Abstract
Heterochromatin is critical for genome integrity, and recent studies have suggested the importance of transcription in heterochromatin for maintaining its silent state. We previously developed a method to generate a large homogeneously staining region (HSR) composed of tandem plasmid sequences in human cells that showed typical heterochromatin characteristics. In this study, we examined transcription in the HSR. We found that transcription of genes downstream to no-inducible SRα promoter was restricted to a few specific points inside the large HSR domain. Furthermore, the HSR localized to either to the surface or to the interior of the nucleolus, where it was more actively transcribed. The perinucleolar or intranucleolar locations were biased to late or early S-phase, and the location depended on either RNA polymerase II/III or I transcription, respectively. Strong activation of the inducible TRE promoter resulted in the reversible loosening of the HSR domain and the appearance of transcripts downstream of not only the TRE promoters, but also the SRα promoters. During this process, detection of HP1α or H3K9Me3 suggested that transcription was activated at many specific points dispersed inside large heterochromatin. The transcriptional rules obtained from studying artificial heterochromatin should be useful for understanding natural heterochromatin.
INTRODUCTION
Heterochromatin is a condensed silent chromatin that is essential for cellular life. It appears constitutively at pericentromeric and telomeric chromosomal regions and in inactivated X chromosomes, as well as conditionally at tissue-specific genes (1,2). Recent studies have revealed an apparently contradictory situation, in which transcription is required for the maintenance of silent heterochromatin (3,4). It was reported that such transcription occurred at specific stages of the cell cycle in yeast (5,6) and mammalian cells (7). The transcript was processed to activate the RNAi mechanism (8,9), which in turn heterochromatinated and silenced the cognate DNA sequence. Interestingly, the inactivated X chromosome in mammalian female cells has been shown to periodically visit the nucleolus in order to maintain the heterochromatic silent state; however, the relationship between nucleolar localization and transcription was not clear (10).
We previously found that a plasmid bearing a mammalian replication initiation region (IR) and a matrix attachment region (MAR) was quite efficiently amplified in human cancer cells (11,12). The amplified plasmid was localized to cytogenetically detectable extrachromosomal double minutes (DMs) or a chromosomal homogeneously staining region (HSR). As the plasmid amplification system appeared to mimic gene amplification during human cell oncogenesis, we utilized the system to uncover the mechanism of gene amplification (13). The novel amplification system was efficiently adapted to the production of recombinant protein (14). Furthermore, it was used to dissect the mammalian replicator sequence in the IR (15), to analyze the interaction between the DNA binding NF-κB or glucocorticoid receptor protein and its cognate sequence (16,17), to reveal the intracellular behavior and entrapment of extrachromosomal DMs into micronuclei (18) and to analyze the transcription of DMs inside micronuclei (19). Because the IR/MAR-plasmid generated a giant HSR composed of plasmid repeats, we also examined the macroscopic folding and replication of the giant HSR in interphase nuclei. We found that the HSR was folded as a giant coiled coil in late S-phase nuclei, and that it was replicated from outside to inside (20).
The HSR generated by the IR/MAR plasmid was composed essentially of plasmid direct repeats with some irregularities (12). It is known that expression from direct repeats is frequently silenced by a mechanism called ‘repeat-induced gene silencing’ (RIGS) (21–23). In actuality, the HSR generated by the IR/MAR plasmid showed many features typical of heterochromatin: (i) it appeared condensed among metaphase spreads stained with DAPI or propidium iodide (PI) (11,12);(ii) it was replicated at the last stage of S-phase when the heterochromatin was replicated (20), (iii) expression levels from genes in the HSR were usually quite low (14); and (iv) the HSR was associated with heterochromatin protein (HP1α; this study) and tri-methylation at Lys 9 of histone H3 (H3K9Me3; this study). In this study, we focused on how this artificial HSR is transcribed in a spatio-temporally regulated manner. From this investigation, we revealed several new features of heterochromatin transcription, which may be applied to a broad spectrum of natural heterochromatin.
MATERIALS AND METHODS
Cell lines and cultures
Human COLO 320DM (CCL 220) neuroendocrine tumor cells were obtained and maintained as described previously (24). The ‘clone 22’ cell line we developed has been described previously (12) namely, clone 22 was obtained by transfection of COLO 320DM cells with a pSFVdhfr plasmid that bears an IR/MAR sequence from the DHFR locus; stable transformants were selected by blasticidine because the plasmid carries a blasticidine resistance (BSR) gene. We showed that the introduced plasmid in clone 22 was amplified and localized in the HSR. For the current study, we established the ‘HSR-CFP’ cell line from COLO 320DM. The HSR in this line can be visualized by its cyan fluorescence, and inducible transcription from the HSR is visible by yellow fluorescence. This line was developed by a similar method that was employed for the establishment of DM-CFP C4C4 cells in our previous paper (19); namely, we first transfected COLO 320DM with the pSV2 ECFP-LacR plasmid (a generous gift from Dr Susan M. Janicki and Dr David L. Spector at the Cold Spring Harbor Laboratory), which expresses a fusion protein composed of the lactose repressor (LacR) and enhanced cyan fluorescence protein (ECFP). We isolated a stable clone in which cyan fluorescence was uniformly detected throughout the nucleus. Into such cells, we co-transfected the pSFVdhfr plasmid and pECMS2β plasmid [a generous gift from Dr Susan M. Janicki and Dr David L. Spector; ref. (25)]. We previously showed that any DNA could be co-amplified by co-transfection with the IR/MAR-bearing plasmid (12,16,19). Therefore, the above plasmids were co-amplified at the HSR; this was visible in the nucleus by the binding of LacR-CFP to the LacO repeat in the pECMS2β sequence. We obtained a clone in which the HSR was visible by cyan fluorescence (‘HSR-CFP’). In HSR-CFP cells, the plasmid sequences were amplified at the HSR, which was confirmed by fluorescence in situ hybridization (FISH) using probes specific to DHFR IR (see ‘Results’ section). We electroporated these cells with pTet-ON or pTet-OFF (Clontech Laboratories Inc., CA) and pMS2-YFP plasmids by using a BioRad ‘Gene Pulser’ at 960 µF, 200 mV. The pECMS2β plasmid bears a TRE promoter that is induced by the rtTA or tTA protein, which is expressed from pTet-ON or oTet-OFF in the presence or absence of Doxycycline (Dox), respectively. RNA transcribed from the TRE promoter has an MS2 target sequence that can be visualized by the binding of the MS2-YFP protein, which is expressed by pMS2-YFP.
For the isolation of transiently transformed cells, we used the ‘MACSelect’ kit (Miltenyi Biotec Inc.) according to the manufacturer's recommended protocol. In short, we electroporated a mixture of pTet-OFF and pMACS-LNGFR plasmid DNA into HSR-CFP cells and cultured the cells for 2 days. The cells expressing LNGFR were tagged by anti-LNGFR beads, which were separated by a magnetic column. From these cells, total DNA and RNA were isolated using TRI reagent (Molecular Research Center Inc.). Quantitative real-time PCR was performed by using the primer pair specific to the BSR coding region and a LightCycler (Roche). Actinomycin D (Sigma) was dissolved in ethanol at 2 mg/ml and was added to the culture at the indicated concentrations for 3 h.
