Abstract
Human DNA polymerase δ (Pol δ4), a key enzyme in chromosomal replication, is a heterotetramer composed of the p125, p50, p68 and p12 subunits. Genotoxic agents such as UV and alkylating chemicals trigger a DNA damage response in which Pol δ4 is converted to a trimer (Pol δ3) by degradation of p12. We show that Pol δ3 has altered enzymatic properties: it is less able to perform translesion synthesis on templates containing base lesions (O6-MeG, 8-oxoG, an abasic site or a thymine-thymine dimer); a greater proofreading activity; an increased exonuclease/polymerase activity ratio; a decreased tendency for the insertion of wrong nucleotides, and for the extension of mismatched primers. Overall, our findings indicate that Pol δ3 exhibits an enhanced ability for the detection of errors in both primers and templates over its parent enzyme. These alterations in Pol δ3 show that p12 plays a major role in Pol δ4 catalytic functions, and provides significant insights into the rationale for the conversion of Pol δ4 to Pol δ3 in the cellular response to DNA damage.
INTRODUCTION
Faithful replication of chromosomal DNA and DNA damage repair are essential for preserving genomic integrity. Replicative DNA polymerases exhibit high fidelity and are able to proofread errors with an intrinsic 3′ to 5′ exonuclease activity (1,2). Structural, kinetic and mechanistic studies have led to models for how replicative DNA polymerases achieve extremely low error rates. This is achieved by the selectivity of the polymerase (pol) activity for nucleotide insertion, and also by proofreading and excision of mismatched nucleotides by the 3′ to 5′ exonuclease (exo) activity (3–5). Shuttling of DNA from the exo to pol catalytic sites is also a key factor in their co-operative actions that lead to efficient proofreading (2,6,7). The balance between exo and pol reaction rates (exo/pol ratio) also plays a key role in proofreading, as alterations in the exo/pol ratio by mutations can either decrease or increase the fidelity of polymerases (8,9).
There are two proofreading DNA polymerases in eukaryotic cells, Pol δ and Pol ε, which are responsible for the replication of genomic DNA (10). Human Pol δ possesses four subunits: p125, which harbors the pol and exo active sites; p50, which is tightly associated with the p125 subunit; p68, which associates with p50; and p12, which binds to both p125 and p50. p12 interacts with both the p50 and p125 subunits, may stabilize Pol δ subunit assembly, and play a role in the interaction of Pol δ with PCNA (11). The latter is a DNA sliding clamp that promotes processive DNA synthesis by Pol δ (10). The maintenance of genomic integrity requires that cells repair DNA damage and mutations. Pol δ also participates in DNA repair as a gap-filling polymerase (10). The functions of Pol δ are thus important not only in replication, but also in understanding how it responds to encounters with DNA damage that can lead to the introduction of mutations.
DNA base damage arises during the course of normal cellular processes or by environmental exposure to genotoxic agents such as UV light, alkylating and oxidizing agents. Two of the most common and extensively studied base lesions are O6-MeG (O6-methylguanine), which is produced by alkylating agents, and 8-oxoG (7,8-dihydro-8-oxoguanine), which is produced by reactive oxygen species. These lesions are highly mutagenic as they are readily bypassed by eukaryotic polymerases (12–14). Bases modified by bulky adducts, cross-linked bases such as thymine-thymine dimers, and abasic sites represent severe blocks to many replicative polymerases (12,15,16).
Mammalian cells respond to DNA damage by a host of defense mechanisms that include activation of cell-cycle checkpoints and DNA repair pathways (17,18). Cells are particularly vulnerable during S-phase, because of the encounter of replisomes with damaged DNA. This can lead to mutations if the replicative polymerases perform translesion (TLS) synthesis and introduce errors, or replication fork stalling if the polymerases are unable to bypass the lesions. Events that lead to prolonged stalling of replication forks result in replication fork collapse, formation of aberrant fork structures, chromosome damage or loss and ultimately cell death (18). To avoid stalling, these lesions can be bypassed by the DNA damage tolerance pathway that requires the ubiquitination of PCNA (19). This pathway allows the bypass of lesions by specialized TLS polymerases that are error-prone, so that bypass carries the risk of introduction of errors, depending on the lesion and the TLS polymerase involved (19,20). TLS synthesis requires a polymerase switch, in which the TLS polymerase exchanges places with Pol δ (or Pol ε), and after limited synthesis past the lesion, again switches with Pol δ to allow resumption of DNA replication (21,22).
Recently, we discovered a novel cellular response to DNA damage, in which exposure of mammalian cells to UV light or alkylating agents such as methyl methane sulfonate leads to the rapid degradation of the p12 subunit of Pol δ. Pol δ is consequently converted from a heterotetramer (Pol δ4) to a trimer lacking the p12 subunit (Pol δ3). This response is dependent on the ATR/Chk1 checkpoint kinases and a functional ubiquitination pathway (23). These observations lead to the hypothesis that the conversion of Pol δ4 to Pol δ3 may contribute to the cellular response to DNA damage, and that this is exerted through alterations in Pol δ activity or behavior (23). Given the number of DNA transactions in which Pol δ may be involved, there are a number of possible outcomes of the alteration in its quaternary structure. In this study, we have tested the hypothesis that Pol δ3 exhibits altered properties when it encounters DNA base lesions. Our studies show that Pol δ3 exhibits a decreased ability for TLS synthesis. Pol δ3 exhibits reduced proclivities for the insertion of mismatched nucleotides, and for the extension of mismatched primer termini. Overall, the behavior of Pol δ3 leads to the conclusions that it is more discriminatory than Pol δ4 in assays that test their abilities to recognize aberrant primer/templates, to insert correctly based-paired nucleotides, and to recognize mismatched primer termini. These findings provide support for the hypothesis that Pol δ3 has altered properties that may be beneficial to the cell when it is subjected to DNA damage.
MATERIALS AND METHODS
Immunoaffinity purification of native Pol δ4 and Pol δ3 from HeLa cells
Hela cells (3 × 108) were divided into two equal portions. One half was treated with UV (20 J/m2) and harvested 4 h later. Pol δ3 and Pol δ4 were isolated from the UV-treated and untreated cells, respectively, by immunoaffinity chromatography on anti-p125 agarose, essentially as described for their isolation from HEK 293T cells (23). Pol δ3 activity in the peak fraction was 45% that of the peak fraction of Pol δ4. The fractions were western blotted for the p125, p50, p68 and p12 subunits to confirm that p12 was absent from the Pol δ3 from the UV-treated cells (23). The amounts of Pol δ4 and Pol δ3 protein in the preparations were estimated based on the specific activities determined for the purified recombinant enzymes.
Expression and purification of human DNA Pol δ
Recombinant human Pol δ4 and Pol δ3 were expressed in Sf9 insect cells and purified by immunoaffinity chromatography on immobilized anti-p125, followed by FPLC on Mono Q ion exchange columns essentially as previously described (11). The preparations of Pol δ4 and Pol δ3 used in these studies had specific activities of 21 000 U/mg protein and 9400 U/mg protein, respectively. Pol δ activity was assayed on poly(dA)/oligo(dT) as described previously (11). The Pol δ trimer lacking the p68 subunit (Pol δ3-p68) was also prepared. Exonuclease-deficient forms of Pol δ4 and Pol δ3 were produced by mutation of D402 of p125 to alanine, and purified as described for the wt enzymes. In mice, the mutation of the cognate residue, D400, to alanine, leads to loss of exonuclease function (24). Protein concentrations of p125 were determined by in-gel analysis with catalase as a protein standard. A baculovirus expressing his-tagged human pol η was a generous gift from Dr. P. Guengerich (Vanderbilt University School of Medicine, Nashville, TN). Pol η was expressed in Sf9 cells and purified on Ni–NTA columns.
