)/SD(I) | 16.7 (5.2)a | 17.8 (4.9)b | 9.6 (3.2)c |
| Redundancy | 5.3 (5.1)a | 3.7 (3.5)b | 2.4 (2.4)c |
| Rmerged (%) | 10 (31)a | 8 (20)b | 10 (26)c |
| Completeness (%) | 99.6 (98.2)a | 99.7 (98.7)b | 99.4 (98.0)c |
| R-factor | 0.1914 | 0.1859 | 0.1667 |
| Rfreee | 0.2357 | 0.2135 | 0.2377 |
| rms bond length deviation (Å) | 0.020 | 0.012 | 0.021 |
| rms bond angle deviation (°) | 2.71 | 1.40 | 2.93 |
| Aerage B factor (Å2) | |||
| Total | 21.283 | 23.294 | 21.598 |
| Main-chain atoms | 18.642 | 21.371 | 20.478 |
| Side-chain atoms and water | 23.657 | 24.934 | 22.646 |
| Ramachandran plot for non-glycine and non-proline residuesf | |||
| % Most favoured regions | 89.2 | 92.0 | 90.1 |
| % Additional allowed regions | 9.7 | 7.6 | 9.1 |
| % Generously allowed regions | 1.1 | 0.4 | 0.9 |
| % Disallowed regions | 0.0 | 0.0 | 0.0 |
aNumbers in parentheses are the corresponding numbers for the highest resolution shell (2.53–2.4 Å).
bNumbers in parentheses are the corresponding numbers for the highest resolution shell (2.42–2.3 Å).
cNumbers in parentheses are the corresponding numbers for the highest resolution shell (2.32–2.2 Å).
dRmerge = ∑hkl | Ii – Im |/∑hkl Im, where Ii and Im are the observed intensity and the mean intensity of related reflections, respectively.
e5% of the total reflections were used for Rfree calculation.
fThe quality of the structure of one molecule in the asymmetric unit was assessed using PROCHECK (30).
TCS recognises two motifs of c11-P: an N-terminal acidic DDD motif and a C-terminal hydrophobic LF motif. The DDD motif is connected to the LF motif by a left-handed conformation (φ/ψ = ∼50°/30°) at Gly6 and a type II β-turn (Gly6 to Leu9). The two conserved glycine residues at positions six and eight accommodate the required positive backbone φ dihedral angles. The DDD motif forms charge-charge interactions with basic charge residues at the C-terminal domain of TCS. In particular, Asp4 of c11-P forms two salt-bridges with Lys173 and Arg174 of TCS, and Asp2 forms a hydrogen bond with Gln169 of TCS (Figure 1C and Table 2). Phe10 in the LF motif of c11-P docks to a hydrophobic pocket of TCS lined by Phe166, Leu188 and Leu215, forming several side chain and main chain interactions between TCS and the C-terminal region of c11-P (Table 2).
Table 2.
Interaction between TCS and the c11-P peptide
| C11-P | Water molecules | TCS | Distance (Å) |
|---|---|---|---|
| Asp 2 OD1 | – | Gln169 NE2 | 3.05 |
| Asp3 O | – | Gln170 NE2 | 3.01 |
| Asp4 OD1 | – | Lys173 NZ | 2.70 |
| Asp4 OD2 | – | Arg174 NH2 | 2.74 |
| Met5 O | – | Gln219 NE2 | 2.97 |
| Met5 O | WAT100 | – | 2.98 |
| – | Gln219 OE1 | 2.88 | |
| – | Ala218 N | 3.06 | |
| Leu9 O | – | Asn236 ND2 | 2.96 |
| Phe10 O | – | Val 232 N | 2.86 |
| Asp11 O | – | Gly231 N | 3.06 |
| Asp11 OD2 | WAT172 | – | 2.56 |
| – | Asn217 ND2 | 3.21 |
Interaction between TCS and the C-terminal residues of c11-P
To confirm that the interaction between TCS and the C-terminal region of c11-P indeed plays a critical role in the TCS–ribosome interaction, we created a TCS variant (V232K/N236D), that was designed to break the interaction. Val232 is located at the mouth of the hydrophobic pocket, where the LF motif of c11-P binds. The substitution with a lysine residue was expected to block the access of P-proteins to the pocket. On the other hand, the N236D substitution removes the hydrogen bond between the amide of Asn236 and the backbone carbonyl of Leu9 of c11-P.