All transfections were performed using the Gene Porter 2 lipofection kit (Genlantis Co. CA), unless otherwise indicated. COLO 320DM and all its derivative cell lines were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum.
FISH and other cytochemical procedures
FISH was performed according to our previously published protocol (19). In brief, to prepare antisense RNA probes, the DNA of the pGEM4 plasmid containing BSR was linearized and transcribed using T7-RNA polymerase and DIG-dUTP. To detect the HSR DNA by FISH, pSFVdhfr plasmid DNA was labeled with biotin as described previously (11).
Cells were washed once with PBS-, cytocentrifuged on poly-l-lysine-coated glass slides (Matsunami Glass Ind. Ltd., Osaka), and then treated with prechilled CSK buffer (100 mM NaCl, 300 mM sucrose, 10 mM PIPES, pH 6.8, 3 mM MgCl2, 1 mM EGTA, 0.5% Triton X-100, 1.2 M phenylmethylsulfonyl fluoride and 1.2 mM vanadyl adenosine) for 30 s on ice. The solution was replaced with 4% paraformaldehyde (PFA) in PBS, and the cells were fixed for 15 min at room temperature. The slides were then washed with PBS and stored in 70% ethanol at 4°C.
To detect specific RNA (RNA FISH), cells on slides were washed once with 2 × SSC and treated with equilibration solution (50% formamide, 2 × SSC) for 15 min at room temperature. The hybridization mixture, which consisted of 20 ng of DIG-labeled probe, 6 µg of salmon sperm DNA, 50% formamide, 10% dextransulfate and 2 × SSC in 15 µl for each slide, was denatured at 75°C for 5 min and then applied to the non-denatured slide. The slide was covered, and hybridization was allowed to occur overnight at 37°C. Stringent washing and detection of the hybridized DIG-labeled probe using an anti-DIG mouse monoclonal antibody (Roche Diagnostics Co.) and a FITC-labeled donkey anti-mouse IgG antibody (Rockland Co.) were performed as previously described (26). To simultaneously detect RNA and DNA, specific RNA was detected as above, and then the signal was fixed by immersing the slide in ethanol/acetic acid (19/1) at –20°C for 3 min. The slide was then rinsed with PBS- and further fixed by 3% PFA in PBS for 10 min at room temperature. The slides were washed with PBS and immersed in methanol/acetic acid (3/1) for 10 min at room temperature. Slide denaturation and hybridization with a denatured biotin-labeled DNA probe were performed as described previously (27). Visualization of the probe was mediated by the use of Alexa 647-conjugated streptavidin (Invitrogen Co.) and a biotinylated goat anti-streptavidin antibody (Vector Co.).
5-bromouridine (BrU; Aldrich), which was freshly dissolved in water at 100 mM each time before the experiment, was added to the culture at a concentration of 2.5 mM for 15 min. The cells were washed with PBS and fixed with 1.75% PFA in PBS for 10 min at room temperature. The slide was washed, permeabilized with 0.5% NP-40 for 10 min, and blocked using ‘Signal Enhancer’ (Molecular Probes Co.). Incorporated BrU was detected by a mouse monoclonal anti-BrdU antibody (Roche Diagnostics Co.), because it cross-reacts with BrU, and a FITC-conjugated donkey anti-mouse IgG antibody (Rockland Inc.). The co-localization of HSR (green signal) and BrU (red signal) among the images was determined by the ‘FV10-ASW’ software equipped to FV1000 confocal microscope (Olympus Co.). 5-Bromo-2′-deoxyuridine (BrdU; Aldrich) or 5-chloro-2′-deoxyuridine (CldU; ICN biomedicals) were freshly dissolved in water and added to the culture at 10 µM for 15 min. The incorporated BrdU or CldU were detected after the FISH procedure by using anti-BrdU mouse monoclonal IgG (Upstate Co.) or rat monoclonal anti-BrdU IgG2a (Oxford Biotechnology). Alternatively, to detect simultaneously with CFP-LacR, the fixed cells were treated for 30 min at 37°C with a mixture containing 5–6 µg/ml mouse monoclonal anti-BrdU antibody (Roche Diagnostics Co.), 30 mM Tris–HCl, pH 8.0, 0.3 mM MgCl2, 0.5 mM mercaptoethanol, 0.5% BSA, 1% BlockAce (Dainippon Pharmaceuticals, Co.), and 6 U/µl RNase-free DNase (Takara, Co.) in PBS. The bound primary antibody was detected by Texas Red-conjugated goat anti-mouse IgG antibody (Southern Biotechnology Inc.). HP1α was detected using a mouse monoclonal anti-HP1β antibody (Upstate Co.) and a Texas red-conjugated goat anti-mouse IgG antibody (Southern Biotechnology). Histone H3 (tri methyl K9)(H3K9Me3) was detected using a rabbit polyclonal anti-H3K9Me3 antibody (Abcam Co.) and a Texas red-conjugated goat anti-rabbit IgG antibody (Vector Co.).
Image acquisition and processing
Images were obtained by using one of the following three microscopes: For the images in Figures 1A, B, F, and 2, 5C, 6C–6D and 7, an epifluorescence microscope (ECLIPSE TE2000-E, Nikon) equipped with an ×63 objective lens (Nikon Plan APO VC NA 1.40) and a CCD camera (DS, Nikon) was used. Images in Figures 2 and 6C and D were deconvoluted using the ‘NIS element’ (Nikon). For the images in Figures 1C and D, 5A and B and 6A and B, a Zeiss confocal system LSM5 Pascal on a Axiovert 200M equipped with a ×63 objective was used. For the images in Figures 1 and I, and 3 and 4, an Olympus FV10-ASW confocal system on FV1000D-IX81 with an ×60 objective (UPLSAPO NA 1.35 × 60 oil) was used. For localization of HSR inside the nucleus, we obtained a z-series of 6 to 20 images with an interval of 0.6 to 1.2 µm. All of the acquired digital images were expressed as pseudo-colors and were merged using Adobe Photoshop CS version 8.0.1 (Adobe Systems Inc.).
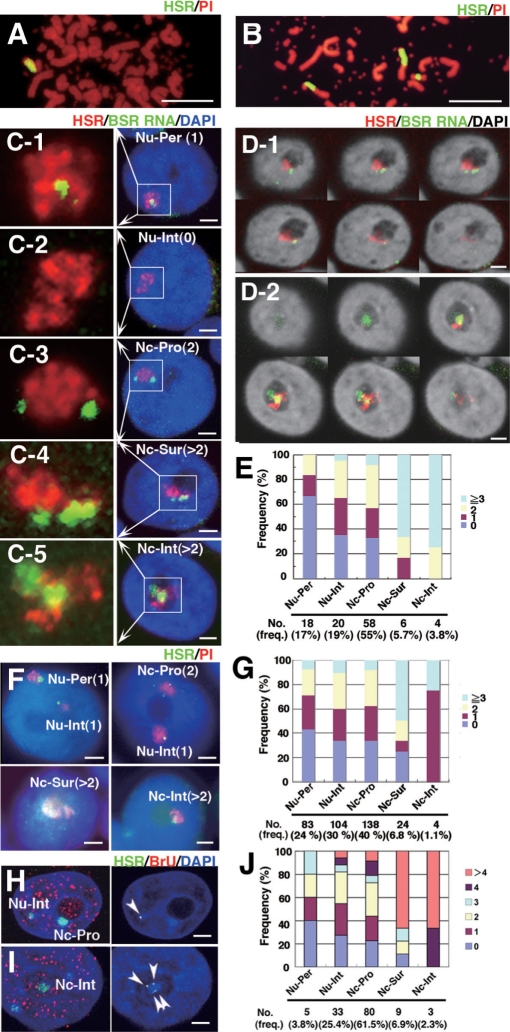
Figure 1.