DNA substrates
Unmodified DNA oligonucleotides were synthesized and PAGE-purified by Integrated DNA Technologies, Inc. (Coralville, IA); modified oligonucleotides were synthesized and RP-HPLC purified by Midland Certified Reagents (Midland, TX). Oligonucleotides were end-labeled with [γ-32P]ATP (5000 Ci/mmol, MP Biochemicals) and T4 polynucleotide kinase (New England Biolabs, MA). The 5′-32P labeled primers were annealed with templates in a ratio of 1:1.2. A 25-mer primer (5′-GCCACTACAGCACCTTGACAGCCAG-3′) was annealed to 40-mer oligonucleotides (5-TCATCGGTCGCATCXCTGGCTGTCAAGGTGCTGTAGTGGC-3′) where X = G, O6-MeG, 8-oxoG, or modified tetrahydrofuran (AP site). The 26-mer primer used for mismatch extension assays was the 25-mer with an additional T at the 3′ end. A 5′-end labeled 24-mer (5′-GCCTCGAGCCAGCCGCAGACGCAG-3′) was used as a ssDNA substrate, and annealed to a 36-mer template (5′-TCGGCGTCCTCGCTGCGTCTGCGGCTGGCTCGAGGC-3′) for use as a dsDNA substrate for assays of 3′ to 5′ exonuclease activity. A 40-mer template containing a cis-syn thymine–thymine dimer at positions 26 and 27 (5′-AAACAACAATGACTTGGGGCTGTCAAGGTGCTGTAGTGTC-3′) was synthesized by TriLink Biotechnologies (San Diego, CA). The unmodified 40-mer and 25-mer, 26-mer, 27-mer primers were: 25-mer, 5′-GACACTACAGCACCTTGACAGCCCC-3′; 26A primer, 5′-GACACTACAGCACCTTGACAGCCCCA-3′; 26G primer, 5′-GACACTACAGCACCTTGACAGCCCCG-3′; 27AA primer, 5′-GACACTACAGCACCTTGACAGCCCCAA-3′; 27GG primer, 5′-GACACTACAGCACCTTGACAGCCCCGG-3′. The bolded nucleotides represent the residues that would be opposite the thymine–thymine dimer.
Pol δ assays on oligonucleotide primer/templates
The 5′-end labeled 25-mer primer annealed to unmodified or modified templates were preincubated for 3 min at room temperature with Pol δ and PCNA. Reactions (20 μl) were initiated by addition of a mixture containing all four dNTPs and MgCl2. Final concentrations of reactants were 500 μM dNTP, 10 mM MgCl2, 50 mM Bis–Tris–HCl pH 6.5, 5 mM dithiothreitol, 200 μg/ml BSA, 30 mM NaCl, 150 nM primer/template, 400 nM PCNA and the indicated amounts of Pol δ. Reactions were performed at 37°C for 5.5 min or the indicated times. Aliquots (8 μl) were quenched with 16 μl of 95% formamide/25 mM EDTA, and heated at 90°C for 3 min. Products were separated by electrophoresis on 16% polyacrylamide gels containing 8 M urea. Radioactive products were visualized by phosphorimaging and analyzed with ImageQuant software (Amersham Biosciences, NJ).
Nucleotide incorporation and mismatched primer extension assays
The 25-/40-mer primer/templates (200 nM) were extended with 5 nM recombinant Pol δ enzymes at 37°C, in the presence of 400 nM PCNA, 10 mM MgCl2 and 500 μM correct (dCTP) or incorrect nucleotides. Other reaction components were the same as for the extension reactions described above. Mismatched primer (T:G) extension assays were carried out under the same conditions, except that 250 nM DNA and 2 nM Pol δ enzymes were used, and only the next correct nucleotide (dGTP) was provided.
3′ to 5′ exonuclease assays
Exonuclease assays were performed in the same reaction buffer as described above for the extension assays. Each reaction contained 4 nM polymerases, 100 nM 24-mer ssDNA or 24-/36-mer dsDNA (partially single stranded); MgCl2 was added to initiate reactions. Reactions were carried out at 22°C. Products were separated by 16% urea denaturing gels and visualized by phosphorimaging. The concentration of dNMP that was generated in order to attain the product distribution found was calculated. To this end, the radioactivity of each band was determined as a fraction of the total radioactivity in its lane, and used to calculate its concentration based on the starting primer concentration. This concentration was multiplied by the number of exonucleolytic cleavages needed to generate such a product length, and the sum of all calculations represents the concentration of dNMP that is generated in order to attain the product distribution found.
RESULTS
Native Pol δ3 produced by UV damage exhibits an increased discrimination against bypass synthesis on templates containing base lesions
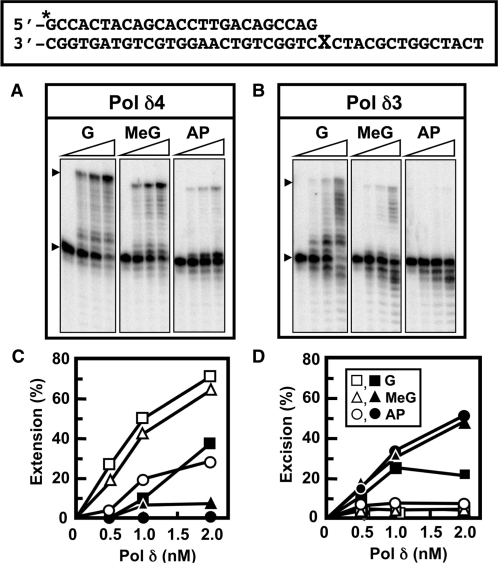
Native forms of Pol δ holoenzyme (Pol δ4) and Pol δ3 were isolated by immunoaffinity chromatography from control and UV-treated HeLa cells, respectively (see ‘Materials and methods’ section). Their abilities to perform TLS synthesis were compared on oligonucleotide primer/templates consisting of a 25-mer end-labeled primer annealed to a 40-mer template that carried O6-MeG or an AP (abasic) site at nt 26 (Figure 1, ‘Materials and methods’ section). We chose O6-MeG as an example of a lesion that is readily bypassed by replicative polymerases in vitro, including Pol δ (25,26). O6-MeG is highly mutagenic because T is often inserted opposite to it by DNA polymerases (13). The AP site was selected as one that represents a severe block to replicative polymerases. Replicative DNA polymerases stall at AP sites after the insertion of a single base, usually A, a phenomenon known as the ‘A’ rule (27).
Figure 1.
Pol δ3 has a reduced ability for the bypass of template lesions when compared to Pol δ4. Pol δ4 and Pol δ3 were prepared by immunoaffinity chromatography from control and UV-treated HeLa cells (see ‘Materials and methods’ section) and assayed for their activities on oligonucleotide primer templates (upper box) containing G, O6-MeG (MeG) or an AP site at position 26 (‘X’). Reactions contained 150 nM 5′-[32P]end-labeled 25-/40-mer primer/template, 400 nM PCNA and 500 μM of each dNTP (see ‘Materials and methods’ section). Products were resolved on 16% denaturing gels and quantitated by phosphorimaging. (A, B) Phosphorimages of the reaction products formed by Pol δ4 and Pol δ3, respectively. For each panel the lanes represent the products formed by 0, 0.5, 1 or 2 nM enzyme. Arrowheads mark the positions of the primer and the full-length 40-mer product. (C, D) Plots of the total amounts of extension products (>25-mer) and excision products (<25-mer), respectively. Products were quantitated and expressed as percentages of the starting primer. Key for symbols [inset, (D)]: template with X = G (squares); O6-MeG (triangles); AP site (circles). Data for Pol δ4 are shown as open symbols and for Pol δ3 as solid symbols.