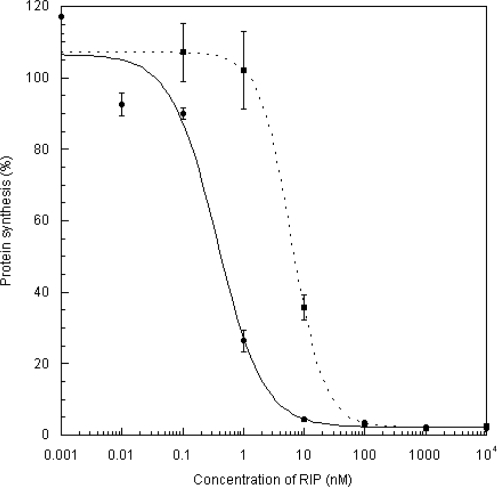
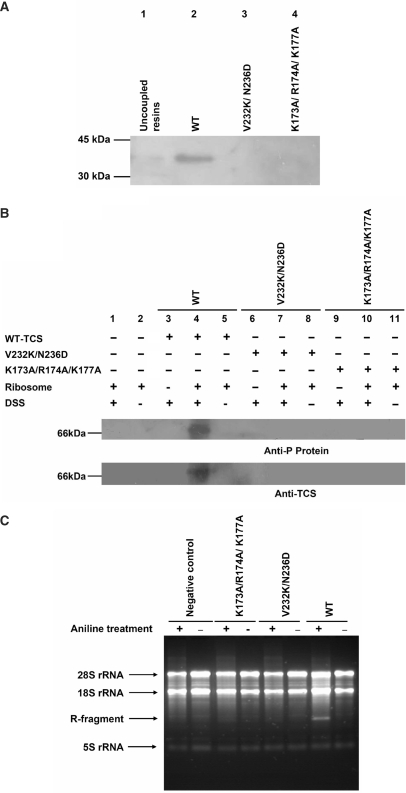
The ribosome-inactivating activity of the V232K/N236D variant was assayed (Figure 2). In agreement with our prediction, the activity in relation to the wild-type protein was reduced by about 17-fold. The experiments assessing the in vitro interaction with ribosome and the cross-linking with ribosomal stalk protein P0 on the 80S rat ribosome showed that neither [V232K/N236D]TCS nor [K173A/R174A/K177A]TCS interact with the ribosome under these specific experimental conditions (Figure 3A and B). The loss of these interactions also drastically reduces the N-glycosidase activity (Figure 3C).
Figure 2.
Assay for the inhibition of protein synthesis. The ability of wild-type and [V232K/N236D]TCS to inhibit the incorporation of [3H]-leucine into the proteins of the nuclease-untreated rabbit reticulocyte lysate was assayed as described (4). Each point denotes the average of an assay done in triplicate. The IC50, the concentration of RIP required to achieve 50% inhibition, was determined by fitting the data to a four-parameter logistic equation. The values of IC50 for wild-type (filled circle) and [V232K/N236D]TCS (filled square) were 0.37 and 6.28 nM, respectively.
Figure 3.