Transcription in HSR was restricted to only few foci, and the number of foci increased at the surface or interior of the nucleolus. Metaphase chromosome spreads prepared from clone 22 cells (A) or HSR-CFP cells (B) were hybridized with a plasmid probe (green), and DNA was counterstained with propidium iodide (PI; red). The clone 22 cells had a single large HSR, whereas the HSR-CFP cells had one small and two large HSRs. Logarithmically growing clone 22 cells (C–E) or HSR-CFP cells (F, G) were fixed with paraformaldehyde (PFA). BSR RNA (green) and HSR DNA (red) were simultaneously detected by nondenaturing and denaturing FISH, respectively, as described in ‘Materials and Methods’ section. DNA was counterstained with DAPI, and its image is shown in blue (C, F, H, I) or gray-scale (D). The nucleolus was identified as a dim region after DAPI staining. The location of the HSR in the nucleus was determined by examining z-series images that were obtained by confocal microscopy. The representative images for the nuclear periphery (Nu-Per, C-1; defined by contact with the nuclear rim), the nuclear interior (Nu-Int, C-2), the nucleolar proximity (Nc-Pro, C-3; defined by contact with the nucleolus), the nucleolar surface (Nc-Sur, C-4; defined by distribution over the nucleolar surface) and the nucleolar interior (Nc-Int, C-5) are shown. BSR RNA was detected as small foci in these HSRs, as well as those in the HSRs of HSR-CFP cells (F, four representative nuclei). The approximate number of foci (in parentheses in each panel) was determined by viewing z-series images. The magnified images for the HSR domain are shown at the left side of each panel C. In D-1 and D-2, the confocal z-series images show the HSR at the surface or interior of the nucleolus, respectively. Such HSRs were associated with many BSR RNA signals. The approximate number of BSR RNA foci was scored for the HSR at each location; these are summarized in panels E (clone 22) and G (HSR-CFP). At the bottom of the graphs, the number and the frequency of HSRs at each location were noted. At each location, the bars show the frequency of HSRs that were associated with indicated number of BSR-RNA foci. Because of technical difficulties, the classification between Nu-Per, Nu-Int and Nc-Pro was not accurate in strict mean, but it provides the relative location of HSR in the nucleoplasm. To detect de novo transcription, the HSR-CFP cells were pulse-treated with BrU for 15 min, and incorporated BrU was detected by red fluorescence (H-J; see ‘Materials and methods’ section). The co-localization of HSR (green) and BrU (red) among the images (left panel of H, I) was shown in gray scale (right panel of H, I). The number of co-localization signals was counted for the HSRs at each location, and it was plotted in J. All bars in images indicate 2 µm, except for A and B (5 µm).
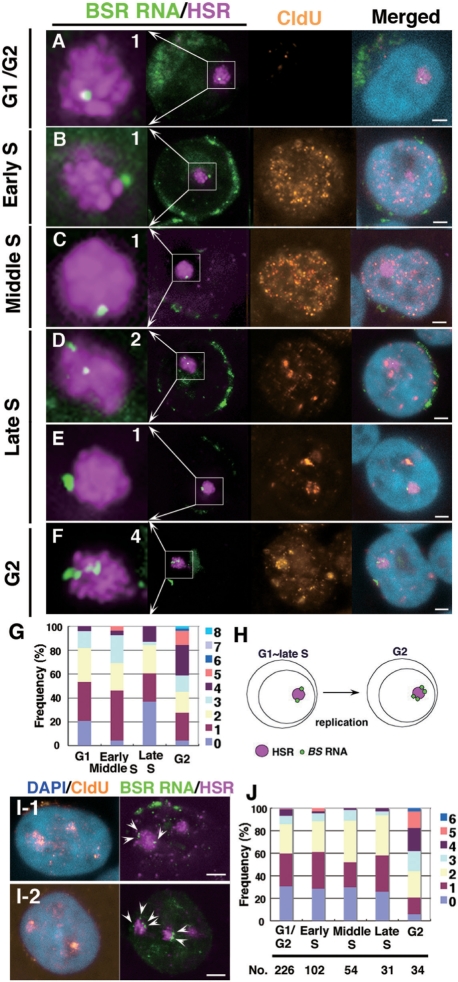
Figure 2.
The number of transcriptional specific points increased after HSR duplication. Logarithmically growing clone 22 cells (A–G) or HSR-CFP cells (I, J) were pulse-labeled (15 min) with CldU and fixed directly with PFA. Both BSR RNA (green) and HSR DNA (magenta) were detected by FISH; CldU (brown) was simultaneously detected by immunofluorescence, and DNA was counterstained with DAPI (blue). According to the CldU pattern in the nucleus, each cell was determined as being in G1/G2 (no signal), or early, middle or late S-phase. G2-phase cells were specifically identified among the cells chased 2 h after the CldU pulse-labeling, and cells showing a CldU pattern typical of late S-phase should be in G2 at the time of harvest. As expected, the HSR, which replicate at the end of S-phase (20), was stained with CldU in these cells (F and I). For such cells, the number of BSR RNA foci per each HSR was scored; these are noted in the left-most enlarged images of A–F. The data obtained for each stage are summarized in the graph (G for clone 22 and J for HSR-CFP cells). We scored more than 50 HSRs for each stage (G) or the numbers indicated in the graph (J). The graph suggested that the number was almost doubled after the duplication of the HSR (H). The green signal surrounding nuclei is BSR RNA transported to the cytoplasm. All bars in images indicate 2 µm.
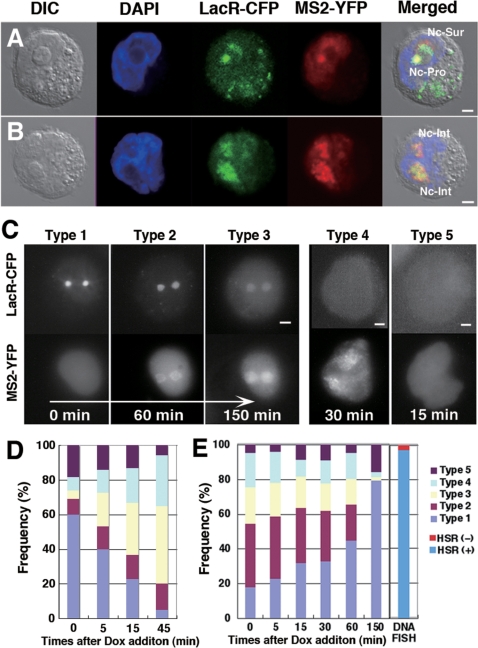
Figure 5.