Pol δ4 readily bypassed the O6-MeG lesion as has been previously observed (26) (Figure 1A and C). Pol δ4 was stalled at the AP site after insertion of a single nt, forming the 26-mer or P+1 (primer+1) product. This behavior is typical of replicative DNA polymerases, including Pol δ, which usually insert an A across the AP site (25,27). Nevertheless, at the highest Pol δ4 concentration (2 nM), ca. 30% of the primer was extended (Figure 1C). In contrast to the unmodified template, Pol δ4 produced a small but detectable increase in exonucleolytic products (smaller than the 25-mer primer) on the O6-MeG and the AP substrates, a result of the stalling of the polymerase and an increased idling reaction (7).
Pol δ3 behaved in a strikingly different manner from Pol δ4, as can be seen from the distribution and amounts of products on the gels. Pol δ3 produced fewer full-length products, and more products of intermediate size than Pol δ4 (Figure 1B). This indicates a less processive behavior or a greater tendency for dissociation from the primer/template. When Pol δ3 encountered the O6-MeG lesion, product formation was decreased (Figure 1B and C). Thus, Pol δ3 exhibits a reduced ability for TLS synthesis past the O6-MeG lesion. Pol δ3 was unable to bypass the AP site at the enzyme concentrations tested. Coupled with this was a very marked increase in exonucleolytic products (<25mer) on all templates by comparison with Pol δ4 (Figure 1B and D). This increase in formation of exonucleolytic products on these model templates is indicative of increased proofreading or idling reactions (28).
These findings are highly significant, as they indicate that Pol δ activity is profoundly altered by the loss of the p12 subunit. Pol δ3 exhibited a markedly increased production of exonucleolytic products, a marked increase in intermediate products that indicated a tendency for stalling, and was less effective in the bypass of template lesions.
TLS synthesis by recombinant Pol δ enzymes
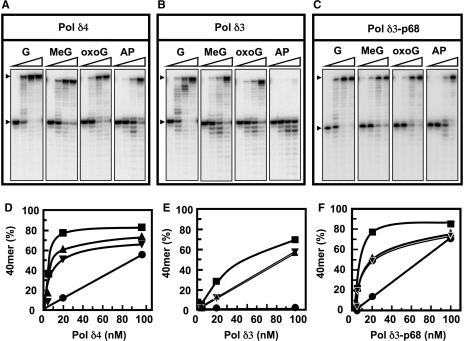
In order to obtain rigorous confirmation of the findings made with the HeLa Pol δ enzymes and to further investigate the alterations in Pol δ3 activity, we turned to the use of recombinant human Pol δ4 and Pol δ3 purified to near-homogeneity (‘Materials and methods’ section). Their activities on lesion-containing templates were examined under the same conditions as for the HeLa enzymes, except that higher enzyme concentrations were used (4, 20 and 100 nM). We also included a template with the 8-oxoG lesion, which is highly mutagenic as it leads to insertion of both C and A to generate G:A mispairs that result in G:T transversion mutations (14,29). 8-oxoG has also been shown to be bypassed by calf thymus Pol δ (30).
Recombinant Pol δ4 and Pol δ3 behaved in the same way as the HeLa enzymes. Pol δ3 produced more exonucleolytic products than Pol δ4 on all the templates (Figure 2A and B). Pol δ3 produced fewer extension products than Pol δ4 on the unmodified and the O6-MeG and 8-oxoG templates, as can be seen from the examination of the products formed at the two lower enzyme concentrations (Figure 2A and B) and also in the graphical plots (Figure 2D and E). With the higher enzyme concentrations, most of the primer was extended, so that changes were less obvious. The amounts of full-length extension products produced by 4 nM Pol δ4 on the unmodified, O6-Me-G, and 8-oxoG templates were 27, 17 and 8.3%, respectively, and those produced by Pol δ3 were 5.9, 2.5 and 2.4%, respectively. The results obtained for the 8-oxoG template were very similar to those obtained for the O6-MeG template. Pol δ3 did not bypass the AP lesion, and the amounts of P+1 or full-length product were negligible even at 100 nM enzyme (Figure 2B and E). In contrast, Pol δ4 was able to extend most of the primer past the AP lesion to the full-length product at 100 nM enzyme concentration (Figure 2A and D).
Figure 2.
Translesion synthesis by recombinant Pol δ. Recombinant Pol δ4, Pol δ3 and the Pol δ trimer lacking the p68 subunit (Pol δ3-p68) were purified to near-homogeneity (see ‘Materials and methods’ section). Translesion synthesis was assayed using 25-/40-mer templates, where nt 26 = G in the unmodified template, and the modified templates where nt 26 = O6-MeG, 8-oxoG or an AP site under the same conditions as described for Figure 1. Polymerase concentrations used were 0, 4, 20 and 100 nM for the four lanes in each panel. (A–C) Phosphorimages of the reaction products formed by Pol δ4, Pol δ3 and Pol δ3-p68, respectively. (D–F) The amounts of full-length 40-mer products produced by Pol δ4, Pol δ3, and Pol δ3-p68, respectively, were plotted as percentage of the starting 25-mer primer against enzyme concentration. The arrowheads mark the positions of the 40-nt full-length product and the 25-nt primer.
We also analyzed the Pol δ trimer that lacks the p68 subunit (Pol δ3-p68). Pol δ3-p68 behaved in a similar manner to Pol δ4 (Figure 2C and F). These data show that the differences in properties of Pol δ3 with regard to TLS synthesis that we observed are specifically associated with absence or presence of the p12 subunit.
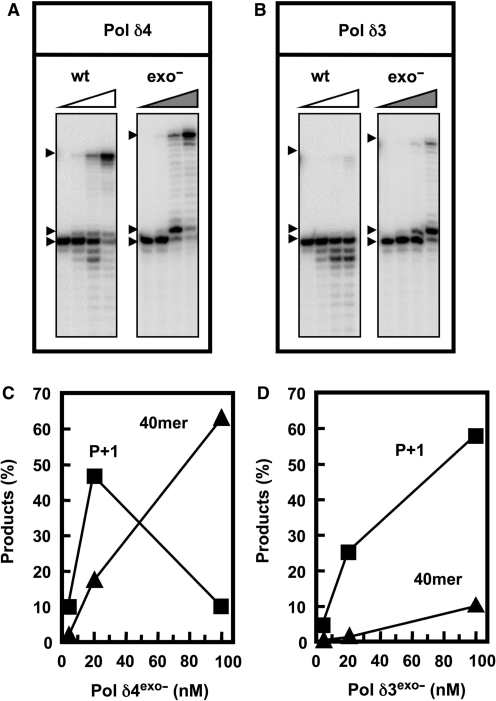
Analysis of the bypass of the AP lesion by exonuclease-deficient mutants of Pol δ4 and Pol δ3
Exonuclease-deficient forms of Pol δ were obtained by mutation of the p125 subunit (see ‘Materials and methods’ section). Examination of the bypass of the AP template by wt and exonuclease-deficient Pol δ4 and Pol δ3 revealed that the exonuclease activity plays a key role in the differences in product formation on the AP template that were observed. There was an increased accumulation of the P+1 product, and a increased formation of the full-length 40-mer by Pol δ4exo− when compared to wt Pol δ4 (Figure 3A). Quantitation of the amounts of P+1 and 40-mer products formed by Pol δ4exo− showed that the amounts of P+1 initially increased, then decreased with increasing enzyme concentration as the accumulating P+1 products were extended to the full-length 40-mer (Figure 3C). At 100 nM enzyme, most of the P+1 product was extended to the full length 40-mer (Figure 3A, and C).