Interaction with the ribosomal stalk for full activity of TCS. (A) Rat 80S ribosome was loaded onto NHS-Sepharose coupled with TCS, [V232K/N236D]TCS or [K173A/R174A/K177A]TCS; the elutants were detected with a Western blot using an anti-P antibody. Ribosomal protein P0 was pull-down by the wild-type TCS (lane 2) but not by TCS variants (lane 3 and 4). (B) Disuccinimidyl suberate (DSS) was used to induce cross-linking between TCS and the ribosome. A protein band at 66 kDa, corresponding to the size of the TCS–P0 complex, was detected by both anti-P and anti-TCS antibodies when the ribosome was cross-linked with wild-type TCS (lane 4) but not with the variants (lane 7 and 10) or negative controls. (C) Depurinated rRNA was hydrolysed by acidic aniline; the 450-bp R-fragments (indicated by an arrow) indicate specific depurination of the 28S rRNA at A4324. Control lanes, in which RNA samples were not treated with aniline, are labeled with the ‘–’ marks. Only wild-type TCS had specific N-glycosidase activity.
The crystal structures of [V232K/N236D]TCS indicated that Asp236 has a similar conformation as the wild-type Asn236. On the other hand, the valine to lysine substitution at residue 232 induces a significant conformational change in the loop composed of residues 230–235. Lysine 232 of the mutant enzyme is flipped towards the solvent surface, most likely because of steric hindrance and a hydrophobic environment that is unfavourable to the lysine residue. This movement generates a structural ‘frameshift’ in the neighbouring residues so that the position of Val 233 is now replaced by Thr 234, Val232 by Val233 and Gly231 by Lys 232. Furthermore, the flipping of Lys 232 drives the backbone of residues 230 and 231 towards the C-terminal end of the c-11 peptide and causes a steric clash. Together with the mutation at residue 236, Asn to Asp, the interactions between TCS and the C-terminal Leu9, Phe10 and Asp11 of c11-P are disrupted (Figure 4).
Figure 4.
Stereo image showing the superimposition of the TCS–c-11P complex and [V232K/N236D]TCS. Note that the side chain of Lys232 is flipped out to the solvent accessible surface. The movement of Lys232-induced changes in the conformation of neighbouring residues; remarkably, the Cα atoms of Ala230 and Gly231 are shifted by 1.98 and 3.43 Å, respectively. The three hydrogen bond interactions that were interrupted by the mutations are indicated. TCS–c-11P complex is coloured in green, and [V232K/N236D]TCS is coloured in white.
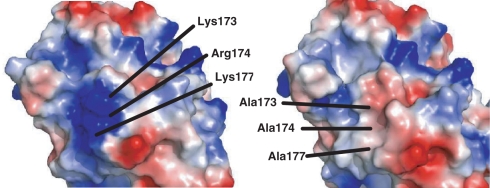
The structural analysis of [K173A/R174A/K177A]TCS indicated that the mutagenesis of the charged cluster to alanine does not change the overall structure of the protein, as the Cα r.m.s. deviation of TCS and [K173A/R174A/K177A]TCS is 0.59 Å. On the other hand, in this triple variant, a patch of surface positive charge is removed (Figure 5), resulting in a reduction of affinity and activity towards the ribosome [Figure 3, ref. (4)].
Figure 5.
Electrostatic surface representation of (A) Wild-type TCS and (B) [K173A/R174A/K177A]TCS, indicating the loss of a patch of surface positive charges in the latter. Positive charge is shown in blue, and negative charge is in red.
DISCUSSION
The acidic proteins P0, P1 and P2 assemble into a pentameric complex, which forms the characteristic protuberance that is termed the ribosomal stalk. In both eukaryotic and prokaryotic ribosomes, this stalk is essential for translation, as it binds a number of initiation and elongation factors (17–19). Through mutagenesis analysis and protein–protein interaction studies, we have shown that TCS interacts with the conserved DDD motif of the C-terminal tail of P2 through K173, R174 and K177 (4). Here, we report the structural evidence for this interaction by elucidating the crystal structure of the TCS–c11-P complex. In this complex, we find that a hydrophobic pocket at the C-terminal domain of TCS also takes part in the interaction, associating with the non-polar residues at the C-terminal tail of the P protein.