The HSR was reversibly decondensed upon transcriptional induction. To logarithmically growing HSR-CFP cells, we co-transfected pMS2-YFP and either pTet-ON (A–D) or pTet-OFF (E) plasmid DNA by electroporation. After 2.5 h, Dox was added to the culture. After 30 min, or the indicated times, cells were observed live (C) or after PFA fixation (A, B, D, E) using confocal microscopy (A, B) or epifluorescence microscopy (C–E). From the LacO sequence-tagged HSR, MS2 sequence-tagged RNA was transcribed after Dox addition in pTet-ON transfected cells. Therefore, HSR was visualized by the binding of LacR-CFP, whereas induced RNA was visualized by the binding of MS2-YFP. On the other hand, transcription was activated in pTet-OFF transfected cells until Dox addition (time 0 in E); after that it was down-regulated. For D and E, more than 150 cells that expressed MS2-YFP were observed at each time point, and the frequencies of Types 1 to 5 were scored. FISH revealed that the HSR-CFP cell line contained about 3.2% of cells that had essentially no HSR, and it was indicated in E. All bars in images indicate 2 µm.
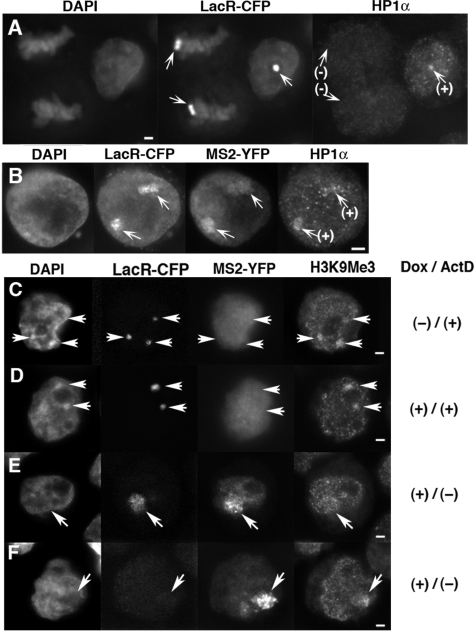
Figure 6.
The heterochromatin marker remained at the decondensed HSR after transcriptional activation. HP1α (A, B) or H3K9Me3 (C–F) was detected by indirect immunofluorescence. (A) In logarithmically growing HSR-CFP cells, the HSR in interphase nuclei (right) coincided with HP1α staining, while the HSR in mitotic cells (left) was not. (B–F) The same cells were co-transfected with pMS2-YFP and pTet-ON, cultured for 3 h in the absence (B, E, F) or presence (C, D) of 2 µg/ml Actinomycin D. During the last 30 min, the cells were treated with Dox except for C. As shown in Figure 5, MS2 sequence-tagged RNA that was visualized by the binding of MS2-YFP was expressed from the HSR visualized by LacR-CFP (B, E, F). The HSR in B and E correspond to Type 3, and the one in F corresponds to Type 4 in Figure 5. The arrows indicate position of the HSRs. All bars in images indicate 2 µm.
Figure 7.
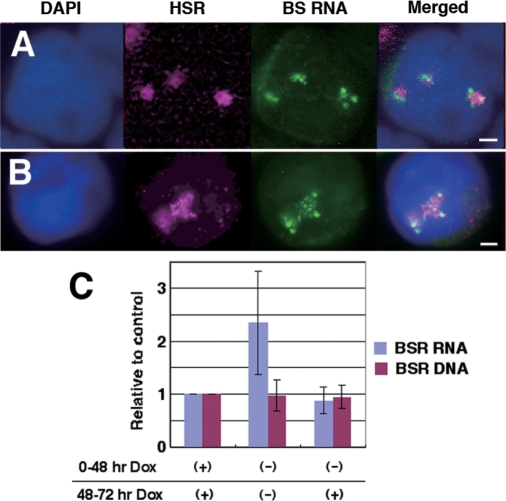
Decondensation of the HSR is accompanied by the activation of a noninducible promoter. (A, B) HSR-CFP cells were co-transfected with pMS2-YFP and pTet-OFF, cultured for 2.5 h in the absence of Dox for 30 min, and fixed. Among these cells, HSR and BSR RNA were simultaneously detected by FISH using different colors. Compared with the experiments that appear in Figures 1 and 2, many BSR RNA signals were associated with the HSR DNA. (C) HSR-CFP cells were co-transfected with pMACS-LNGFR and pTet-OFF and cultured for 2 days in the presence or absence of Dox, as indicated. Cells expressing LNGFR were isolated by the MACSelect system. From these cells, total RNA and DNA were extracted, and the amount of BSR sequence was determined by real-time PCR. The graph was generated from three independent experiments; error bars represent standard deviations. All bars in images indicate 2 µm.
RESULTS
Transcription in the HSR was restricted to only a few foci, and the number of foci increased at surface or inside of the nucleolus
In order to analyze transcription in the HSR, we utilized two clonal cell lines that we developed, i.e. ‘clone 22’ and ‘HSR-CFP’ cells (see ‘Materials and Methods’ section). The cells of clone 22 bear a long HSR at chromosome arm (Figure 1A). By using this cell line, we previously showed that the HSR was a few tens of megabase-pairs in length; that it consisted solely of plasmid sequences, most of which were arranged as direct repeats (12); and that how it was folded and replicated in the nucleus (20). Now, we examined the expression of the BSR genes, which are tandemly arranged in the HSR, and whose expression is driven by noninducible SRα promoters. For this, we simultaneously detected the HSR DNA and BSR RNA in different colors using FISH. As a result, we found that the RNA transcript, if it was detected, always appeared as small foci in the HSR subchromosomal domain (Figure 1C), which implied the presence of specific points for the transcription in the long and homogeneous HSR. We also found that some HSRs were spread along the surface of the nucleolus (Figure 1C-4 and D-1) or were located inside the nucleolus (Figure 1C-5 and D-2). At such locations, the HSRs appeared to be more actively transcribed as many BSR RNA foci were detected. To clarify this point, we obtained the frequencies of HSRs bearing various numbers of BSR RNA foci in relation to their nuclear position (Figure 1E): HSRs inside the nucleolus (Nc-Int) or at the nucleolar surface (Nc-Sur) were associated with markedly higher numbers of RNA foci, compared with the HSRs at the nucleoplasm (Nu-Per, Nu-Int, or Nc-Pro; Figure 1C-1–C3).
Similar observations were also obtained by using another cell line (HSR-CFP cells). This cell line was also constructed from human COLO 320DM cells; however, the HSR was composed of a mixture of two co-transfected plasmid sequences (see ‘Materials and Methods’ section). Furthermore, the cells contained one small and two large HSRs in a single cell (Figure 1B). In the following experiment, we took into account only the two large HSRs and neglected the small one. For such HSRs, transcription was restricted to a small number of foci, although the number was larger if the HSRs were located at the surface of the nucleolus (Figure 1F and G). In these cells, the HSR in the nucleolar interior was not associated with many RNA foci; however, the frequency of HSRs with more than three foci was markedly higher. Therefore, from the data obtained from the two different cell strains, we concluded that the HSR was more actively transcribed if it was located either at the surface or in the interior of the nucleolus.