Figure 3.
Bypass synthesis on AP containing templates by wt and exonuclease-deficient forms of Pol δ. The recombinant Pol δ4 and Pol δ3 enzymes containing the D402A mutants in the p125 subunit were used (see ‘Materials and methods’ section). These exonuclease-deficient forms of Pol δ were assayed for their abilities to bypass the AP lesion as described in Figure 2; enzyme concentrations were 0, 4, 20 and 100 nM. (A, B) Phosphorimages of the reaction products produced by Pol δ4exo– and Pol δ3exo–, respectively. Arrowheads show the position of the 40-mer, the P+1 product (26-mer) and the primer (25-mer). (C, D) The amounts of the P+1 (squares) and full-length extension products (triangles) were plotted against enzyme concentrations for Pol δ4exo– and Pol δ3exo–, respectively.
In the case of Pol δ3, very little P+1 product was seen with the wt enzyme. Loss of the exonuclease activity now allowed the accumulation of the P+1 product, and a small amount of full-length product was now observed (Figure 3B). Quantitation of the P+1 and 40-mer products formed by Pol δ3exo− showed that P+1 levels continued to rise with increasing enzyme concentration, to a level of ca. 60% of the starting primer at 100 nM enzyme (Figure 3D). Thus, apparent absence of the P+1 product with the wild-type Pol δ3 can be attributed to its more rapid removal by the exonuclease activity. In addition, since Pol δ3 exo− was much less able to extend the P+1 product than Pol δ4 exo– (Figure 3D) so it can be concluded that there is an altered behavior of the polymerase activity per se. Thus, the analyses of the exonuclease-deficient mutants provide evidence that the altered behavior of Pol δ3 when it encounters base lesions are due to alterations in the polymerase activity, in addition to the observed increase in proofreading.
The experiments described above provide an analysis of the accumulation of products generated by the combined actions of the polymerase and exonuclease activities as a function of enzyme concentration. In order to examine the rates of formation of extended primers we carried out time-course experiments and determined the rates of formation of extended primers for Pol δ4 and Pol δ3 as well for their exonuclease-deficient forms (Table 1). The data were normalized to the activity of each enzyme on the unmodified template. This analysis allowed for a comparison of the activities of Pol δ4 and Pol δ3 on the O6-MeG and 8-oxoG templates. Pol δ4 exhibited lower rates of activity on these templates by comparison with the unmodified template, both in terms of total product formation and formation of the full-length 40-mer (Table 1, first column). Pol δ3 also exhibited lower rates with the O6-MeG and 8-oxoG templates when compared to the unmodified template, but in both instances the rates were significantly slower by comparison with the data for Pol δ4. These results indicate that Pol δ3 bypasses these two template lesions more slowly than does Pol δ4.
Table 1.
Relative Rates of Primer Extension by Pol δ4 and Pol δ3a
| Product | Pol δ4 wt | Pol δ3 wt | Pol δ4exo– | Pol δ3exo– |
|---|---|---|---|---|
| G template: | ||||
| >25mer | 100 | 100 | 100 | 100 |
| 40mer | 47 | 26 | 41 | 21 |
| O6-MeG template: | ||||
| >25mer | 54 | 30 | 65 | 54 |
| 40mer | 28 | 10 | 25 | 9 |
| 8-oxoG template: | ||||
| >25mer | 56 | 35 | 57 | 40 |
| 40mer | 31 | 8 | 24 | 7 |
| AP template: | ||||
| >25mer | 28 | 6.5 | 42 | 21 |
| 40mer | 4.6 | 0.36 | 6.1 | 0.6 |
| P+1 | 16 | 3.3 | 30 | 18 |
aThe Pol δ enzymes (2 nM) were incubated with the template/primers for 1, 2 and 4 min (Materials and Methods section) and the rates of primer extension were determined for total extension products (>25mer) and the full-length product (40mer). The amounts of extension product were plotted against time. The plots were linear between 1 and 4 min and rates were determined by linear regression analysis (R2 values were >0.99 in almost all cases and the lowest was 0.90.) The rates obtained for each enzyme were normalized to the rate on the unmodified template.
Loss of the exonuclease activity did not greatly affect the relative rates for bypass of the O6-MeG and 8-oxoG templates by Pol δ4 (Table 1, first and third columns). A similar observation was made when Pol δ3 was compared to Pol δ3exo– (Table 1, second and fourth columns), except for the O6-MeG template where the rate was almost doubled for Pol δ3exo–. These findings were surprising, given the evidence for increased levels of exonucleolytic products by Pol δ3, so that one might have expected greater increases. However, in this type of assay, these rates do not directly measure total nucleotide incorporation but conversion of the primer to extended products. Also, these experiments may point to alterations in the polymerase activities that in turn could also alter proofreading rates. A more detailed kinetic analysis of the activities on these templates will be required to determine the true rates of nucleotide incorporation.
For the AP template, we examined the rates of formation of the P+1 product as well as for the total and full-length products. The formation of the full-length product by Pol δ3 was 16-fold slower than by Pol δ4, and the formation of the P+1 product was 5-fold slower (Table 1). This is consistent with the data of Figure 3, which shows that Pol δ3 does not readily bypass the AP lesion or accumulate the P+1 product. The rates of formation of products by Pol δ4exo– were faster than those of Pol δ4, by ca. 2-fold (Table 1, columns 1 and 3). In the case of Pol δ3exo,– similar increases in rates were observed by comparison to Pol δ3, except for formation of the P+1 product which was increased by >5-fold. These data support the view that faster removal of the P+1 product by the exonuclease is an important factor in the low accumulation of the P+1 product by Pol δ3 when compared to Pol δ4 (Figure 3).
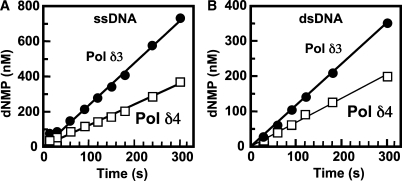
Pol δ3 has a higher exonuclease activity than Pol δ4
As already noted, a striking difference in Pol δ3 is the evidence for increased proofreading activity, as evidenced by the increased levels of exonucleolytic products. We compared the exonuclease activities of Pol δ3 and Pol δ4 by determining their activities under steady-state conditions on ssDNA and dsDNA oligonucleotide substrates. Product formation time courses with ssDNA and dsDNA substrates revealed that Pol δ3 has a 1.9-fold higher exonuclease specific activity compared to Pol δ4 on a ssDNA substrate (Figure 4A) and a 1.8-fold higher specific activity on a dsDNA substrate (Figure 4B). These findings show for the first time that loss of the p12 subunit affects the exonuclease activity of the Pol δ. In addition, assays of polymerase activity in the standard assay on poly(dA)/oligo(dT) template/primer shows that Pol δ4 has approximately twice the specific activity of Pol δ3 (see ‘Materials and methods’ section). Thus, Pol δ3 exhibits an increase in the ratio of exonuclease to polymerase ratio of ca. 4-fold compared to Pol δ4.
Figure 4.