In the human ribosomal P proteins, the DDD motif is present in P1 and P2 but is replaced by DED in P0. This indicates that, the second Asp in this motif may not be important for the interaction. Consistent with this, the TCS–c11-P structure shows that the side chain of the second Asp (D3) in c11-P points toward the solvent and does not take part in the interaction with TCS. Since the other amino acids in c11-P are identical in the three P proteins, our observation of the interaction between TCS and c11-P may be applied to all of them.
On the surface of the ribosome, the negative electrostatic potential that is contributed by the phosphodiester backbone and the solvent-exposed acidic patches of ribosomal proteins covers much of the ribosomal surface. Recently, it has been found that the electrostatic interaction between ribotoxin and the ribosome leads to an unusually high catalytic efficiency; it is hypothesised that ribotoxins may bind to several sites on the ribosome and diffuse to the SRL (20,21). Our structure of the complex also confirmed that charge-charge interactions are crucial for TCS to interact with the P protein. The present finding does not exclude the possibility that TCS may interact with other ribosomal proteins, although we have not found such interactions by yeast two-hybrid screening (22). By passing rat liver extract through a TCS-sepharose column, a group has recently observed that TCS binds to ribosomal protein L10a with high affinity; whether this interaction contributes to the activity of TCS has not been determined (23). By superimposing the TCS-SRL model on the eFE2-SRL model, we have previously found that the three basic residues (K173, R174 and K177) of TCS are in close proximity to the P-protein binding site of eEF2 (4). Also, a model of the mammalian 60S ribosome, with the structure of P0 including up to the first Asp in the c11-P sequence, has just been released (24). The distance between the targeted adenine (A4565) on SRL and P0 was found to be about 50 Å, and the distance between the concerned adenine and L10a was about 140 Å. This observation further supports the argument that the binding of TCS to P proteins would make the TCS readily accessible to the SRL. Since we have shown that the interaction between TCS and the P protein is needed for the full activity of TCS, this interaction should be crucial for TCS to act on the targeted adenine.
Pokeweed anti-viral protein, a type I RIP distinctly related to TCS and active on prokaryotic and eukaryotic ribosomes, has been found to interact with ribosomal protein L3 (25) and does not need the C-terminus of the P protein in Trypaosoma cruzi ribosomes for full activity (26). On the other hand, a number of other type I and type II RIPs have been shown to access the ribosome through interactions with P proteins. With cross-linking experiments, the interaction between RTA and P0 of human lung carcinoma cells has been observed (27). Also, NSBr-saporin has been cross-linked to a 30 kDa ribosomal protein, presumably P0, from the 60S yeast ribosome (28), and several positively charged residues in the C-terminal region of SO6 have been protected from chemical modification in a SO6-ribosome complex (12). Pepocin has been shown to require P proteins for the depurination of rRNA (29). Recently, it has also been found that the C-terminal 11-aa of the P protein interacts directly with SLT-1A and RTA (5), thus providing independent support to our structural observations of TCS and the c-11 peptide.