It was possible that the BSR RNA FISH signal may reflect some kind of structural RNA that was associated with HSR domain. Therefore, we examined whether the RNA FISH signals reflect de novo transcription or not. Thus, we simultaneously detected the HSR and the sites of transcription in HSR-CFP cells (Figure 1H–J). We pulse-labeled the newly transcribed RNA by treating the cells with BrU, and detected it by immunofluorescence in red. Co-localization of CFP and red signals indicated the site of de novo transcription in HSR. Such sites were also detected as foci, and the numbers of which at each nuclear locations were counted. The result (Figure 1J) was very similar to the result from the RNA FISH (Figure 1E and G); namely, many foci were associated with the HSR at the nucleolus compared with the ones at the nucleoplasm. Therefore, it suggested that the RNA FISH signal reflects the site of de novo transcription.
The number of transcriptional points in the HSR appeared to double after HSR duplication
The above experiments suggested the presence of specific points for transcription in the large, homogeneous HSR. Therefore, we next addressed whether the number of points varies during progression of the cell cycle. We determined the cellcycle stage of individual cells by detecting pulse-incorporated CldU in cells in which BSR RNA and HSR DNA were simultaneously detected by FISH (Figure 2A–F). As we reported previously (20), the HSR was replicated at the last stage of S-phase, because the HSRs overlapped with the CldU signal in nuclei bearing a CldU pattern typical of late S-phase (Figure 2E). Interestingly, the number of BSR RNA foci in each HSR was almost constant during G1 to late S-phase, whereas it increased to approximately double at G2-phase (Figure 2G). A similar experiment on a different cell line (HSR-CFP cells) resulted in almost the same result (Figure 2I and J). These results suggested that the transcriptional specific points in the HSR appeared to have doubled after HSR duplication (Figure 2H).
Nucleolar visits by the HSR are biased to specific cell-cycle stages
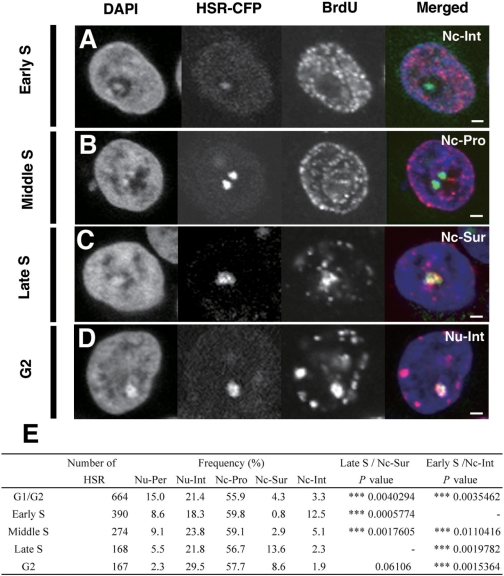
We have shown above that the HSR sometimes visited the nucleolus, and the HSR was actively transcribed there. We next examined the relationship between the cell-cycle stage and the localization of the HSR. For this, we simultaneously visualized the HSR and pulse-incorporated BrdU in logarithmically growing HSR-CFP cells (Figure 3A–D). For individual cells, we noted both the localization of the HSR, which was determined by a z-series of confocal images, and the cell cycle stage, which was determined by the BrdU signal pattern. We found that HSR localization at the nucleolus was not restricted to a specific cell-cycle stage. However, a summary of the results (Figure 3E) indicated that the nucleolar interior was frequented often at early S, whereas the nucleolar surface was frequented at late S-phase. Therefore, although HSR nucleolar localization was not restricted to certain stages of the cell cycle, we concluded that it was biased to specific stages. The bias was small, thus it did not appear as increase in terms of RNA-FISH foci during S-phase (Figure 2G and J), but it was statistically significant (Figure 3E). Notably, the most biased stage was different between the interior (early S) and surface (late S) of the nucleoli. Our data suggest that such localizations have different biological implications. This is consistent with the following result and will be discussed below.
Figure 3.
Nucleolar visits by the HSR are biased to specific cell-cycle stages. The logarithmically growing HSR-CFP cells were pulse-labeled by BrdU for 15 min, harvested, and fixed (A–C). Alternatively, cells were further chased in fresh medium for 2 h before the harvest (D). The incorporated BrdU was detected by immunofluorescence and was viewed simultaneously with HSRs that were visualized by binding of the LacR-CFP protein. The cell-cycle stages of individual cells were determined by the BrdU distribution pattern. G2-phase cells were identified among cells chased 2 h after the BrdU pulse, as in Figure 2. The representative images for each cell-cycle stage are shown in A–D. The localization of HSRs (for abbreviations, see the legend to Figure 1), as well as the cell-cycle stages, were determined by confocal z-series images. These data are summarized and shown in E. In the table, the number of HSRs examined at each cell cycle stage (number of HSRs) and the percentage of the HSR at each location (frequency) was noted. The significances of the Nc-Sur at Late S or the Nc-Int at Early S among the other cell-cycle stages were analyzed by the analysis of variance (ANOVA), and the results were noted as P-values. All bars in images indicate 2 µm.
Nucleolar visitation by the HSR is transcription dependent
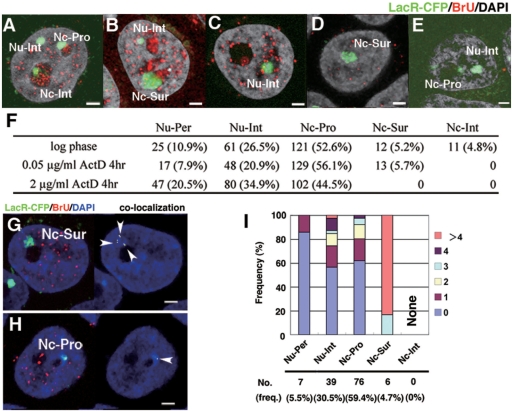
Because we have shown that the nucleolar localization of the HSR was associated with a higher level of transcription, we next addressed whether or not transcription itself was required for the nucleolar localization. Thus, we examined the effect of transcription inhibition on nucleolar localization by treating HSR-CFP cells with actinomycin D and monitoring overall transcription by pulse-treating the cells with BrU. In the logarithmically growing cells without drug (Figure 4A and B), BrU signal was detected heavily in the nucleoli reflecting RNA polymerase I transcription of the rDNA. The BrU signal was also scattered throughout the nucleoplasm, most of which should reflect RNA polymerase II transcription. When the cells were treated with 0.05 µg/ml actinomycin D, a concentration that usually inhibits only RNA polymerase I, the nucleolar BrU signal was completely diminished, while the nucleoplasmic signal remained (Figure 4C and D). Under that condition, the HSR at the nucleolar interior completely disappeared, while the one at the nucleolar surface remained (Figure 4F). Treatment of the cells with a higher concentration of actinomycin D resulted in the complete disappearance of the nuclear BrU signal, reflecting the complete inhibition of all types of RNA polymerases (Figure 4E). Under that condition, the morphology of the nucleolus appeared to be intact, and the HSRs at both the nucleolar surface and the interior disappeared. These results suggested that the localization at the nucleolar interior requires polymerase I transcription at the nucleolus, while localization at the nucleolar surface requires polymerase II or III. On the other hand, HSR at the nucleoplasm or the surface of nucleolus coincided with BrU signal in the logarithmically growing (Figure 1H and I) and the 0.05 µg/ml actinomycin D-treated (Figure 4G and H) cells. In the latter cells, much larger number of transcription foci were associated with the HSR at the nucleolar surface compared with the one at the nucleoplasm (Figure 4G and I), as seen in the logarithmically growing cells (Figure 1J). These data suggested that the HSR was transcribed by polymerase II or III at those locations.