The 3′ to 5′ exonuclease activities of Pol δ4 and Pol δ3. (A, B) Time courses for the exonuclease activities assayed on a ssDNA substrate (5′-[32P]end-labeled 24-nt oligonucleotide), and a 24-/36-mer dsDNA substrate, respectively (see ‘Materials and methods’ section). Data for Pol δ4 are shown as open squares and for Pol δ3 as solid circles. Reactions contained 4 nM polymerases, 100 nM 24-mer ssDNA or 24-/36-mer dsDNA; MgCl2 was added to initiate the reactions. The concentration of dNMP that was released in order to attain the product distribution found was calculated (see ‘Materials and methods’ section). The specific activities (apparent kcat) of the 3′ to 5′ exonuclease activities were determined from the slopes of the curves divided by the enzyme concentration (4 nM); these were 0.53 s–1 and 0.26 s–1, respectively, for Pol δ3 and Pol δ4 activity on ssDNA, and 0.30 s–1 and 0.17 s–1, respectively, on the dsDNA substrate.
Pol δ3 is less likely than Pol δ4 to insert an incorrectly base-paired nucleotide
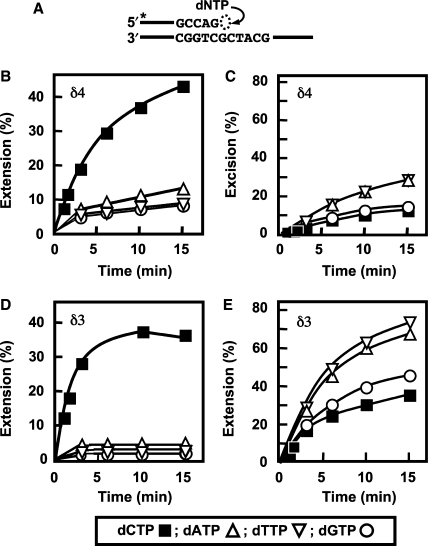
The behavior of Pol δ3 when it attempts to synthesize DNA past template lesions suggested that Pol δ3 might also be less likely to insert wrong nucleotides than Pol δ4. The tendencies of Pol δ4 and Pol δ3 to incorporate the wrong nucleotides (misincorporation) were compared in the single nucleotide incorporation assay (Figure 5A). The unmodified 25-mer/40-mer primer/template was used, and the correct (dCTP) and incorrect nucleotides (dATP, dTTP and dGTP) were supplied individually at a high concentration of 500 μM (see ‘Materials and methods’ section). The amounts of extension and excision products formed by Pol δ4 (Figure 5B and C) and Pol δ3 (Figure 5D and E) were determined. Pol δ3 produced far less products by insertion of the wrong nucleotides (dATP, dTTP and dGTP) than Pol δ4 (cf. Figure 5B and D). In parallel, Pol δ3 produced more excision products than Pol δ4 (cf. Figure 5C and E). These results indicate that Pol δ3 is less likely than Pol δ4 to insert an incorrectly base paired nucleotide.
Figure 5.
Comparison of the misinsertion of wrong nucleotides by Pol δ4 and Pol δ3 in the single nucleotide incorporation assay. (A) Diagram of the assay for single nucleotide incorporation. The abilities of Pol δ (5 nM each) to extend the unmodified 25-/40-mer primer/template (200 nM) (Figure 1) in the presence of 400 nM PCNA and 500 μM correct (dCTP) or incorrect nucleotides were examined (see ‘Materials and methods’ section). Reactions were performed for 0, 2, 4, 6, 10 and 15 min. Products were resolved by denaturing gel electrophoresis followed by phosphorimaging and quantitation. (B, C) Extension products and excision products, respectively, for Pol δ4. (D, E) Extension and excision products, respectively, for Pol δ3. Symbols (bottom box): dCTP, solid squares; dTTP, open inverted triangles; dGTP, open circles; dATP, open triangles. Amounts of products were expressed as the percentages of the initial primer amounts.
Comparison of the exonuclease-deficient forms of Pol δ4 and Pol δ3 in the single nucleotide incorporation assay
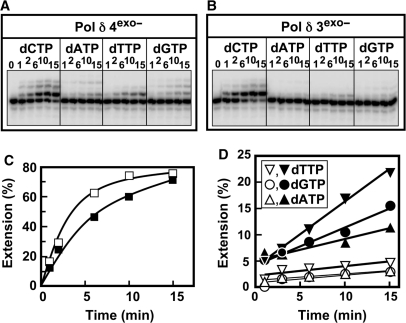
The observed behaviors of Pol δ3 on templates containing lesions and in the single nucleotide assay could be due to a more active proofreading activity. The latter does not necessarily imply an increased intrinsic 3′ to 5′exonuclease activity, since decreases in insertion rates at the pol site can form a kinetic barrier that favors translocation of the primer from the pol to the exo site (5,7). We therefore compared the Pol δ4 and Pol δ3 exonuclease-deficient mutants in the single nucleotide incorporation assay (Figure 6A and B). Quantitation of the amounts of misincorporation products show that Pol δ3exo– has a reduced tendency to insert the wrong nucleotide when compared to Pol δ4exo– (Figure 6D). This suggests that changes in the polymerase activity of Pol δ3 may play a role in its greater discrimination against insertion of mismatched nucleotides.
Figure 6.
Comparison of the misinsertion of wrong nucleotides by Pol δ4exo– and Pol δ3exo– in the single nucleotide incorporation assay. The exonuclease-deficient forms of Pol δ4 and Pol δ3 were used (see ‘Materials and methods’ section). Assays for single nucleotide incorporation were performed as described in Figure 5. (A, B) Phosphorimages of the gels for the reaction products formed by Pol δ4exo– and Pol δ3exo–, respectively. (C) Quantitation of the amounts of extension products formed expressed as % of the primer converted for the correct nucleotide (dCTP). Data for Pol δ4exo– are shown by the solid squares and data for Pol δ3exo– are shown as open squares. (D) Data for insertion of the wrong nucleotides: dTTP, inverted triangles; dGTP, circles; dATP, triangles. Data for Pol δ4exo– are shown by the solid symbols and data for Pol δ3exo– are shown as open symbols.
Pol δ3 is less likely to extend a mismatched primer than Pol δ4
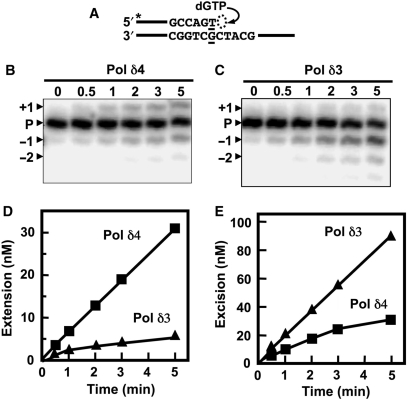
We next compared the ability of Pol δ4 and Pol δ3 to extend a primer in which the 3′nucleotide is mismatched, i.e. does not form a Watson–Crick base pair with the template. In this ‘mismatch extension’ assay, the same template used above was annealed to a 26-mer primer with an additional 3′ T, such that a T:G mismatch was formed, and only dGTP, which could correctly base pair with the next template base, was provided (Figure 7A). Pol δ3 had a decreased rate of extension of the mismatched primer (Figure 7D) and an increased rate of formation of excision products when compared to Pol δ4 (Figure 7E). Analysis of the slopes of the linear portions of the time courses for incorporation showed that Pol δ3 extended the mismatched primer 6.5-fold more slowly and degraded the mismatched primer 3.1-fold faster than Pol δ4 (Figure 7D and E). Thus, the ratio of excision/extension rates by Pol δ3 can be calculated to be 19-fold greater than that of Pol δ4. These findings show that Pol δ3 is less likely to extend a mismatched primer, and the altered balance of excision and extension product formation is in keeping with an interpretation that it is more active in proofreading.
Figure 7.