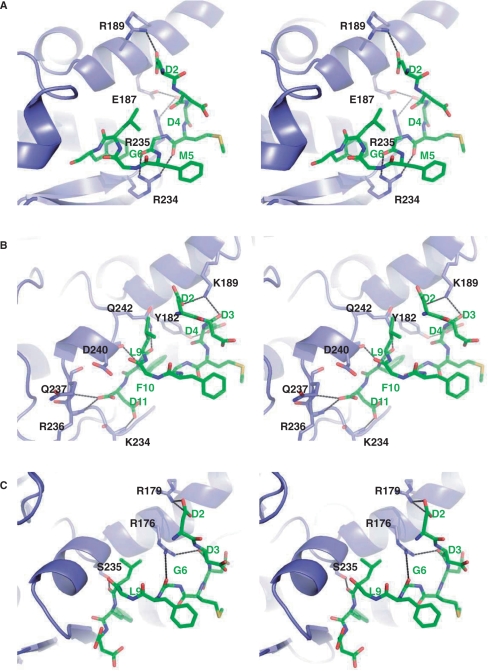
Using the TCS–c11-P complex as a reference and docking the c11-P peptide to RTA, SO6 and SLT-1A, we have found that the C-terminal domains of these RIPs share a common binding surface with the c11-peptide. The N-terminal charged residues of c11-P, in particular the side chains of Asp2 and Asp4 and the backbone oxygen of Asp3, form hydrogen bonds with the side chains of an α-helix of the RIP (Figure 6 and Table 3). Such hydrophilic interaction involving the N-terminal Asp appears to be crucial, as a C-7 peptide (MGFGLFD) did not interact with TCS, RTA or SLT-1A (4,5). Similar to the case of TCS–c11-P, there is a well-defined hydrophobic pocket in RTA, SO6 and SLT-1A for the insertion of Phe 10 of the c-11P. In RTA, SO6 and SLT-1A, we have also found potential hydrogen bonds between them and residues in the C-terminal region of c11-P (Table 3). The additional interactions are attributed by the extended two stranded β-sheet in both TCS (residues 213–225) and RTA (residues 230–242) or loop in SO6 (residues 227–236). Such interactions are absent in SLT-A of which the C-terminal 80 residues fold in a completely different conformation. The potential interaction of C-11 peptide with PAP, which has not been shown to interact with P proteins, was also examined (Supplementary Figure 1). In the N-terminal region of c11-P, only Asp4 is hydrogen bonded to the PAP (Supplementary Table 1). Hence, the importance of the N-terminal Asp residues that was demonstrated in other RIP–c11-P complexes is not well supported in the PAP–c11-P model. Also, a clear molecular interaction surface on PAP, in particular a hydrophobic cleft for Phe 10 of c11-P, is absent, suggesting that the interaction between PAP and c11-P, if it occurs, may not be as strong or specific as that shown for TCS, RTA, SO6 and SLT-1A.
Figure 6.
Stereo images of the molecular docking of c11-P peptide to RTA, SO6 and SLT-1A. (A) RTA, (B) SO6 and (C) SLT-1A. c11-P is shown in ball and stick representation and in green colour. Potential hydrogen bond interactions between the RIPs and the c-11 peptide are indicated.
Table 3.
Possible hydrogen bonds formed between the c11-P peptide and RTA, SO6 or SLT-1A
| c11-P | RTA | Distance (Å) |
| Asp 2 OD2 | Arg 189 NH1 | 3.24 |
| Asp 4 OD1 | Glu 187 OE2 | 3.23 |
| Asp 4 OD1 | Arg 235 NH2 | 3.50 |
| Met 5 O | Arg 234 NH2 | 3.19 |
| Gly 6 O | Arg 234 NH1 | 2.87 |
| Asp 11 OD1 | Ser 241 N | 3.36 |
| c11-P | SO6 | Distance (Å) |
| Asp 2 OD1 | Lys 189 NZ | 3.61 |
| Asp 3 O | Lys 189 NZ | 2.88 |
| Asp 4 OD1 | Tyr 182 OH | 3.06 |
| Leu 9 O | Gln 242 NE2 | 2.92 |
| Phe 10 N | Asp 240 O | 2.79 |
| Asp 11 OD1 | Lys 234 N | 2.97 |
| Asp 11 OXT | Arg 236 N | 2.98 |
| Asp 11 OXT | Gln 237 N | 3.20 |
| c11-P | SLT-1A | Distance (Å) |
| Asp 2 OD1 | Arg 179 NH2 | 3.42 |
| Asp 2 OD2 | Arg 179 NH2 | 3.43 |
| Asp 3 O | Arg 176 NH2 | 3.10 |
| Gly 6 O | Arg 176 NH1 | 3.54 |
| Leu 9 O | Ser 235 OH | 2.67 |
In conclusion, we have provided structural evidence of the interaction between TCS and the consensus C-terminal motif of ribosomal stalk proteins. This interaction is required for TCS to exert its full activity and may be generalised to other RIPs.