Figure 4.
Nucleolar visits by the HSR are transcription-dependent. Logarithmically growing HSR-CFP cells were treated with 0 µg/ml (A, B), 0.05 µg/ml (C, D, G–I), or 2 µg/ml (E) of actinomycin D for 3 h and harvested. During the last 15 min before the harvest, BrU was added to all cultures. Incorporated BrU was detected by red fluorescence, and DNA was counterstained by DAPI (gray in A–E, blue in G, H). The slide was viewed by confocal microscopy, and the representative z-section images are shown (A–E, G, H). The location of the HSR, which was visualized by binding of LacR-CFP (pseudo-colored in green), was determined as in Figure 1, and is noted in the panels. The number of HSRs at each location as well as their percent of the total (in parentheses) is summarized in F. The co-localization of HSR (green) and BrU (red) among the images (left panel of G, H) was determined, and it was shown in right panel (indicated by arrowheads). The number of co-localization signals was counted for HSRs at each location, as in Figure 1J, and it was plotted in I. All bars in images indicate 2 µm.
The HSR was reversibly decondensed upon transcriptional induction
We next examined induced transcription in the HSR in live cells. For this, we introduced a mixture of pMS2-YFP DNA and either pTet-ON or pTet-OFF DNA to logarithmically growing HSR-CFP cells by electroporation. After 2.5 h, Doxycyclin (Dox) was added to the culture. In the pTet-ON-transfected cells, MS2 sequence-tagged RNA that was visualized by the binding of the MS2-YFP fusion protein appeared from the LacO sequence-tagged HSR that was visualized by the binding of LacR-CFP. Such visualization appeared to be independent from the HSR location, and the expression in the HSR located at the surface or interior of the nucleoli might be visualized (Figure 5A and B). We first chased a live cell to learn the chronological order of patterns of HSRs relative to RNAs. Before induction, the HSR visualized by LacR-CFP appeared as a compact sphere (Figure 5C, Type 1 cell). Once Dox was added, RNA visualized by the MS2-YFP protein appeared (Figure 5C, Type 2 cell). The HSR in Type 2 cell was still a compact sphere. Then, the HSR was loosened, and transcription proceeded further (Figure 5C, Type 3 cell). There were also cells in which the HSR was invisible, although expressed RNA was visible (Figure 5C, Type 4 cell), or both HSR and RNA were invisible, while the MS2-YFP protein was transiently expressed in the cell (Figure 5C, Type 5 cell). The frequency of these types of cells among the cells expressing MS2-YFP (Figure 5D) suggested that the cells underwent the transition from Type 1 to Type 4 in this order. It implied that the HSR was decondensed while it was transcribed, and transcription appeared to initiate throughout the HSR, which was in contrast to BSR gene expression observed from the HSR (Figures 1 and 2). Type 4 cells may represent a situation where the HSR was too decondensed and might not be visible any more. Such an idea was in line with the observation that the transcribed RNA was spread widely in Type 4 cells suggesting that the HSR might also be spread widely. Alternatively, it is also possible that extensive transcription prevents binding of LacR-CFP to its target sequence. On the other hand, FISH revealed that the HSR-CFP cell line contained about 3.2% of cells that had essentially no HSR (Figure 5E). Because the frequency was smaller than that of Type 5, the presence of Type 5 cells cannot be simply explained by cells without a HSR. It may be possible that Type 4 cells might be converted to Type 5 by stopping MS2 RNA transcription.
In pTet-OFF-introduced cells, the HSR was already decondensed at the time of Dox addition (Figure 5E), because transcription from the TRE promoter was already induced during a 2.5-h incubation after the electroporation of pTet-OFF. Addition of Dox to the culture resulted in the cessation of transcription and the rapid condensation of the HSR, because the frequency of Type 1 increased while the frequency of Types 2 and 3 decreased. These results showed that the HSR underwent reversible decondensation/recondensation upon transcriptional activation/cessation.
The heterochromatin marker remained at the decondensed HSR after transcriptional activation
As described above, the HSR used in this study showed several features typical to heterochromatin. Therefore, it was reasonable to observe that the HP-1α protein, which usually associates with constitutive heterochromatin, was detected at HSRs in interphase nuclei (Figure 6A). On the other hand, HP1α was not detected at HSRs in mitotic cells (Figure 6A), which was consistent with the report that HP1α is released from heterochomatin during mitosis (28,29). We have shown above that the HSRs underwent reversible decondensation upon transcriptional activation. We now found that the HP1α signal was associated with the decondensed HSR that was expressing RNA (Figure 6B). The signal almost entirely or partially overlapped with the HSR domain that appeared in lower or upper part of this panel, respectively. On the other hand, H3K9Me3 is another heterochomatin marker, and it was heavily detected at the HSR that was not transcriptionally activated (Figure 6C and D). Upon transcriptional activation, we found H3K9Me3 was still associated with the decondensed HSR that was expressing RNA. Interestingly, the distribution of H3K9Me3 inside the HSR was not homogeneous but patchy. It suggested that loss of the heterochromatin trait was partial, and it will be discussed.
Decondensation of HSR activated noninducible promoters located in cis
We next addressed the effect of HSR decondensation on the noninducible SRα promoters that drive the BSR genes in the HSR. We transfected pTet-OFF into HSR-CFP cells, cultured in the absence of Dox to activate the TRE promoter, and simultaneously detected HSR DNA and BSR RNA. We used FISH to detect HSR DNA, because BSR RNA was detectable only by FISH, which involved a hybridization step and dimmed the LacR-CFP fluorescence. As a result, we found that most HSRs had many BSR RNA signals (Figure 7A and B), which was in contrast with that observed previously (Figures 1 and 2). To examine this quantitatively, we isolated the cells that transiently expressed the electroporated DNA by a MACS cell separation system (see ‘Materials and Methods’ section). From the isolated cells, we extracted total DNA and total RNA and measured the amount of BSR sequence by real-time PCR. The result (Figure 7C) showed that the level of BSR RNA after the induction of the TRE promoter was more than two times higher than the level without induction, which suggested that the decondensation of the HSR by transcriptional activation of the TRE promoter resulted in activation of the noninducible SRα promoter located in cis. This can be explained if the silent BSR genes in the compact HSR domain became accessible to the protein required for its transcription.