Comparison of the extension of a mismatched primer terminus by Pol δ4 and Pol δ3 (A) Diagram of the assay for mismatch extension. The abilities of Pol δ4 and Pol δ3 to extend a 26-/40-mer primer/template in which the 3′ nt of the primer forms a T:G mismatch (underlined) with the template was examined. The reactions contained 2 nM Pol δ4 or Pol δ3, 400 nM PCNA, 250 nM 26-/40-mer and 500 μM dGTP. Only the correct next nucleotide (dGTP) was added to the reactions. Reactions were carried out for 0, 0.5, 1, 2, 3 and 5 min. (see ‘Materials and methods’ section). (B, C) Phosphorimages of the reaction products formed by Pol δ4 and Pol δ3, respectively. The positions of the 26-mer primer (‘P’), the 27-mer mismatch extension product (+1), the exonucleolytic 25-mer (–1) and 24-mer (–2) products are shown by the arrowheads. (D, E) The amounts of mismatch extension (27-mer) and excision products (>26mer), respectively, that were formed. Product formation was expressed as concentrations formed in the assay in nanomoles, based on the initial concentration of the primer. Data for Pol δ4 are shown as squares and for Pol δ3 as triangles.
Comparison of the exonuclease-deficient forms of Pol δ4 and Pol δ3 in the mismatch extension assay
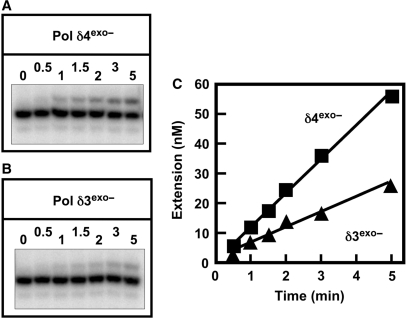
The exo− forms of Pol δ were examined in the mismatch extension assay (Figure 8A and B). Pol δ3exo− extended the mismatched primer terminus at about half the rate of Pol δ4exo– (Figure 8C). This result demonstrates that polymerase activity of Pol δ3 is less likely than Pol δ4 to extend a mismatched primer terminus. These findings show that alterations in polymerase activity per se are involved in the reduced tendency of Pol δ3 for mismatch extension, and are consistent with the results obtained in the single nucleotide insertion assay (Figure 6).
Figure 8.
Comparison of the extension of a mismatched primer terminus by Pol δ4exo– and Pol δ3exo–. Assays for mismatch extension were performed as in Figure 7. (A, B) Phosphorimages of the reaction products formed by Pol δ4exo– and Pol δ3exo–, respectively. (C) Mismatch extension products (27 nt) formed by Pol δ4exo– and Pol δ3exo– were quantitated and are shown as squares and triangles, respectively.
Activities of Pol δ4 and Pol δ3 on a template containing a thymine–thymine dimer
The activities of Pol δ4 and Pol δ3 were compared on a 25-mer primer annealed to a 40-mer template containing a thymine–thymine CPD (cyclobutane pyrimidine dimer), the major lesion produced by UV damage (20). Pol η was also assayed for comparison, as it readily bypasses thymine–thymine dimers, with a preference for the insertion of two As opposite the lesion (20,31,32). Pol η readily bypassed the lesion (Figure 9A, two left panels). Pol δ4 was able to insert a single nucleotide to form the 26-mer, but only weakly extended the primer to the full-length product (Figure 9A). Thus, the CPD forms a stronger barrier to Pol δ4 than does the AP lesion (cf. Figure 2A). Pol δ3 did not produce significant amounts of product on the CPD template (Figure 9A). Both Pol δ4 and Pol δ3 produced exonucleolytic products when blocked at the CPD lesion, with Pol δ3 producing more products than Pol δ4 (Figure 9A).
Figure 9.
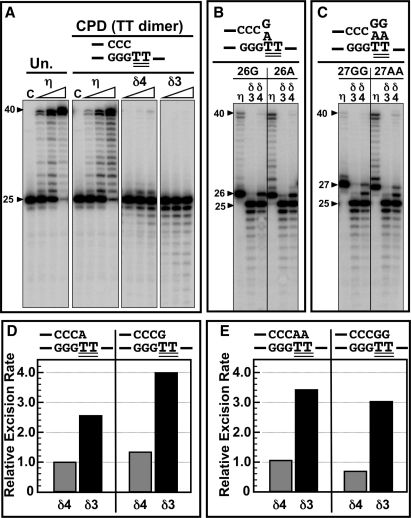
Translesion synthesis by Pol δ4 and Pol δ3 on a template containing a thymine–thymine dimer. The abilities of Pol δ4 and Pol δ3 to extend primers on a template containing a thymine–thymine dimer (CPD) were examined. The template used was a 40-mer containing a thymine–thymine dimer at nts 26 and 27. The primers used were a 25-mer primer, two 26-mer primers that had an added A (‘26A’) or G (‘26G’) at position 26, and two 27-mer primers that had two added As (27AA’) or Gs (‘27GG’) (see ‘Materials and methods’ section). (A) Phosphorimage of the reaction products formed on a template containing a thymine–thymine dimer. DNA polymerases (4, 20 and 100 nM) were incubated with 500 μM dNTPs, 400-nM PCNA, 100-nM 25-mer end-labeled primer annealed to the unmodified (‘Un’) template or with the CPD template as indicated. Reactions were incubated at 37°C for 15 min. The sequence at the primer terminus is shown, where the two double-underlined TT residues represent the thymine–thymine dimer. From left to right: Pol η activity on the unmodified template (‘Un’), followed by Pol η, Pol δ4 and Pol δ3 activities on the CPD template. Lane ‘C’ represents controls in which no enzyme was added. (B) Pol η, Pol δ4 and Pol δ3 were assayed as in A at 20-nM concentration using the 26G or 26A primers annealed to the 40-mer template containing the thymine–thymine dimer as shown above the phosphorimage. Assays were performed for 5 min. (C). Pol η, Pol δ4 or Pol δ3 were assayed as in B using the 27GG or 27AA primers annealed to the 40-mer template containing the thymine–thymine dimer as shown above the phosphorimage. (D) The rate of exonucleolytic cleavage of the 26A and 26G primers by Pol δ4 and Pol δ3 were assayed as in (B). Pol δ4 and Pol δ3 were incubated with the 26A or 26G primer annealed to the thymine–thymine dimer template as in (B), and the reactions carried out for 0, 1, 2, 4, 6 and 10 min. The amounts of the 26-mers remaining at each time point was determined, were plotted against time and fitted into a single exponential decay equation to obtain the rate constant of 26-mer disappearance. The values were normalized to the rate for Pol δ4 activity on the 26A primer. Values for Pol δ4 are shown by the shaded bars and for Pol δ3 by the solid bars. (E). The rate of exonucleolytic cleavage of the 27AA and 27GG primers annealed to the 40-mer template containing a CPD lesion by Pol δ4 and Pol δ3 were assayed and plotted as described in D. The values were normalized to the rate for Pol δ4 activity on the 26A primer.