Accession codes
Coordinates of TCS–C11-P complex, the V232K/N236D and the K173A/R174A/K177A variants have been deposited with PDB accession codes 2JDL, 2JJR and 2VS6, respectively.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by grants (CUHK4301/03M and CUHK4606/06M) from the Research Grants Council of Hong Kong SAR. Funding for open access charge: Department of Biochemistry, Faculty of Science, The Chinese University of Hong Kong.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Tchorzewski M. The acidic ribosomal P proteins. Int. J. Biochem. Cell Biol. 2002;34:911–915. doi: 10.1016/s1357-2725(02)00012-2. [DOI] [PubMed] [Google Scholar]
- 2.Helgstrand M, Mandava CS, Mulder FAA, Liljas A, Sanyal S, Akke M. The ribosomal stalk binds to translation factors IF2, EF-Tu, EF-G and RF3 via a conserved region of the L12 C-terminal domain. J. Mol. Biol. 2007;365:468–479. doi: 10.1016/j.jmb.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 3.Shaw PC, Lee KM, Wong KB. Recent advances in trichosanthin, a ribosome-inactivating protein with multiple pharmacological properties. Toxicon. 2005;45:683–689. doi: 10.1016/j.toxicon.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 4.Chan DSB, Chu LO, Lee KM, Too PHM, Ma KW, Sze KH, Zhu G, Shaw PC, Wong KB. Interaction between trichosanthin, a ribosome-inactivating protein, and the ribosomal stalk protein P2 by chemical shift perturbation and mutagenesis analyses. Nucleic Acids Res. 2007;35:1660–1672. doi: 10.1093/nar/gkm065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCluskey AJ, Poon GMK, Bolewska-Pedyczak E, Srikumar T, Jeram SM, Raught B, Gariépy J. The catalytic subunit of Shiga-like toxin 1 interacts with ribosomal stalk proteins and is inhibited by their conserved C-terminal domain. J. Mol. Biol. 2008;378:375–386. doi: 10.1016/j.jmb.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 6.Leslie AGW. 1992. Joint CCP4+ ESF-EAMCB Newsletter on Protein Crystallography, No. 26. [Google Scholar]
- 7.Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 8.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 9.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve W, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 10.McRee DE. XtalView/Xfit – A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 11.Mlsna D, Monzingo AF, Katzin BJ, Ernst S, Robertus JD. Structure of recombinant ricin A chain at 2.3 Å. Protein Sci. 1993;2:429–435. doi: 10.1002/pro.5560020315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Savino C, Federici L, Ippoliti R, Lendaro E, Tsemoglou D. The crystal structure of saporin SO6 from Saponaria officinalis and its interaction with the ribosome. FEBS Lett. 2000;470:239–243. doi: 10.1016/s0014-5793(00)01325-9. [DOI] [PubMed] [Google Scholar]
- 13.Fraser ME, Chernaia MM, Kozlov YV, James MN. Crystal structure of the holotoxin from Shigella dysenteriae at 2.5 Å resolution. Nat. Struct. Biol. 1994;1:59–64. doi: 10.1038/nsb0194-59. [DOI] [PubMed] [Google Scholar]
- 14.Kurinov IV, Rajamohan F, Venkatachalam TK, Uckun FM. X-ray crystallographic analysis of the structural basis for the interaction of pokeweed antiviral protein with guanine residues of ribosomal RNA. Protein Sci. 1999;8:2399–2405. doi: 10.1110/ps.8.11.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holm L, Sander C. Alignment of three-dimensional protein structures: network server for database searching. Methods Enzymol. 1996;266:653–662. doi: 10.1016/s0076-6879(96)66041-8. [DOI] [PubMed] [Google Scholar]