DISCUSSION
By utilizing a long and homogeneous HSR made of an artificial plasmid array, we have obtained several findings regarding transcription in heterochromatin. First, the noninducible SRα promoter in the HSR was active at only a few specific points, as specific RNA appeared as foci in the HSR domain (Figures 1 and 2), and RNA polymerase II appeared to be responsible for such transcription (Figure 4G–I). We tentatively assumed that SRα promoter drove the BSR RNA transcription. However, it is possible that the RNA was transcribed from another sequence, because recent paper suggests multiple transcription start sites from heterochromatic satellite DNA sequences (7). It may be related to our previous finding that the generation of HSR from the IR/MAR plasmid was affected by the transcription that was not driven by the plasmid-encoded promoter (15). In any case, our data suggested that HSR was transcribed at several specific points by RNA polymerase II/III (Figure 4G–I). As described in the ‘Introduction’ section, many reports addressed the role of RNA polymerase II transcription in heterochromatin to maintain its silenced state. However, detection of specific transcription points in transgene array by in situ detection of transcribed RNA has not been reported. One of the reasons for this should be that making large and homogeneous transgene array had been very difficult until our establishment of IR/MAR plasmid-mediated gene amplification technology. If short transgene array was used, in situ detection would resulted in only overlapping signals for DNA and RNA. Lu et al. (7) detected the specific transcript from the pericentric satellite repeat by RNA FISH. They showed that the RNA signal appeared as foci at the surface of the pericentric heterochromatin, which appears similar to the result shown in this report. Thus, our argument that the heterochromatin was transcribed at a specific point may be a general feature of heterochromatin. The HSR used in this study was composed of a homogeneous plasmid array (12). Therefore, the transcriptional specific point may not appear genetically, and some kind of epigenetic mechanism may determine the location of this point. Our results showed that the number of points appeared to double after replication of the HSR itself. One idea is that the specific point for transcription arises at the junction between the heterochromatin and the normal chromosome arm. Identifying the origin and nature of the specific points in the heterochromatin should be an important future task.
We have shown that the HSR might localize at the nucleolar surface or the nucleolar interior, and that the HSRs at such locations were transcribed more actively than the one at the nucleoplasm. In general, the perinucleolar region is a well-known heterochromatin-rich nuclear domain, and many kinds of constitutive or facultative heterochromatin reside there. For example, a recent report showed that the inactivated X chromosome in female somatic cells periodically visited the surface of the nucleoli during mid- to late S-phase (10). Furthermore, it was previously suggested that a small HSR, which was made of transgenes using a different method from that used in this study, were in close proximity to the nucleoli in S-phase cells (30). Consistent with these reports, our current study showed that the HSR at the nucleolar surface appeared most frequently during late S-phase. It has been shown previously that the ‘perinucleolar component (PNC)’ was transcribed by RNA polymerase II or III, and it was suggested that the PNC might be involved in some kind of RNA metabolism (31). This is consistent with our result that RNA polymerase II/III transcription was required for the HSR to locate at the nucleolar surface. As we have described, many recent reports underscore the important role of RNA polymerase II transcription for heterochromatin maintenance. Therefore, transcription at the nucleolar surface might have an implication for heterochromatin maintenance. Localization of satellite DNA inside the nucleolus has been reported repeatedly, although the physiological implication of this observation was not known (32–34). We have shown that localization of the HSR to the nucleolar interior was most frequent during early S-phase, and it required RNA polymerase I transcription. Therefore, the physiological implication of HSR localization to the interior of the nucleolus may be different from that at the nucleolar surface, because the timing of localization as well as the responsible RNA polymerase was different. Therefore, clarifying the physiological meaning of this localization will be an important future task.
If an inducible promoter in the HSR was strongly activated, the entire HSR was loosened, and transcription appeared throughout the HSR. This reproduces the findings of previous reports, in which a transgene array was decondensed by the binding of the VP16 acidic activation domain (AAD) to the target sequences in the array (35). These authors also showed that the same domain fused to rtTA, which was expressed from the pTet-ON plasmid used in this study, also decondensed the target chromatin (25,36). We further showed in this paper that the process was reversible. On the other hand, an apparent discrepancy arose because the above paper showed that HP1α was depleted from the array upon induction (25), whereas our data showed that it at least partially remained at the HSR. However, this might be explained by differences in the size of the constructs used in the two studies. They used an array composed of 200 copies of pECMS2β plasmid sequences (25), whereas the HSR in HSR-CFP cells contained a few thousands copies of plasmid sequence. On the other hand, we have shown that the H3K9Me3 signal remained at the HSR as a patchy appearance (Figure 6C–F). Therefore, our data on HP1α and H3K9Me3 most likely suggests that there still remained heterochromatin in our large HSR even after the loosening of entire HSR domain. Inside such HSR, transcription was activated at many specific points, where the heterochromatin trait may be removed as suggested before (25). The idea was consistent with data in Figure 7 namely, during the induction of TRE promoter, the noninducible promoters in the HSR also were activated. This suggests that the promoter might be activated if the chromatin was loosened and if the transcription factors were accessible. Importantly, the transcript appeared as many discrete foci, which suggests activation of many transcriptional specific points. On the other hand, transcript detected by MS2-YFP-binding appeared homogeneously in the HSR domain (Figures 5 and 6), which may contradict with above idea. However, high background of this detection method might smoothen the patchy appearance. Taken together, upon transcriptional activation, the HSR appeared to be loosened and transcribed at many specific points inside large heterochromatin. How and why such specific points may arise will be an important aspect of future work on this topic.
FUNDING
A Grant-in-Aid for Scientific Research (B) (17370002 to N.S.) from the Japan Society for the Promotion of Science; and a Grant-in-Aid for Scientific Research on Priority Areas—Nuclear dynamics (19038016 to N.S.) from the Ministry of Education, Science, Sports and Culture of Japan. Funding for open access charge: Ministry of Education, Science, Sports and Culture of Japan (19038016).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr. Susan M. Janicki and Dr. David L. Spector at Cold Spring Harbor Laboratory for their kind gifts of pECMS2β pLacR-CFP, and pMS2-YFP.