Next, we used 26-mer primers with an added A or G, and 27-mer primers with two As or Gs added to the 25-mer. These primers would present paired or mispaired primer termini opposite the 5′ and 3′ thymidines of the CPD. Pol η poorly extends primers with bases other than A opposite the 3′ and the 5′ thymidines (32). These primers were used to assess the comparative rates at which Pol δ4 and Pol δ3 degraded these primers, since it has been suggested that Pol δ may function to provide extrinsic exonucleolytic proofreading for Pol η (33,34). Pol δ3 degraded most of the 26-mer to the 25-mer, while much less cleavage was evident with Pol δ4 (Figure 9B). Neither primer was significantly extended by either enzyme. Similar results were found with the 27-mer primers, where most of the 27-mer was degraded to the 25-mer by Pol δ3, while only partial degradation was observed with Pol δ4 (Figure 9C), and neither Pol δ4 nor Pol δ3 extended the primers. In order to compare the rates of exonucleolytic cleavage of the 26-mer and 27-mer primers, time course experiments were performed, and the relative rates of exonucleolytic cleavage of the primers were determined (Figure 9D and E). Pol δ3 degraded the mismatched 5′ G more rapidly than A (Figure 9D), but did not discriminate between the 27-mer primers (Figure 9E). In all cases, Pol δ3 cleaved the primers between 2- and 4-fold faster than Pol δ4. Thus, with these substrates, Pol δ3 was more efficient than Pol δ4 at removal of bases that were opposite the CPD lesion.
DISCUSSION
Our studies provide new information regarding the properties of human Pol δ3. Pol δ3 has a reduced ability for TLS bypass synthesis, and exhibits an increased proofreading activity, as indicated by the increased production of exonucleolytic products. Analyses using the single nucleotide incorporation assays showed that Pol δ3 is less likely to insert an incorrect nucleotide and is less likely to extend a mismatched primer. Pol δ3 exhibited a higher ratio of exonuclease to polymerase activity than Pol δ4 that is consistent with an increased proofreading activity. The importance of the exonuclease activity is clearly observed in the products formed on the AP template, where significant levels of the P+1 product were only observed when the exonuclease activity of Pol δ3 was inactivated. Analyses of the exo- mutants showed that alterations the polymerase activity of Pol δ3 are also involved. These analyses indicate that Pol δ3 is altered in such a way that it displays attributes of increased fidelity (in a biochemical context). These biochemical properties imply that Pol δ3 may have the ability to synthesize DNA with a greater fidelity than Pol δ4. This possibility needs to be confirmed by experimental approaches that will allow direct comparison of the error rates of Pol δ4 and Pol δ3, both by in vitro and in vivo approaches. The loss of p12 in vivo can be regarded as an example of enzyme regulation by alteration of quaternary structure, and the increased ability of Pol δ3 to detect DNA damage can be viewed as a gain of function over Pol δ4. The present findings that Pol δ3 exhibits properties consistent with a greater ability to recognize template lesions, and a reduced proclivity for misincorporation and mismatch extension, provides prima facie support for the hypothesis that the conversion of Pol δ4 to Pol δ3 represents an adaptation that could be beneficial to cells undergoing genotoxic insult.
There are several possible explanations for the mechanistic basis for the changes that were observed in Pol δ3. Our findings that Pol δ3 exhibits altered steady-state ratios of exo/pol activity indicate that the loss of p12 must alter the catalytic site of the p125 subunit. We found that Pol δ3 exhibited an increased steady exonuclease activity, and an increased exonuclease/polymerase ratio, which would be expected to favor proofreading activity. However, the increased exonucleolytic behavior of Pol δ3 could be induced by changes in the functioning of the polymerase catalytic site alone. This possibility is supported by our analyses of the exonuclease-deficient mutants, which show that Pol δ3exo– exhibits lowered rates of misincorporation and mismatch extension. This would result in an increased kinetic barrier, i.e. an increased probability for translocation of DNA from the pol to the exo sites (4,5,7). There could also be alterations in the rates of translocation between the pol and exo sites, or changes in the kinetic properties of the exonuclease itself (4,5,7). Further studies using detailed kinetic analyses will be required to determine the mechanistic basis for these alterations. Taken together, our findings suggest that Pol δ3 exhibits an increased ability to discriminate against aberrant primer/template structures. Crystallographic studies of replicative polymerase interactions with a variety of primer/template lesions, in particular those of the RB69 polymerase, have provided a greater understanding of how aberrant primer/template structures interact with polymerase catalytic sites (12,35). From a structural perspective, we can infer that loss of p12 may alter the geometry of the Pol δ active site, or alter its ability to undergo the large conformational changes that take place during catalysis (3–5). It is noted that p12 interacts with both the p125 and p50 subunits that form the core enzyme (11).
There are a number of potential implications of the alterations that we observed in Pol δ3 besides the obvious possibility that the potential for error introduction may be reduced. Pol δ participates in a number of DNA transactions that include both DNA replication and repair processes which may also be impacted when its properties are altered. Reduction in activity and or processivity of Pol δ may be relevant to recent evidence for an elongation checkpoint that involves slowing of replication fork progression following DNA damage by UV or alkylating agents (36,37). The mechanism for this is unknown, but it is relevant that Pol δ3 exhibits an impaired activity when assayed on a singly primed ssM13 DNA in an assay which also requires PCNA, its clamp-loader RFC, and RPA (23).
The increased tendency for stalling by Pol δ3 may be relevant to two processes that are involved in the DNA damage response. Polymerase stalling on encounter with DNA lesions results in formation of ssDNA that triggers ATR activation (38,39), and activation of the DNA damage tolerance response (19). An increased tendency to stall at template lesions may promote the activation of checkpoint responses. Current models for TLS bypass involve a polymerase switch between a TLS polymerase and Pol δ that takes place on mono-ubiquitinated PCNA (40), and this may involve Pol δ3 rather than Pol δ4. The polymerase switch between yeast Pol δ and Pol η has been shown to require the stalling of Pol δ (41).
The increase in the exonucleolytic behavior of Pol δ3 may be relevant to its proposed functions as an extrinsic proofreading exonuclease for the error prone TLS polymerases, e.g. in the bypass of thymine–thymine dimers by yeast Pol δ and Pol η (33,34). The increased exonuclease activity of Pol δ, its increased idling behavior on encounter with base lesions (Figure 1), and its greater tendency for excision of bases opposite AP and CPD lesions (Figures 2 and 9) would represent positive adaptations for the purpose of extrinsic proofreading.
In summary, our studies provide evidence that the p12 subunit plays a significant role in the catalytic functions of Pol δ. Pol δ3 possesses altered properties that provide support for the hypothesis that the conversion of Pol δ4 to Pol δ3 may represent an adaptive response that contributes to the DNA damage response. Our findings also raise issues whose further investigation may add to our understanding of the DNA damage response.
FUNDING
The National Institutes of Health (GM31973 and ES014737 to M.Y.W.T.L., AI052395 to D.N.F.). Funding for open access charge: National Institutes of Health (GM31973).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr. Maurice J. Bessman for reading the manuscript and for his helpful advice.