- 16.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 17.Bargis-Surgey P, Lavergne JP, Gonzalo P, Vard C, Filhol-Cochet O, Reboud JP. Interaction of elongation factor eEF-2 with ribosomal P proteins. Eur. J. Biochem. 1999;262:606–611. doi: 10.1046/j.1432-1327.1999.00434.x. [DOI] [PubMed] [Google Scholar]
- 18.Datta PP, Sharma MR, Qi L, Frank J, Agrawal RK. Interaction of the G' domain of elongation factor G and the C-terminal domain of ribosomal protein L7/L12 during translocation as revealed by Cryo-EM. Mol. Cell. 2005;20:723–731. doi: 10.1016/j.molcel.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 19.Helgstrand M, Mandava CS, Mulder FAA, Liljas A, Sanyal S, Akke M. The ribosomal stalk binds to translation factors IF2, EF-Tu, EF-G and RF3 via a conserved region of the L12 C-terminal domain. J. Mol. Biol. 2007;365:468–479. doi: 10.1016/j.jmb.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 20.Korennykh AV, Piccirilli JA, Correll CC. The electrostatic character of the ribosomal surface enables extraordinarily rapid target location by ribotoxins. Nat. Struct. Mol. Biol. 2006;13:436–443. doi: 10.1038/nsmb1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korennykh AV, Correll CC, Piccirilli JA. Evidence for the importance of electrostatics in the function of two distinct families of ribosome inactivating toxins. RNA. 2007;13:1391–1396. doi: 10.1261/rna.619707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan SH, Hung FSJ, Chan DSB, Shaw PC. Trichosanthin interacts with acidic ribosomal proteins P0 and P1 and mitotic checkpoint protein MAD2B. Eur. J. Biochem. 2001;268:2107–2112. doi: 10.1046/j.1432-1327.2001.02091.x. [DOI] [PubMed] [Google Scholar]
- 23.Xia X, Hou F, Li J, Nie H. Ribosomal protein L10a, a bridge between trichosanthin and the ribosome. Biochem. Biophys. Res. Commun. 2005;336:281–286. doi: 10.1016/j.bbrc.2005.08.074. [DOI] [PubMed] [Google Scholar]
- 24.Chandramouli P, Topf M, Ménétret J.-F, Eswar N, Cannone JJ, Gutell RR, Sali A, Akey CW. Structure of the mammalian 80S ribosome at 8.7Å. Structure. 2008;16:535–548. doi: 10.1016/j.str.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hudak KA, Dinman JD, Tumer NE. Pokeweed antiviral protein accesses ribosomes by binding to L3. J. Biol. Chem. 1999;274:3859–3864. doi: 10.1074/jbc.274.6.3859. [DOI] [PubMed] [Google Scholar]
- 26.Ayub MJ, Smulski CR, Ma KW, Levin MJ, Shaw PC, Wong KB. The C-terminal end of P proteins mediates ribosome inactivation by trichosanthin but does not affect the pokeweek antiviral protein activity. Biochem. Biophys. Res. Commun. 2008;369:314–319. doi: 10.1016/j.bbrc.2008.01.170. [DOI] [PubMed] [Google Scholar]
- 27.Vater CA, Bartle LM, Leszyk JD, Lambert JM, Goldmacher VS. Ricin A chain can be chemically cross-linked to the mammalian ribosomal proteins L9 and L10e. J. Biol. Chem. 1995;270:12933–12940. doi: 10.1074/jbc.270.21.12933. [DOI] [PubMed] [Google Scholar]
- 28.Ippoliti R, Lendaro E, Bellelli A, Brunori M. A ribosomal protein is specifically recognized by saporin, a plant toxin which inhibits protein synthesis. FEBS Lett. 1992;298:145–148. doi: 10.1016/0014-5793(92)80042-f. [DOI] [PubMed] [Google Scholar]
- 29.Uchiumi T, Honma S, Endo Y, Hachimori A. Ribosomal proteins at the stalk region modulate functional rRNA structures in the GTPase center. J. Biol. Chem. 2002;277:41401–41409. doi: 10.1074/jbc.M207424200. [DOI] [PubMed] [Google Scholar]
- 30.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.