REFERENCES
- 1.Dillon N, Festenstein R. Unravelling heterochromatin: competition between positive and negative factors regulates accessibility. Trends Genet. 2002;18:252–258. doi: 10.1016/s0168-9525(02)02648-3. [DOI] [PubMed] [Google Scholar]
- 2.Trojer P, Reinberg D. Facultative heterochromatin: is there a distinctive molecular signature? Mol. Cell. 2007;28:1–13. doi: 10.1016/j.molcel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 3.Huisinga KL, Brower-Toland B, Elgin SC. The contradictory definitions of heterochromatin: transcription and silencing. Chromosoma. 2006;115:110–122. doi: 10.1007/s00412-006-0052-x. [DOI] [PubMed] [Google Scholar]
- 4.Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 6.Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA interference guides histone modification during the S phase of chromosomal replication. Curr. Biol. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu J, Gilbert DM. Proliferation-dependent and cell cycle regulated transcription of mouse pericentric heterochromatin. J. Cell Biol. 2007;179:411–421. doi: 10.1083/jcb.200706176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, Nakayama T, Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- 9.Ho CY, Murnane JP, Yeung AK, Ng HK, Lo AW. Telomeres acquire distinct heterochromatin characteristics during siRNA-induced RNA interference in mouse cells. Curr. Biol. 2008;18:183–187. doi: 10.1016/j.cub.2007.12.059. [DOI] [PubMed] [Google Scholar]
- 10.Zhang LF, Huynh KD, Lee JT. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell. 2007;129:693–706. doi: 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]
- 11.Shimizu N, Miura Y, Sakamoto Y, Tsutsui K. Plasmids with a mammalian replication origin and a matrix attachment region initiate the event similar to gene amplification. Cancer Res. 2001;61:6987–6990. [PubMed] [Google Scholar]
- 12.Shimizu N, Hashizume T, Shingaki K, Kawamoto JK. Amplification of plasmids containing a mammalian replication initiation region is mediated by controllable conflict between replication and transcription. Cancer Res. 2003;63:5281–5290. [PubMed] [Google Scholar]
- 13.Shimizu N, Shingaki K, Kaneko-Sasaguri Y, Hashizume T, Kanda T. When, where and how the bridge breaks: anaphase bridge breakage plays a crucial role in gene amplification and HSR generation. Exp. Cell Res. 2005;302:233–243. doi: 10.1016/j.yexcr.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Shimizu N, Hanada N, Utani K, Sekiguchi N. Interconversion of intra- and extra-chromosomal sites of gene amplification by modulation of gene expression and DNA methylation. J. Cell. Biochem. 2007;102:515–529. doi: 10.1002/jcb.21313. [DOI] [PubMed] [Google Scholar]
- 15.Hashizume T, Shimizu N. Dissection of mammalian replicators by a novel plasmid stability assay. J. Cell. Biochem. 2007;101:552–565. doi: 10.1002/jcb.21210. [DOI] [PubMed] [Google Scholar]
- 16.Bosisio D, Marazzi I, Agresti A, Shimizu N, Bianchi ME, Natoli G. A hyper-dynamic equilibrium between promoter-bound and nucleoplasmic dimers controls NF-kappaB-dependent gene activity. EMBO J. 2006;25:798–810. doi: 10.1038/sj.emboj.7600977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diefenbacher M, Sekula S, Heilbock C, Maier JV, Litfin M, van Dam H, Castellazzi M, Herrlich P, Kassel O. Restriction to Fos family members of Trip6-dependent coactivation and glucocorticoid receptor-dependent trans-repression of activator protein-1. Mol. Endocrinol. 2008;22:1767–1780. doi: 10.1210/me.2007-0574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimizu N, Misaka N, Utani K. Nonselective DNA damage induced by a replication inhibitor results in the selective elimination of extrachromosomal double minutes from human cancer cells. Genes, Chrom., Cancer. 2007;46:865–874. doi: 10.1002/gcc.20473. [DOI] [PubMed] [Google Scholar]
- 19.Utani K, Kawamoto JK, Shimizu N. Micronuclei bearing acentric extrachromosomal chromatin are transcriptionally competent and may perturb the cancer cell phenotype. Mol. Cancer Res. 2007;5:695–704. doi: 10.1158/1541-7786.MCR-07-0031. [DOI] [PubMed] [Google Scholar]
- 20.Shimizu N, Shingaki K. Macroscopic folding and replication of the homogeneously staining region in late S phase leads to the appearance of replication bands in mitotic chromosomes. J. Cell Sci. 2004;117:5303–5312. doi: 10.1242/jcs.01414. [DOI] [PubMed] [Google Scholar]
- 21.Garrick D, Fiering S, Martin DI, Whitelaw E. Repeat-induced gene silencing in mammals. Nat. Genet. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- 22.Henikoff S. Conspiracy of silence among repeated transgenes. Bioessays. 1998;20:532–535. doi: 10.1002/(SICI)1521-1878(199807)20:7<532::AID-BIES3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 23.McBurney MW, Mai T, Yang X, Jardine K. Evidence for repeat-induced gene silencing in cultured Mammalian cells: inactivation of tandem repeats of transfected genes. Exp. Cell Res. 2002;274:1–8. doi: 10.1006/excr.2001.5443. [DOI] [PubMed] [Google Scholar]
- 24.Shimizu N, Kanda T, Wahl GM. Selective capture of acentric fragments by micronuclei provides a rapid method for purifying extrachromosomally amplified DNA. Nat. Genet. 1996;12:65–71. doi: 10.1038/ng0196-65. [DOI] [PubMed] [Google Scholar]
- 25.Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, et al. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimizu N, Ochi T, Itonaga K. Replication timing of amplified genetic regions relates to intranuclear localization but not to genetic activity or G/R band. Exp. Cell Res. 2001;268:201–210. doi: 10.1006/excr.2001.5286. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka T, Shimizu N. Induced detachment of acentric chromatin from mitotic chromosomes leads to their cytoplasmic localization at G1 and the micronucleation by lamin reorganization at S phase. J. Cell Sci. 2000;113:697–707. doi: 10.1242/jcs.113.4.697. [DOI] [PubMed] [Google Scholar]
- 28.Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 29.Terada Y. Aurora-B/AIM-1 regulates the dynamic behavior of HP1alpha at the G2-M transition. Mol. Boil. Cell. 2006;17:3232–3241. doi: 10.1091/mbc.E05-09-0906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li G, Sudlow G, Belmont AS. Interphase cell cycle dynamics of a late-replicating, heterochromatic homogeneously staining region: precise choreography of condensation/decon densation and nuclear positioning. J. Cell Biol. 1998;140:975–989. doi: 10.1083/jcb.140.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang S, Deerinck TJ, Ellisman MH, Spector DL. The perinucleolar compartment and transcription. J. Cell Biol. 1998;143:35–47. doi: 10.1083/jcb.143.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rae MM, Franke WW. The interphase distribution of satellite DNA-containing heterochromatin in mouse nuclei. Chromosoma. 1972;39:443–456. doi: 10.1007/BF00326177. [DOI] [PubMed] [Google Scholar]
- 33.Jacob J, Gillies K, Macleod D, Jones KW. Molecular hybridization of mouse satellite DNA-complementary RNA in ultrathin sections prepared for electron microscopy. J. Cell Sci. 1974;14:253–261. doi: 10.1242/jcs.14.2.253. [DOI] [PubMed] [Google Scholar]
- 34.Demirtas H, Candemir Z, Cucer N, Imamoglu N, Donmez H, Bokesoy I. Essay on the nucleoli survey by the alpha- and beta-satellite DNA probes of the acrocentric chromosomes in mitogen-stimulated human lymphocytes. Annales de genetique. 2000;43:61–68. doi: 10.1016/s0003-3995(00)01014-5. [DOI] [PubMed] [Google Scholar]
- 35.Tumbar T, Sudlow G, Belmont AS. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J. Cell Biol. 1999;145:1341–1354. doi: 10.1083/jcb.145.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsukamoto T, Hashiguchi N, Janicki SM, Tumbar T, Belmont AS, Spector DL. Visualization of gene activity in living cells. Nat. Cell Biol. 2000;2:871–878. doi: 10.1038/35046510. [DOI] [PubMed] [Google Scholar]