REFERENCES
- 1.Brutlag D, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. 36. A proofreading function for the 3′ to 5′ exonuclease activity in deoxyribonucleic acid polymerases. J. Biol. Chem. 1972;247:241–248. [PubMed] [Google Scholar]
- 2.Johnson A, O'Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 3.Joyce CM, Steitz TA. Function and structure relationships in DNA polymerases. Annu. Rev. Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 4.Kunkel TA, Bebenek K. DNA replication fidelity. Annu. Rev. Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 5.Johnson KA. Conformational coupling in DNA polymerase fidelity. Annu. Rev. Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 6.Goodman MF, Gore WC, Muzyczka N, Bessman MJ. Studies on the biochemical basis of spontaneous mutation. III. Rate model for DNA polymerase-effected nucleotide misincorporation. J. Mol. Biol. 1974;88:243–235. doi: 10.1016/0022-2836(74)90492-6. [DOI] [PubMed] [Google Scholar]
- 7.Khare V, Eckert KA. The proofreading 3′ - 5′ exonuclease activity of DNA polymerases: a kinetic barrier to translesion DNA synthesis. Mutat. Res. 2002;510:45–54. doi: 10.1016/s0027-5107(02)00251-8. [DOI] [PubMed] [Google Scholar]
- 8.Muzyczka N, Poland RL, Bessman MJ. Studies on the biochemical basis of spontaneous mutation. I. A comparison of the deoxyribonucleic acid polymerases of mutator, antimutator, and wild type strains of bacteriophage T4. J. Biol. Chem. 1972;247:7116–7122. [PubMed] [Google Scholar]
- 9.Reha-Krantz LJ. Regulation of DNA polymerase exonucleolytic proofreading activity: studies of bacteriophage T4 “antimutator” DNA polymerases. Genetics. 1998;148:1551–1557. doi: 10.1093/genetics/148.4.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garg P, Burgers PM. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 2005;40:115–128. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- 11.Li H, Xie B, Zhou Y, Rahmeh A, Trusa S, Zhang S, Gao Y, Lee EY, Lee MY. Functional roles of p12, the fourth subunit of human DNA polymerase delta. J. Biol. Chem. 2006;281:14748–14755. doi: 10.1074/jbc.M600322200. [DOI] [PubMed] [Google Scholar]
- 12.Hogg M, Wallace SS, Doublie S. Bumps in the road: how replicative DNA polymerases see DNA damage. Curr. Opin. Struct. Biol. 2005;15:86–93. doi: 10.1016/j.sbi.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 13.Warren JJ, Forsberg LJ, Beese LS. The structural basis for the mutagenicity of O(6)-methyl-guanine lesions. Proc. Natl Acad. Sci. USA. 2006;103:19701–19706. doi: 10.1073/pnas.0609580103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu GW, Ober M, Carell T, Beese LS. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature. 2004;431:217–221. doi: 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- 15.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 16.Beard WA, Prasad R, Wilson SH. Activities and mechanism of DNA polymerase beta. Methods Enzymol. 2006;408:91–107. doi: 10.1016/S0076-6879(06)08007-4. [DOI] [PubMed] [Google Scholar]
- 17.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol. Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 18.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 19.Andersen PL, Xu F, Xiao W. Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res. 2008;18:162–173. doi: 10.1038/cr.2007.114. [DOI] [PubMed] [Google Scholar]
- 20.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 21.Friedberg EC, Lehmann AR, Fuchs RP. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol. Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 22.Lehmann AR. Translesion synthesis in mammalian cells. Exp. Cell Res. 2006;312:2673–2676. doi: 10.1016/j.yexcr.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 23.Zhang S, Zhou Y, Trusa S, Meng X, Lee EY, Lee MY. A novel DNA damage response: rapid degradation of the p12 subunit of dna polymerase delta. J. Biol. Chem. 2007;282:15330–15340. doi: 10.1074/jbc.M610356200. [DOI] [PubMed] [Google Scholar]
- 24.Goldsby RE, Hays LE, Chen X, Olmsted EA, Slayton WB, Spangrude GJ, Preston BD. High incidence of epithelial cancers in mice deficient for DNA polymerase delta proofreading. Proc. Natl Acad. Sci. USA. 2002;99:15560–15565. doi: 10.1073/pnas.232340999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mozzherin DJ, Shibutani S, Tan CK, Downey KM, Fisher PA. Proliferating cell nuclear antigen promotes DNA synthesis past template lesions by mammalian DNA polymerase delta. Proc. Natl Acad. Sci. USA. 1997;94:6126–6131. doi: 10.1073/pnas.94.12.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi JY, Chowdhury G, Zang H, Angel KC, Vu CC, Peterson LA, Guengerich FP. Translesion synthesis across O6-alkylguanine DNA adducts by recombinant human DNA polymerases. J. Biol. Chem. 2006;281:38244–38256. doi: 10.1074/jbc.M608369200. [DOI] [PubMed] [Google Scholar]
- 27.Taylor JS. New structural and mechanistic insight into the A-rule and the instructional and non-instructional behavior of DNA photoproducts and other lesions. Mutat. Res. 2002;510:55–70. doi: 10.1016/s0027-5107(02)00252-x. [DOI] [PubMed] [Google Scholar]
- 28.Khare V, Eckert KA. The 3′ - 5′ exonuclease of T4 DNA polymerase removes premutagenic alkyl mispairs and contributes to futile cycling at O6-methylguanine lesions. J. Biol. Chem. 2001;276:24286–24292. doi: 10.1074/jbc.M011025200. [DOI] [PubMed] [Google Scholar]
- 29.Michaels ML, Miller JH. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J. Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Einolf HJ, Guengerich FP. Fidelity of nucleotide insertion at 8-oxo-7,8-dihydroguanine by mammalian DNA polymerase delta. Steady-state and pre-steady-state kinetic analysis. J. Biol. Chem. 2001;276:3764–3771. doi: 10.1074/jbc.M006696200. [DOI] [PubMed] [Google Scholar]
- 31.McCulloch SD, Kokoska RJ, Masutani C, Iwai S, Hanaoka F, Kunkel TA. Preferential cis-syn thymine dimer bypass by DNA polymerase eta occurs with biased fidelity. Nature. 2004;428:97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- 32.Washington MT, Johnson RE, Prakash L, Prakash S. Accuracy of lesion bypass by yeast and human DNA polymerase eta. Proc. Natl Acad. Sci. USA. 2001;98:8355–8360. doi: 10.1073/pnas.121007298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCulloch SD, Kokoska RJ, Chilkova O, Welch CM, Johansson E, Burgers PM, Kunkel TA. Enzymatic switching for efficient and accurate translesion DNA replication. Nucleic Acids Res. 2004;32:4665–4675. doi: 10.1093/nar/gkh777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nick McElhinny SA, Pavlov YI, Kunkel TA. Evidence for extrinsic exonucleolytic proofreading. Cell Cycle. 2006;5:958–962. doi: 10.4161/cc.5.9.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hogg M, Wallace SS, Doublie S. Crystallographic snapshots of a replicative DNA polymerase encountering an abasic site. EMBO J. 2004;23:1483–1493. doi: 10.1038/sj.emboj.7600150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merrick CJ, Jackson D, Diffley JF. Visualization of altered replication dynamics after DNA damage in human cells. J. Biol. Chem. 2004;279:20067–20075. doi: 10.1074/jbc.M400022200. [DOI] [PubMed] [Google Scholar]
- 37.Conti C, Seiler JA, Pommier Y. The mammalian DNA replication elongation checkpoint: implication of Chk1 and relationship with origin firing as determined by single DNA molecule and single cell analyses. Cell Cycle. 2007;6:2760–2767. doi: 10.4161/cc.6.22.4932. [DOI] [PubMed] [Google Scholar]
- 38.Paulsen RD, Cimprich KA. The ATR pathway: fine-tuning the fork. DNA Repair (Amst) 2007;6:953–966. doi: 10.1016/j.dnarep.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 39.Andreassen PR, Ho GP, D’Andrea AD. DNA damage responses and their many interactions with the replication fork. Carcinogenesis. 2006;27:883–892. doi: 10.1093/carcin/bgi319. [DOI] [PubMed] [Google Scholar]
- 40.Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 41.Zhuang Z, Johnson RE, Haracska L, Prakash L, Prakash S, Benkovic SJ. Regulation of polymerase exchange between Pol eta and Pol delta by monoubiquitination of PCNA and the movement of DNA polymerase holoenzyme. Proc. Natl Acad. Sci. USA. 2008;105:5361–5366. doi: 10.1073/pnas.0801310105. [DOI] [PMC free article] [PubMed] [Google Scholar]